Abstract
Regulatory measures and public concerns regarding bisphenol A (BPA) have led to its replacement by structural analogues, such as BPAF, BPAP, BPB, BPF, BPP, BPS, and BPZ. However, these alternatives are under surveillance for potential endocrine disruption, particularly during the critical period of fetal development. Despite their structural analogies, these BPs differ greatly in their placental transport efficiency. For predicting the fetal exposure of this important class of emerging contaminants, quantitative structure-activity relationship (QSAR) studies were developed to model and predict the placental clearance indices (CI). The most usual input parameters were molecular descriptors obtained by modelling, but for bisphenols (BPs) with structural similarities or heteroatoms such as sulfur, these descriptors do not contrast greatly. This study evaluated and compared the capacity of QSAR models based either on molecular or chromatographic descriptors or a combination of both to predict the placental passage of BPs. These chromatographic descriptors include both the retention mechanism and the peak shape on columns that reflect specific molecular interactions between solute and stationary and mobile phases and are characteristic of the molecular structure of BPs. The chromatographic peak shape such as the asymmetry and tailing factors had more influence on predicting the placental passage than the usual retention parameters. Furthermore, the QSAR model, having the best prediction capacity, was obtained with the chromatographic descriptors alone and met the criteria of internal and cross validation. These QSAR models are crucial for predicting the fetal exposure of this important class of emerging contaminants.
1. Introduction
Bisphenol A (BPA), which was widely used in everyday products, has been prohibited in food packaging in several countries because of its endocrine disrupting properties. As a result, structurally similar phenolic compounds such as BPAF, BPAP, BPB, BPF, BPP, BPS, and BPZ (Figure 1) have gradually replaced BPA in the polymer industry. Because of the wide use of these analogues in food packaging and personal care products [1,2,3], human exposure to these compounds is ubiquitous, as demonstrated by their detection in human urine [1,4,5]. These analogues may display endocrine disrupting properties similar to those of BPA [6,7,8,9,10] particularly during the critical period of pregnancy [11,12,13]. In vivo studies performed in pregnant sheep showed that, similar to BPA, BPS and BPF can also cross the placenta [14,15,16]. Recently, ex vivo human placental perfusion carried out to assess the placental transfer of 15 bisphenols (BPs) has shown that despite their structural similarities, the efficiency of placental transport differs greatly among BPs, as reflected in the large range of their clearance indices, from 0.065 to 0.842 [17]. Indeed, while BP4-4, BPAP, BPE, BPF, 3-3BPA, BPB, and BPA cross the placenta by passive diffusion [14], the materno-fetal placental transfer of BPS and BPFL is very limited and may involve membrane efflux transporters [17,18].
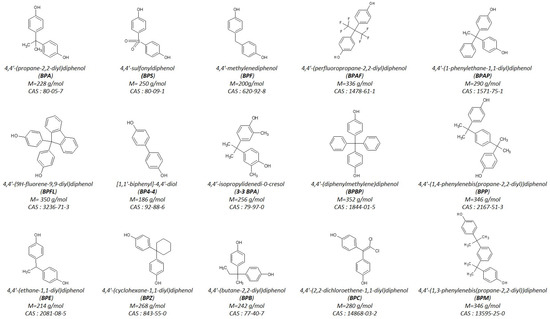
Figure 1.
Molecular structure of the fifteen bisphenols with their respective name, abbreviation, molecular weight, and CAS number.
The physicochemical properties of the molecules, such as molecular weight, degree of ionization (pKa), and lipophilicity (LogP), are considered possible factors that determine the extent of placental passage and the mechanism of transfer [19]. In this context, several studies have predicted the maternal-fetal transfer rates of many drugs or organic compounds through quantitative-structure activity relationship (QSAR) modelling [20,21,22,23]. These models are mostly based on molecular descriptors in order to establish a relationship between the biological activities or the toxicological responses of molecules and their molecular, physicochemical, and structural properties [24]. However, these numerical descriptors are calculated from the two- and/or three-dimensional structures of molecules and do not take into account the dynamic interactions with their biological targets [25]. For BP analogues, the physicochemical parameters generally involved in placental transfer (pKa within 8.5–9, LogP within 2–7, and molecular weight (MW) below 600 Da) would imply that their potential passive diffusion across the placenta would be high [17,20,21,22]. It is, therefore, anticipated that the placental transfer efficiency of these structurally related BPs will be hardly predicted by descriptors based on molecular structure alone. In many cases, experimental descriptors such as chromatographic retention have been added to molecular ones to improve the QSAR models [26,27]. Indeed, the chromatographic retention depends on the interaction of the compound of interest, i.e., its structure, with the stationary phase and the mobile phase. Thus, chromatographic retention of a molecule depends strongly on both its physicochemical properties and its environment. To the best of our knowledge, few studies have investigated the use of chromatographic descriptors alone in QSAR modelling, and all of them have only considered retention parameters [28,29,30].
The aim of the present study was to evaluate the reliability of the QSAR model based on chromatographic descriptors compared to classical molecular descriptors to predict the placental clearance of BPs, using the placental clearance indices determined in a previous study [17] as the biological output. To that end, the set of 15 selected BPs was eluted on thirteen chromatographic columns using two elution solvents. We then built a QSAR model based either on classical molecular descriptors of the fifteen bisphenols or on chromatographic descriptors, such as retention and peak shape parameters, or on both molecular and chromatographic descriptors. Their performances for predicting the BP placental transport efficiency were verified by cross validation procedures. This QSAR approach, by allowing the prediction of a key parameter, i.e., placental clearance, should provide new insight into screening of this important class of emerging BPs and assessing the hazards of BPA substitution in a vulnerable developing fetus.
2. Results
2.1. Data Collection
2.1.1. Clearance Indices
Clearance indices (CI) of the 15 BPs perfused in cocktail were determined on five placentae using the ex vivo human perfused placenta model described in our previous study [17], as shown in Table 1. CI data were used as outcomes due to their ability to eliminate inter-placental variation by standardizing BP clearance to the reference compound antipyrine. These BPs were classified into two groups according to their mechanism of placental passage, by passive diffusion for seven BPs (similar to antipyrine) or the limited transport for eight BPs [17]. This data set obtained in the same experimental conditions was relatively distributed in a wide range of placental CI and is, thus far, suited for the QSAR modelling of the ability of these newly-introduced BPs to cross the placenta [22]. Moreover, the use of individual data of CI instead of the mean CI of both experimental and biological variabilities needs to be taken into account and, thus, improves the predictive capacity of the model [31].

Table 1.
Mean ± standard deviation (SD) of CI of the 15 studied BPs (seven BPs with transfer rates that did not differ significantly from that of antipyrine and eight BPs with a significantly lower transfer rate than that of antipyrine) evaluated on five placentae. The clearance index corresponds to the ratio of BP-studied placental transfer rate divided by the antipyrine transfer rate [17].
