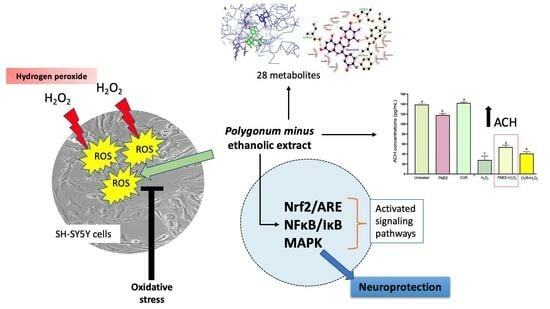
Ethanolic Extract of Polygonum minus Protects Differentiated Human Neuroblastoma Cells (SH-SY5Y) against H2O2-Induced Oxidative Stress
Abstract
:1. Introduction
2. Results and Discussion
2.1. LC–MS/MS Analysis
2.2. Phase Contrast Microscopy and Immunocytochemistry Confirmed Neuronal Marker β-Tubulin III Expression
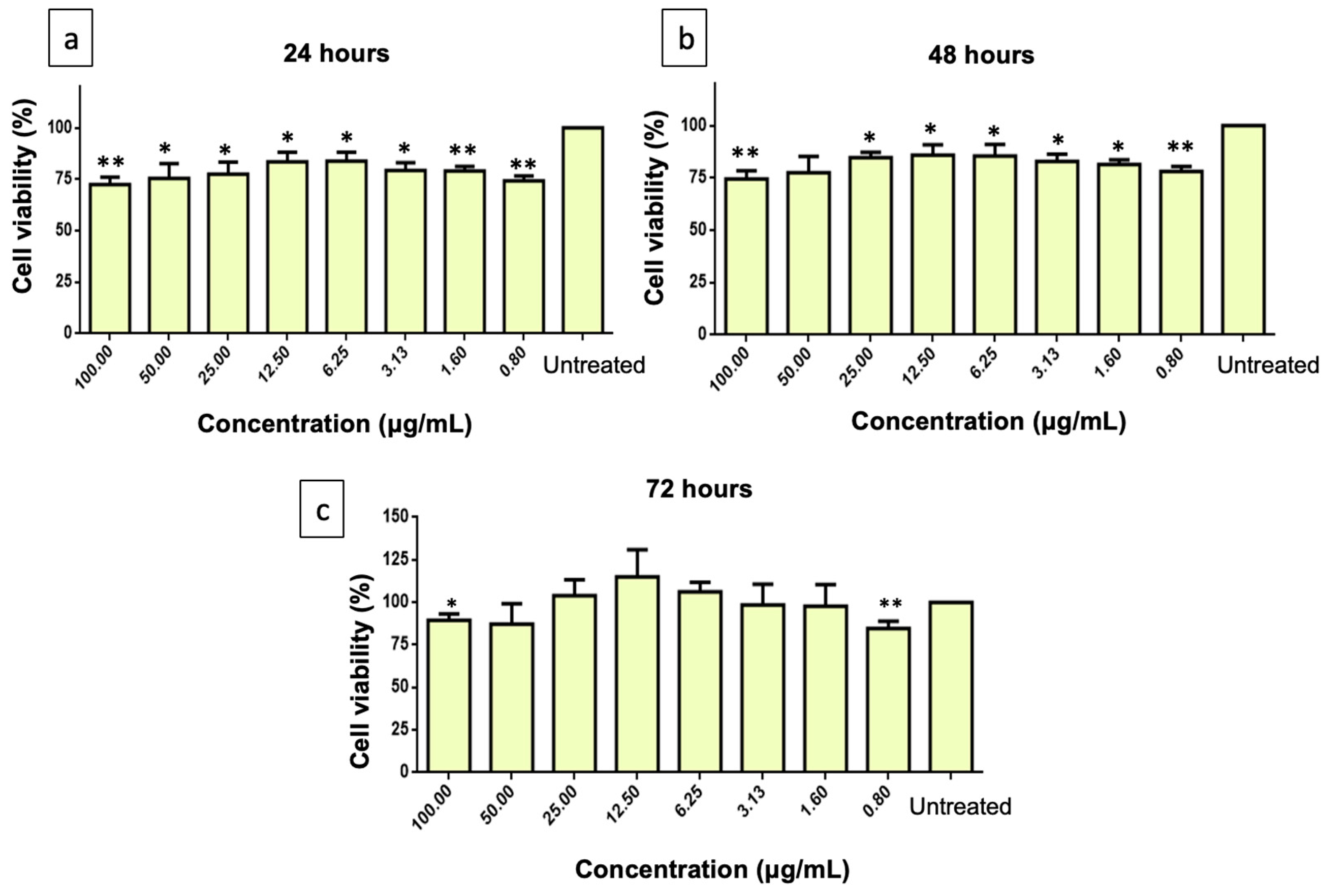
2.3. Cytotoxicity Effect of PMEE and H2O2 on SH-SY5Y Viability
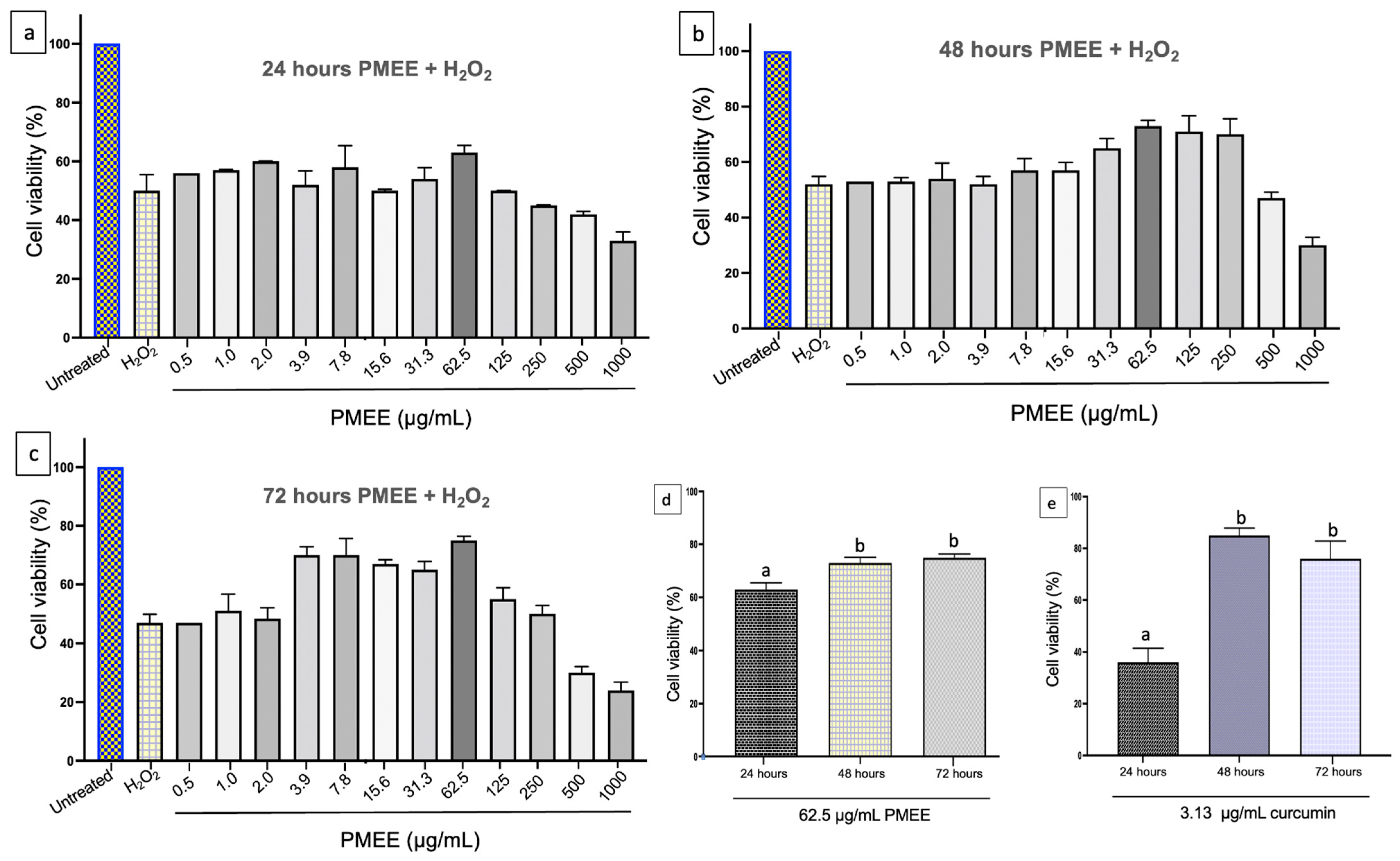
2.4. Neuroprotective Effect of PMEE against H2O2-Induced Cytotoxicity
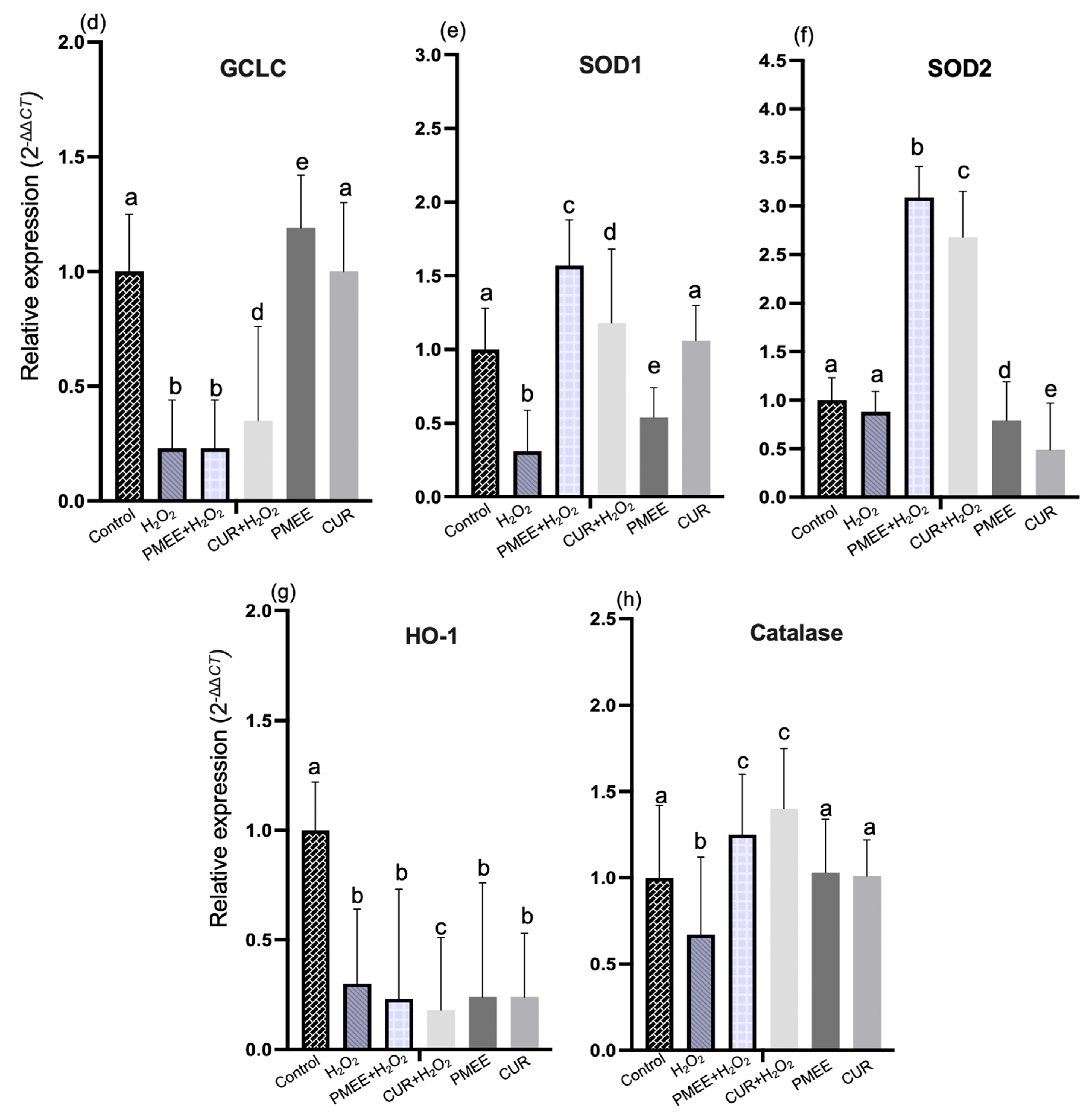
2.5. PMEE Pre-Treatment Influenced Gene Expressions in Nrf2/ARE Pathway
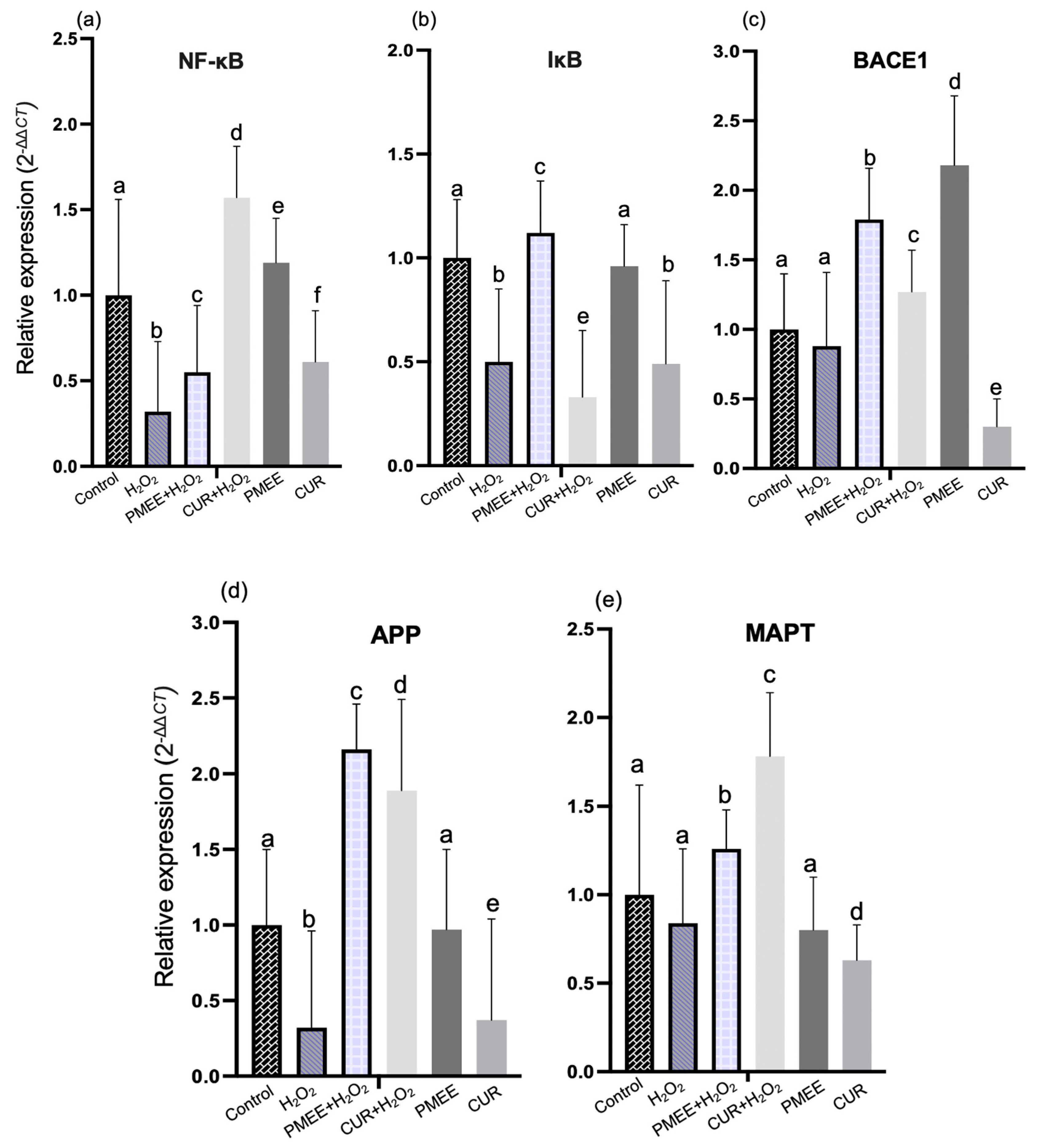
2.6. PMEE Pre-Treatment Influenced Gene Expressions in NF-κB/IκB Pathway
2.7. PMEE Pre-Treatment Influenced Gene Expressions in the MAPK Pathway
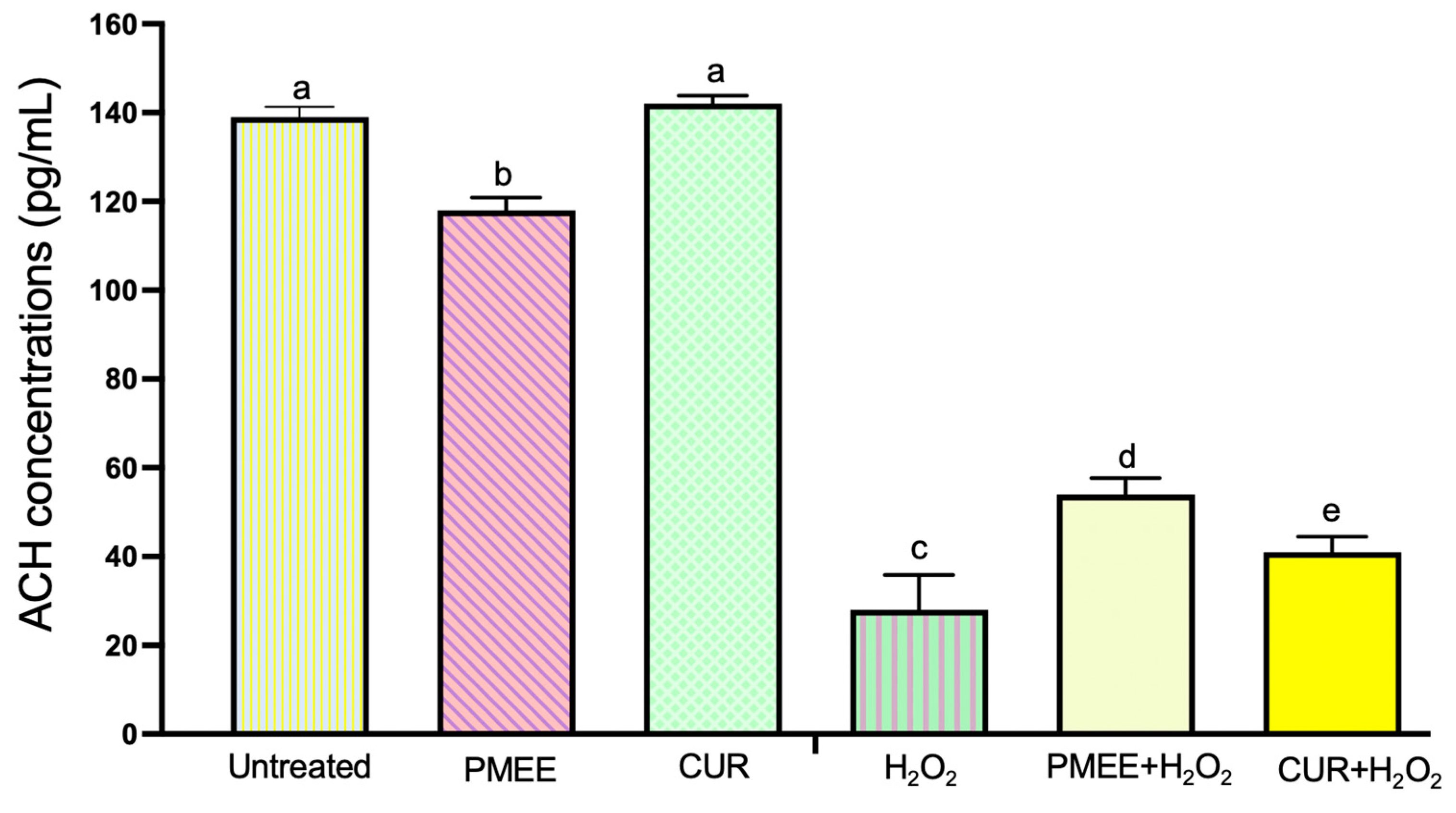
2.8. PMEE Pre-Treatment Increased the Expression of Acetylcholine (ACH) in SH-SY5Y Differentiated Cells
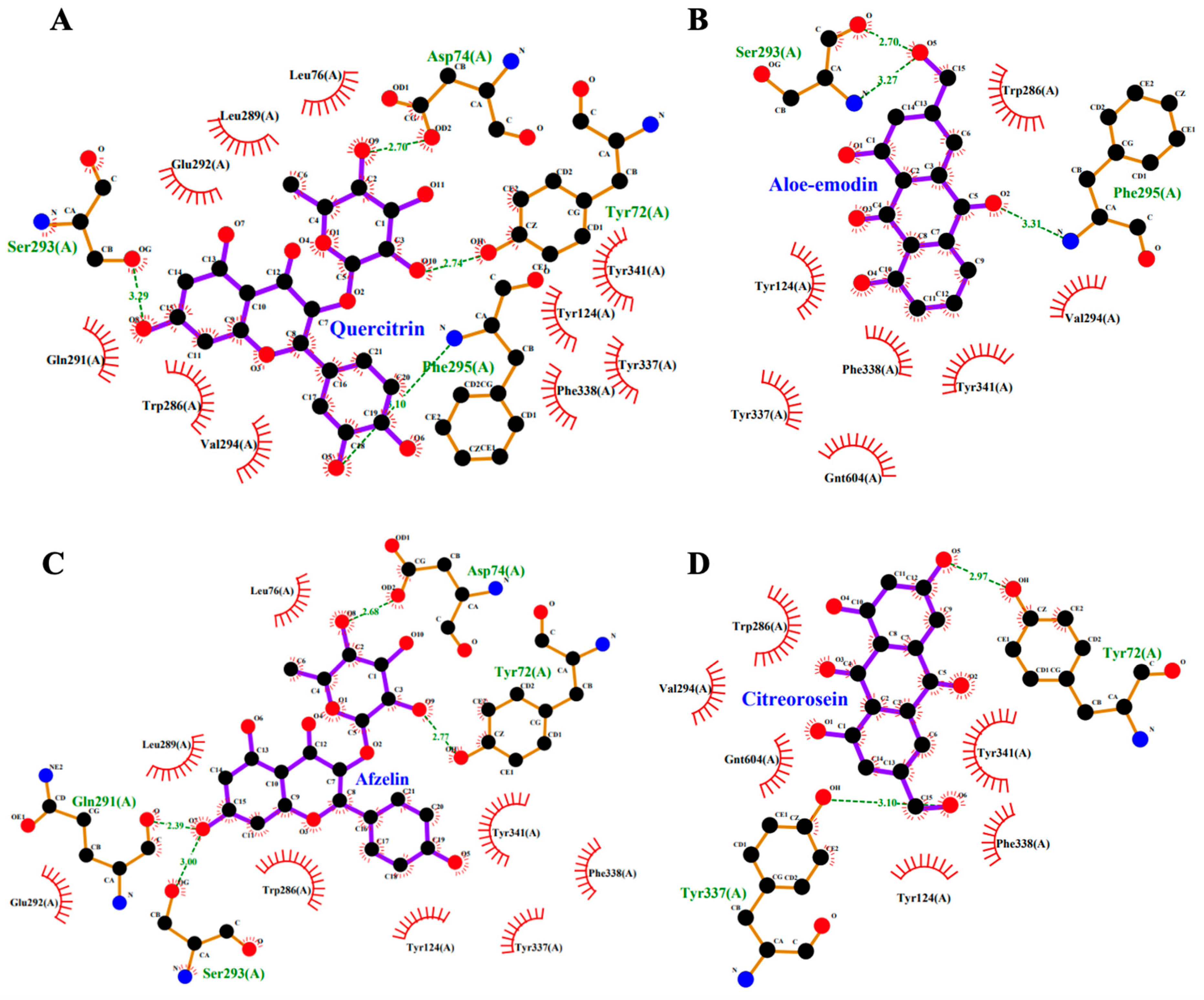
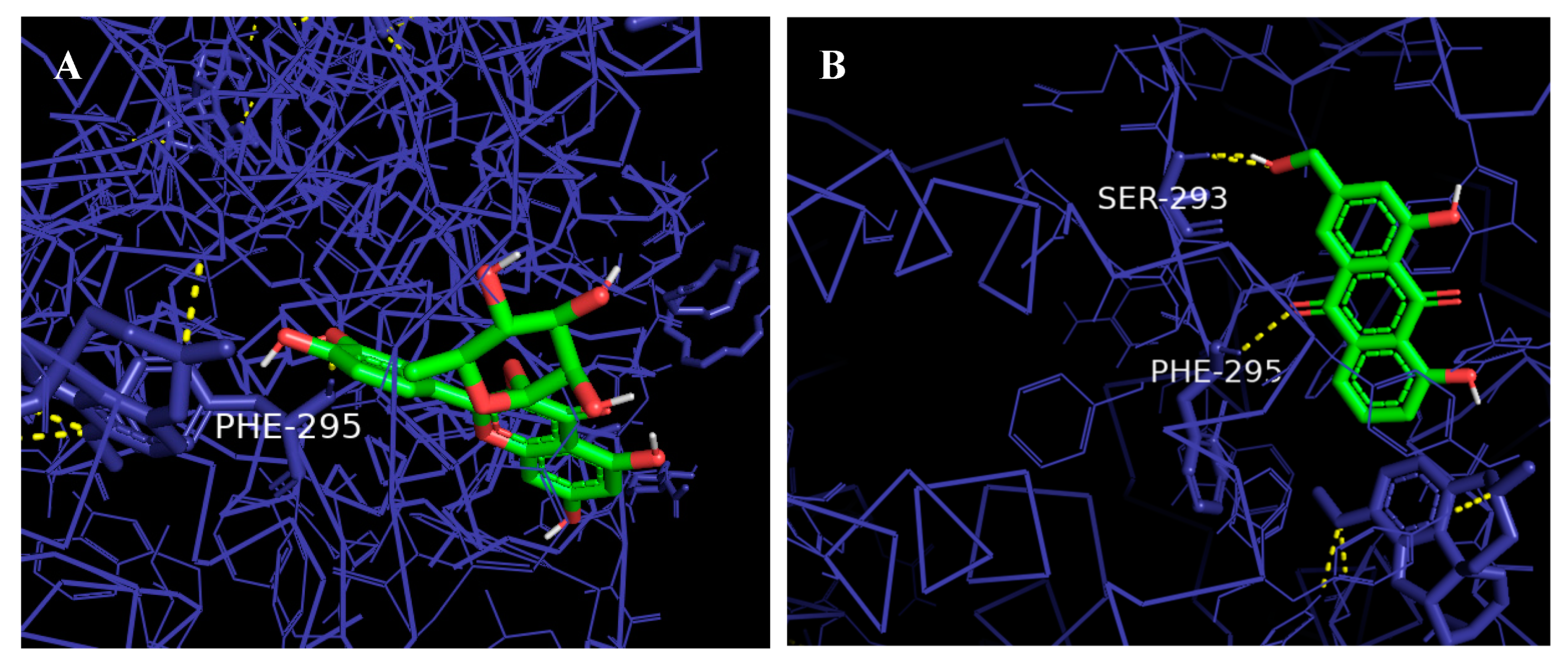
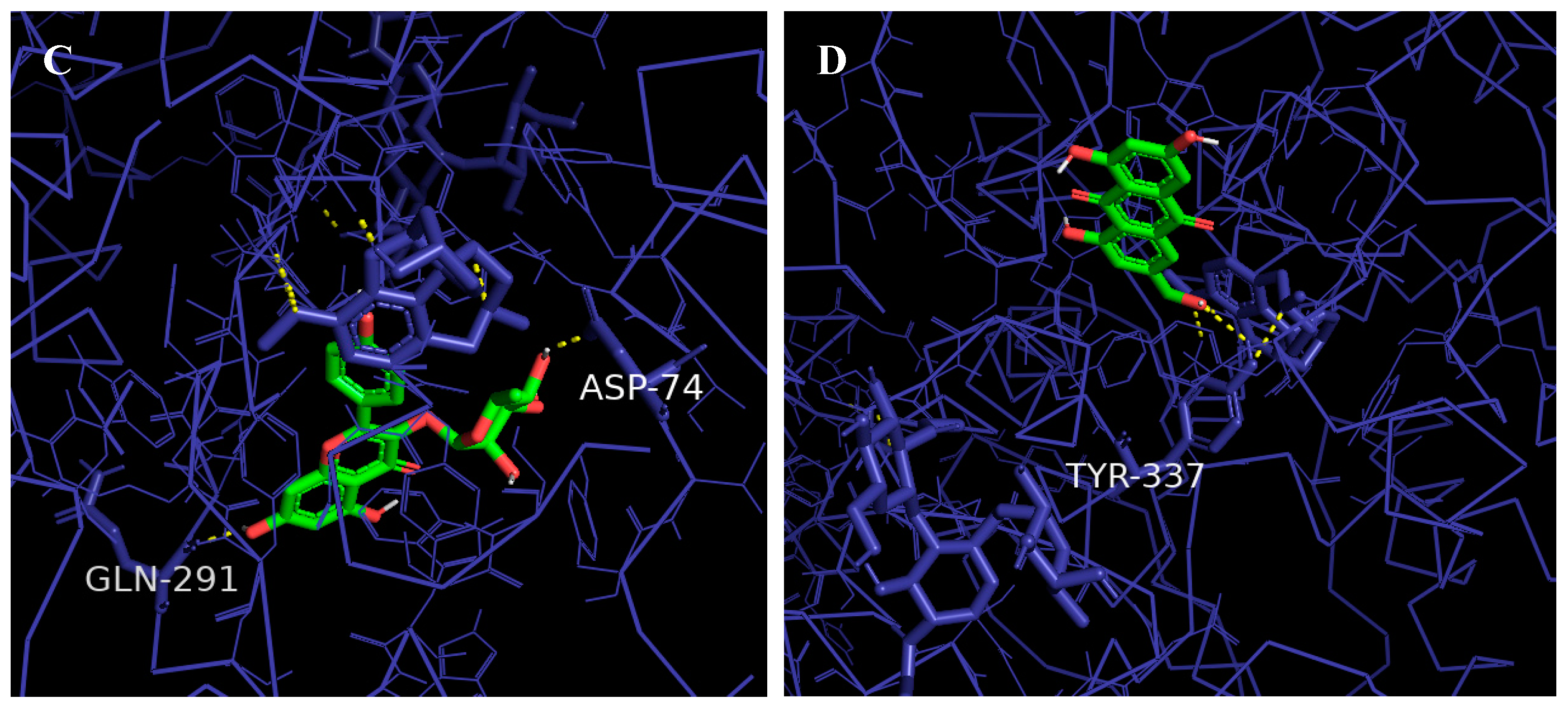
2.9. Molecular Docking
3. Materials and Methods
3.1. Plant Collection and Preparation of Ethanolic Extract
3.2. Liquid Chromatography–Mass Spectrometry (LC–MS/MS)
3.3. Cell Lines and Cell Cultures
3.4. Neuronal Differentiation of SH-SY5Y Cells
3.5. Immunocytochemistry (ICC) Assay
3.6. Cytotoxicity of PMEE on the SH-SY5Y Cells
3.7. Neuroprotection of PMEE on the SH-SY5Y Cells
3.8. PMEE Pre-Treatment and H2O2 Exposure
3.9. Gene Expression Study of PMEE-Treated SH-SY5Y Cells
3.10. Acetylcholine (ACH) Enzyme-Linked Immunosorbent Assay (ELISA)
3.11. Molecular Docking
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative Stress: A Key Modulator in Neurodegenerative Diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef] [PubMed]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, M.; Anil Kumar, N.V.; Zucca, P.; Varoni, E.M.; Dini, L.; Panzarini, E.; Rajkovic, J.; Tsouh Fokou, P.V.; Azzini, E.; Peluso, I.; et al. Lifestyle, Oxidative Stress, and Antioxidants: Back and Forth in the Pathophysiology of Chronic Diseases. Front. Physiol. 2020, 11, 694. [Google Scholar] [CrossRef] [PubMed]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef]
- Kurutas, E.B. The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: Current state. Nutr. J. 2016, 15, 71. [Google Scholar] [CrossRef]
- Ahmad, R.; Rosandy, A.R.; Sahidin, I.; Ab Ghani, N.S.; Noor, N.M.; Baharum, S.N. Bioassay Analysis and Molecular Docking Study Revealed the Potential Medicinal Activities of Active Compounds Polygonumins B, C and D from Polygonum minus (Persicaria minor). Plants 2023, 12, 59. [Google Scholar] [CrossRef]
