Modes of Action of a Novel c-MYC Inhibiting 1,2,4-Oxadiazole Derivative in Leukemia and Breast Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Growth Inhibition Assay
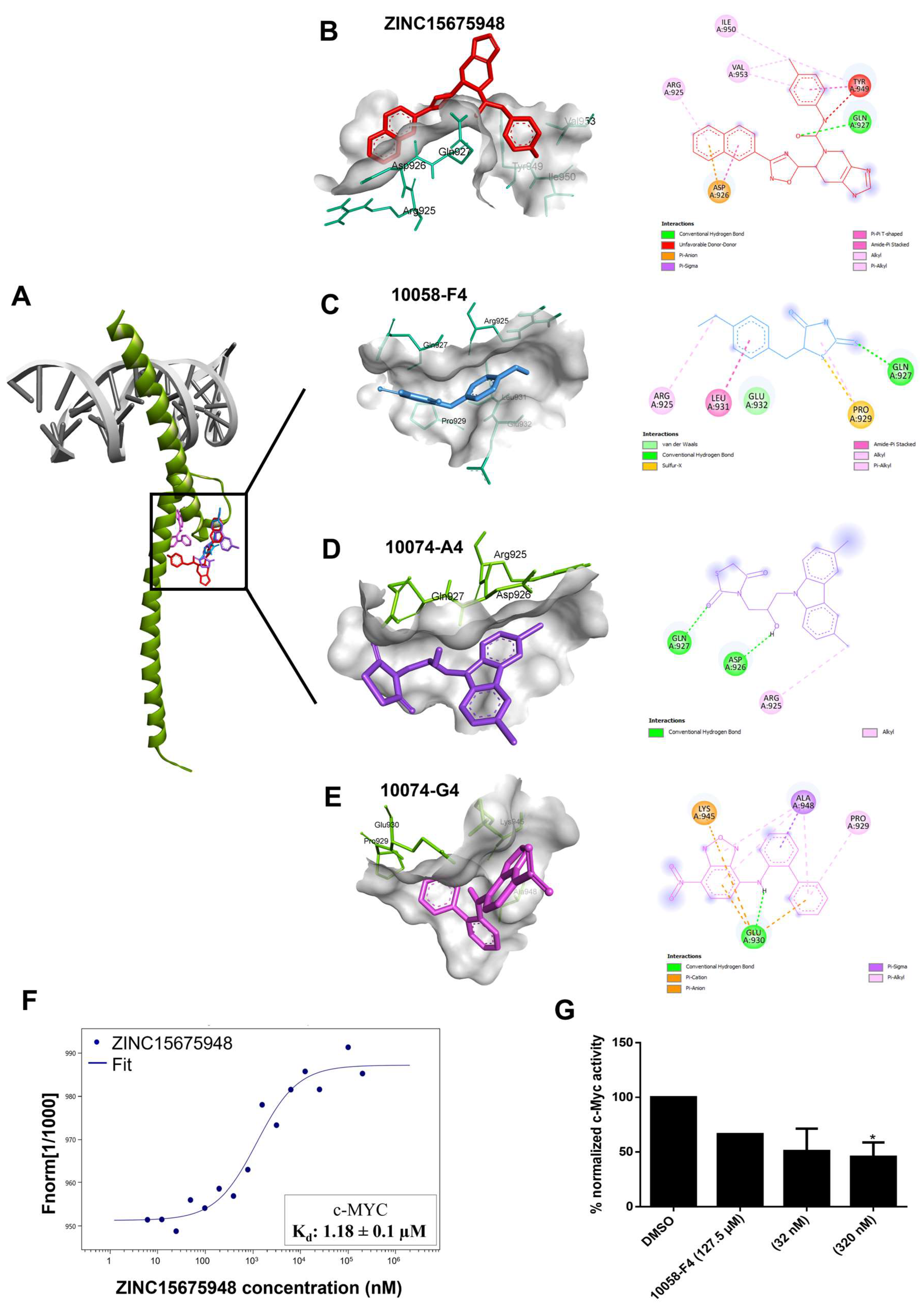
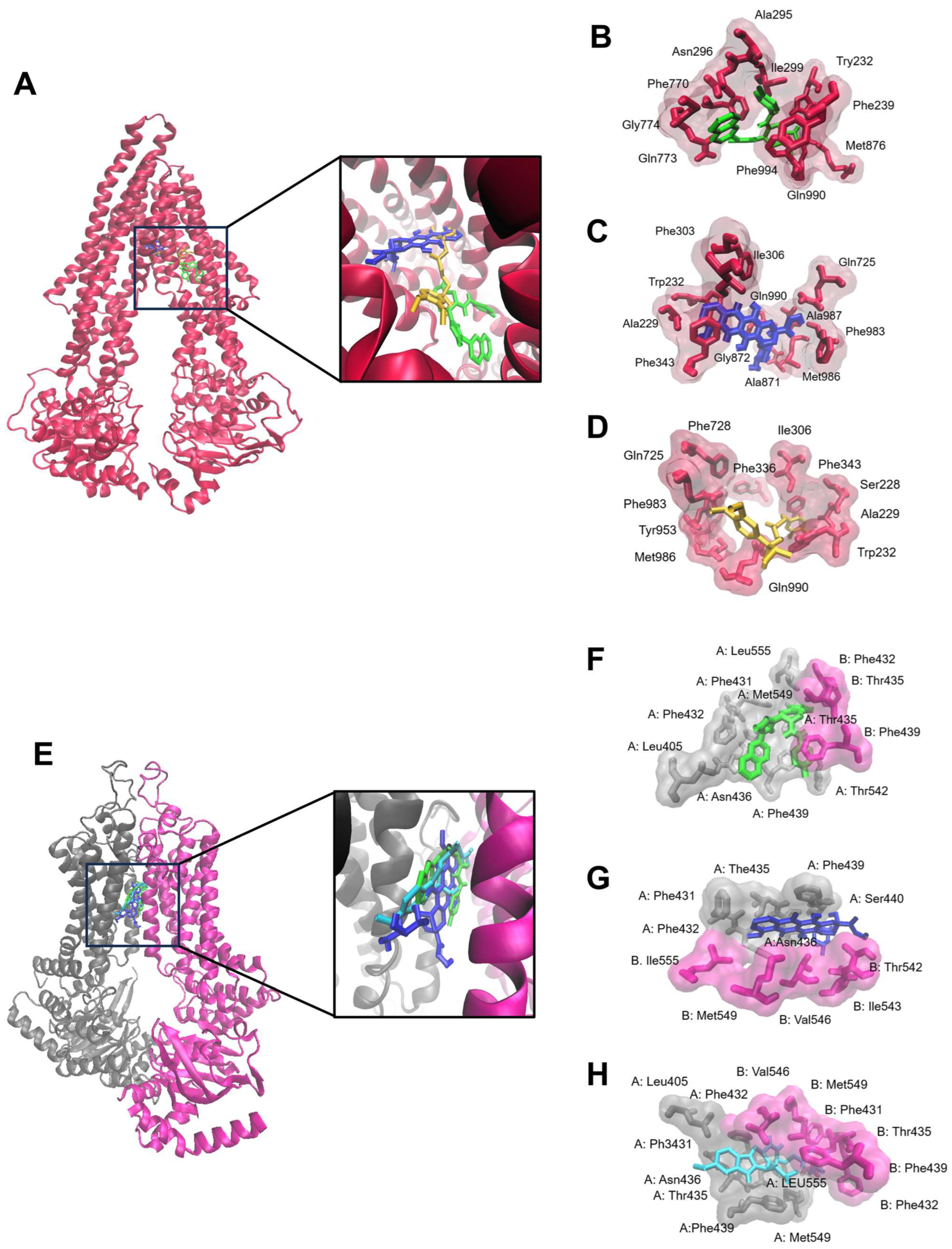
2.2. Molecular Docking
2.3. Microscale Thermophoresis
2.4. c-MYC Reporter Assay
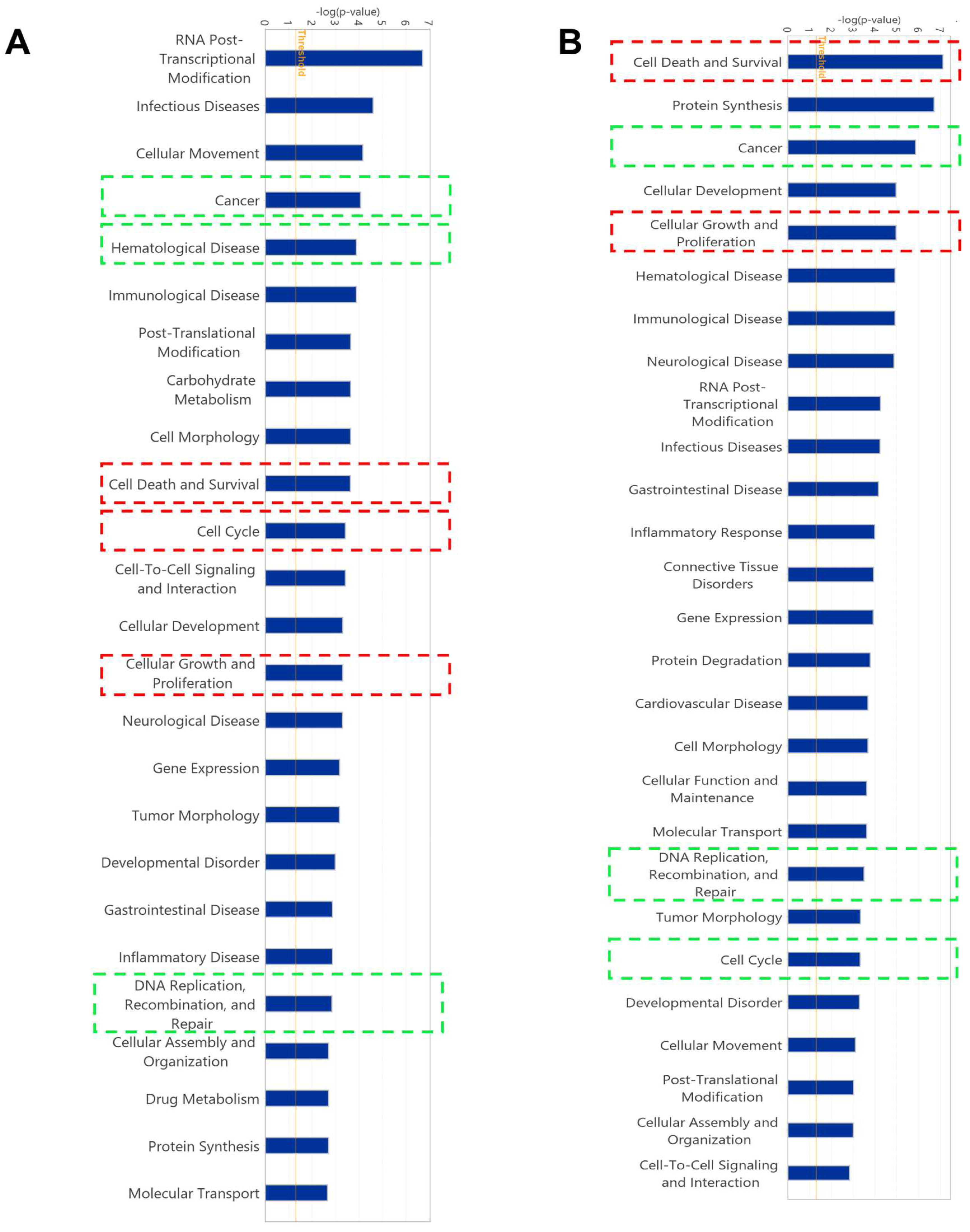
2.5. Gene Expression Profile of Cell Lines Using Microarray Analyses
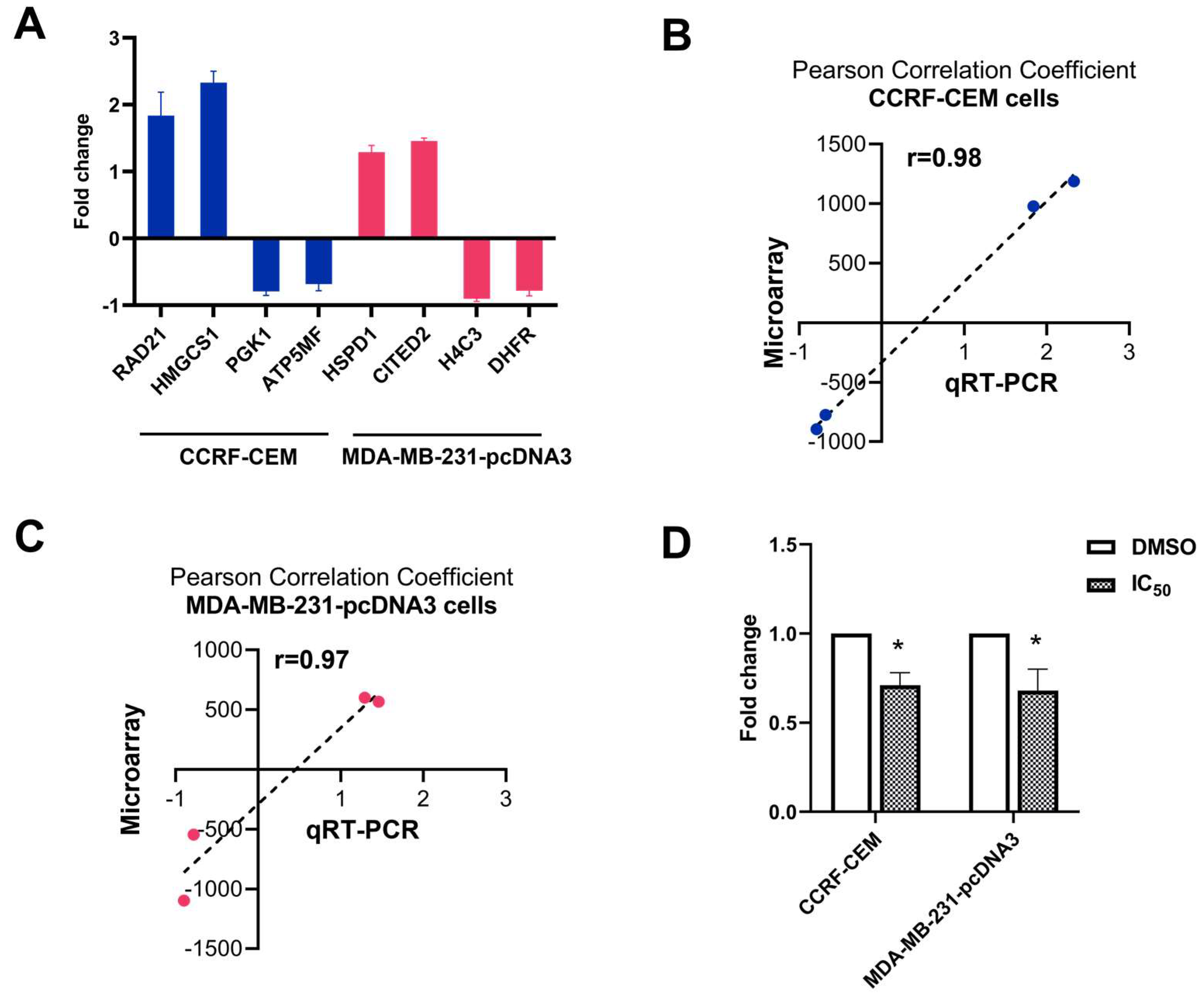
2.6. Quantitative Reverse Transcription PCR (qRT-PCR)
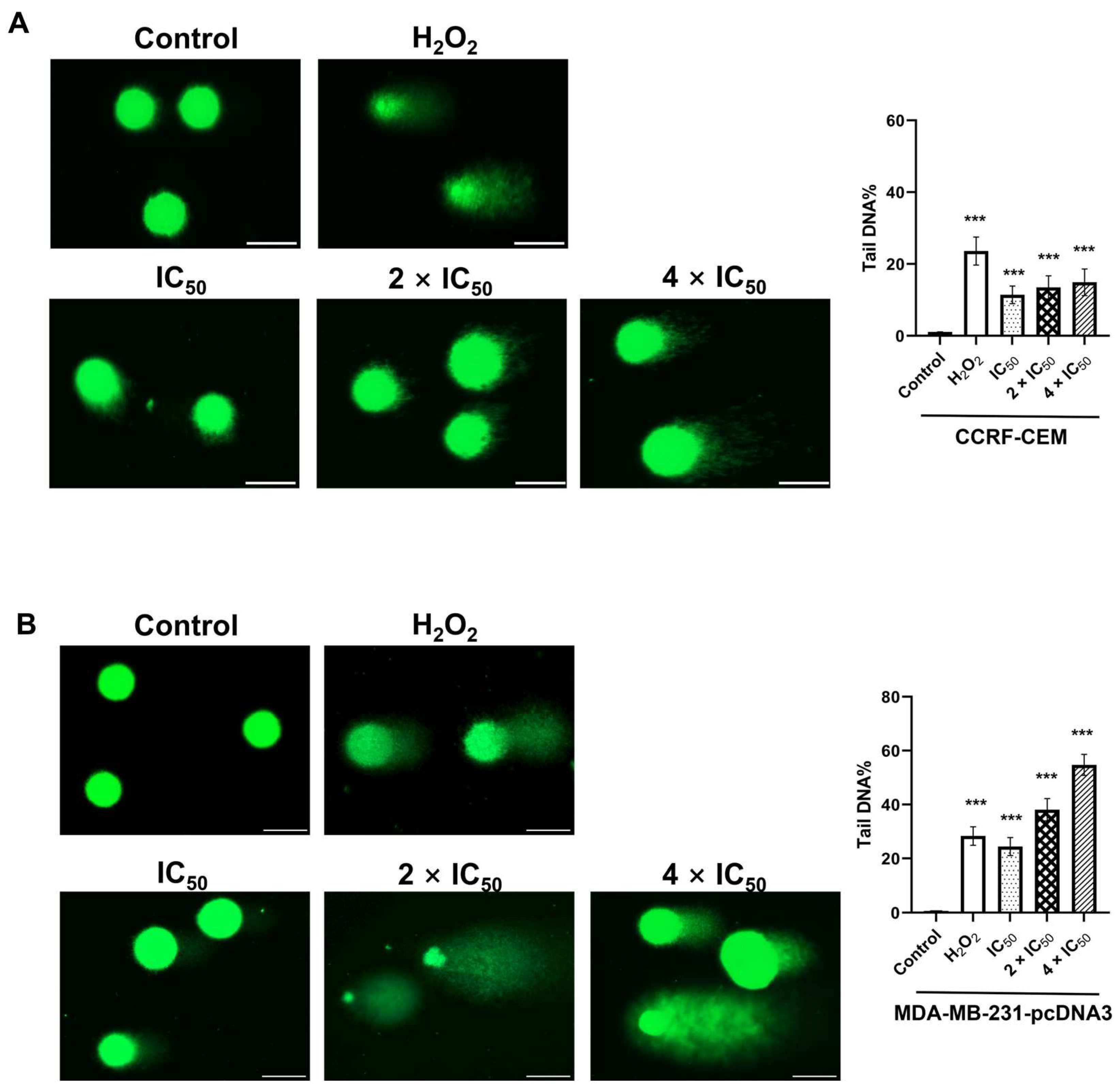
2.7. Single Cell Gel Electrophoresis (Alkaline Comet Assay)
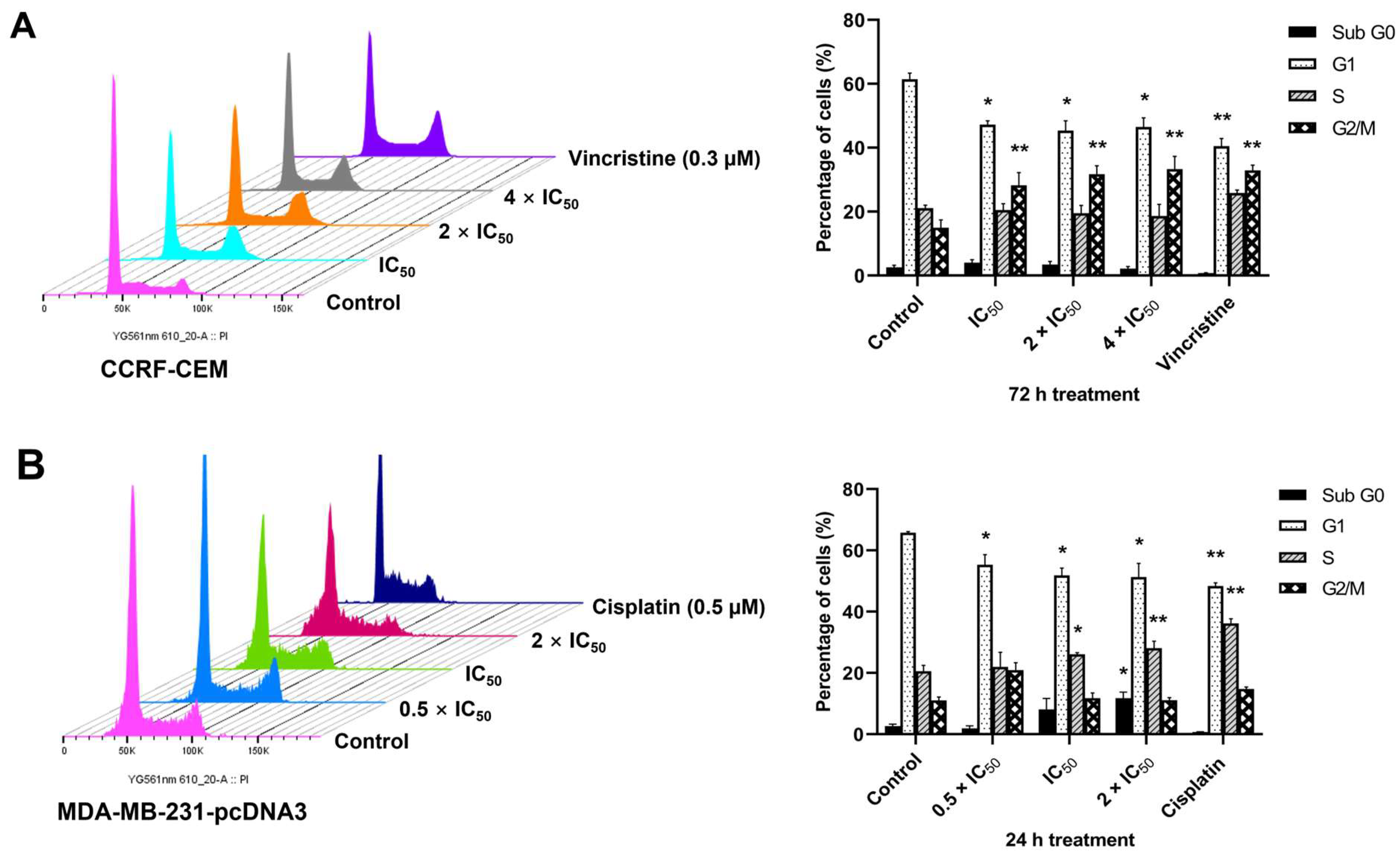
2.8. Cell Cycle Arrest
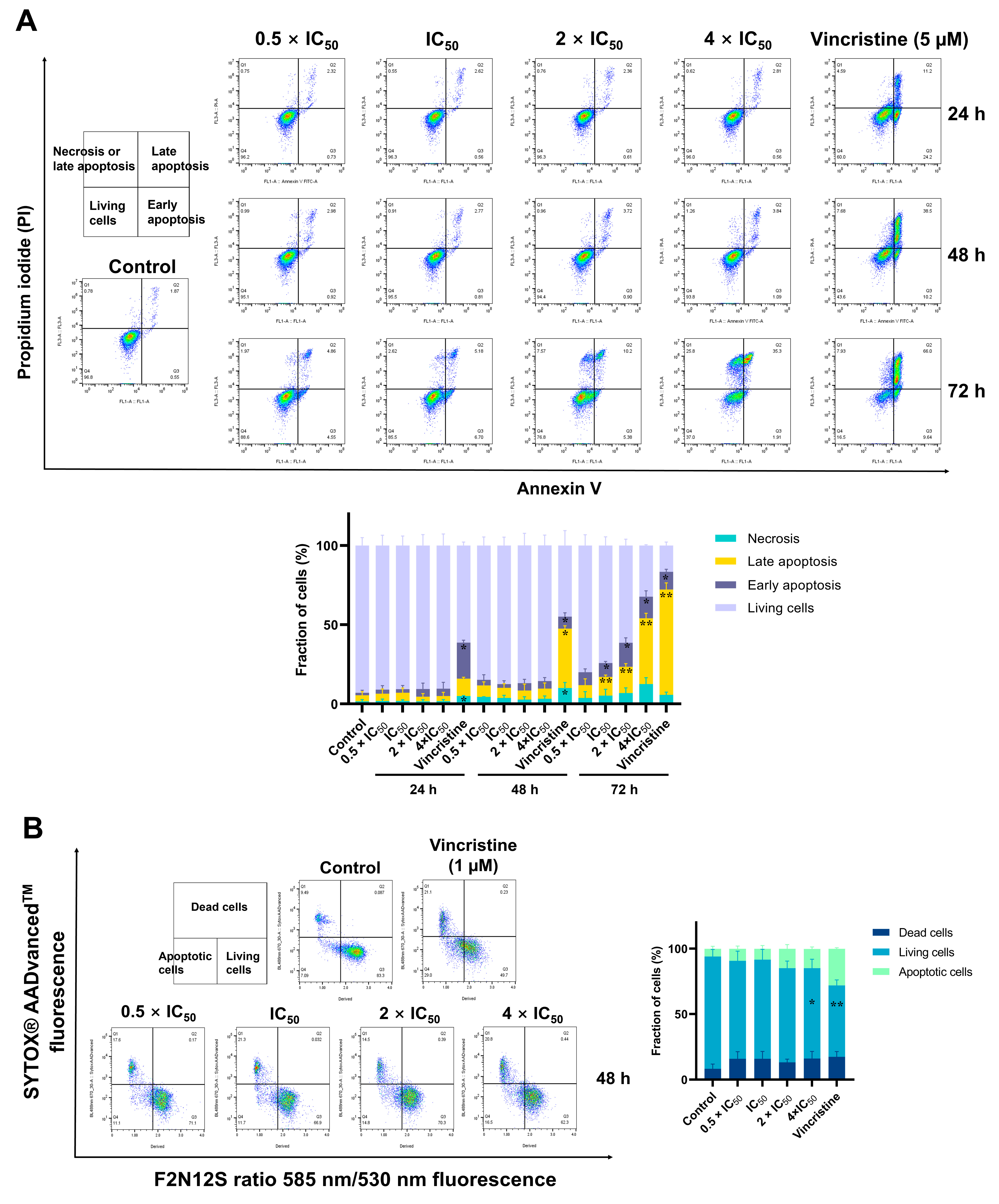
2.9. Detection of Apoptosis
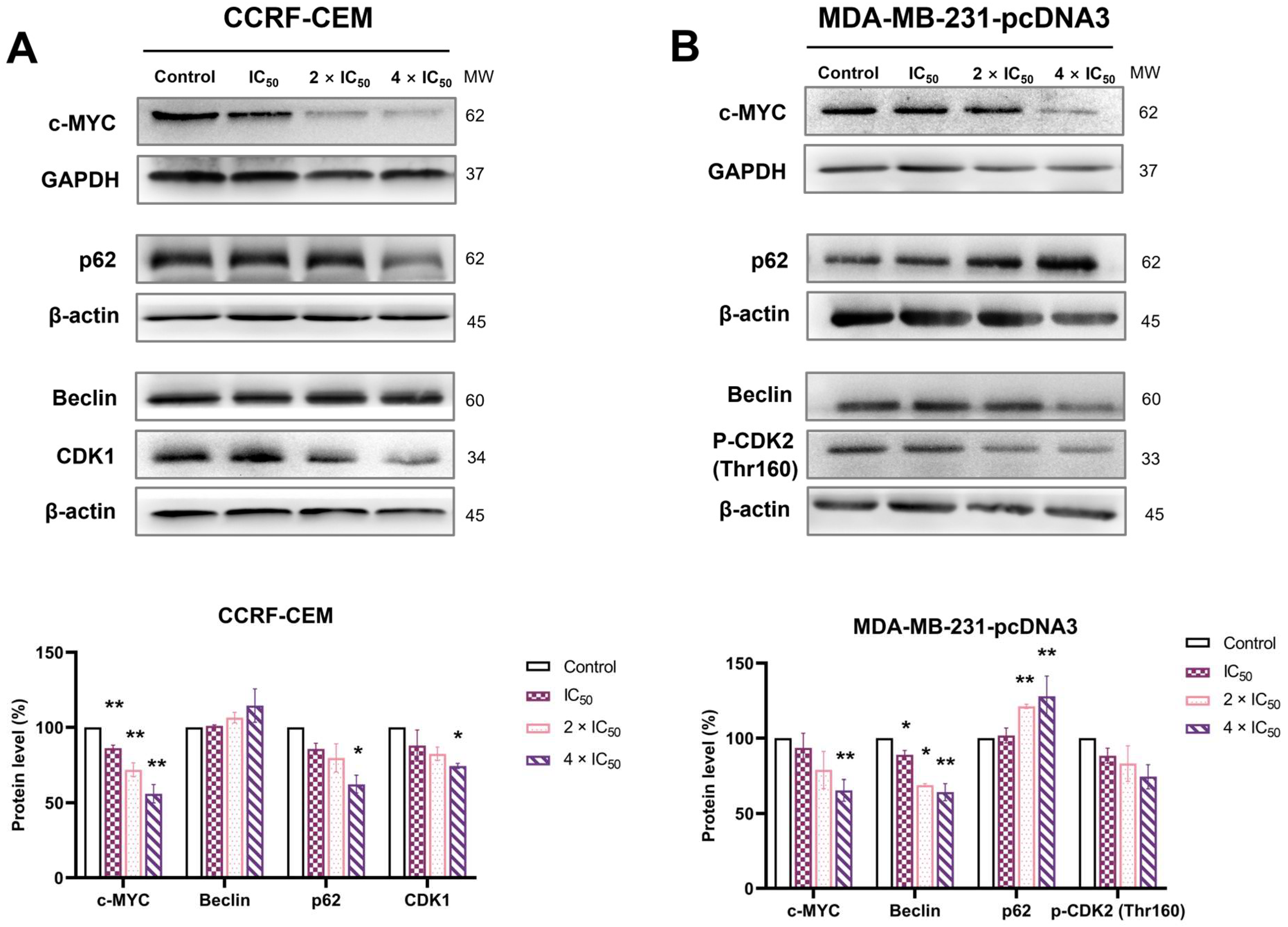
2.10. Western Blotting
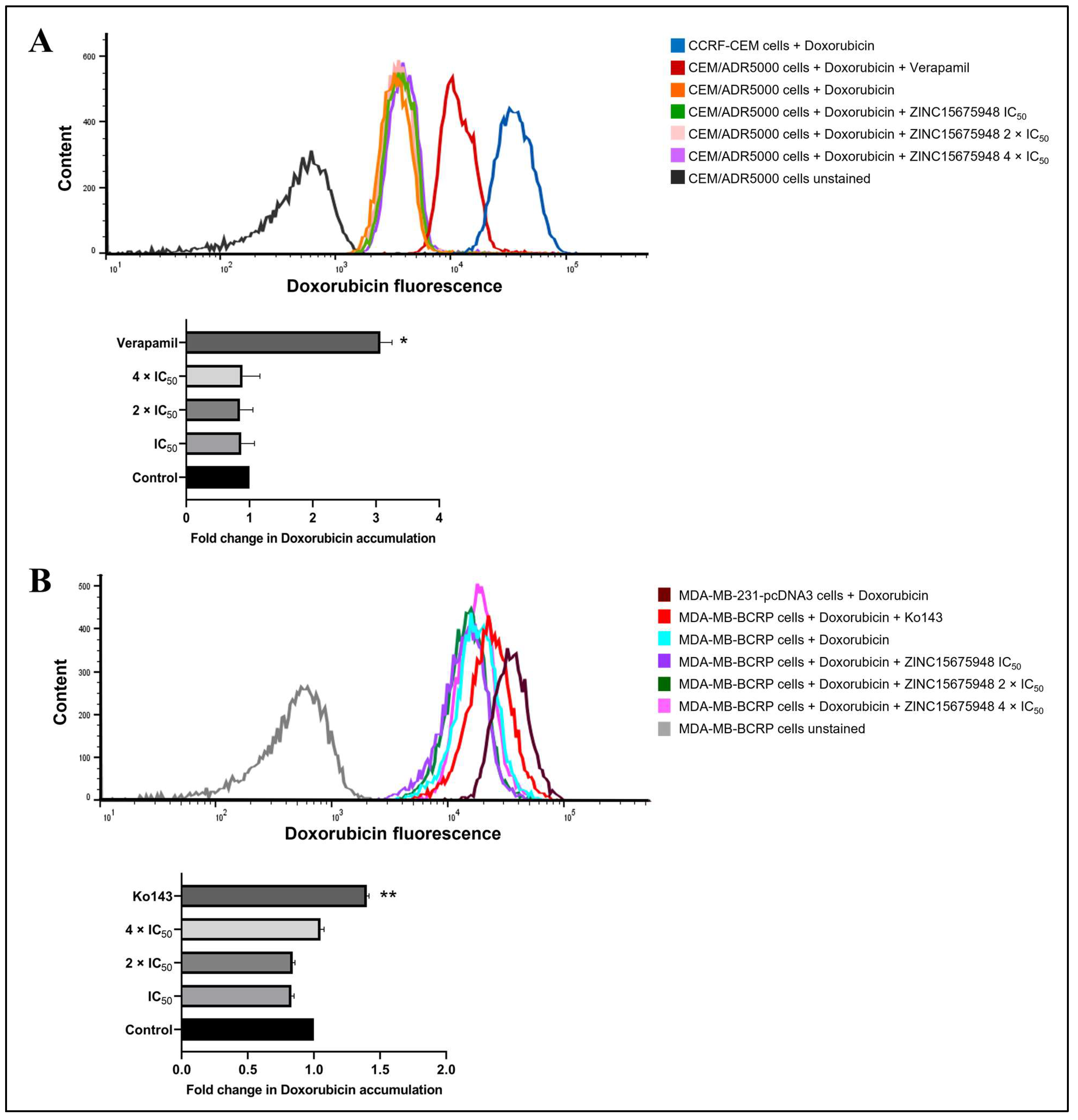
2.11. Doxorubicin Uptake Assay
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Cell Culture
4.3. Growth Inhibition Assay
4.4. Molecular Docking
4.5. Microscale Thermophoresis
4.6. c-MYC Reporter Assay
4.7. Gene Expression Profiles
4.8. Pathway Analysis of Microarray Data
4.9. Quantitative Real-Time Reverse Transcription PCR
4.10. Single Cell Gel Electrophoresis (Alkaline Comet Assay)
4.11. Cell Cycle Arrest
4.12. Detection of Apoptosis in Suspension Cells
4.13. Detection of Apoptosis in Adherent Cells
4.14. Protein Analyses by SDS-PAGE and Immunoblotting
4.15. Doxorubicin Uptake Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Bishop, J.M. Molecular themes in oncogenesis. Cell 1991, 64, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, C.; Resetca, D.; Redel, C.; Lin, P.; MacDonald, A.S.; Ciaccio, R.; Kenney, T.M.G.; Wei, Y.; Andrews, D.W.; Sunnerhagen, M.; et al. MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer 2021, 21, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Albihn, A.; Johnsen, J.I.; Henriksson, M.A. MYC in oncogenesis and as a target for cancer therapies. Adv. Cancer Res. 2010, 107, 163–224. [Google Scholar] [CrossRef]
- Sammak, S.; Hamdani, N.; Gorrec, F.; Allen, M.D.; Freund, S.M.V.; Bycroft, M.; Zinzalla, G. Crystal structures and nuclear magnetic resonance studies of the apo form of the c-MYC: MAX bHLHZip complex reveal a helical basic region in the absence of DNA. Biochemistry 2019, 58, 3144–3154. [Google Scholar] [CrossRef]
- Duffy, M.J.; O’Grady, S.; Tang, M.; Crown, J. MYC as a target for cancer treatment. Cancer Treat. Rev. 2021, 94, 102154. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef]
