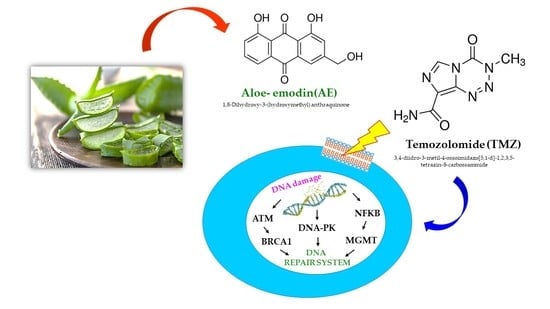
Aloe-Emodin Overcomes Anti-Cancer Drug Resistance to Temozolomide and Prevents Colony Formation and Migration in Primary Human Glioblastoma Cell Lines NULU and ZAR
Abstract
1. Introduction
2. Results
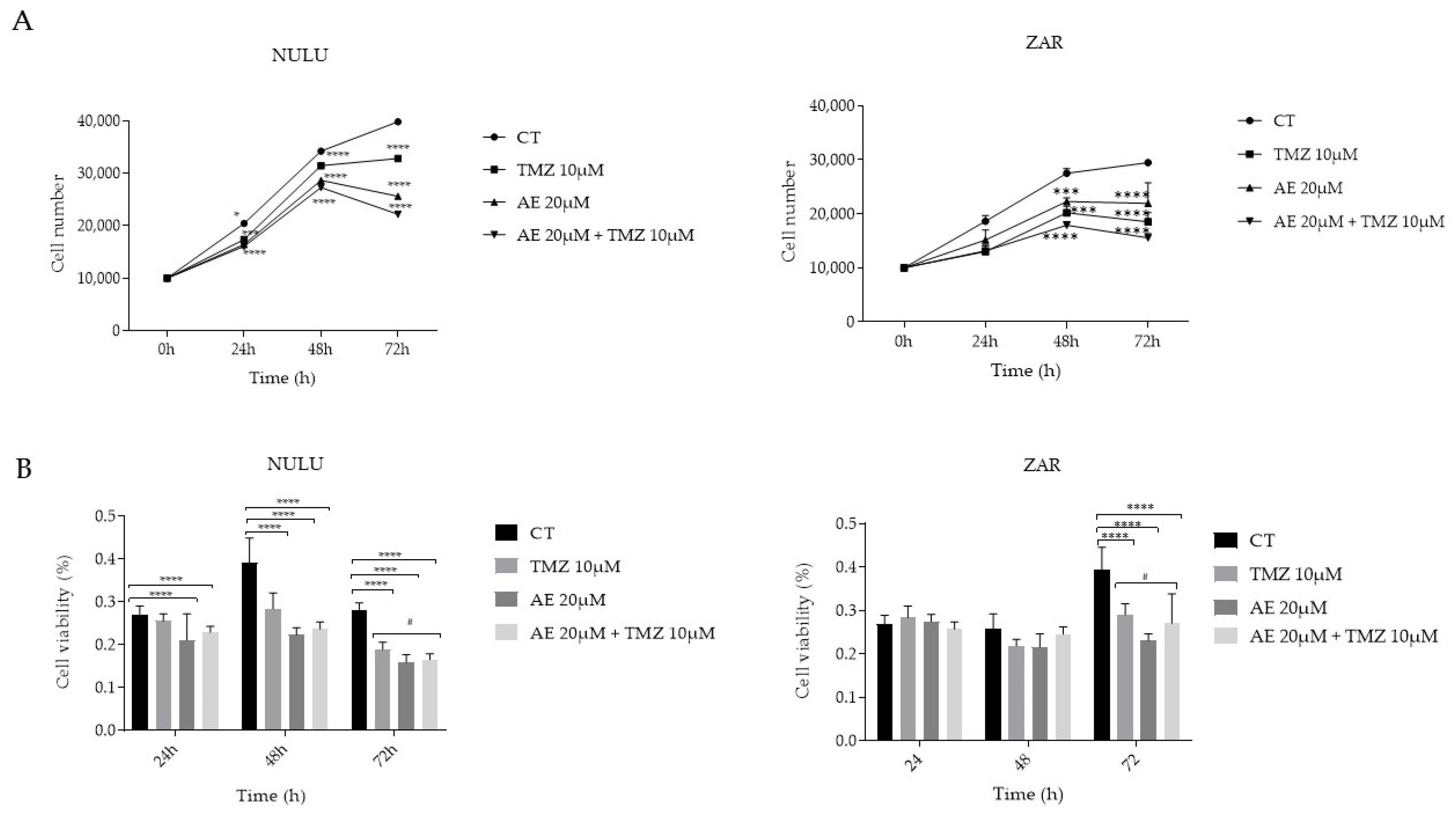
2.1. Impact of AE on Primary GBM Cell Lines NULU and ZAR, Both as a Single Agent and in Combination with TMZ
2.2. Analysis of the Expression of the MGMT Protein in the NULU and ZAR Primary GBM Lines Treated with TMZ and AE Alone/in Combination
2.3. Analysis of the Expression of the Transcription Factor NF-κB in Primary GBM Lines Exposed to Treatment with AE, TMZ, and in Combination
2.4. NULU mRNA Levels of BRCA1, ATM, and DNA-PK after AE and TMZ Treatment
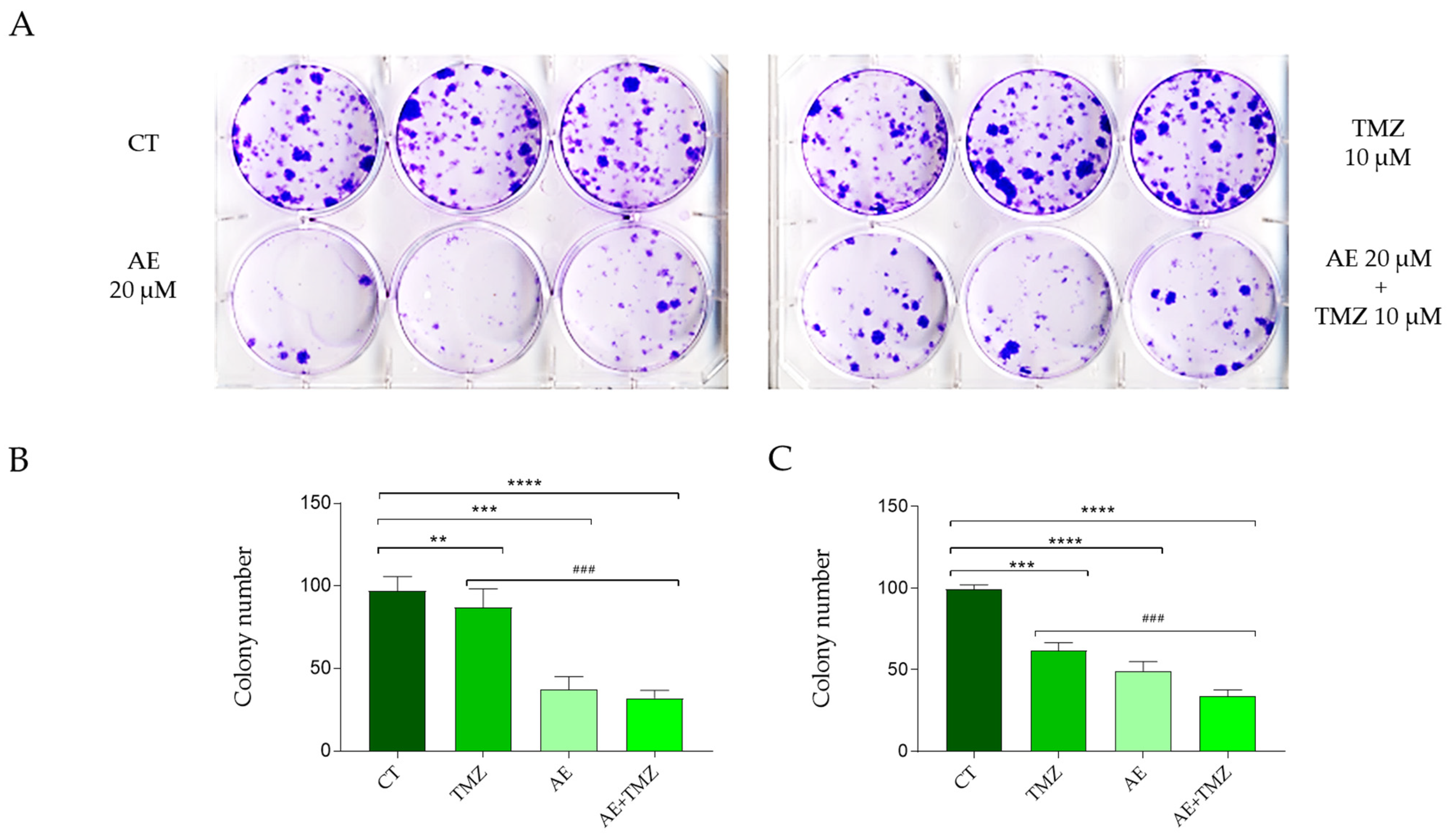
2.5. The Effects of TMZ and AE Adjuvant Therapy on the Clonogenic Capacity of the Primary GBM Lines NULU and ZAR
2.6. Scratch Test to Assess the Effectiveness of the Combination of AE and TMZ in Preventing the Migration of the Primary GBM Lines NULU and ZAR
3. Discussion
4. Methods
4.1. Cell Culture
4.2. Cell Counts
4.3. Cell Viability Assay (MTT)
4.4. Clonogenic Assay
4.5. Wound Healing Assay
4.6. Western Blot
4.7. Reverse Transcription of RNA
4.8. Statistical Analysis
4.9. Determination of the Half-Maximal Inhibitory Concentration (IC50) of AE in ZAR Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ciceroni, C.; Bonelli, M.; Mastrantoni, E.; Niccolini, C.; Laurenza, M.; Larocca, L.M.; Pallini, R.; Traficante, A.; Spinsanti, P.; Ricci-Vitiani, L.; et al. Type-3 metabotropic glutamate receptors regulate chemoresistance in glioma stem cells, and their levels are inversely related to survival in patients with malignant gliomas. Cell Death Differ. 2013, 20, 396–407. [Google Scholar] [CrossRef] [PubMed]