2.1.2. Molecular Descriptors
In total, 50 molecular descriptors were calculated for each BP with different algorithms available in the ChemDraw Pro 17.1 software. Among these 50 descriptors, 16 concern physicochemical properties (e.g., pKa, LogP), 14 are topological descriptors (e.g., Balaban index, polar surface), and 20 are involved with thermodynamic and electronic properties.
2.1.3. Chromatographic Descriptors
Figure 2 shows that the retention and the elution order of BPAF, BPAP, and 3-3BPA on the C18 column change according to the elution solvent, whether AcN or MeOH, (Figure 2) and the BPS peak shape is larger with MeOH than with AcN. Peak asymmetry, tailing factor, peak width (5%), and k′ vs. BPA were determined with the two organic solvents and for the 13 columns, resulting in a total of 104 descriptors per BP (Supplementary Materials Table S2).
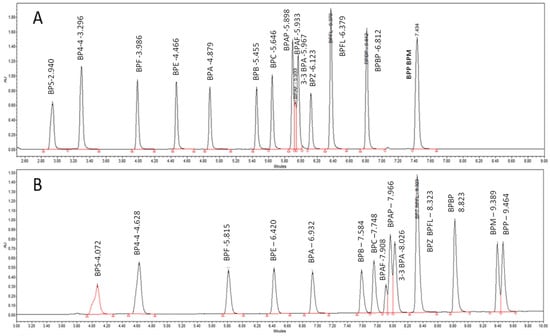
Figure 2.
Chromatograms of 15 studied BPs eluted on a BEH C18 column with an AcN/H2O gradient elution (A) and with an MeOH/H2O gradient elution (B).
2.2. QSAR Modelling
First, the ability of the models with molecular descriptors obtained in silico to predict the BP CI was investigated. Secondly, the chromatographic descriptors were added to the molecular descriptors to assess whether this combination could improve the prediction ability of the model. Finally, a model using only chromatographic descriptors was developed and compared to those using molecular descriptors.
2.2.1. Molecular Descriptors in the QSAR Model for Predicting Placental Passage
Among the 50 normalized molecular descriptors (Supplementary Materials Table S1), eleven descriptors gave a weight greater than twice the median of the weights using artificial neural network (ANN) learning and were selected as significant descriptors in the prediction of CI. Three of them were physicochemical descriptors (partition coefficients: LP2 and CD2, and a melting point CPP10), two were steric descriptors (Connolly Molecular Area CPS2, Connolly Solvent Excluded Volume CPS3), and two were topologic descriptors (Cluster count MT2 and Polar Surface Area MT5). The other four were involved in electrostatic and thermodynamic interactions (heat of formation DE1, total energy DE2, cosmo area DE5, and ionization potential DE7). Then, using these eleven descriptors, twenty-eight combinations were selected to build the model, considering low pair correlations among the variables (determination coefficient < 0.5). Five of these combinations had a regression R² > 0.7 and significant parameters in the multilinear regression (MLR). The final model, with significant and not correlated descriptors, was obtained with the following four molecular descriptors: LogP (LP2), Polar Surface Area (MT5), the heat of formation (DE1), and the total energy (DE2), and by using Equation (1) with a weighting by 1/Y:
CI = 0.46 + 0.23 × LP2 − 0.08 × MT5 − 0.30 × DE1 + 0.41 × DE2
Regression-adjusted CI and leverage are reported in Supplementary Materials Table S3. The criterion of the regression and of the cross validation are acceptable with a mean coefficient of regression determination (R2) of 0.77 and mean cross-validated correlation coefficient (Q2) higher than 0.70, respectively. However, the mean correlation coefficient of leave-many-out (LMO) validation (QLMO²) and mean correlation coefficient of concordance (CCC) for the three data sets were 0.57 and 0.64, respectively, which did not fulfill the recommended criteria for validating the predictive ability of this model (i.e., QLMO² > 0.6 and CCC > 0.85, Table 2).

Table 2.
Statistical results of the QSAR for the three selected models built with either molecular, chromatographic, or combined descriptors.
Moreover, the slope of the LMO validation curve [Ypred = f(mean(Yobs))] is significantly different than 1 (Figure 3A), and for most of the BPs of the test sets, the predicted CI are underestimated by the model except for BPZ (Figure 4A), for which its predicted CI were twice as high as its observed CI (Figure 4A). The results suggest that these in-silico calculated descriptors alone are not robust enough for predicting the clearance index of a new BP.
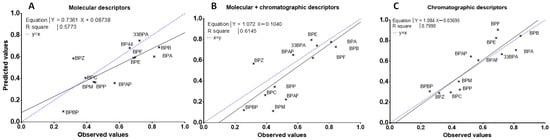
Figure 3.
LMO validation curves of the mean observed placental CI of BPs and predicted for the three test data sets by multilinear regression based on molecular (A), both molecular and chromatographic (B), or chromatographic (C) descriptors. The dotted line represents a perfect fit with the equation CIobs = CIpred and the straight line is the representation of CIpred = a × CIobs + b where a and b are the coefficients of the corresponding linear regression model.
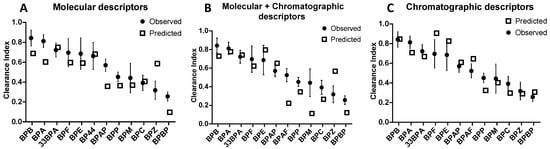
Figure 4.
Mean ± SD observed CI determined for the BPs on five perfused human placentae compared to the CI predicted by QSAR models based on molecular descriptors (A), molecular combined with chromatographic descriptors (B), and chromatographic descriptors (C). The placental CI of some BPs cannot be predicted because they are pivotal in building the QSAR model (BPAF, BPFL, BPS in (A), BP44, BPFL, BPS in (B) and (C)).
2.2.2. Combining Chromatographic Descriptors and Molecular Descriptors in the QSAR Model for Predicting Placental Passage
The 104 chromatographic descriptors were combined with the 50 molecular descriptors. Thirty-eight descriptors from these 154 descriptors were selected by ANN (Table 3), leading to 341 combinations of descriptors without correlation. Interestingly, only five molecular descriptors included in the previous molecular QSAR modelling (MT2, MT5, DE1, DE2, and DE5) were also significant in this present model, whereas the molecular descriptors related to LogP were not selected by ANN using molecular and chromatographic descriptors. The retention factors related to BPA were first selected by ANN (k′ vs. BPA, #5), but were progressively removed from the multilinear regression models because of their strong correlations with the molecular and peak shape descriptors. After the MLR assay, 50 models were chosen for their good fitting and the significance of their parameters. However, only a few of them met the LMO validation criteria.

Table 3.
Molecular and chromatographic descriptors selected to build QSAR for predicting placental CI, i.e., with weight in the artificial neural networks higher than twice the median of all descriptors’ weight.