- Llombart, V.; Mansour, M.R. Therapeutic targeting of “undruggable” MYC. eBioMedicine 2022, 75, 103756. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Sears, R.C. The life cycle of C-myc: From synthesis to degradation. Cell Cycle 2004, 3, 1133–1137. [Google Scholar] [CrossRef]
- Dingar, D.; Tu, W.B.; Resetca, D.; Lourenco, C.; Tamachi, A.; De Melo, J.; Houlahan, K.E.; Kalkat, M.; Chan, P.K.; Boutros, P.C.; et al. MYC dephosphorylation by the PP1/PNUTS phosphatase complex regulates chromatin binding and protein stability. Nat. Commun. 2018, 9, 3502. [Google Scholar] [CrossRef] [PubMed]
- Fallah, Y.; Brundage, J.; Allegakoen, P.; Shajahan-Haq, A.N. MYC-driven pathways in breast cancer subtypes. Biomolecules 2017, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Boxer, L.M.; Dang, C.V. Translocations involving c-myc and c-myc function. Oncogene 2001, 20, 5595–5610. [Google Scholar] [CrossRef] [PubMed]
- Felsher, D.W.; Bishop, J.M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 1999, 4, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Hassan, A.; Khan, A.H.; Saleem, F.; Ahmad, H.; Khan, K.M. A patent review of pharmaceutical and therapeutic applications of oxadiazole derivatives for the treatment of chronic diseases (2013–2021). Expert Opin. Ther. Pat. 2022, 32, 969–1001. [Google Scholar] [CrossRef]
- Camci, M.; Karali, N. Bioisosterism: 1,2,4-oxadiazole rings. ChemMedChem 2023, 18, e202200638. [Google Scholar] [CrossRef]
- Atmaram, U.A.; Roopan, S.M. Biological activity of oxadiazole and thiadiazole derivatives. Appl. Microbiol. Biotechnol. 2022, 106, 3489–3505. [Google Scholar] [CrossRef]
- Dhameliya, T.M.; Chudasma, S.J.; Patel, T.M.; Dave, B.P. A review on synthetic account of 1,2,4-oxadiazoles as anti-infective agents. Mol. Divers. 2022, 26, 2967–2980. [Google Scholar] [CrossRef]
- Carbone, M.; Li, Y.; Irace, C.; Mollo, E.; Castelluccio, F.; Di Pascale, A.; Cimino, G.; Santamaria, R.; Guo, Y.-W.; Gavagnin, M. Structure and cytotoxicity of phidianidines A and B: First finding of 1,2,4-oxadiazole system in a marine natural product. Org. Lett. 2011, 13, 2516–2519. [Google Scholar] [CrossRef] [PubMed]
- Labriere, C.; Elumalai, V.; Staffansson, J.; Cervin, G.; Le Norcy, T.; Denardou, H.; Réhel, K.; Moodie, L.W.K.; Hellio, C.; Pavia, H.; et al. Phidianidine A and synthetic analogues as naturally inspired marine antifoulants. J. Nat. Prod. 2020, 83, 3413–3423. [Google Scholar] [CrossRef]
- Shamsi, F.; Hasan, P.; Queen, A.; Hussain, A.; Khan, P.; Zeya, B.; King, H.M.; Rana, S.; Garrison, J.; Alajmi, M.F.; et al. Synthesis and SAR studies of novel 1,2,4-oxadiazole-sulfonamide based compounds as potential anticancer agents for colorectal cancer therapy. Bioorg. Chem. 2020, 98, 103754. [Google Scholar] [CrossRef]