- de Almeida Sassi, F.; Lunardi Brunetto, A.; Schwartsmann, G.; Roesler, R.; Abujamra, A.L. Glioma revisited: From neurogenesis and cancer stem cells to the epigenetic regulation of the niche. J. Oncol. 2012, 2012, 537861. [Google Scholar] [CrossRef] [PubMed]
- Ohgaki, H.; Kleihues, P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005, 109, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Surawicz, T.S.; Davis, F.; Freels, S.; Laws, E.R.; Menck, H.R. Brain tumor survival: Results from the National Cancer Data Base. J. Neurooncol. 1998, 40, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Weber, D.C. The role of radio- and chemotherapy in glioblastoma. Onkologie 2005, 28, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Holland, E.C. Astrocyte differentiation states and glioma formation. Cancer J. 2003, 9, 72–81. [Google Scholar] [CrossRef]
- Oliva, M.A.; Staffieri, S.; Castaldo, S.; Giangaspero, F.; Esposito, V.; Arcella, A. Characterization of primary glioma cell lines derived from the patients according to 2016 CNS tumour WHO classification and comparison with their parental tumours. J. Neurooncol. 2021, 151, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Kawaguchi, A.; Yamanaka, R. Promising Prognosis Marker Candidates on the Status of Epithelial-Mesenchymal Transition and Glioma Stem Cells in Glioblastoma. Cells 2019, 8, 1312. [Google Scholar] [CrossRef]
- Butle, M.; Pongo, L.; Su, Y.T.; Xi, L.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 2020, 6, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Binabaj, M.M.; Bahrami, A.; ShahidSales, S.; Joodi, M.; Joudi Mashhad, M.; Hassanian, S.M.; Anvari, K.; Avan, A. The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J. Cell Physiol. 2018, 233, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Pistollato, F.; Bremer-Hoffmann, S.; Basso, G.; Cano, S.S.; Elio, I.; Vergara, M.M.; Giampieri, F.; Battino, M. Targeting Glioblastoma with the Use of Phytocompounds and Nanoparticles. Target Oncol. 