Revealing the Control Mechanisms of pH on the Solution Properties of Chitin via Single-Molecule Studies
Abstract
:1. Introduction
2. Results and Discussion
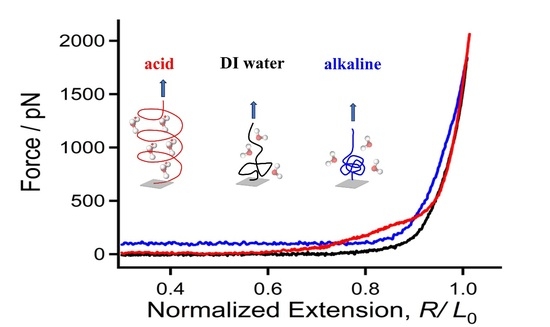
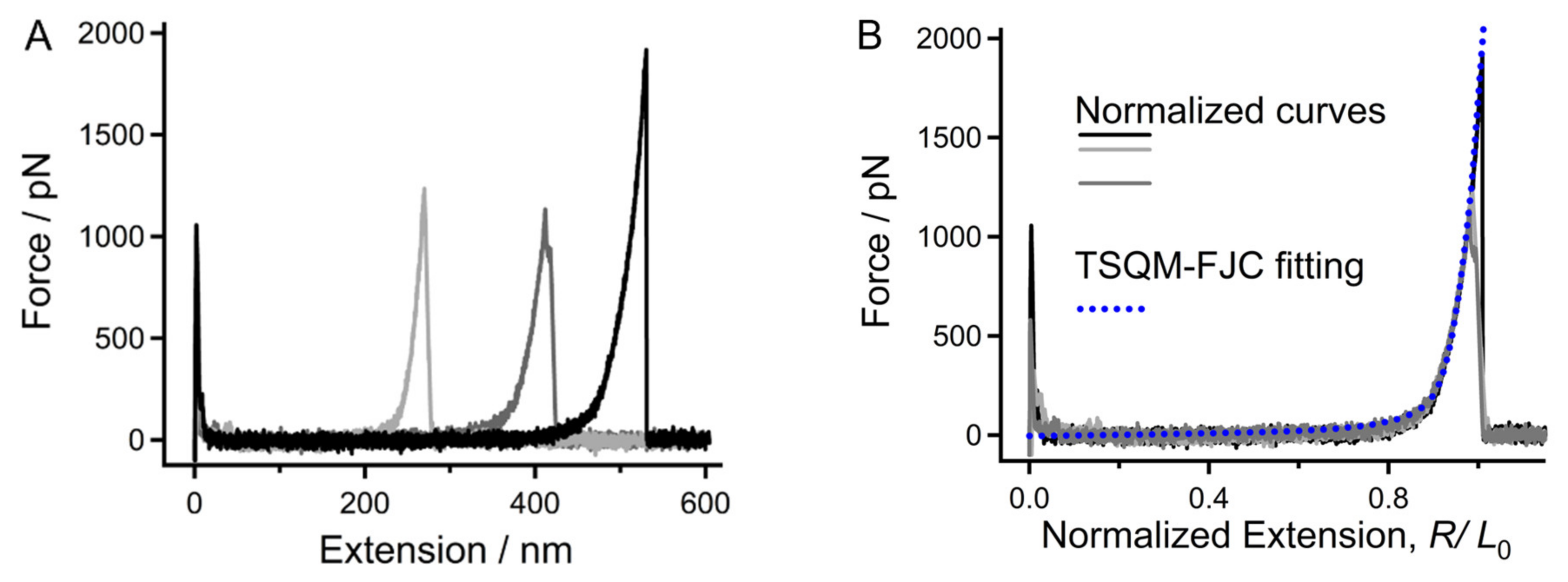
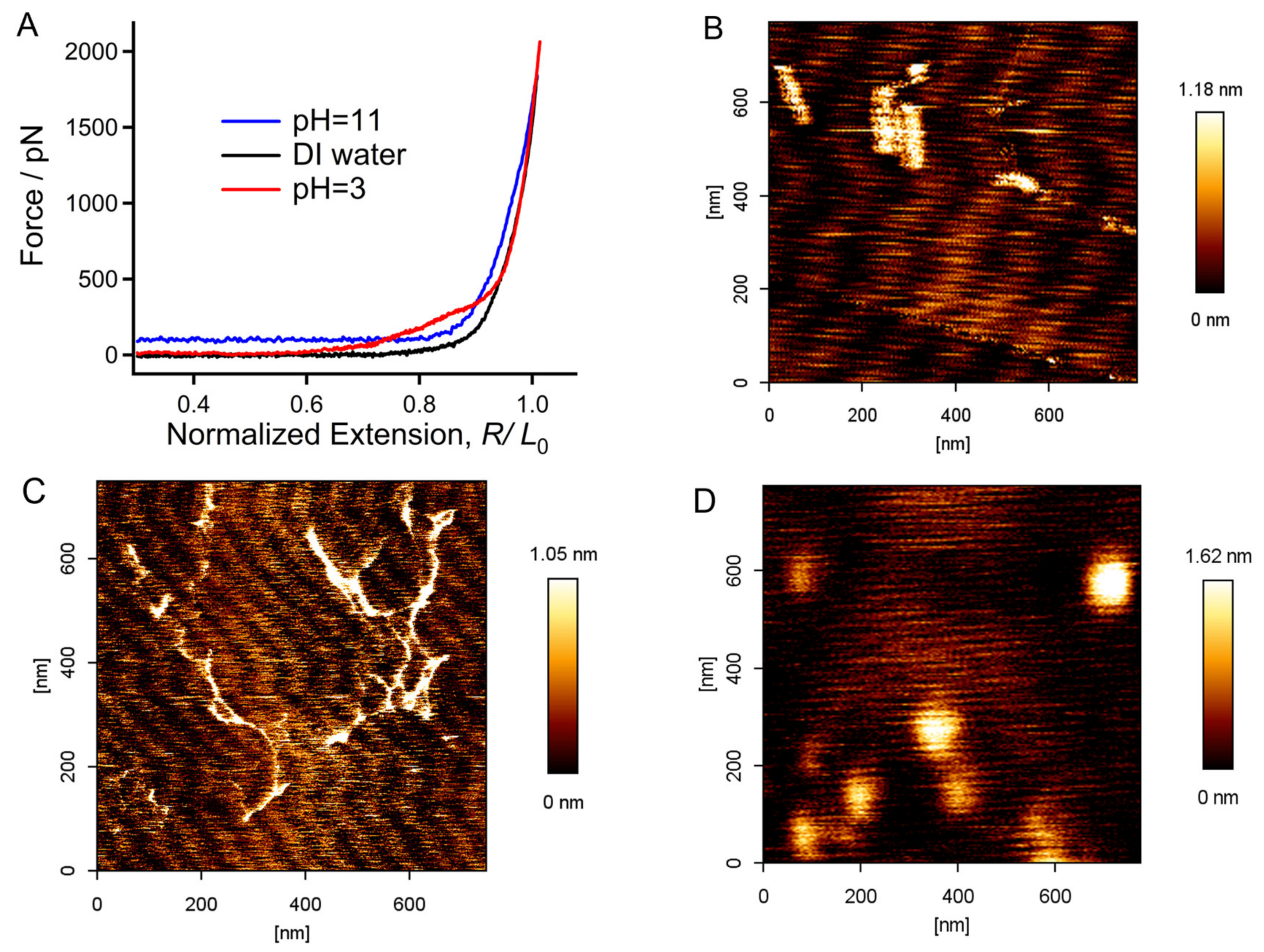
2.1. Molecular Mechanical Properties of Chitin in DI Water
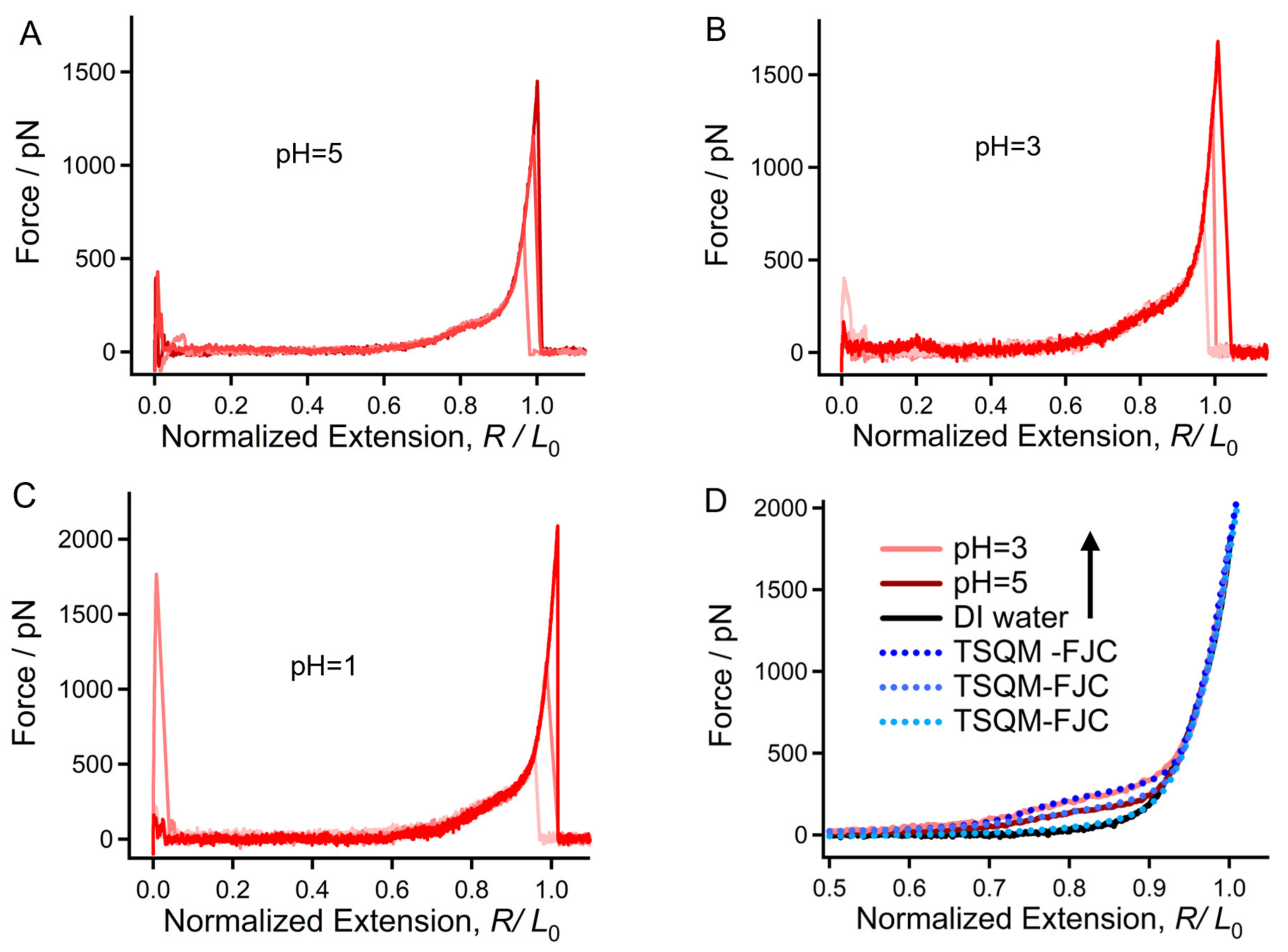
2.2. Surprising Enhancement of Single-Molecule Elasticity Due to Strong Hydratability in Acid
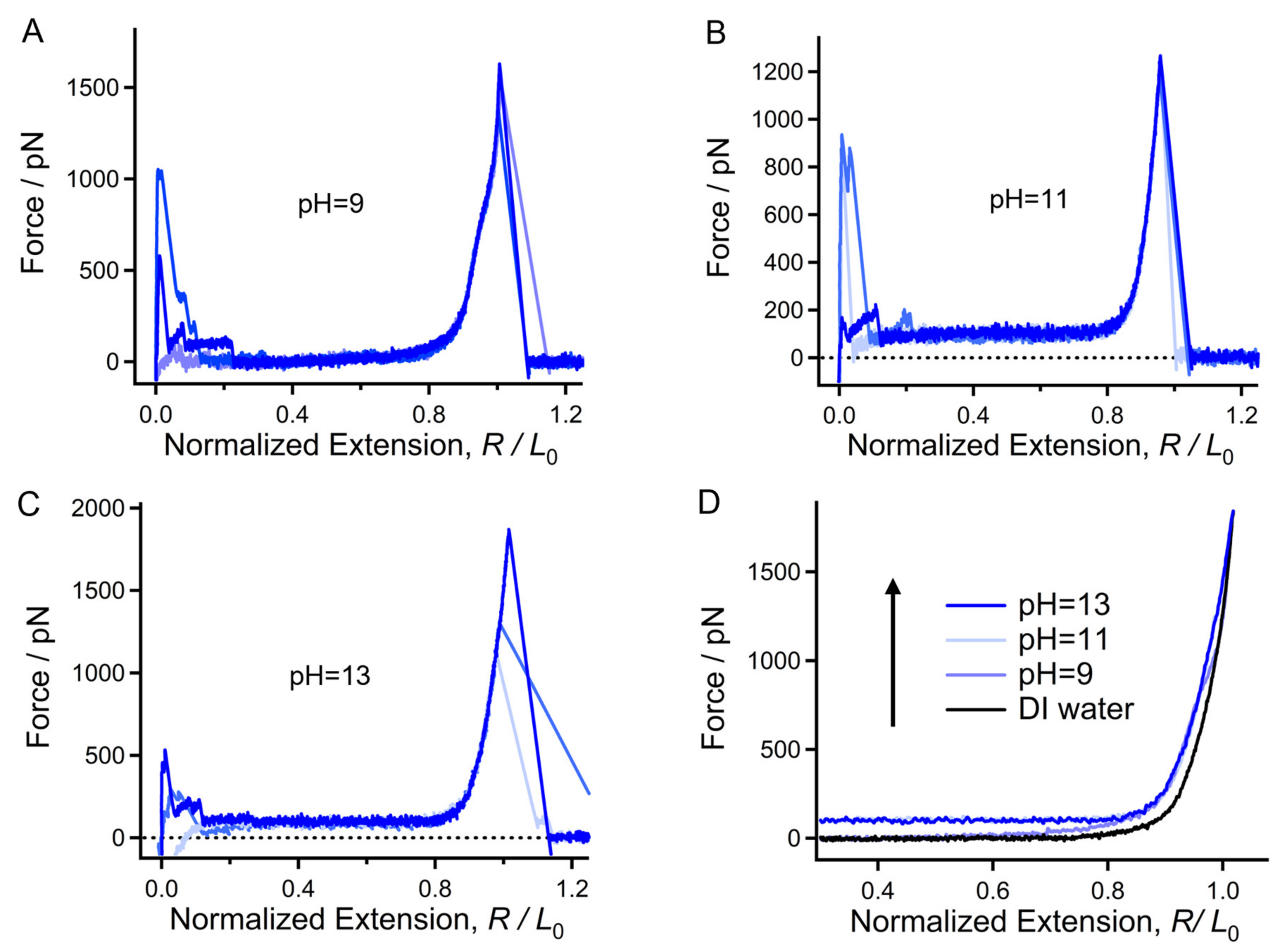
2.3. Mechanical Properties of Chitin Chains in Alkaline Environments
2.4. Origin of the Mechanical Deviation of Chitin in Acid, DI Water, and Alkali Medium in Relation to the Aspect of Molecular Morphology
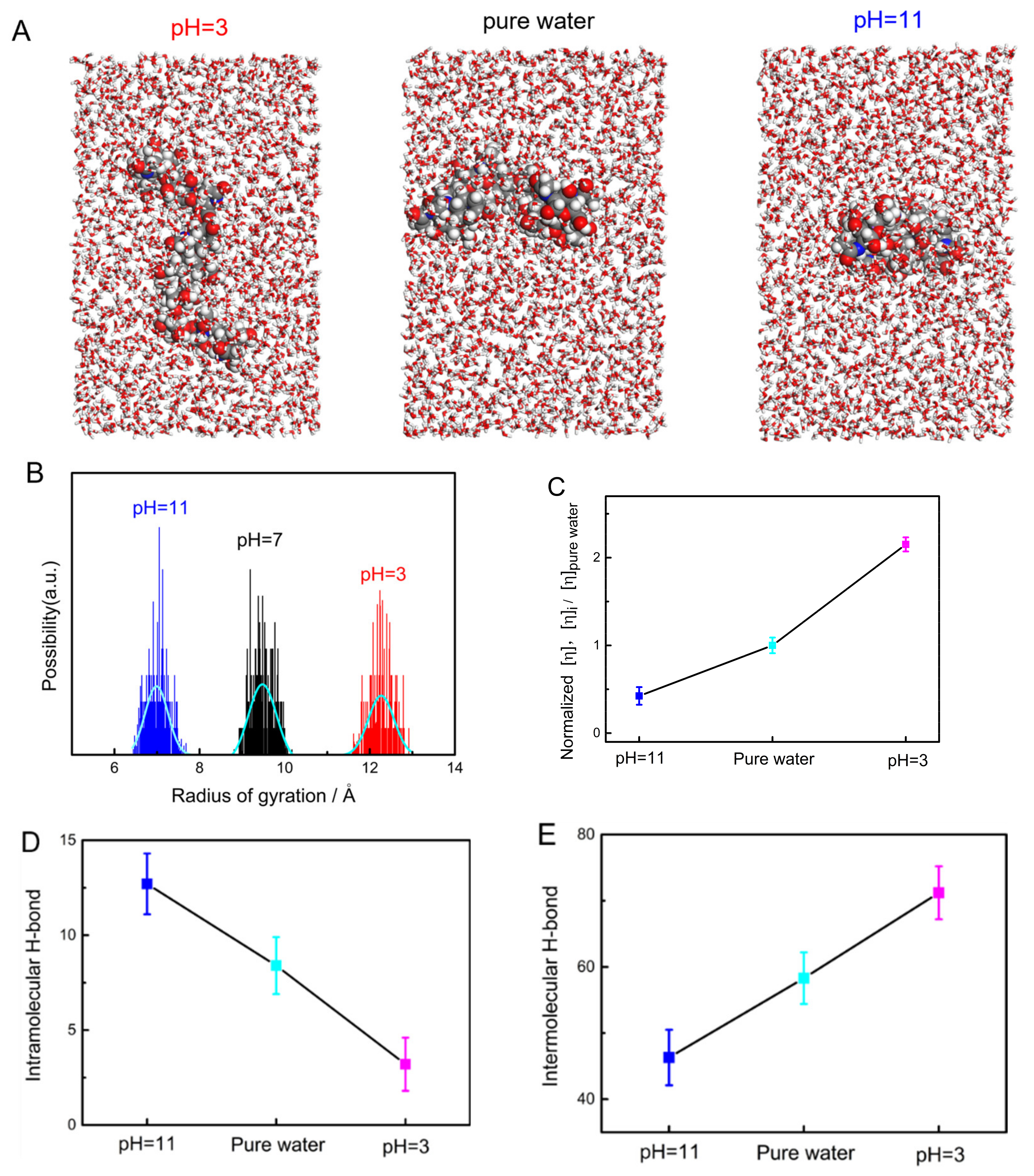
2.5. Analysis of the Single-Molecule Physical Properties of Chitin via MD Simulations
2.6. The Intramolecular and Intermolecular H-Bonds of a Chitin Chain in Solution
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Details of SMFS
3.3. Details of AFM Imaging
3.4. Details of MD Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Chen, C.; Li, D.; Yano, H.; Abe, K. Insect cuticle-mimetic hydrogels with high mechanical properties achieved via the combination of chitin nanofiber and gelatin. J. Agric. Food Chem. 2019, 67, 5571–5578. [Google Scholar] [CrossRef]
- Ma, M.; Zhong, Y.; Jiang, X. Thermosensitive and pH-responsive tannin-containing hydroxypropyl chitin hydrogel with long-lasting antibacterial activity for wound healing. Carbohydr. Polym. 2020, 236, 116096. [Google Scholar] [CrossRef]
- Cho, Y.-W.; Cho, Y.-N.; Chung, S.-H.; Yoo, G.; Ko, S.-W. Water-soluble chitin as a wound healing accelerator. Biomaterials 1999, 20, 2139–2145. [Google Scholar] [CrossRef]
- Izumi, R.; Komada, S.; Ochi, K.; Karasawa, L.; Osaki, T.; Murahata, Y.; Tsuka, T.; Imagawa, T.; Itoh, N.; Okamoto, Y. Favorable effects of superficially deacetylated chitin nanofibrils on the wound healing process. Carbohydr. Polym. 2015, 123, 461–467. [Google Scholar] [CrossRef]