- Caneschi, W.; Enes, K.B.; Carvalho de Mendonça, C.; de Souza Fernandes, F.; Miguel, F.B.; da Silva Martins, J.; Le Hyaric, M.; Pinho, R.R.; Duarte, L.M.; Leal de Oliveira, M.A.; et al. Synthesis and anticancer evaluation of new lipophilic 1,2,4 and 1,3,4-oxadiazoles. Eur. J. Med. Chem. 2019, 165, 18–30. [Google Scholar] [CrossRef]
- Mohamed, M.F.A.; Marzouk, A.A.; Nafady, A.; El-Gamal, D.A.; Allam, R.M.; Abuo-Rahma, G.E.-D.A.; El Subbagh, H.I.; Moustafa, A.H. Design, synthesis and molecular modeling of novel aryl carboximidamides and 3-aryl-1,2,4-oxadiazoles derived from indomethacin as potent anti-inflammatory iNOS/PGE2 inhibitors. Bioorg. Chem. 2020, 105, 104439. [Google Scholar] [CrossRef]
- Il’in, M.V.; Sysoeva, A.A.; Bolotin, D.S.; Novikov, A.S.; Suslonov, V.V.; Rogacheva, E.V.; Kraeva, L.A.; Kukushkin, V.Y. Aminonitrones as highly reactive bifunctional synthons. An expedient one-pot route to 5-amino-1,2,4-triazoles and 5-amino-1,2,4-oxadiazoles—Potential antimicrobials targeting multi-drug resistant bacteria. New J. Chem. 2019, 43, 17358–17366. [Google Scholar] [CrossRef]
- Kim, J.; Shin, J.S.; Ahn, S.; Han, S.B.; Jung, Y.-S. 3-Aryl-1,2,4-oxadiazole derivatives active against human rhinovirus. ACS Med. Chem. Lett. 2018, 9, 667–672. [Google Scholar] [CrossRef]
- Dos Santos Filho, J.M.; de Queiroz, E.S.D.M.A.; Macedo, T.S.; Teixeira, H.M.P.; Moreira, D.R.M.; Challal, S.; Wolfender, J.L.; Queiroz, E.F.; Soares, M.B.P. Conjugation of N-acylhydrazone and 1,2,4-oxadiazole leads to the identification of active antimalarial agents. Bioorg. Med. Chem. 2016, 24, 5693–5701. [Google Scholar] [CrossRef]
- Mohammad, B.D.; Baig, M.S.; Bhandari, N.; Siddiqui, F.A.; Khan, S.L.; Ahmad, Z.; Khan, F.S.; Tagde, P.; Jeandet, P. Heterocyclic compounds as dipeptidyl peptidase-IV inhibitors with special emphasis on oxadiazoles as potent anti-diabetic agents. Molecules 2022, 27, 6001. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, T.; Chen, S.; Wu, M.; Han, J.; Li, Z. Design and synthesis of 3-(4-pyridyl)-5-(4-sulfamido-phenyl)-1,2,4-oxadiazole derivatives as novel GSK-3β inhibitors and evaluation of their potential as multifunctional anti-Alzheimer agents. Eur. J. Med. Chem. 2021, 209, 112874. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.B.; Fizazi, K.; Miller, K.; Higano, C.; Moul, J.W.; Akaza, H.; Morris, T.; McIntosh, S.; Pemberton, K.; Gleave, M. Phase 3, randomized, placebo-controlled study of zibotentan (ZD4054) in patients with castration-resistant prostate cancer metastatic to bone. Cancer 2012, 118, 5709–5718. [Google Scholar] [CrossRef] [PubMed]