2016, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Fu, J.; Yin, X.; Cao, S.; Li, X.; Lin, L.; Huyiligeqi; Ni, J. Emodin: A Review of its Pharmacology, Toxicity and Pharmacokinetics. Phytother. Res. 2016, 30, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, G.K.; Mudiam, M.K.R.; Vashishtha, V.M.; Raisuddin, S.; Das, M. Activity-guided chemo toxic profiling of Cassia occidentalis (CO) seeds: Detection of toxic compounds in body fluids of CO-exposed patients and experimental rats. Chem. Res. Toxicol. 2015, 28, 1120–1132. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, X.H.; Yu, X.; Cao, F.; Cai, X.; Chen, P.; Li, M.; Feng, Y.; Li, H.; Wang, X. Pharmacokinetics of Anthraquinones from Medicinal Plants. Front. Pharmacol. 2021, 12, 638993. [Google Scholar]
- Stompor-Gorący, M. The Health Benefits of Emodin, a Natural Anthraquinone Derived from Rhubarb—A Summary Update. Int. J. Mol. Sci. 2021, 22, 9522. [Google Scholar] [CrossRef] [PubMed]
- Arcella, A.; Oliva, M.A.; Staffieri, S.; Sanchez, M.; Madonna, M.; Riozzi, B.; Esposito, V.; Giangaspero, F.; Frati, L. Effects of aloe emodin on U87MG glioblastoma cell growth: In vitro and in vivo study. Environ. Toxicol. 2018, 33, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Aasland, D.; Reich, T.R.; Tomicic, M.T.; Switzeny, O.J.; Kaina, B.; Christmann, M. Repair gene O6-methylguanine-DNA methyltransferase is controlled by SP1 and up-regulated by glucocorticoids, but not by temozolomide and radiation. J. Neurochem. 2018, 144, 139–151. [Google Scholar] [CrossRef]
- Hu, Y.H.; Jiao, B.H.; Wang, C.Y.; Wu, J.L. Regulation of temozolomide resistance in glioma cells via the RIP2/NF-κB/MGMT pathway. CNS Neurosci. Ther. 2021, 27, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Ferri, A.; Stagni, V.; Barilà, D. Targeting the DNA Damage Response to Overcome Cancer Drug Resistance in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 4910. [Google Scholar] [CrossRef] [PubMed]