- Seimandi, G.; Álvarez, N.; Stegmayer, M.I.; Fernández, L.; Ruiz, V.; Favaro, M.A.; Derita, M. An Update on Phytochemicals and Pharmacological Activities of the Genus Persicaria and Polygonum. Molecules 2021, 26, 5956. [Google Scholar] [CrossRef]
- Vikram, P.; Chiruvella, K.K.; Ripain, I.H.A.; Arifullah, M. A recent review on phytochemical constituents and medicinal properties of kesum (Polygonum minus Huds.). Asian Pac. J. Trop. Biomed. 2014, 4, 430–435. [Google Scholar] [CrossRef]
- Hamid, A.A.; Aminuddin, A.; Yunus, M.H.M.; Murthy, J.K.; Hui, C.K.; Ugusman, A. Antioxidative and anti-inflammatory activities of Polygonum minus: A review of literature. Rev. Cardiovasc. Med. 2020, 21, 275–288. [Google Scholar] [CrossRef]
- Maizura, M.; Abdullah, A.; Mustapha, W. Total phenolic content and antioxidant activity of kesum (Polygonum minus), ginger (Zingiber officinale) and turmeric (Curcuma longa) extract. Int. Food Res. J. 2011, 18, 526–531. [Google Scholar]
- Nadzirah, A.S.; Rusop, M.; Noriham, A. A review of potential of antioxidant properties using Polygonum minus. Adv. Mater. Res. 2014, 832, 659–664. [Google Scholar] [CrossRef]
- Abdullah, M.Z.; Mohd Ali, J.; Abolmaesoomi, M.; Abdul-Rahman, P.S.; Hashim, O.H. Anti-proliferative, in vitro antioxidant, and cellular antioxidant activities of the leaf extracts from Polygonum minus Huds: Effects of solvent polarity. Int. J. Food Prop. 2017, 20, 846–862. [Google Scholar] [CrossRef]
- Wasman, S.Q.; Mahmood, A.A.; Salehhuddin, H.; Zahra, A.A.; Salmah, I. Cytoprotective activities of Polygonum minus aqueous leaf extract on ethanol-induced gastric ulcer in rats. J. Med. Plants Res. 2010, 4, 2658–2665. [Google Scholar] [CrossRef]
- Christapher, P.; Parasuraman, S.; Christina, J.; Asmawi, M.Z.; Vikneswaran, M. Review on Polygonum minus. Huds, a commonly used food additive in Southeast Asia. Pharmacogn. Res. 2015, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- George, A.; Ng, C.P.; O’Callaghan, M.; Jensen, G.S.; Wong, H.J. In vitro and ex-vivo cellular antioxidant protection and cognitive enhancing effects of an extract of Polygonum minus Huds (LineminusTM) demonstrated in a Barnes Maze animal model for memory and learning. BMC Complement. Altern. Med. 2014, 14, 161. [Google Scholar] [CrossRef]
- Almey, A.; Khan, A.J.; Zahir, S.; Suleiman, M.; Aisyah, M.R.; Rahim, K. Total phenolic content and primary antioxidant activity of methanolic and ethanolic extracts of aromatic plants’ leaves. Int. Food Res. J. 2010, 17, 1077–1084. [Google Scholar]
- Faujan, H.; Noriham, A.; Norrakiah, B. Antioxidative Activities of Water Extracts of Some Malaysian Herbs 61. ASEAN Food J. 2007, 14, 61–68. [Google Scholar]