- Engebraaten, O.; Vollan, H.K.M.; Børresen-Dale, A.L. Triple-negative breast cancer and the need for new therapeutic targets. Am. J. Pathol. 2013, 183, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pan, S.; Xie, T.; Liu, H. MYC in T-cell acute lymphoblastic leukemia: Functional implications and targeted strategies. Blood Sci. 2021, 3, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, M.F.; Dawood, M.; Mahmoud, N.; Elbadawi, M.; Sugimoto, Y.; Klauck, S.M.; Mohamed, N.; Efferth, T. 2α-Hydroxyalantolactone from Pulicaria undulata: Activity against multidrug-resistant tumor cells and modes of action. Phytomedicine 2021, 81, 153409. [Google Scholar] [CrossRef]
- Beaulieu, M.E.; Soucek, L. Finding MYCure. Mol. Cell. Oncol. 2019, 6, e1618178. [Google Scholar] [CrossRef]
- Whitfield, J.R.; Soucek, L. The long journey to bring a Myc inhibitor to the clinic. J. Cell Biol. 2021, 220, e202103090. [Google Scholar] [CrossRef]
- Ghobrial, A.; Flick, N.; Daly, R.; Hoffman, M.; Milcarek, C. ELL2 influences transcription elongation, splicing, Ig secretion and growth. J. Mucosal Immunol. Res. 2019, 3, 112. [Google Scholar]
- Chen, Y.; Zhou, C.; Ji, W.; Mei, Z.; Hu, B.; Zhang, W.; Zhang, D.; Wang, J.; Liu, X.; Ouyang, G.; et al. ELL targets c-Myc for proteasomal degradation and suppresses tumour growth. Nat. Commun. 2016, 7, 11057. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Yap, J.L.; Wang, H.; Hu, A.; Chauhan, J.; Jung, K.Y.; Gharavi, R.B.; Prochownik, E.V.; Fletcher, S. Pharmacophore identification of c-Myc inhibitor 10074-G5. Bioorg. Med. Chem. Lett. 2013, 23, 370–374. [Google Scholar] [CrossRef]
- Hammoudeh, D.I.; Follis, A.V.; Prochownik, E.V.; Metallo, S.J. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 2009, 131, 7390–7401. [Google Scholar] [CrossRef]
- Massó-Vallés, D.; Soucek, L. Blocking Myc to treat cancer: Reflecting on two decades of omomyc. Cells 2020, 9, 883. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Jain, A.D.; Truica, M.I.; Izquierdo-Ferrer, J.; Anker, J.F.; Lysy, B.; Sagar, V.; Luan, Y.; Chalmers, Z.R.; Unno, K.; et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell 2019, 36, 483–497.e15. [Google Scholar] [CrossRef] [PubMed]
- Boike, L.; Cioffi, A.G.; Majewski, F.C.; Co, J.; Henning, N.J.; Jones, M.D.; Liu, G.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; et al. Discovery of a functional covalent ligand targeting an intrinsically disordered cysteine within MYC. Cell Chem. Biol. 2021, 28, 4–13.e17. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Saha, P.; Das, T.; Dash, J. Target guided synthesis using DNA nano-templates for selectively assembling a G-quadruplex binding c-MYC inhibitor. Nat. Commun. 2017, 8, 16103. [Google Scholar] [CrossRef]