- McCarthy-Leo, C.; Darwiche, F.; Tainsky, M.A. DNA Repair Mechanisms, Protein Interactions and Therapeutic Targeting of the MRN Complex. Cancers 2022, 14, 5278. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Bai, C.; Xie, D.; Ma, T.; Zhou, P.K. DNA-PKcs: A Multi-Faceted Player in DNA Damage Response. Front. Genet. 2020, 11, 607428. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, M.; Ouchi, T. Regulation of ATM/DNA-PKcs Phosphorylation by BRCA1-Associated BAAT1. Genes Cancer 2010, 1, 1211–1214. [Google Scholar] [CrossRef]
- Munshi, A.; Hobbs, M.; Meyn, R.E. Clonogenic cell survival assay. Methods Mol. Med. 2005, 110, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Dymova, M.A.; Kuligina, E.V.; Richter, V.A. Molecular Mechanisms of Drug Resistance in Glioblastoma. Int. J. Mol. Sci. 2021, 22, 6385. [Google Scholar] [CrossRef] [PubMed]
- Suboj, P.; Babykutty, S.; Valiyaparambil Gopi, D.R.; Nair, R.S.; Srinivas, P.; Gopala, S. Aloe emodin inhibits colon cancer cell migration/angiogenesis by downregulating MMP-2/9, RhoB and VEGF via reduced DNA binding activity of NF-κB. Eur. J. Pharm. Sci. 2012, 45, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.M.; Xiao, B.X.; Liu, Q.; Zhang, S.; Liu, D.H.; Gong, Z.H. Anticancer effect of aloe-emodin on cervical cancer cells involves G2/M arrest and induction of differentiation. Acta Pharmacol. Sin. 2007, 28, 1991–1995. [Google Scholar] [CrossRef]
- Rouse, J.; Jackson, S.P. Interfaces between the detection, signaling, and repair of DNA damage. Science 2002, 297, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Amundson, S.A.; Fornace, A.J. Gene expression profiles for monitoring radiation exposure. Radiat. Prot. Dosim. 2001, 97, 11–16. [Google Scholar] [CrossRef]