- Kähkönen, M.P.; Hopia, A.I.; Vuorela, H.J.; Rauha, J.-P.; Pihlaja, K.; Kujala, T.S.; Heinonen, M. Antioxidant Activity of Plant Extracts Containing Phenolic Compounds. J. Agric. Food Chem. 1999, 47, 3954–3962. [Google Scholar] [CrossRef]
- Ahmad, R.; Sahidin, I.; Taher, M.; Low, C.; Noor, N.M.; Sillapachaiyaporn, C.; Chuchawankul, S.; Sarachana, T.; Tencomnao, T.; Iskandar, F.; et al. Polygonumins A, a newly isolated compound from the stem of Polygonum minus Huds with potential medicinal activities. Sci. Rep. 2018, 8, 4202. [Google Scholar] [CrossRef]
- Yaacob, K.B. Kesom Oil-A Natural Source of Aliphatic Aldehydes. Perfum. Flavor. 1987, 12, 28–30. [Google Scholar]
- Baharum, S.N.; Bunawan, H.; Ghani, M.A.; Wan Mustapha, W.A.; Noor, N.M. Analysis of the Chemical Composition of the Essential Oil of Polygonum minus Huds. Using Two-Dimensional Gas Chromatography-Time-of-Flight Mass Spectrometry (GC-TOF MS). Molecules 2010, 15, 7006. [Google Scholar] [CrossRef] [PubMed]
- Ruijters, E.J.B.; Weseler, A.R.; Kicken, C.; Haenen, G.R.M.M.; Bast, A. The flavanol (-)-epicatechin and its metabolites protect against oxidative stress in primary endothelial cells via a direct antioxidant effect. Eur. J. Pharmacol. 2013, 715, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Chen, J.; Xing, Y. Eupatilin prevents H2O2-induced oxidative stress and apoptosis in human retinal pigment epithelial cells. Biomed. Pharmacother. 2017, 85, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yang, W.; He, Z.; Guo, J.; Yang, X.; Wang, R.; Li, H. Kaempferol Alleviates Oxidative Stress and Apoptosis Through Mitochondria-dependent Pathway During Lung Ischemia-Reperfusion Injury. Front. Pharmacol. 2021, 12, 624402. [Google Scholar] [CrossRef]
- Shojaee, M.S.; Moeenfard, M.; Farhoosh, R. Kinetics and stoichiometry of gallic acid and methyl gallate in scavenging DPPH radical as affected by the reaction solvent. Sci. Rep. 2022, 12, 8765. [Google Scholar] [CrossRef]
- Khan, F.A.; Maalik, A.; Murtaza, G. Inhibitory mechanism against oxidative stress of caffeic acid. J. Food Drug Anal. 2016, 24, 695–702. [Google Scholar] [CrossRef]
- Medeiros, D.L.; Lima, E.T.G.; Silva, J.C.; Medeiros, M.A.; Pinheiro, E.B.F. Rhamnetin: A review of its pharmacology and toxicity. J. Pharm. Pharmacol. 2022, 74, 793–799. [Google Scholar] [CrossRef]
- Dai, X.; Ding, Y.; Zhang, Z.; Cai, X.; Li, Y. Quercetin and quercitrin protect against cytokine-induced injuries in RINm5F β-cells via the mitochondrial pathway and NF-κB signaling. Int. J. Mol. Med. 2013, 31, 265–271. [Google Scholar] [CrossRef]