- Michel, J.; Cuchillo, R. The impact of small molecule binding on the energy landscape of the intrinsically disordered protein C-myc. PLoS ONE 2012, 7, e41070. [Google Scholar] [CrossRef]
- Follis, A.V.; Hammoudeh, D.I.; Wang, H.; Prochownik, E.V.; Metallo, S.J. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem. Biol. 2008, 15, 1149–1155. [Google Scholar] [CrossRef]
- Farrell, A.S.; Sears, R.C. MYC degradation. Cold Spring Harb. Perspect. Med. 2014, 4, a014365. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, X.-X.; Sears, R.C.; Dai, M.-S. Writing and erasing MYC ubiquitination and SUMOylation. Genes Dis. 2019, 6, 359–371. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Yang, C. Evaluating In Vitro DNA Damage Using Comet Assay. J. Vis. Exp. 2017, 128, e56450. [Google Scholar] [CrossRef]
- Goga, A.; Yang, D.; Tward, A.D.; Morgan, D.O.; Bishop, J.M. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat. Med. 2007, 13, 820–827. [Google Scholar] [CrossRef]
- Gu, Y.; Rosenblatt, J.; Morgan, D.O. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992, 11, 3995–4005. [Google Scholar] [CrossRef]
- Kurbegovic, A.; Trudel, M. The master regulators Myc and p53 cellular signaling and functions in polycystic kidney disease. Cell. Signal. 2020, 71, 109594. [Google Scholar] [CrossRef] [PubMed]
- Kelly, G.L.; Grabow, S.; Glaser, S.P.; Fitzsimmons, L.; Aubrey, B.J.; Okamoto, T.; Valente, L.J.; Robati, M.; Tai, L.; Fairlie, W.D.; et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev. 2014, 28, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. p62 links the autophagy pathway and the ubiqutin–proteasome system upon ubiquitinated protein degradation. Cell Mol. Biol. Lett. 2016, 21, 29. [Google Scholar] [CrossRef] [PubMed]
- Elbadawi, M.; Boulos, J.C.; Dawood, M.; Zhou, M.; Gul, W.; ElSohly, M.A.; Klauck, S.M.; Efferth, T. The novel artemisinin dimer isoniazide ELI-XXIII-98-2 induces c-MYC inhibition, DNA damage, and autophagy in leukemia cells. Pharmaceutics 2023, 15, 1107. [Google Scholar] [CrossRef]
- Li, W.; Zhang, H.; Assaraf, Y.G.; Zhao, K.; Xu, X.; Xie, J.; Yang, D.H.; Chen, Z.S. Overcoming ABC transporter-mediated multidrug resistance: Molecular mechanisms and novel therapeutic drug strategies. Drug Resist. Updates 2016, 27, 14–29. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Tiwari, A.K.; Sodani, K.; Dai, C.L.; Ashby, C.R., Jr.; Chen, Z.S. Revisiting the ABCs of multidrug resistance in cancer chemotherapy. Curr. Pharm. Biotechnol. 2011, 12, 570–594. [Google Scholar] [CrossRef]
- Doyle, L.; Ross, D.D. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2). Oncogene 2003, 22, 7340–7358. [Google Scholar] [CrossRef]