Furthermore, the influence of BPs (leverage) on the regression differed according to the type of descriptors used. Indeed, BPAF, which had a strong influence using molecular descriptors, was no longer leveraged but was replaced by BP44, resulting in a change in the arbitrary choice of BPs in the test set 1 for LMO validation [32]. The final model included the following parameters: Connolly Solvent Excluded Volume (CPS3), cluster count (MT2), and LUMO energy (DE9) as molecular descriptors, and shape peak parameters (width #2, asymmetry #7, and tailing factor #8) on the biphenyl and BEH C8 column with AcN elution (RCBA8 and C8A2) and on the PFP column with MeOH as the organic modifier (PFPM2 and PFPM7). The equation of the multilinear regression, assigning a weight 1/Y, was:
CI = 0.48 − 0.03 × CPS3 − 0.23 × MT2 + 0.33 × DE9 − 0.25 × C8A2 + 0.37 ×
RCBA8 − 0.27 × PFPM2 + 0.23 × PFPM7
RCBA8 − 0.27 × PFPM2 + 0.23 × PFPM7
Regression-adjusted CI and leverage are reported in Supplementary Materials Table S3. The R² of the regression was 0.85, the Q² of the cross validation was 0.82, and the Bayesian information criterion (BIC) was lower than that of the model with molecular descriptors. However, the QLMO² of the LMO validation was lower than 0.5 (Table 2).
As shown in Figure 3B, the correlation curve (straight line) was systematically below the perfect curve (dot line) and the predicted CI for the BPs having low CI were misestimated (Figure 4B). The combination of chromatographic and molecular descriptors was not able to improve the prediction capacity of the QSAR model based solely on molecular descriptors.
2.2.3. Chromatographic Descriptors in the QSAR Model for Predicting Placental Passage
Among the 104 descriptors experimentally obtained by liquid chromatography, the ANN selected 17 chromatographic descriptors with weights higher than twice the median of all descriptors’ weight (Table 4). Eight chromatographic descriptors were also significant in the QSAR model based on both molecular and chromatographic descriptors (Table 3). Twenty-four models based on uncorrelated descriptors were identified, resulting in the selection of eight models with significant descriptors for predicting CI.

Table 4.
Chromatographic descriptors with weight in the artificial neural networks higher than twice the median of all descriptors’ weight and selected to build the QSAR model for predicting placental passage.
The final model was obtained using six chromatographic descriptors with a weighting by 1/Y according to the following Equation (3):
CI = 0.48 − 0.66 × T3A2 − 0.05 × PFPA7 + 0.07 × CC18A7 − 0.50 × C8M7 − 0.14 × PFPM7 + 0.38 × CNM8
Regression-adjusted CI and leverage of Equation (3) are reported in Supplementary Materials Table S3. All of these descriptors referred to peak shape parameters, and half were eluted with AcN and half with MeOH. Three of them were eluted on a hydrophobic phase (HSS T3, C18, and C8), two on a fluorophenyl phase (PFP), and one on a phase containing a polar group (CN). The asymmetry on the PFP column with MeOH elution (PFPM7) was the only descriptor in common with the multilinear regression based on these molecular and chromatographic descriptors (Equation (2)). This model has the lowest BIC value and the R² of the regression and the Q² of the cross-validation were 0.84 and 0.81, respectively. Moreover, the LMO validation values (QLMO² = 0.73 and CCC = 0.81) almost met the recommended criteria (Table 2), [33]. The Spearman’s correlation coefficient (R² spearman) was higher than 0.8 (Table 2), attesting to the ability of the model to classify CI according to the BPs. In addition, the mean square error of the calibration (RMSEC), cross-validation (RMSECV), and prediction (RMSEP) were low, between 0.11 and 0.13 (Table 2). The correlation curve (straight line, Ypred = f(mean(Yobs))) was close to the perfect line (dotted line, Figure 3C) with an intercept not significantly different from 0 and a slope close to 1. The predicted CI of BPs were grossly well estimated, with predicted CI included in standard deviation intervals of experimented values, except for BPE, BPF, and BPAF, which were slightly overestimated, and BPP, which was slightly underestimated (Figure 4C).
3. Discussion
This work aimed at developing a predictive model of placental transfer for newly emerging BPs. This approach was based on QSAR modeling of the CI of 15 BPs previously evaluated using ex vivo human placental perfusion [17]. BPs were selected because of their occurrence in foodstuffs [34,35], their production volume [36], and the diversity of their structures. In the present study, we have shown that a QSAR model based on chromatographic parameters provides more reliable predictions of BP placental transfer than QSAR models based on molecular descriptors alone or in combination with chromatographic descriptors.
The classical QSAR model based on molecular descriptors (Equation (1)) showed that physicochemical, topological, thermodynamic, and electronic parameters (LogP, total energy, polar surface area, and heat of formation) were able to influence the placental passage of this family of emerging BPs. Among physicochemical properties, the molecular weight and the lipophilicity are known to be key factors determining the passage across placenta [37,38,39]. The molecular weights of all BPs were below 600 Da and would not impact their passage across the placenta. Moreover, in a QSAR methodology using multivariate data analysis to model the placental passage of structurally diverse drugs and chemicals, the polar surface area and lipophilicity proved to be the most important factors in the model [21]. A QSAR model using multilinear analysis identified molecular weight, polarity, and also the heat of formation as important parameters to predict the materno-fetal transport of organohalogen compounds [20]. Whatever the importance of these factors, our results showed that a QSAR model based on molecular descriptors alone cannot provide a robust prediction of the placental transport for this class of emerging BPs, even if the ranking of their CI can be determined with a relatively good confidence level. It means that the placental transfer efficiency of these structurally related BPs cannot be predicted solely from their physicochemical properties determined in silico.
Chromatographic techniques offer the possibility of taking the interactions of the molecules with different columns in several conditions (pH, solvent) into account, and then better characterizing the molecular interactions with the biological system. That is the reason why chromatographic descriptors were combined with molecular ones in the QSAR models [26,27,28,29,30,40,41,42]. The most commonly used chromatographic descriptors are the retention factors that can be determined on several orthogonal stationary phases with different pH and solvents [26,28,30]. Indeed, many new reverse stationary phases were developed to extend the selectivity of chromatographic retention, involving many interactions with the column during elution [43]. The use of these experimental tools might be of great interest for QSAR modelling because these chromatographic parameters integrate several physicochemical properties of the compound. In this study, we used 13 chromatographic columns containing different stationary phases and two elution solvents, MeOH and AcN, involving different interactions with the 3D structure of the chemical.
Indeed, the organic modifier used in the gradient mode influences the fluidity of the alkyl chains of the stationary phases and then the mechanisms of molecular interactions between the analytes and the stationary phases [44], leading to modification of the chromatographic parameters. In our experiment, the use of MeOH as an organic modifier in the gradient mode favors the H-bonding and π-π interactions, while ion-dipole interactions are masked. Conversely, the use of AcN, due to the interactions of its cyano function with the stationary phase, limits the π-π interactions [45] and promotes dipole-dipole interactions [46]. The chromatographic behavior of the chemical is described not only by the retention factors but also by the peak shape parameters, such as asymmetry, tailing factors, or width at 5% [47,48], and can be linked to specific molecular descriptors [44].