- Xu, H.; Fang, Z.; Tian, W.; Wang, Y.; Ye, Q.; Zhang, L.; Cai, J. Green fabrication of amphiphilic quaternized β-chitin derivatives with excellent biocompatibility and antibacterial activities for wound healing. Adv. Mater. 2018, 30, 1801100. [Google Scholar] [CrossRef]
- Liao, J.; Hou, B.; Huang, H. Preparation, properties and drug controlled release of chitin-based hydrogels: An updated review. Carbohydr. Polym. 2022, 283, 119177. [Google Scholar] [CrossRef]
- Fan, Z.; Wang, L.; Qin, Y.; Li, P. Activity of chitin/chitosan/chitosan oligosaccharide against plant pathogenic nematodes and potential modes of application in agriculture: A review. Carbohydr. Polym. 2023, 306, 120592. [Google Scholar] [CrossRef]
- Shen, X.; Shamshina, J.L.; Berton, P.; Gurau, G.; Rogers, R.D. Hydrogels based on cellulose and chitin: Fabrication, properties, and applications. Green Chem. 2016, 18, 53–75. [Google Scholar] [CrossRef]
- Yu, Y.; Gim, S.; Kim, D.; Arnon, Z.A.; Gazit, E.; Seeberger, P.H.; Delbianco, M. Oligosaccharides self-assemble and show intrinsic optical properties. J. Am. Chem. Soc. 2019, 141, 4833–4838. [Google Scholar] [CrossRef]
- Xue, C.; You, J.; Zhang, H.; Xiong, S.; Yin, T.; Huang, Q. Capacity of myofibrillar protein to adsorb characteristic fishy-odor compounds: Effects of concentration, temperature, ionic strength, pH and yeast glucan addition. Food Chem. 2021, 363, 130304. [Google Scholar] [CrossRef]
- Walker, G.M.; Basso, T.O. Mitigating stress in industrial yeasts. Fungal Biol. 2020, 124, 387–397. [Google Scholar] [CrossRef]
- Abdel Latef, A.A.H.; Mostofa, M.G.; Rahman, M.M.; Abdel-Farid, I.B.; Tran, L.-S.P. Extracts from yeast and carrot roots enhance maize performance under seawater-induced salt stress by altering physio-biochemical characteristics of stressed plants. J. Plant Growth Regul. 2019, 38, 966–979. [Google Scholar] [CrossRef]