- Silva, R.; Vilas-Boas, V.; Carmo, H.; Dinis-Oliveira, R.J.; Carvalho, F.; de Lourdes Bastos, M.; Remião, F. Modulation of P-glycoprotein efflux pump: Induction and activation as a therapeutic strategy. Pharmacol. Ther. 2015, 149, 1–123. [Google Scholar] [CrossRef]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef]
- Modi, A.; Roy, D.; Sharma, S.; Vishnoi, J.R.; Pareek, P.; Elhence, P.; Sharma, P.; Purohit, P. ABC transporters in breast cancer: Their roles in multidrug resistance and beyond. J. Drug Target. 2022, 30, 927–947. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.D.; Handley, M.D.; Gottesman, M.M. Is resistance useless? Multidrug resistance and collateral sensitivity. Trends Pharmacol. Sci. 2009, 30, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T.; Konkimalla, V.B.; Wang, Y.F.; Sauerbrey, A.; Meinhardt, S.; Zintl, F.; Mattern, J.; Volm, M. Prediction of broad spectrum resistance of tumors towards anticancer drugs. Clin. Cancer Res. 2008, 14, 2405–2412. [Google Scholar] [CrossRef]
- Saeed, M.E.M.; Boulos, J.C.; Elhaboub, G.; Rigano, D.; Saab, A.; Loizzo, M.R.; Hassan, L.E.A.; Sugimoto, Y.; Piacente, S.; Tundis, R.; et al. Cytotoxicity of cucurbitacin E from Citrullus colocynthis against multidrug-resistant cancer cells. Phytomedicine 2019, 62, 152945. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.E.M.; Mahmoud, N.; Sugimoto, Y.; Efferth, T.; Abdel-Aziz, H. Molecular determinants of sensitivity or resistance of cancer cells toward sanguinarine. Front. Pharmacol. 2018, 9, 136. [Google Scholar] [CrossRef]
- Doyle, L.A.; Yang, W.; Abruzzo, L.V.; Krogmann, T.; Gao, Y.; Rishi, A.K.; Ross, D.D. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. USA 1998, 95, 15665–15670. [Google Scholar] [CrossRef]
- Kadioglu, O.; Cao, J.; Kosyakova, N.; Mrasek, K.; Liehr, T.; Efferth, T. Genomic and transcriptomic profiling of resistant CEM/ADR-5000 and sensitive CCRF-CEM leukaemia cells for unravelling the full complexity of multi-factorial multidrug resistance. Sci. Rep. 2016, 6, 36754. [Google Scholar] [CrossRef]
- Efferth, T.; Sauerbrey, A.; Olbrich, A.; Gebhart, E.; Rauch, P.; Weber, H.O.; Hengstler, J.G.; Halatsch, M.E.; Volm, M.; Tew, K.D.; et al. Molecular modes of action of artesunate in tumor cell lines. Mol. Pharmacol. 2003, 64, 382–394. [Google Scholar] [CrossRef]
- Kimmig, A.; Gekeler, V.; Neumann, M.; Frese, G.; Handgretinger, R.; Kardos, G.; Diddens, H.; Niethammer, D. Susceptibility of multidrug-resistant human leukemia cell lines to human interleukin 2-activated killer cells. Cancer Res. 1990, 50, 6793–6799. [Google Scholar]