- Kwon, Y.K.; Choi, S.J.; Kim, C.R.; Kim, J.K.; Kim, Y.-J.; Choi, J.H.; Song, S.-W.; Kim, C.-J.; Park, G.G.; Park, C.-S.; et al. Antioxidant and cognitive-enhancing activities of Arctium lappa L. roots in Aβ1-42-induced mouse model. Appl. Biol. Chem. 2016, 59, 553–565. [Google Scholar] [CrossRef]
- Cimini, A.; Ardini, M.; Gentile, R.; Giansanti, F.; Benedetti, E.; Cristiano, L.; Fidoamore, A.; Scotti, S.; Panella, G.; Angelucci, F.; et al. A peroxiredoxin-based proteinaceous scaffold for the growth and differentiation of neuronal cells and tumour stem cells in the absence of prodifferentiation agents. J. Tissue Eng. Regen. Med. 2016, 11, 2462–2470. [Google Scholar] [CrossRef]
- Jaafaru, M.S.; Nordin, N.; Shaari, K.; Rosli, R.; Abdull Razis, A.F. Isothiocyanate from Moringa oleifera seeds mitigates hydrogen peroxide-induced cytotoxicity and preserved morphological features of human neuronal cells. PLoS ONE 2018, 13, e0196403. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.; Ismail, M.; Imam, M.U.; Azmi, N.H.; Fathy, S.F.; Foo, J.B.; Abu Bakar, M.F. Mechanistic basis for protection of differentiated SH-SY5Y cells by oryzanol-rich fraction against hydrogen peroxide-induced neurotoxicity. BMC Complement. Altern. Med. 2014, 14, 467. [Google Scholar] [CrossRef] [PubMed]
- Qader, S.W.; Abdulla, M.A.; Chua, L.S.; Najim, N.; Zain, M.M.; Hamdan, S. Antioxidant, total phenolic content and cytotoxicity evaluation of selected Malaysian plants. Molecules 2011, 16, 3433–3443. [Google Scholar] [CrossRef]
- Mohd Ghazali, M.A.; Al-Naqeb, G.; Krishnan Selvarajan, K.; Hazizul Hasan, M.; Adam, A. Apoptosis Induction by Polygonum minus is Related to Antioxidant Capacity, Alterations in Expression of Apoptotic-Related Genes, and S-Phase Cell Cycle Arrest in HepG2 Cell Line. Biomed. Res. Int. 2014, 2014, 539607. [Google Scholar] [CrossRef]
- Zhao, X.; Fang, J.; Li, S.; Gaur, U.; Xing, X.; Wang, H.; Zheng, W. Artemisinin Attenuated Hydrogen Peroxide (H2O2)-Induced Oxidative Injury in SH-SY5Y and Hippocampal Neurons via the Activation of AMPK Pathway. Int. J. Mol. Sci. 2019, 20, 2680. [Google Scholar] [CrossRef]
- Shen, B.; Truong, J.; Helliwell, R.; Govindaraghavan, S.; Sucher, N.J. An in Vitro Study of Neuroprotective Properties of Traditional Chinese Herbal Medicines Thought to Promote Healthy Ageing and Longevity. BMC Complement. Altern. Med. 2013, 13, 373. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Song, M.Y.; Kim, E.H. Role of Oxidative Stress and Nrf2/KEAP1 Signaling in Colorectal Cancer: Mechanisms and Therapeutic Perspectives with Phytochemicals. Antioxidants 2021, 10, 743. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.R.; Reindl, K.M. Glutathione S-Transferases in Cancer. Antioxidants 2021, 10, 701. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Yan, L.J.; Jana, C.K.; Das, N. Role of Catalase in Oxidative Stress- and Age-Associated Degenerative Diseases. Oxidative Med. Cell. Longev. 2019, 2019, 9613090. [Google Scholar] [CrossRef] [PubMed]