Our modelling approach (Equation (2)) showed that the Connolly Solvent Excluded Volume, which is the volume enclosed in the molecule, had a weak influence on the placental passage prediction. The topological descriptor, cluster count, which reflects the complexity of the molecule [49], had a negative contribution to the CI of bisphenols. Inversely, the LUMO energy had a positive contribution. Our results are consistent with a previous QSAR model [20] showing that electronic descriptors are important parameters that may predict the ability of the molecule to cross the placenta. Surprisingly, although the goodness of fit is better, the contribution of the chromatographic descriptors does not improve the global predictive ability of our model based on the molecular descriptors. These results are not consistent with a previous study showing that the nonlinear QSAR model of prediction of gastrointestinal absorption was improved by adding the measured chromatographic descriptors to the molecular ones [27]. This discrepancy could be attributed to the QSAR model used, which was linear in our study and nonlinear in the Deconinck study, and to differences in the chromatographic parameters used in our QSAR model, which not only include the retention factors but also the peak shape parameters such as asymmetry, tailing factor, and peak width. These chromatographic parameters evaluated in the same experimental conditions reflect, in part, the mass transfer kinetics and thermodynamics of the analytes between the mobile and the stationary phase [50]. The chromatographic stationary phase selected in our model mainly involved polarizability (RCB) and π-π interactions (PFP) of the analytes. Thus, the implementation of these chromatographic descriptors led to a combination of uncorrelated descriptors having a great influence on the prediction of placental passage, which was quite different from those selected in the two QSAR model based either on molecular or chromatographic parameters. A significant influence of the thermodynamic properties of the BPs at the expense of the lipophilicity parameters is observed. In fact, the evaluation of the relationship between the molecular and the chromatographic descriptors selected in the two chromatography-based QSAR models through the ANN learning showed that the selected chromatographic descriptors are a combination of molecular descriptors selected to describe CI. Nevertheless, the four chromatographic descriptors presented in the model based on molecular and chromatographic descriptors are not directly related to LogP, unlike the chromatographic descriptors selected by the chromatography-based QSAR (Supplementary Materials Table S4).
When we developed a QSAR model based on chromatographic descriptors alone to predict the passage of BPs across the placenta, the main chromatographic descriptors selected by the ANNs involved the peak shape, such as asymmetry, width, and tailing factor, as in the previous model. More specifically, the most important descriptors in the validated model (Equation (3)) are the width peak on the HSS T3 column with AcN as the eluent, followed by the asymmetry peak on the BEH C8 column with MeOH elution and the tailing factor on the HSS CN column with MeOH elution. Several studies have shown that residual silanols can increase the tailing factor of basic or polar compounds due to hydrogen bonding between oxygen and hydroxyl groups or ion exchange [51]. The HSS CN stationary phase is not end capped; thus, the silanols are accessible to the eluted compounds and the CN group favors the retention of polar compounds due to dipole interactions. The importance of this descriptor could reflect the involvement of H bond interactions in the transplacental passage, which was highlighted in several QSAR studies [21,22,23]. Moreover, the predictive capacity of the model obtained with the multilinear regression and based on these chromatographic descriptors alone has been verified by an extensive internal validation procedure. Therefore, chromatographic descriptors appear to be more relevant than molecular descriptors alone or in combination with chromatographic descriptors for predicting the passage of BPs across the placental barrier [33,52,53].
The main limitation of this study is the restricted number of BPs included in the models, which did not allow for an external validation, i.e., predicting the CI of BPs not included in the QSAR development process. Consequently, leave-many-out validation was performed with eleven BPs in the training set and four in the test set, and this validation was repeated three times with three training and test sets. Moreover, unlike what is usually practiced in QSAR modelling, our training sets contained five repeatedly measured CI for each bisphenol, and it was previously shown that the incorporation of experimental errors improves the predictive ability of the QSAR model [31]. Moreover, it is important to consider that BPs with a high leverage value (as BPS and BPFL) must be included in the training set to reinforce the model, and their prediction should be considered unreliable [32,54]. Indeed, whatever the QSAR model may be, BPS and BPFL have a strong influence on the regression, which explains the poor prediction of their CI when one of them was not included in the training dataset. Interestingly, the placental CI of these two BPs were significantly lower than those of the other BPs [17], and they are structurally different in terms of lipophilicity for BPS and thermodynamic for BPFL. Regarding their chromatographic behavior, BPS and BPFL showed inverse asymmetric parameters, especially on the HSS PFP column with MeOH elution, reflecting the involvement of both H-bonds and π-π interactions. In the same way, BP44, which has a significant influence on the prediction of placental passage (high leverage) in the QSAR model based on chromatographic descriptors, was excluded for prediction even if its CI could be predicted with good accuracy using QSAR based only on molecular descriptors. Conversely, BPAF could not be included in the test set in the QSAR model based only on molecular descriptors because of the great difference in its thermodynamic properties, which is related to its fluorine atoms, whereas its CI could be predicted with the QSAR model based on chromatographic descriptors. Thus, the inclusion of new compounds containing more heteroatoms in the model would extend the range of values of molecular and chromatographic descriptors, and thus improve the ability of the QSAR model to predict the CL of BPs [55].
Further investigations including the quantitative structure-retention relationship studies (QSRR) are required to link the molecular properties and the chromatographic behavior of these BPs [44,56] and, thus, to better understand the interactions involved in the placental passage of BPs.
4. Materials and Methods
4.1. Data Set
4.1.1. Compounds
Bisphenol S (BPS) (purity ≥ 98%), Bisphenol A (BPA) (purity ≥ 99%), Bisphenol E (BPE) (purity ≥ 98%), 2,2-Bis(4-hydroxy-3-methylphenyl)propane (3-3BPA) (purity ≥ 97%), Bisphenol B (BPB) (purity ≥ 98%), Bis(4-hydroxyphenyl)-2,2-dichloroethylene (BPC) (purity ≥ 98%), Bisphenol BP (BPBP)(purity ≥ 98%), Bisphenol F (BPF) (purity ≥ 98%), Bisphenol FL (BPFL) (purity ≥ 97%), Bisphenol Z (BPZ) (purity ≥ 98%), 4,4′-Dihydroxybiphenyl (BP4-4) (purity ≥ 97%), Bisphenol AP (BPAP) (purity ≥ 99%), Bisphenol AF (BPAF) (purity ≥ 97%), Bisphenol P (BPP) (purity ≥ 98%), Bisphenol M (BPM) (purity ≥ 99%) were purchased from Sigma-Aldrich (Saint Louis, MO, USA).