- Abdelfatah, S.; Böckers, M.; Asensio, M.; Kadioglu, O.; Klinger, A.; Fleischer, E.; Efferth, T. Isopetasin and S-isopetasin as novel P-glycoprotein inhibitors against multidrug-resistant cancer cells. Phytomedicine 2021, 86, 153196. [Google Scholar] [CrossRef]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Yin, X.; Giap, C.; Lazo, J.S.; Prochownik, E.V. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 2003, 22, 6151–6159. [Google Scholar] [CrossRef]
- Kallio, M.A.; Tuimala, J.T.; Hupponen, T.; Klemelä, P.; Gentile, M.; Scheinin, I.; Koski, M.; Käki, J.; Korpelainen, E.I. Chipster: User-friendly analysis software for microarray and other high-throughput data. BMC Genom. 2011, 12, 507. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Boulos, J.C.; Klauck, S.M.; Efferth, T. The cardiac glycoside ZINC253504760 induces parthanatos-type cell death and G2/M arrest via downregulation of MEK1/2 phosphorylation in leukemia cells. Cell Biol. Toxicol. 2023, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Benhusein, G.M.; Mutch, E.; Aburawi, S.; Williams, F.M. Genotoxic effect induced by hydrogen peroxide in human hepatoma cells using comet assay. Libyan J. Med. 2010, 5, 4637. [Google Scholar] [CrossRef]
- Gyori, B.M.; Venkatachalam, G.; Thiagarajan, P.S.; Hsu, D.; Clement, M.V. OpenComet: An automated tool for comet assay image analysis. Redox Biol. 2014, 2, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R. The comet assay for DNA damage and repair. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Marfell, B.J.; Scott, A.P.; Waterhouse, N.J. Quantitation of apoptosis and necrosis by annexin V binding, propidium iodide uptake, and flow cytometry. Cold Spring Harb. Protoc. 2016, 2016, 953–957. [Google Scholar] [CrossRef] [PubMed]
- Shynkar, V.V.; Klymchenko, A.S.; Kunzelmann, C.; Duportail, G.; Muller, C.D.; Demchenko, A.P.; Freyssinet, J.-M.; Mely, Y. Fluorescent biomembrane probe for ratiometric detection of apoptosis. J. Am. Chem. Soc. 2007, 129, 2187–2193. [Google Scholar] [CrossRef] [PubMed]
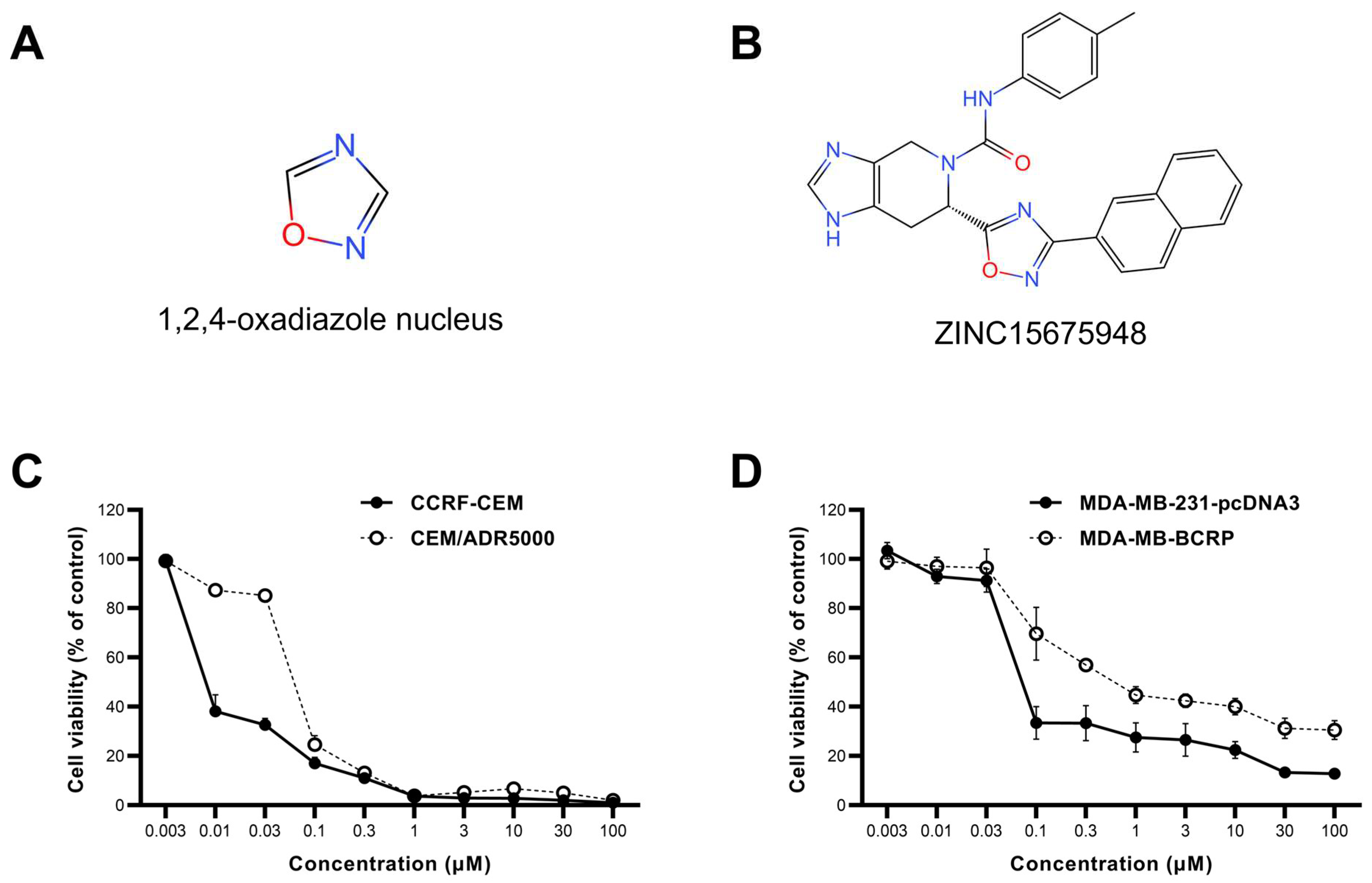

| Cell Lines | IC50 (µM) | Degree of Resistance |
|---|---|---|
| CCRF-CEM | 0.008 ± 0.001 | 8.37 |
| CEM/ADR5000 | 0.071 ± 0.002 | |
| MDA-MB-231-pcDNA3 | 0.08 ± 0.004 | 9 |
| MDA-MB-BCRP | 0.72 ± 0.07 |
| Compound | Lowest Binding Energy (kcal/mol) | pKi (µM) | Amino Acids Interactions (Residues in H-Bond Bolded) |
|---|---|---|---|
| ZINC15675948 | −9.91 | 0.055 | Arg924, Asp926, Gln927, Tyr949, Ile950, Val953 |
| 10058-F4 | −4.92 | 247.03 ± 1.5 | Arg925, Gln927, Pro929, Leu931, Glu932 |
| 10074-A4 | −6.42 ± 0.01 | 19.53 ± 0.30 | Arg925, Asp926, Gln927 |
| 10074-G5 | −6.96 ± 0.01 | 7.93 ± 0.13 | Pro929, Pro930, Lys945, Ala948 |