- Nijboer, C.H.A.; Heijnen, C.J.; Groenendaal, F.; May, M.J.; Van Bel, F.; Kavelaars, A. Strong Neuroprotection by Inhibition of NF-κB after Neonatal Hypoxia-Ischemia Involves Apoptotic Mechanisms but is Independent of Cytokines. Stroke 2008, 39, 2129–2137. [Google Scholar] [CrossRef]
- Chen, C.H.; Zhou, W.; Liu, S.; Deng, Y.; Cai, F.; Tone, M.; Tone, Y.; Tong, Y.; Song, W. Increased NF-κB signalling up-regulates BACE1 expression and its therapeutic potential in Alzheimer’s disease. Int. J. Neuropsychopharmacol. 2012, 15, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.L.; Webb, M.S.; Copik, A.J.; Wang, Y.; Johnson, B.H.; Kumar, R.; Thompson, E.B. p38 Mitogen-Activated Protein Kinase (MAPK) Is a Key Mediator in Glucocorticoid-Induced Apoptosis of Lymphoid Cells: Correlation between p38 MAPK Activation and Site-Specific Phosphorylation of the Human Glucocorticoid Receptor at Serine 211. Mol. Endocrinol. 2005, 19, 1569–1583. [Google Scholar] [CrossRef] [PubMed]
- Cipriani, R.; Chara, J.C.; Rodríguez-Antigüedad, A.; Matute, C. FTY720 attenuates excitotoxicity and neuroinflammation. J. Neuroinflamm. 2015, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Majd, S.; Power, J.H.T.; Koblar, S.A.; Grantham, H.J.M. Early glycogen synthase kinase-3β and protein phosphatase 2A independent tau dephosphorylation during global brain ischaemia and reperfusion following cardiac arrest and the role of the adenosine monophosphate kinase pathway. Eur. J. Neurosci. 2016, 44, 1987–1997. [Google Scholar] [CrossRef]
- Sangodkar, J.; Farrington, C.C.; McClinch, K.; Galsky, M.D.; Kastrinsky, D.B.; Narla, G. All roads lead to PP2A: Exploiting the therapeutic potential of this phosphatase. FEBS J. 2016, 283, 1004–1024. [Google Scholar] [CrossRef]
- Tan, X.L.; Wright, D.K.; Liu, S.; Hovens, C.; O’Brien, T.J.; Shultz, S.R. Sodium selenate, a protein phosphatase 2A activator, mitigates hyperphosphorylated tau and improves repeated mild traumatic brain injury outcomes. Neuropharmacology 2016, 108, 382–393. [Google Scholar] [CrossRef]
- Işık, M.; Beydemir, Ş. AChE mRNA expression as a possible novel biomarker for the diagnosis of coronary artery disease and Alzheimer’s disease, and its association with oxidative stress. Arch. Physiol. Biochem. 2019, 128, 352–359. [Google Scholar] [CrossRef]