4.1.2. Molecular Descriptors
The 3D structures of these bisphenols were minimized by MMFF94 (Merck Molecular Force Field) using a semi-empirical molecular orbital package, MOPAC application, in Chem 3D Ultra software (V17.1, Perkin Elmer informatics). An RMS gradient of 0.10 was used to minimize energy with a maximum of 1000 iterations. Electrostatic (such as dipole moment, HOMO and LUMO energy, etc.), thermodynamic (such as dipole heat of formation, Gibbs free energy, etc.), steric (such as ovality, Connolly accessible area, etc.), topologic (such as polar surface area, shape attribute, etc.), and physicochemical (such as molecular weight, pKa, LogP, etc.) descriptors were calculated. The set of molecular descriptors calculated for each studied BP is detailed in the Supplementary Materials Table S1.
4.1.3. Chromatographic Descriptors
In most of the BP chromatographic studies, the BPs are eluted on a C18 column with an H2O/AcN (Acetonitrile) gradient [57,58]. Therefore, the separation of the 15 BPs was optimized using the conventional U-HPLC BEH C18 stationary phase with acetonitrile as the organic modifier (Figure 2A). Next, the same solvent percentage, flow rate, and time were applied to thirteen chromatographic columns (Table 5), either with AcN or with MeOH (methanol), in order to compare the retention and peak shape of 15 BPs in all of these chromatographic conditions. The chromatographic descriptors were calculated from the chromatographic parameters determined by the analysis of the chromatogram of each BP and injected alone in the aqueous gradient mode on 13 analytical stationary phases listed in Table 5, using two solvents as the organic modifier: methanol and acetonitrile.

Table 5.
Analytical columns used for determining the chromatographic descriptors with their abbreviation, dimension, and selectivity [48,59].
Each BP solution was prepared in (H2O/MeOH: 50/50) at a concentration of 0.01 mg/mL. Ten microliters of each solution were injected on an Acquity UPLC system with UV detection set at 210 nm (Waters, Milford, MA, USA). The column temperature was set at 40 °C, and the BPs were eluted at 0.3 mL/min using the linear gradient mode with two organic solvents (MeOH or AcN) and water: pump A (H2O)/pump B (MeOH or AcN): t(0→10 min) 84% A→5% A; t(10→15 min) 5% A. The retention time, peak asymmetry (4.4%), tailing factor, and peak width (5%) were determined with Empower Suitability System software (Waters®). A specific retention factor k′ vs. BPA (Equation (4)) relative to BPA was calculated by Equation (4) to normalize all retentions:
k′ vs. BPA = (trBP − trBPA)/trBPA
Descriptors corresponding to peak asymmetry, tailing factor, peak width, and retention factor were identified as #7, #8, #2, and #5, respectively (Supplementary Materials Table S2).
4.2. QSAR Modelling
4.2.1. Variable Selection and Multi-Linear Regression
QSAR models were performed on RStudio software (Version 1.2.5001). The descriptor values were centered and scaled to the unit standard deviation of each descriptor. An artificial neural network (ANN) was created using the R package neural net (version: 1.44.2) with a sigmoid activation function and a backpropagation learning algorithm (code in Supplementary Materials). The ANNs were adjusted by assigning a weight to each descriptor to minimize error between predicted and experimental clearance indices using a sigmoid activation function and backpropagation learning algorithm [60]. The ANN was structured with an input layer connected to each descriptor and an output layer linked to the clearance indices (CI) for each BP. The stepmax and threshold were set at 100,000 and 0.005, respectively. After the learning cycles, descriptors with a weight (absolute value) greater than twice the median of all descriptor weights were considered significant. To avoid collinearities between descriptors, a correlation matrix was calculated with the significant descriptors. When a correlation (R2 greater than 0.5) between two descriptors was observed, two new matrices were created, one containing the first descriptor and the other containing the other descriptor. This operation was carried out until a matrix containing uncorrelated descriptors was obtained; these descriptors were then used to build the final QSAR model. A multilinear regression (MLR) was then performed between the selected descriptors and CI for each QSAR model using Rstudio software. The regression was tested with three weightings: 1, 1/Y, and 1/Y² with Y = observed clearance index, and residuals and leverages were plotted to evaluate, by visual inspection, the quality of the regression and to choose the weighting. The models with R-squared greater than 0.7 were selected for further development. For these models, if the descriptors were not significant (p < 0.05), sub-models were built by subtracting the non-significant descriptors one by one, and multilinear regressions were performed until all descriptors were significant. When too many descriptors were not significant to build the sub-models manually, the stepwise method was used to obtain the best model simplification by Akaike information criterion (AIC) [61]. The goodness of fit of each model was compared using the Bayesan information criterion calculated from each of the multilinear regressions with the RStudio software.
4.2.2. Data Splitting
Clearance indices for fifteen BPs were determined on five placentae, corresponding to a total of 75 values. This data set was divided into a training set (55 values, 11 BPs used for model development) and a test set of 20 values corresponding to 4 BPs randomly selected for model predictive assessment. Three prediction data sets were constructed to allow a balanced distribution of the different structures of the BPs. Each BP was allocated to one test set except BPs with high leverage in the multilinear regression model [54,62], i.e., BPS and BPFL for the model using molecular and chromatographic descriptors, BPAF for the model using only molecular descriptors, and BP44 for the model using only chromatographic descriptors. Consequently, data set 1 included BP44, BPB, 3-3BPA, and BPZ in the test set for the validation of the model based on molecular descriptors, and data set 1 included BPAF, BPB, 3-3BPA, and BPZ for the models based on chromatographic descriptors or on both types of descriptors. Data set 2 included BPP, BPM, BPAP, and BPBP in the test set, and data set 3 included BPE, BPA, BPF, and BPC in the test set. The training set was used for multilinear regression, and the corresponding test set was used for the predictive ability validation for the three data sets separately.
4.2.3. Validation
Multilinear regression models were validated with the three data sets. For each data set, the training set was used to configure regression and for cross-validation. The CI of the four BPs included in the test set were predicted by data set regression. Test set predictions were used for leave-many-out validation (LMO) according to the criteria described for external validation [33,53,55,63,64]. Multilinear regression was estimated by R2 (coefficient of regression determination, Equation (5)) and the root mean square error of calibration (RMSEC, Equation (6)). The robustness of the QSAR model was validated by the determination of Q2 (cross-validated correlation coefficient, Equation (7)) and the root mean square error of cross validation (RMSECV, Equation (8)). The predictive quality of the model was estimated with Q2LMO (Equation (9)): the correlation coefficient of LMO validation, the root mean square error of prediction (RMSEP, Equation (10)), the correlation coefficient of concordance (CCC, Equation (11)), and the Pearson squared correlation coefficient between predicted and experimental values. Finally, the Spearman’s correlation squared coefficient was determined to assess the model’s ranking capability [63]. The accepted criteria are R² > 0.65, Q2 > 0.5, QLMO² > 0.65 [52,53], and CCC > 0.85 [33]. Additionally, RMSEC, RMSECV, and RMSEP should be close and as low as possible [53].
where y(i): experimental data values of the training set, ŷc(i): multilinear-regression predicted values, ӯ: average of the experimental values of the training set, ŷv(i): cross-validation predicted values, ye(i): experimental data values of the test set, ŷe(i): test predicted values, ӯe: average of the experimental values of the test set, : average of the predicted values of the LMO validation, I: number of samples in the training set, A: number of descriptors, nEXT: number of descriptors in the test set, nTR: number of descriptors in the training set.