- Suryawanshi, N.; Eswari, J.S. Chitin from seafood waste: Particle swarm optimization and neural network study for the improved chitinase production. J. Chem. Technol. Biotechnol. 2022, 97, 509–519. [Google Scholar] [CrossRef]
- Sherrington, S.L.; Sorsby, E.; Mahtey, N.; Kumwenda, P.; Lenardon, M.D.; Brown, I.; Ballou, E.R.; MacCallum, D.M.; Hall, R.A. Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 2017, 13, e1006403. [Google Scholar] [CrossRef] [PubMed]
- Jayakumar, R.; Nair, A.; Rejinold, N.S.; Maya, S.; Nair, S. Doxorubicin-loaded pH-responsive chitin nanogels for drug delivery to cancer cells. Carbohydr. Polym. 2012, 87, 2352–2356. [Google Scholar] [CrossRef]
- Valdivieso, H.; Durán, A.; Roncero, C.J.E. Chitin synthases in yeast and fungi. Exs 1999, 87, 55–69. [Google Scholar]
- Cabib, E.; Arroyo, J. How carbohydrates sculpt cells: Chemical control of morphogenesis in the yeast cell wall. Nat. Rev. Microbiol. 2013, 11, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Nakajima, I.; Ohno, T. Development of a new method for evaluation of yeast vitality by measuring intracellular pH. J. Am. Soc. Brew. Chem. 1994, 52, 5–8. [Google Scholar] [CrossRef]
- Heilmann, C.J.; Sorgo, A.G.; Mohammadi, S.; Sosinska, G.J.; de Koster, C.G.; Brul, S.; de Koning, L.J.; Klis, F.M. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot. Cell 2013, 12, 254–264. [Google Scholar] [CrossRef]
- Jayakumar, R.; Prabaharan, M.; Nair, S.; Tokura, S.; Tamura, H.; Selvamurugan, N. Novel carboxymethyl derivatives of chitin and chitosan materials and their biomedical applications. Prog. Mater. Sci 2010, 55, 675–709. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, P.; Lv, X.; Liang, Y. Insight into the structure-function relationships of the solubility of chitin/chitosan in natural deep eutectic solvents. Mater. Today Commun. 2021, 27, 102374. [Google Scholar] [CrossRef]