| Oligo Name | Type | Sequence 5′–3′ |
|---|---|---|
| GAPDH | forward | AGAAAATCTGGCACCACACC |
| reverse | GGGGTGTTGAAGGTCTCAAA | |
| BRCA1 | forward | ACTGCAGCCAGCCACAGGTA |
| reverse | TGACCAGGACAGTAGAAGGA | |
| ATM | forward | TTTACCTAACTGTGAGCTGTCTCCAT |
| reverse | ACTTCCGTAAGGCATCGTAACAC | |
| DNA-PK | forward | CCAGCTCTTACGCTCTGATATG |
| reverse | CAAACGCATGCCCAAAGTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staffieri, S.; Russo, V.; Oliva, M.A.; Alborghetti, M.; Russo, M.; Arcella, A. Aloe-Emodin Overcomes Anti-Cancer Drug Resistance to Temozolomide and Prevents Colony Formation and Migration in Primary Human Glioblastoma Cell Lines NULU and ZAR. Molecules 2023, 28, 6024. https://doi.org/10.3390/molecules28166024
Staffieri S, Russo V, Oliva MA, Alborghetti M, Russo M, Arcella A. Aloe-Emodin Overcomes Anti-Cancer Drug Resistance to Temozolomide and Prevents Colony Formation and Migration in Primary Human Glioblastoma Cell Lines NULU and ZAR. Molecules. 2023; 28(16):6024. https://doi.org/10.3390/molecules28166024
Chicago/Turabian StyleStaffieri, Sabrina, Veronica Russo, Maria Antonietta Oliva, Marika Alborghetti, Miriam Russo, and Antonietta Arcella. 2023. "Aloe-Emodin Overcomes Anti-Cancer Drug Resistance to Temozolomide and Prevents Colony Formation and Migration in Primary Human Glioblastoma Cell Lines NULU and ZAR" Molecules 28, no. 16: 6024. https://doi.org/10.3390/molecules28166024
APA StyleStaffieri, S., Russo, V., Oliva, M. A., Alborghetti, M., Russo, M., & Arcella, A. (2023). Aloe-Emodin Overcomes Anti-Cancer Drug Resistance to Temozolomide and Prevents Colony Formation and Migration in Primary Human Glioblastoma Cell Lines NULU and ZAR. Molecules, 28(16), 6024. https://doi.org/10.3390/molecules28166024