- Attar, U.A.; Ghane, S.G. In Vitro Antioxidant, Antidiabetic, Antiacetylcholine Esterase, Anticancer Activities and RP-HPLC Analysis of Phenolics from the Wild Bottle Gourd (Lagenaria siceraria (Molina) Standl.). S. Afr. J. Bot. 2019, 125, 360–370. [Google Scholar] [CrossRef]
- Khan, H.; Maryal; Amin, S.; Kamal, M.A.; Patel, S. Flavonoids as acetylcholinesterase inhibitors: Current therapeutic standing and future prospects. Biomed. Pharmacother. 2018, 101, 860–870. [Google Scholar] [CrossRef]
- Ahmad, R.; Baharum, S.; Bunawan, H.; Lee, M.; Mohd Noor, N.; Rohani, E.R.; Ilias, N.; Zin, N.M. Volatile profiling of aromatic traditional medicinal plant, Polygonum minus in different tissues and its biological activities. Molecules 2014, 19, 19220–19242. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz-Ozden, T.; Nasabi, N.T.; Hasbal-Celikok, G.; Kocyigit, M.; Özhan, G. Antioxidant, Anti-Acetylcholinesterase, and Anticancer Activities of Four Polygonum Species from Istanbul. Int. Food Res. J. 2021, 28, 1298–1309. [Google Scholar] [CrossRef]
- Bingol, M.N.; Bursal, E. LC-MS/MS Analysis of Phenolic Compounds and In Vitro Antioxidant Potential of Stachys lavandulifolia Vahl. var. brachydon Boiss. Int. Lett. Nat. Sci. 2018, 72, 28–36. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
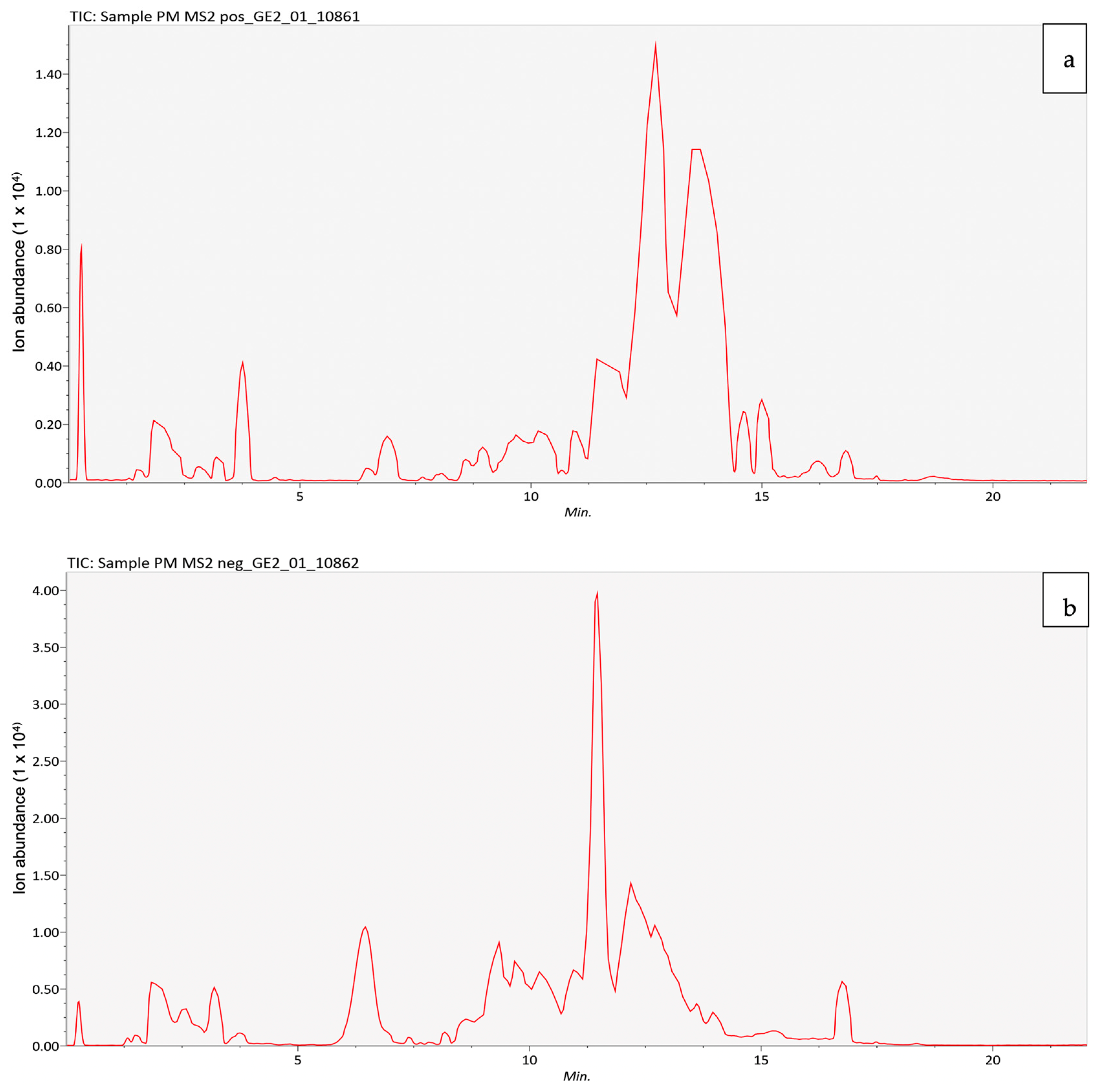





| No | Identified Compounds | Retention Time (min) | Molecular Formula | Molecular Weight (g/mol) |
|---|---|---|---|---|
| 1 | Quinic acid | 2.540 | C7H12O6 | 191.06 |
| 2 | Gallic acid | 6.456 | C7H6O5 | 169.01 |
| 3 | 2,3-Dihydroxybenzoic acid | 8.170 | C7H6O4 | 153.02 |
| 4 | (−)-epicatechin | 8.686 | C15H14O6 | 291.09 |
| 5 | 5-Hydroxy-2-penten-1-yl-3-oxocyclopentyl acetic acid | 8.753 | C12H18O4 | 227.13 |
| 6 | Caffeic acids | 9.142 | C9H8O4 | 179.04 |
| 7 | Loliolide | 9.506 | C11H16O3 | 197.12 |
| 8 | Quercitrin | 9.673 | C21H20O11 | 449.11 |
| 9 | Naphtho[2,3-b]furan-9(4H)-1,4,8-bis(acetyloxy)-4a,5,6,7,8,8a-hexahydro-3,4a,5-trimethyl-, (4S,4aR,5S,8S,8aS) | 9.823 | C19H24O6 | 331.15 |
| 10 | Carboxymethyl-cyclohexanecarboxylic acid | 9.961 | C9H14O4 | 185.08 |
| 11 | Afzelin | 10.073 | C21H20O10 | 433.11 |
| 12 | 6-Hydroxy-3-isopropylidene-4a,5-dimethyl-4,4a,5,6,7,8-hexahydro-2(3H)-naphthalenone | 10.157 | C15H22O2 | 235.17 |
| 13 | Feruloyltyramine | 10.157 | C18H19NO4 | 314.14 |
| 14 | Daphnetin | 10.362 | C9H6O4 | 177.02 |
| 15 | Esculetin | 10.423 | C9H6O4 | 179.03 |
| 16 | Citreorosein | 10.910 | C15H10O6 | 287.05 |
| 17 | Quercetin | 11.428 | C15H10O7 | 303.05 |
| 18 | Aloe-emodin | 12.250 | C15H10O5 | 271.06 |
| 19 | Kaempferol | 12.250 | C15H10O6 | 287.05 |
| 20 | Rhamnetin | 12.250 | C16H12O7 | 317.06 |
| 21 | (2Z)-4,6-dihydroxy-2-[(4-hydroxy-3,5-dimethoxyphenyl)methylidene]-1-benzofuran-3-1 | 12.516 | C17H14O7 | 331.08 |