- Hou, F.; He, L.; Ma, X.; Wang, D.; Ding, T.; Ye, X.; Liu, D. Ultrasound enhanced the binding ability of chitinase onto chitin: From an AFM insight. Ultrason. Sonochem. 2020, 67, 105117. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vallet, A.; Tian, H.; Rodriguez-Moreno, L.; Valkenburg, D.-J.; Saleem-Batcha, R.; Wawra, S.; Kombrink, A.; Verhage, L.; de Jonge, R.; van Esse, H.P. A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PLoS Pathog. 2020, 16, e1008652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.M.; Jaroniec, J.; Lee, G.; Marszalek, P.E. Direct detection of inter-residue hydrogen bonds in polysaccharides by single-molecule force spectroscopy. Angew. Chem. Int. Edit. 2005, 44, 2723–2727. [Google Scholar] [CrossRef] [PubMed]
- Pfitzner, E.; Wachauf, C.; Kilchherr, F.; Pelz, B.; Shih, W.M.; Rief, M.; Dietz, H. Rigid DNA beams for high-resolution single-molecule mechanics. Angew. Chem. Int. Edit. 2013, 52, 7766–7771. [Google Scholar] [CrossRef]
- Li, H.; Rief, M.; Oesterhelt, F.; Gaub, H.E.; Zhang, X.; Shen, J. Single-molecule force spectroscopy on polysaccharides by AFM–nanomechanical fingerprint of α-(1, 4)-linked polysaccharides. Chem. Phys. Lett. 1999, 305, 197–201. [Google Scholar] [CrossRef]
- Cai, W.; Bullerjahn, J.T.; Lallemang, M.; Kroy, K.; Balzer, B.N.; Hugel, T. Angle-dependent strength of a single chemical bond by stereographic force spectroscopy. Chem. Sci. 2022, 13, 5734–5740. [Google Scholar] [CrossRef]
- Cai, W.; Jäger, M.; Bullerjahn, J.T.; Hugel, T.; Wolf, S.; Balzer, B.N. Anisotropic Friction in a Ligand-Protein Complex. Nano Lett. 2023, 23, 4111–4119. [Google Scholar] [CrossRef]
- Cai, W.; Trefs, J.L.; Hugel, T.; Balzer, B.N. Anisotropy of π–π Stacking as Basis for Superlubricity. ACS Mater. Lett. 2022, 5, 172–179. [Google Scholar] [CrossRef]
- Lei, H.; Zhang, J.; Li, Y.; Wang, X.; Qin, M.; Wang, W.; Cao, Y. Histidine-specific bioconjugation for single-molecule force spectroscopy. ACS Nano 2022, 16, 15440–15449. [Google Scholar] [CrossRef]