5. Conclusions
In this study, three QSAR models based either on molecular or chromatographic descriptors alone or a combination of molecular and chromatographic descriptors were developed, and their ability to predict the placental transfer of BPs was compared. The model based on chromatographic descriptors alone showed a better predictive ability. To the best of our knowledge, this study is the first to implement new sets of chromatographic peak shape parameters in addition to the classical retention factors in QSAR modelling. These parameters allow a better prediction of placental transfer of BPs than molecular descriptors. This model may be a useful tool for a rapid screening and subsequent removal of endocrine-active emerging BPs with a high fetal exposure potential. Further investigation by means of QSRR is needed to establish a relationship between the chromatographic behaviors of BPs in the diverse separation systems and the chemical structure to better understand the interactions involved in the placental passage of BPs. Moreover, this new concept deserves to be applied to other larger molecular families as well as to other physiological processes, such as absorption or metabolism.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28020500/s1, Table S1: Molecular descriptors used in QSAR development. Table S2: Chromatographic descriptors used in QSAR development. Table S3: Predicted clearance indices for the 15 bisphenols determined using three QSAR models based on either molecular descriptors, a combination of molecular and chromatographic descriptors or chromatographic descriptors. Table S4: Molecular descriptors significantly describing the chromatographic descriptors selected in QSAR models based either on chromatographic parameters alone or both chromatographic and molecular.
Author Contributions
C.A.G.: Conceptualization, Methodology, Formal analysis, Investigation, Writing—Original draft preparation. M.Z.L.: Conceptualization, Methodology, Investigation, Writing—Original draft preparation, Supervision. M.C.: Investigation, Formal analysis. J.-C.G.: Investigation, Methodology. N.P.-H.: Conceptualization, Investigation, Supervision, Draft Reviewing. V.G.: Conceptualization, Investigation, Draft Reviewing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Molecular and chromatographic descriptors for the 15 bisphenols are described in the Excel file Supplementary Materials.
Acknowledgments
The authors thank A. Huesca and F. Ramon-Portugal for their assistance and involvement in the model constructions.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Sample Availability
Samples of the BPs compounds are available from the authors.
References
- Chen, D.; Kannan, K.; Tan, H.; Zheng, Z.; Feng, Y.-L.; Wu, Y.; Widelka, M. Bisphenol Analogues Other Than BPA: Environmental Occurrence, Human Exposure, and Toxicity—A Review. Environ. Sci. Technol. 2016, 50, 5438–5453. [Google Scholar] [CrossRef] [PubMed]
- González, N.; Marquès, M.; Cunha, S.C.; Fernandes, J.O.; Domingo, J.L.; Nadal, M. Biomonitoring of Co-Exposure to Bisphenols by Consumers of Canned Foodstuffs. Environ. Int. 2020, 140, 105760. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Kannan, K. A Survey of Alkylphenols, Bisphenols, and Triclosan in Personal Care Products from China and the United States. Arch. Environ. Contam. Toxicol. 2014, 67, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Karrer, C.; Andreassen, M.; von Goetz, N.; Sonnet, F.; Sakhi, A.K.; Hungerbühler, K.; Dirven, H.; Husøy, T. The EuroMix Human Biomonitoring Study: Source-to-Dose Modeling of Cumulative and Aggregate Exposure for the Bisphenols BPA, BPS, and BPF and Comparison with Measured Urinary Levels. Environ. Int. 2020, 136, 105397. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Quan, Q.; Zhang, M.; Zhang, N.; Zhang, W.; Zhan, M.; Xu, W.; Lu, L.; Fan, J.; Wang, Q. Occurrence of Bisphenol A and Its Alternatives in Paired Urine and Indoor Dust from Chinese University Students: Implications for Human Exposure. Chemosphere 2020, 247, 125987. [Google Scholar] [CrossRef]
- Le Fol, V.; Aït-Aïssa, S.; Sonavane, M.; Porcher, J.-M.; Balaguer, P.; Cravedi, J.-P.; Zalko, D.; Brion, F. In Vitro and in Vivo Estrogenic Activity of BPA, BPF and BPS in Zebrafish-Specific Assays. Ecotoxicol. Environ. Saf. 2017, 142, 150–156. [Google Scholar] [CrossRef]
- Moreman, J.; Lee, O.; Trznadel, M.; David, A.; Kudoh, T.; Tyler, C.R. Acute Toxicity, Teratogenic, and Estrogenic Effects of Bisphenol A and Its Alternative Replacements Bisphenol S, Bisphenol F, and Bisphenol AF in Zebrafish Embryo-Larvae. Environ. Sci. Technol. 2017, 51, 12796–12805. [Google Scholar] [CrossRef]
- Pelch, K.; Wignall, J.A.; Goldstone, A.E.; Ross, P.K.; Blain, R.B.; Shapiro, A.J.; Holmgren, S.D.; Hsieh, J.-H.; Svoboda, D.; Auerbach, S.S.; et al. A Scoping Review of the Health and Toxicological Activity of Bisphenol A (BPA) Structural Analogues and Functional Alternatives. Toxicology 2019, 424, 152235. [Google Scholar] [CrossRef]
- Rosenmai, A.K.; Dybdahl, M.; Pedersen, M.; van Vugt-Lussenburg, B.M.A.; Wedebye, E.B.; Taxvig, C.; Vinggaard, A.M. Are Structural Analogues to Bisphenol A Safe Alternatives? Toxicol. Sci. 2014, 139, 35–47. [Google Scholar] [CrossRef]
- Siracusa, J.S.; Yin, L.; Measel, E.; Liang, S.; Yu, X. Effects of Bisphenol A and Its Analogs on Reproductive Health: A Mini Review. Reprod. Toxicol. 2018, 79, 96–123. [Google Scholar] [CrossRef]
- Kaimal, A.; Al Mansi, M.H.; Dagher, J.B.; Pope, C.; Varghese, M.G.; Rudi, T.B.; Almond, A.E.; Cagle, L.A.; Beyene, H.K.; Bradford, W.T.; et al. Prenatal Exposure to Bisphenols Affects Pregnancy Outcomes and Offspring Development in Rats. Chemosphere 2021, 276, 130118. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Sekulovski, N.; MacLean, J.A.; Whorton, A.; Hayashi, K. Prenatal Exposure to Bisphenol A Analogues on Female Reproductive Functions in Mice. Toxicol. Sci. 2019, 168, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Tucker, D.K.; Hayes Bouknight, S.; Brar, S.S.; Kissling, G.E.; Fenton, S.E. Evaluation of Prenatal Exposure to Bisphenol Analogues on Development and Long-Term Health of the Mammary Gland in Female Mice. Environ. Health Perspect. 2018, 126, 087003. [Google Scholar] [CrossRef] [PubMed]