| 22 | Eupatilin | 12.516 | C18H16O7 | 345.09 |
| 23 | Corynoxeine | 12.971 | C22H26N2O4 | 383.20 |
| 24 | Valerenic acid | 13.062 | C15H22O2 | 233.16 |
| 25 | alpha-Cyperone | 13.154 | C15H22O | 219.17 |
| 26 | Dibutylphthalate | 13.304 | C16H22O4 | 279.16 |
| 27 | (2Z)-2-[(E)-6-(hydroxymethyl)-2,4,8,10-tetramethyldodec-2-enylidene]-4-methylpentanedioic acid | 13.847 | C23H40O5 | 419.27 |
| 28 | Prespatane | 14.260 | C15H24 | 205.19 |
| No. | PMEE’s Identified Compounds | Binding Affinity to AChE (kcal/mol) |
|---|---|---|
| 1. | Quercitrin | −9.5 |
| 2. | Aloe-emodin | −9.4 |
| 3. | Afzelin | −9.3 |
| 4. | Citreorosein | −9.0 |
| 5. | alpha-Cyperone | −8.7 |
| 6. | Quercetin | −8.7 |
| 7. | Kaempferol | −8.6 |
| 8. | Rhamnetin | −8.6 |
| 9. | (2Z)-4,6-dihydroxy-2-[(4-hydroxy-3,5-dimethoxyphenyl) methylidene]-1-benzofuran-3-1 | −8.5 |
| 10. | 6-Hydroxy-3-isopropylidene-4a,5-dimethyl-4,4a,5,6,7,8- hexahydro-2(3H)-naphthalenone | −8.3 |
| 11. | Eupatilin | −8.3 |
| 12. | Feruloyltyramine | −8.0 |
| 13. | Naphtho[2,3-b]furan-9(4H)-1,4,8-bis(acetyloxy)-4a,5,6,7,8,8a- hexahydro-3,4a,5-trimethyl-, (4S,4aR,5S,8S,8aS) | −7.8 |
| 14. | (−)-epicatechin | −7.7 |
| 15. | Esculetin | −7.5 |
| 16. | Prespatane | −7.5 |
| 17. | (2Z)-2-[(E)-6-(hydroxymethyl)-2,4,8,10- tetramethyldodec-2-enylidene]-4-methylpentanedioic acid | −7.3 |
| 18. | Daphnetin | −7.3 |
| 19. | Valerenic acid | −7.3 |
| 20. | Caffeic acids | −7.0 |
| 21. | Corynoxeine | −7.0 |
| 22. | Dibutylphthalate | −7.0 |
| 23. | Loliolide | −6.6 |
| 24. | 5-Hydroxy-2-penten-1-yl-3-oxocyclopentyl acetic acid | −6.5 |
| 25. | Carboxymethyl-cyclohexanecarboxylic acid | −6.5 |
| 26. | Gallic acid | −6.5 |
| 27. | 2,3-Dihydroxybenzoic acid | −6.2 |
| 28. | Quinic acid | −5.8 |
| Gene and Accession No | Forward Primer | Reverse Primer |
|---|---|---|
| AKT [NM_005465.4] | AGGTGACACTATAGAATA AGACATTAAATTTCCTCGAA | GTACGACTCACTATAGGG AATCCTCATCATATTTTTCAGGT |
| APP [NM_000484.3] | AGGTGACACTATAGAATA CTGTGGCAGACTGAACATGC | GTACGACTCACTATAGGG ATCACCAACTAAGCAGCGGTA |
| BACE1 [NM_012104.4] | AGGTGACACTATAGAATA CGAGCTGGATTATGGT | GTACGACTCACTATAGGG AGGAGAGGGAGCTTGG |
| Catalase [NM_001752.3] | AGGTGACACTATAGAATA AGAAATCCTCAGACACATCT | GTACGACTCACTATAGGG AATGTCATGACCTGGATGTAA |
| GCLC [NM_001498.3] | AGGTGACACTATAGAATA ATGAAGCAATAAACAAGCAC | GTACGACTCACTATAGGG ATGGAATGTCACCTGGAG |
| GST [NM_015917.2] | AGGTGACACTATAGAATA ATACATGGCAAATGACTTAAA | GTACGACTCACTATAGGG ATGATGTCTTCATTCCTTGAC |
| HO-1 [NM_002133.2] | AGGTGACACTATAGAATA ACTGCGTTCCTGCTCAACAT | GTACGACTCACTATAGGG AGGGCAGAATCTTGCACTTTGT |
| IκB [NM_020529.2] | AGGTGACACTATAGAATA CTGCAGCAGACTCCAC | GTACGACTCACTATAGGG AGGGTATTTCCTCGAAAGT |