- Milles, L.F.; Gaub, H.E. Extreme mechanical stability in protein complexes. Curr. Opin. Struct. Biol. 2020, 60, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Di, W.; Li, Y.; Huang, W.; Wang, X.; Qin, M.; Wang, W.; Cao, Y. Mg2+-Dependent High Mechanical Anisotropy of Three-Way-Junction pRNA as Revealed by Single-Molecule Force Spectroscopy. Angew. Chem. Int. Ed. 2017, 56, 9376–9380. [Google Scholar] [CrossRef]
- Müller, D.J.; Gaub, H.E. Membrane proteins scrambling through a folding landscape. Science 2017, 355, 907–908. [Google Scholar] [CrossRef] [PubMed]
- Baumann, F.; Bauer, M.S.; Milles, L.F.; Alexandrovich, A.; Gaub, H.E.; Pippig, D.A. Monovalent Strep-Tactin for strong and site-specific tethering in nanospectroscopy. Nat. Nanotechnol. 2016, 11, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Juan, M.L.; Righini, M.; Quidant, R. Plasmon nano-optical tweezers. Nat. Photonics 2011, 5, 349–356. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Luo, Z.; Cui, S. Environment-dependent single-chain mechanics of synthetic polymers and biomacromolecules by atomic force microscopy-based single-molecule force spectroscopy and the implications for advanced polymer materials. Chem. Soc. Rev. 2020, 49, 2799–2827. [Google Scholar] [CrossRef]
- Hospital, A.; Goñi, J.R.; Orozco, M.; Gelpí, J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. 2015, 19, 37–47. [Google Scholar]
- Qian, L.; Guo, X.; Zhang, K.; Yu, M. Effects of hydrogen bonds on the single-chain mechanics of chitin. Phys. Chem. Chem. 2022, 24, 24535–24541. [Google Scholar] [CrossRef]
- Yang, P.; Song, Y.; Feng, W.; Zhang, W. Unfolding of a single polymer chain from the single crystal by air-phase single-molecule force spectroscopy: Toward better force precision and more accurate description of molecular behaviors. Macromolecules 2018, 51, 7052–7060. [Google Scholar] [CrossRef]