- Corbel, T.; Gayrard, V.; Puel, S.; Lacroix, M.Z.; Berrebi, A.; Gil, S.; Viguié, C.; Toutain, P.-L.; Picard-Hagen, N. Bidirectional Placental Transfer of Bisphenol A and Its Main Metabolite, Bisphenol A-Glucuronide, in the Isolated Perfused Human Placenta. Reprod. Toxicol. 2014, 47, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Gingrich, J.; Pu, Y.; Ehrhardt, R.; Karthikraj, R.; Kannan, K.; Veiga-Lopez, A. Toxicokinetics of Bisphenol A, Bisphenol S, and Bisphenol F in a Pregnancy Sheep Model. Chemosphere 2019, 220, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Grandin, F.C.; Lacroix, M.Z.; Gayrard, V.; Gauderat, G.; Mila, H.; Toutain, P.-L.; Picard-Hagen, N. Bisphenol S Instead of Bisphenol A: Toxicokinetic Investigations in the Ovine Materno-Feto-Placental Unit. Environ. Int. 2018, 120, 584–592. [Google Scholar] [CrossRef]
- Gély, C.A.; Lacroix, M.Z.; Morin, M.; Vayssière, C.; Gayrard, V.; Picard-Hagen, N. Comparison of the Materno-Fetal Transfer of Fifteen Structurally Related Bisphenol Analogues Using an Ex Vivo Human Placental Perfusion Model. Chemosphere 2021, 276, 130213. [Google Scholar] [CrossRef]
- Grandin, F.C.; Lacroix, M.Z.; Gayrard, V.; Viguié, C.; Mila, H.; de Place, A.; Vayssière, C.; Morin, M.; Corbett, J.; Gayrard, C.; et al. Is Bisphenol S a Safer Alternative to Bisphenol A in Terms of Potential Fetal Exposure? Placental Transfer across the Perfused Human Placenta. Chemosphere 2019, 221, 471–478. [Google Scholar] [CrossRef]
- Pacifici, G.M.; Nottoli, R. Placental Transfer of Drugs Administered to the Mother. Clin. Pharmacokinet. 1995, 28, 235–269. [Google Scholar] [CrossRef]
- Eguchi, A.; Hanazato, M.; Suzuki, N.; Matsuno, Y.; Todaka, E.; Mori, C. Maternal–Fetal Transfer Rates of PCBs, OCPs, PBDEs, and Dioxin-like Compounds Predicted through Quantitative Structure–Activity Relationship Modeling. Environ. Sci. Pollut. Res. 2018, 25, 7212–7222. [Google Scholar] [CrossRef]
- Giaginis, C.; Zira, A.; Theocharis, S.; Tsantili-Kakoulidou, A. Application of Quantitative Structure–Activity Relationships for Modeling Drug and Chemical Transport across the Human Placenta Barrier: A Multivariate Data Analysis Approach. J. Appl. Toxicol. 2009, 29, 724–733. [Google Scholar] [CrossRef]
- Hewitt, M.; Madden, J.C.; Rowe, P.H.; Cronin, M.T.D. Structure-Based Modelling in Reproductive Toxicology: (Q)SARs for the Placental Barrier. SAR QSAR Environ. Res. 2007, 18, 57–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-H.; Xia, Z.-N.; Yan, L.; Liu, S.-S. Prediction of Placental Barrier Permeability: A Model Based on Partial Least Squares Variable Selection Procedure. Molecules 2015, 20, 8270–8286. [Google Scholar] [CrossRef] [PubMed]
- Danishuddin; Khan, A.U. Descriptors and Their Selection Methods in QSAR Analysis: Paradigm for Drug Design. Drug Discov. Today 2016, 21, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Kaliszan, R. QSRR: Quantitative Structure-(Chromatographic) Retention Relationships. Chem. Rev. 2007, 107, 3212–3246. [Google Scholar] [CrossRef]
- Apostolov, S.; Vastag, G.; Mrdjan, G.; Nakomčić, J.; Stojiljković, I. Chromatographic Descriptors in QSAR Study of Barbiturates. J. Liq. Chromatogr. Relat. Technol. 2019, 42, 194–203. [Google Scholar] [CrossRef]
- Deconinck, E.; Ates, H.; Callebaut, N.; Van Gyseghem, E.; Vander Heyden, Y. Evaluation of Chromatographic Descriptors for the Prediction of Gastro-Intestinal Absorption of Drugs. J. Chromatogr. A 2007, 1138, 190–202. [Google Scholar] [CrossRef]
- Jevrić, L.R.; Podunavac-Kuzmanović, S.O.; Švarc-Gajić, J.V.; Kovačević, S.Z.; Jovanović, B.Ž. RP-HPTLC Retention Data in Correlation with the In-Silico ADME Properties of a Series of s-Triazine Derivatives. Iran. J. Pharm. Res. 2014, 13, 1203–1211. [Google Scholar]
- Koba, M.; Bączek, T.; Marszałł, M.P. Importance of Retention Data from Affinity and Reverse-Phase High-Performance Liquid Chromatography on Antitumor Activity Prediction of Imidazoacridinones Using QSAR Strategy. J. Pharm. Biomed. Anal. 2012, 64–65, 87–93. [Google Scholar] [CrossRef]
- Perisic-Janjic, N.; Kaliszan, R.; Wiczling, P.; Milosevic, N.; Uscumlic, G.; Banjac, N. Reversed-Phase TLC and HPLC Retention Data in Correlation Studies with in Silico Molecular Descriptors and Druglikeness Properties of Newly Synthesized Anticonvulsant Succinimide Derivatives. Mol. Pharm. 2011, 8, 555–563. [Google Scholar] [CrossRef]
- Wenlock, M.C.; Carlsson, L.A. How Experimental Errors Influence Drug Metabolism and Pharmacokinetic QSAR/QSPR Models. J. Chem. Inf. Model. 2015, 55, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Golbraikh, A.; Tropsha, A. Predictive QSAR Modeling Based on Diversity Sampling of Experimental Datasets for the Training and Test Set Selection. J. Comput. Aided Mol. Des. 2002, 16, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Chirico, N.; Gramatica, P. Real External Predictivity of QSAR Models. Part 2. New Intercomparable Thresholds for Different Validation Criteria and the Need for Scatter Plot Inspection. J. Chem. Inf. Model. 2012, 52, 2044–2058. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Casero, N.; Lunar, L.; Rubio, S. Analytical Methods for the Determination of Mixtures of Bisphenols and Derivatives in Human and Environmental Exposure Sources and Biological Fluids. A Review. Anal. Chim. Acta 2016, 908, 22–53. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Kannan, K. Concentrations and Profiles of Bisphenol A and Other Bisphenol Analogues in Foodstuffs from the United States and Their Implications for Human Exposure. J. Agric. Food Chem. 2013, 61, 4655–4662. [Google Scholar] [CrossRef] [PubMed]
- European Chemical Agency ECHA-Information on Registered Substances. Available online: https://echa.europa.eu/fr/information-on-chemicals/registered-substances (accessed on 30 November 2022).