| JNK [NM_001323327.1] | AGGTGACACTATAGAATA AAGGAAAACGTGGATTTATG | GTACGACTCACTATAGGG ACCAGCATATTTAGGTCTGTT |
| MAPT [NM_001123066.3] | AGGTGACACTATAGAATA CCCAGATCTGAGAGAGGT | GTACGACTCACTATAGGG ACTTATTAATTATCTGCACCTTCC |
| MKP1 [NM_004417.3] | AGGTGACACTATAGAATA AGAAGAACCAAATACCTCAA | GTACGACTCACTATAGGG ACAGGTCATAAATAATCAGCA |
| NF-κB [NM_002908.3] | AGGTGACACTATAGAATA CGTTTTAGATACAAATGTGAAG | GTACGACTCACTATAGGG ACACTTTTCCTTTTCCATAAT |
| NQO1 [NM_000903.2] | AGGTGACACTATAGAATA CTGCGAACTTTCAGTATCC | GTACGACTCACTATAGGG AGAAGGGTCCTTTGTCATAC |
| Nrf2 [NM_006164.4] | AGGTGACACTATAGAATA TCGCAAACAACTCTTTATCT | GTACGACTCACTATAGGG AAGAGGAGGTCTCCGTTA |
| p38 [NM_001315.2] | AGGTGACACTATAGAATA TGAGCTGAAGATTCTGGA | GTACGACTCACTATAGGG ATGTCAGACGCATAATCTG |
| PP5 [NM_006247.3] | AGGTGACACTATAGAATA CAAGGACTACGAGAACGCCA | GTACGACTCACTATAGGGA GCTTCACCTTGACCACCGTC |
| PP2A [NM_002715.3] | AGGTGACACTATAGAATA CCGCCATTACAGAGAG | GTACGACTCACTATAGGGA AGGATTTCTTTAGCCTTCT |
| SOD1 [NM_000454.4] | AGGTGACACTATAGAATA AAGTACAAAGACAGGAAACG | GTACGACTCACTATAGGGA TGACAAGTTTAATACCCATCT |
| SOD2 [NM_000636.3] | AGGTGACACTATAGAATA ACAACAGGCCTTATTCC | GTACGACTCACTATAGGGA AGAGCTTAACATACTCAGCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayuti, N.H.; Zulkefli, N.; Tan, J.K.; Saad, N.; Baharum, S.N.; Hamezah, H.S.; Bunawan, H.; Ahmed, Q.U.; Parveen, H.; Mukhtar, S.; et al. Ethanolic Extract of Polygonum minus Protects Differentiated Human Neuroblastoma Cells (SH-SY5Y) against H2O2-Induced Oxidative Stress. Molecules 2023, 28, 6726. https://doi.org/10.3390/molecules28186726
Sayuti NH, Zulkefli N, Tan JK, Saad N, Baharum SN, Hamezah HS, Bunawan H, Ahmed QU, Parveen H, Mukhtar S, et al. Ethanolic Extract of Polygonum minus Protects Differentiated Human Neuroblastoma Cells (SH-SY5Y) against H2O2-Induced Oxidative Stress. Molecules. 2023; 28(18):6726. https://doi.org/10.3390/molecules28186726
Chicago/Turabian StyleSayuti, Nor Hafiza, Nabilah Zulkefli, Jen Kit Tan, Norazalina Saad, Syarul Nataqain Baharum, Hamizah Shahirah Hamezah, Hamidun Bunawan, Qamar Uddin Ahmed, Humaira Parveen, Sayeed Mukhtar, and et al. 2023. "Ethanolic Extract of Polygonum minus Protects Differentiated Human Neuroblastoma Cells (SH-SY5Y) against H2O2-Induced Oxidative Stress" Molecules 28, no. 18: 6726. https://doi.org/10.3390/molecules28186726
APA StyleSayuti, N. H., Zulkefli, N., Tan, J. K., Saad, N., Baharum, S. N., Hamezah, H. S., Bunawan, H., Ahmed, Q. U., Parveen, H., Mukhtar, S., Alsharif, M. A., & Sarian, M. N. (2023). Ethanolic Extract of Polygonum minus Protects Differentiated Human Neuroblastoma Cells (SH-SY5Y) against H2O2-Induced Oxidative Stress. Molecules, 28(18), 6726. https://doi.org/10.3390/molecules28186726