- Zhang, T.; Mahgsoudy-Louyeh, S.; Tittmann, B.; Cosgrove, D.J. Visualization of the nanoscale pattern of recently-deposited cellulose microfibrils and matrix materials in never-dried primary walls of the onion epidermis. Cellulose 2014, 21, 853–862. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Z.; Bao, Y.; Lu, S.; Gong, Z.; Qian, H.-J.; Lu, Z.-Y.; Cui, S. Nanoscopic Characterization Reveals that Bulk Amorphous Elementary Boron Is Composed of a Ladder-like Polymer with B4 as the Structural Unit. ACS Nano 2023, 17, 10958–10964. [Google Scholar] [CrossRef] [PubMed]
- Lostao, A.; Lim, K.; Pallarés, M.C.; Ptak, A.; Marcuello, C. Recent advances in sensing the inter-biomolecular interactions at the nanoscale–A comprehensive review of AFM-based force spectroscopy. Int. J. Biol. Macromol. 2023, 238, 124089. [Google Scholar] [CrossRef]
- Edwards, D.T.; LeBlanc, M.-A.; Perkins, T.T. Modulation of a protein-folding landscape revealed by AFM-based force spectroscopy notwithstanding instrumental limitations. Proc. Natl. Acad. Sci. USA 2021, 118, e2015728118. [Google Scholar] [CrossRef]
- Yi, Y.; Xu, H.; Wang, L.; Cao, W.; Zhang, X. A New Dynamic Covalent Bond of Se—N: Towards Controlled Self-Assembly and Disassembly. Chem. Eur. J. 2013, 19, 9506–9510. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Lu, S.; Wei, J.; Cui, S. Single-chain polymer models incorporating the effects of side groups: An approach to general polymer models. Macromolecules 2019, 52, 7324–7330. [Google Scholar] [CrossRef]
- Bao, Y.; Qian, H.-J.; Lu, Z.-Y.; Cui, S. The unexpected flexibility of natural cellulose at a single-chain level and its implications to the design of nano materials. Nanoscale 2014, 6, 13421–13424. [Google Scholar] [CrossRef]
- Qian, L.; Cai, W.; Xu, D.; Bao, Y.; Lu, Z.-Y.; Cui, S. Single-molecule studies reveal that water is a special solvent for amylose and natural cellulose. Macromolecules 2019, 52, 5006–5013. [Google Scholar] [CrossRef]