| Protein | Compound | Lowest Binding Energy (kcal/mol) | pKi (µM) | Amino Acids Interactions (Residues in H-Bond Bolded) |
|---|---|---|---|---|
| P-gp | ZINC15675948 | −10.55 ± 0.24 | 0.07 ± 0.01 | Trp232, Phe239, Ala295, Asn296, Ile299, Phe770, Gln773, Gly774, Met876, Gln990, Phe994 |
| Doxorubicin | −6.42 ± 0.05 | 147.35 ± 72.62 | Ala229, Trp232, Phe303, Ile306, Phe343, Gln725, Ala871, Gly872, Phe983, Met986, Ala987, Gln990 | |
| Verapamil | −7.61 ± 0.31 | 3.0 ± 1.46 | Ser228, Ala229, Trp232, Ile306, Phe336, Phe343, Gln725, Phe728, Tyr953, Phe983, Met986, Gln990 | |
| BCRP | ZINC15675948 | −11.49 ± 0.01 | 0.007 ± 0.004 | A: Leu405, Phe431, Phe432, Thr435, Asn436, Phe439, Thr542, Met549, Leu555 B: Phe432, Thr435, Phe439 |
| Doxorubicin | −7.00 ± 0.49 | 122.79 ± 10.96 | A: Phe431, Phe432, The435, Asn436, Phe439, Ser440 B: The542, Ile 543, Val546, Met549, Ile555 | |
| Ko143 | −10.24 ± 0.19 | 0.03 ± 0.01 | A: Leu405, Phe431, Phe432, Thr435, Asn436, Phe439, Met549, Leu555 B: Phe431, Phe32, Thr435, Phe439, Val546, Met549 |
| Gene Symbol | Forward Primer | Reverse Primer |
|---|---|---|
| RAD21 | GAGTCAGCTATGCCTCCACC | TGGAGGTTCTTCTGGGGGAA |
| HMGCS1 | CTTTCGTGGCTCACTCCCTT | GTTTCCTCCTTCGGGCACA |
| PGK1 | TGTGTGGAATGGTCCTGTGG | TGGCTTTCACCACCTCATCC |
| ATP5MF | CGGACACCAGGACTCCAAAA | GGACTGAAGTCCCGCATCAA |
| CITED2 | GGCGAAGCTGGGGAATAACA | AATCAGCCCTCCTCATCCTG |
| HSPD1 | GCCGCCCCGCAGAAAT | AAGCCCGAGTGAGATGAGGA |
| H4C3 | CAGGGCATTACAAAACCGGC | GTGCTCCGTATAGGTGACGG |
| DHFR | GCCACCGCTCAGGAATGAAT | AGGTTGTGGTCATTCTCTGGAA |
| c-MYC | ACACTAACATCCCACGCTCTG | CTCGCTAAGGCTGGGGAAAG |
| GAPDH | ATGAATGGGCAGCCGTTAGG | AGCATCACCCGGAGGAGAAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, M.; Boulos, J.C.; Omer, E.A.; Klauck, S.M.; Efferth, T. Modes of Action of a Novel c-MYC Inhibiting 1,2,4-Oxadiazole Derivative in Leukemia and Breast Cancer Cells. Molecules 2023, 28, 5658. https://doi.org/10.3390/molecules28155658
Zhou M, Boulos JC, Omer EA, Klauck SM, Efferth T. Modes of Action of a Novel c-MYC Inhibiting 1,2,4-Oxadiazole Derivative in Leukemia and Breast Cancer Cells. Molecules. 2023; 28(15):5658. https://doi.org/10.3390/molecules28155658
Chicago/Turabian StyleZhou, Min, Joelle C. Boulos, Ejlal A. Omer, Sabine M. Klauck, and Thomas Efferth. 2023. "Modes of Action of a Novel c-MYC Inhibiting 1,2,4-Oxadiazole Derivative in Leukemia and Breast Cancer Cells" Molecules 28, no. 15: 5658. https://doi.org/10.3390/molecules28155658
APA StyleZhou, M., Boulos, J. C., Omer, E. A., Klauck, S. M., & Efferth, T. (2023). Modes of Action of a Novel c-MYC Inhibiting 1,2,4-Oxadiazole Derivative in Leukemia and Breast Cancer Cells. Molecules, 28(15), 5658. https://doi.org/10.3390/molecules28155658