- Audus, K.L. Controlling Drug Delivery across the Placenta. Eur. J. Pharm. Sci. 1999, 8, 161–165. [Google Scholar] [CrossRef][Green Version]
- Evain-Brion, D.; Berveiller, P.; Gil, S. Le passage transplacentaire des médicaments. Therapies 2014, 69, 3–11. [Google Scholar] [CrossRef]
- Syme, M.R.; Paxton, J.W.; Keelan, J.A. Drug Transfer and Metabolism by the Human Placenta. Clin. Pharmacokinet. 2004, 43, 487–514. [Google Scholar] [CrossRef]
- Gao, H.; Huang, H.; Zheng, A.; Yu, N.; Li, N. Determination of Quantitative Retention-Activity Relationships between Pharmacokinetic Parameters and Biological Effectiveness Fingerprints of Salvia Miltiorrhiza Constituents Using Biopartitioning and Microemulsion High-Performance Liquid Chromatography. J. Chromatogr. B 2017, 1067, 10–17. [Google Scholar] [CrossRef]
- Giaginis, C.; Tsantili-Kakoulidou, A. Quantitative Structure–Retention Relationships as Useful Tool to Characterize Chromatographic Systems and Their Potential to Simulate Biological Processes. Chromatographia 2013, 76, 211–226. [Google Scholar] [CrossRef]
- Quiñones-Torrelo, C.; Sagrado, S.; Villanueva-Camañas, R.M.; Medina-Hernández, M.J. Opioid Analgetics Retention-Pharmacologic Activity Models Using Biopartitioning Micellar Chromatography. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 766, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Lesellier, E.; West, C. Σpider Diagram: A Universal and Versatile Approach for System Comparison and Classification. Part 2: Stationary Phase Properties. J. Chromatogr. A 2018, 1574, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Lafossas, C.; Benoit-Marquié, F.; Garrigues, J.C. Analysis of the Retention of Tetracyclines on Reversed-Phase Columns: Chemometrics, Design of Experiments and Quantitative Structure-Property Relationship (QSPR) Study for Interpretation and Optimization. Talanta 2019, 198, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Fazio, S.; Munch, D.; Drumm, P. Impact of Methanol and Acetonitrile on Separations Based on π–π Interactions with a Reversed-Phase Phenyl Column. J. Chromatogr. A 2005, 1097, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Le Mapihan, K.; Vial, J.; Jardy, A. Reversed-Phase Liquid Chromatography Column Testing and Classification: Physicochemical Interpretation Based on a Wide Set of Stationary Phases. J. Chromatogr. A 2007, 1144, 183–196. [Google Scholar] [CrossRef]
- Ravisankar, P.; Anusha, S.; Supriya, K.; Kumar, U.A. Fundamental Chromatographic Parameters. Int. J. Pharm. Sci. Rev. Res. 2020, 55, 46. [Google Scholar]
- Waters Acquity UPLC Colums. Available online: https://www.waters.com/webassets/cms/library/docs/720001140en.pdf (accessed on 9 February 2022).
- Todeschini, R.; Consonni, V. Methods and Principles in Medicinal Chemistry. In Handbook of Molecular Descriptors, 1st ed.; WILEY-VCH Verlag GmbH: Weinheim, Germany, 2000; ISBN 978-3-527-29913-3. [Google Scholar]
- Miyabe, K.; Guiochon, G. Measurement of the Parameters of the Mass Transfer Kinetics in High Performance Liquid Chromatography. J. Sep. Sci. 2003, 26, 155–173. [Google Scholar] [CrossRef]
- Nawrocki, J. The Silanol Group and Its Role in Liquid Chromatography. J. Chromatogr. A 1997, 779, 29–71. [Google Scholar] [CrossRef]
- Chirico, N.; Gramatica, P. Real External Predictivity of QSAR Models: How to Evaluate It? Comparison of Different Validation Criteria and Proposal of Using the Concordance Correlation Coefficient. J. Chem. Inf. Model. 2011, 51, 2320–2335. [Google Scholar] [CrossRef]
- Gramatica, P.; Sangion, A. A Historical Excursus on the Statistical Validation Parameters for QSAR Models: A Clarification Concerning Metrics and Terminology. J. Chem. Inf. Model. 2016, 56, 1127–1131. [Google Scholar] [CrossRef]
- Gramatica, P. Principles of QSAR Models Validation: Internal and External. QSAR Comb. Sci. 2007, 26, 694–701. [Google Scholar] [CrossRef]
- Tropsha, A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010, 29, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Kaliszan, R. Quantitative Structure-Retention Relationships Applied to Reversed-Phase High-Performance Liquid Chromatography. J. Chromatogr. A 1993, 656, 417–435. [Google Scholar] [CrossRef]
- Rocha, B.A.; da Costa, B.R.B.; de Albuquerque, N.C.P.; de Oliveira, A.R.M.; Souza, J.M.O.; Al-Tameemi, M.; Campiglia, A.D.; Barbosa, F., Jr. A Fast Method for Bisphenol A and Six Analogues (S, F, Z, P, AF, AP) Determination in Urine Samples Based on Dispersive Liquid-Liquid Microextraction and Liquid Chromatography-Tandem Mass Spectrometry. Talanta 2016, 154, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Kramer, J.P.; Calafat, A.M.; Ye, X. Automated On-Line Column-Switching High Performance Liquid Chromatography Isotope Dilution Tandem Mass Spectrometry Method for the Quantification of Bisphenol A, Bisphenol F, Bisphenol S, and 11 Other Phenols in Urine. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 944, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Restek Raptor Stationary Phase: Biphenyl. Available online: https://www.restek.com/globalassets/pdfs/literature/gnbr1891c-unv.pdf (accessed on 9 February 2022).
- Günther, F.; Fritsch, S. Neuralnet: Training of Neural Networks. R J. 2010, 2, 30–38. [Google Scholar] [CrossRef]
- Ripley, B.; Venables, B.; Bates, D.M.; Hornik, K.; Gebhardt, A.; Firth, D. R Package “Mass” 7.3-54. Available online: https://rdocumentation.org/packages/MASS/versions/7.3-54 (accessed on 30 November 2022).
- Hebbali, A. Olsrr|Rsquared Academy. Available online: https://olsrr.rsquaredacademy.com/ (accessed on 30 November 2022).
- Alexander, D.L.J.; Tropsha, A.; Winkler, D.A. Beware of R(2): Simple, Unambiguous Assessment of the Prediction Accuracy of QSAR and QSPR Models. J. Chem. Inf. Model. 2015, 55, 1316–1322. [Google Scholar] [CrossRef]
- Ferreira, M.M.C. Multivariate QSAR. J. Braz. Chem. Soc. 2002, 13, 742–753. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).