- Milião, G.L.; de Souza Soares, L.; Balbino, D.F.; Barbosa, É.d.A.A.; Bressan, G.C.; de Carvalho Teixeira, A.V.N.; dos Reis Coimbra, J.S.; de Oliveira, E.B. pH influence on the mechanisms of interaction between chitosan and ovalbumin: A multi-spectroscopic approach. Food Hydrocoll. 2022, 123, 107137. [Google Scholar] [CrossRef]
- Cao, N.; Zhao, Y.; Chen, H.; Huang, J.; Yu, M.; Bao, Y.; Wang, D.; Cui, S. Poly (ethylene glycol) becomes a supra-polyelectrolyte by capturing hydronium ions in water. Macromolecules 2022, 55, 4656–4664. [Google Scholar] [CrossRef]
- Huang, J.; Zhong, Y.; Zhang, L.; Cai, J. Distinctive viewpoint on the rapid dissolution mechanism of α-chitin in aqueous potassium hydroxide–urea solution at low temperatures. Macromolecules 2020, 53, 5588–5598. [Google Scholar] [CrossRef]
- Zhang, S.; Qian, H.J.; Liu, Z.H.; Ju, H.Y.; Lu, Z.Y.; Zhang, H.M.; Chi, L.F.; Cui, S.X. Towards unveiling the exact molecular structure of amorphous red phosphorus by single-molecule studies. Angew. Chem. Int. Ed. 2019, 58, 1659–1663. [Google Scholar] [CrossRef]
- Bao, Y.; Qian, H.J.; Lu, Z.Y.; Cui, S. Revealing the hydrophobicity of natural cellulose by single-molecule experiments. Macromolecules 2015, 48, 3685–3690. [Google Scholar] [CrossRef]
- Yamamoto, E.; Akimoto, T.; Mitsutake, A.; Metzler, R. Universal relation between instantaneous diffusivity and radius of gyration of proteins in aqueous solution. Phys. Rev. Lett. 2021, 126, 128101. [Google Scholar] [CrossRef] [PubMed]
- Jayakumar, R.; Prabaharan, M.; Kumar, P.S.; Nair, S.; Tamura, H. Biomaterials based on chitin and chitosan in wound dressing applications. Biotechnol. Adv. 2011, 29, 322–337. [Google Scholar] [CrossRef]
- Hao, L.T.; Park, S.; Choy, S.; Kim, Y.-M.; Lee, S.-W.; Ok, Y.S.; Koo, J.M.; Hwang, S.Y.; Hwang, D.S.; Park, J. Strong, multifaceted guanidinium-based adhesion of bioorganic nanoparticles to wet biological tissue. JACS Au 2021, 1, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Pele, K.G.; Amaveda, H.; Mora, M.; Marcuello, C.; Lostao, A.; Alamán-Díez, P.; Pérez-Huertas, S.; Ángeles Pérez, M.; García-Aznar, J.M.; García-Gareta, E. Hydrocolloids of Egg White and Gelatin as a Platform for Hydrogel-Based Tissue Engineering. Gels 2023, 9, 505. [Google Scholar] [CrossRef] [PubMed]
- Köhler, M.; Karner, A.; Leitner, M.; Hytönen, V.P.; Kulomaa, M.; Hinterdorfer, P.; Ebner, A. pH-dependent deformations of the energy landscape of avidin-like proteins investigated by single molecule force spectroscopy. Molecules 2014, 19, 12531–12546. [Google Scholar] [CrossRef]
- Zhang, F.; Gong, Z.; Cai, W.; Qian, H.-J.; Lu, Z.-Y.; Cui, S. Single-chain mechanics of cis-1, 4-polyisoprene and polysulfide. Polymer 2022, 240, 124473. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Yu, M.; Zhang, G.; He, G.; Ji, Y.; Dong, J.; Zheng, H.; Qian, L. Revealing the Control Mechanisms of pH on the Solution Properties of Chitin via Single-Molecule Studies. Molecules 2023, 28, 6769. https://doi.org/10.3390/molecules28196769
Zhang S, Yu M, Zhang G, He G, Ji Y, Dong J, Zheng H, Qian L. Revealing the Control Mechanisms of pH on the Solution Properties of Chitin via Single-Molecule Studies. Molecules. 2023; 28(19):6769. https://doi.org/10.3390/molecules28196769
Chicago/Turabian StyleZhang, Song, Miao Yu, Guoqiang Zhang, Guanmei He, Yunxu Ji, Juan Dong, Huayan Zheng, and Lu Qian. 2023. "Revealing the Control Mechanisms of pH on the Solution Properties of Chitin via Single-Molecule Studies" Molecules 28, no. 19: 6769. https://doi.org/10.3390/molecules28196769
APA StyleZhang, S., Yu, M., Zhang, G., He, G., Ji, Y., Dong, J., Zheng, H., & Qian, L. (2023). Revealing the Control Mechanisms of pH on the Solution Properties of Chitin via Single-Molecule Studies. Molecules, 28(19), 6769. https://doi.org/10.3390/molecules28196769