1. Introduction
Free radicals are generated within the body during normal metabolic activities and are markedly involved in many signaling pathways that control development and maintain cellular homeostasis [
1,
2]. However, the overproduction of these free radicals from both endo/exogenous sources has a great impact on humans in the etiology of various diseases [
3]. The body possesses defense mechanisms against antioxidant nutrients and enzymes that arrest the damaging properties of the free radicals. However, continuous exposure to chemicals and contaminants may increase the amount of free radicals in the body beyond its ability to control and cause irreversible oxidative damage [
4].
Therefore, antioxidants with free radical scavenging potential may be relevant in the therapeutic and prevention of diseases where free radicals are implicated. In addition to natural antioxidants such as vitamin C, vitamin E and carotenoid flavonoids, a number of cheaper and effective synthetic antioxidants have been prepared in research laboratories with promising antioxidant potential even higher than natural antioxidants in some cases [
5].
An important class of compounds with excellent antioxidant potential appeared in the form of Schiff base transition metal complexes [
6,
7,
8,
9]. The azomethine (-HC=N) linkage of Schiff bases provides an opportunity for remarkable biological activities [
10]. The lone pair of electrons on the nitrogen atom of the azomethine group has good chelating ability on Schiff bases, especially in combination with other donor atoms close to the azomethine group, and can result in the formation of stable transition metal complexes with simple synthesis and versatility and has a wide range of applications [
7].
In addition to their antioxidant potential, Schiff base transition metal complexes have also shown encouraging binding abilities with DNA [
6,
8], a primary target for most chemotherapeutic agents. Bio-essential transition metals such as copper, cobalt and nickel complexes have shown their efficiencies as potential DNA binders and anticancer agents [
9,
10], thus offering a safe and more effective alternative to the available platinum-based drugs with higher toxicity, tumor resistance and severe side effects [
10].
Keeping in view the importance of first-row transition metal Schiff base complexes, in the present study, synthesis, characterization, antioxidant, DNA binding activity and molecular docking studies of newly synthesized Schiff base ligand and its Co(II), Ni(II), Cu(II) and Zn(II) complexes were evaluated.
2. Results and Discussion
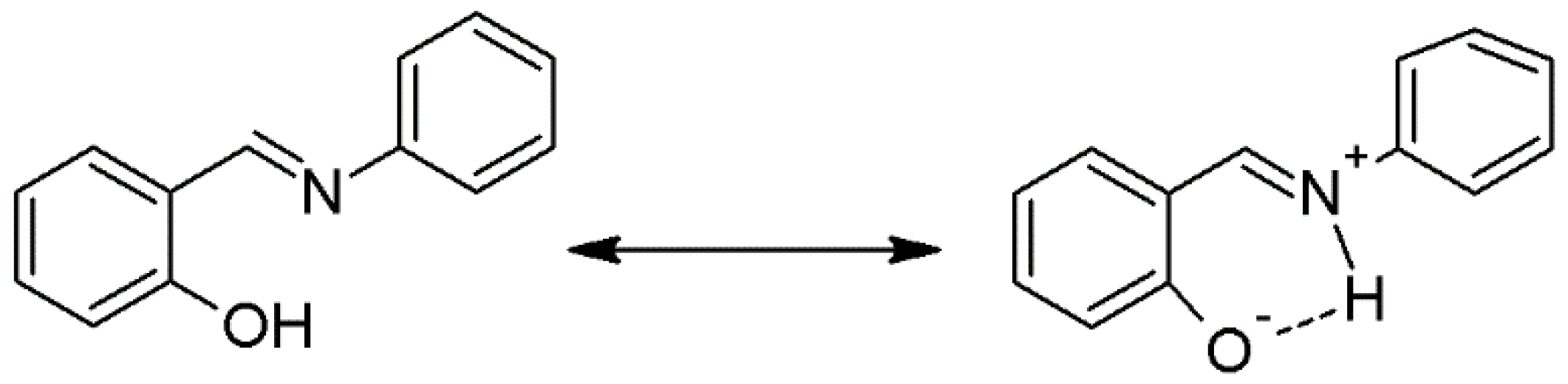
The Schiff base ligand 2-[(phenylimino)methyl]phenol (H-pimp) was synthesized by the condensation reaction of salicylaldehyde with aniline and characterized through different spectroscopic and analytical techniques. The infrared spectral data of the ligand (H-pimp) revealed a peak around 3200 cm
−1 assigned to the NH stretch. The peak is weakly observed because of the possible intramolecular hydrogen bonding as shown in
Scheme 1.
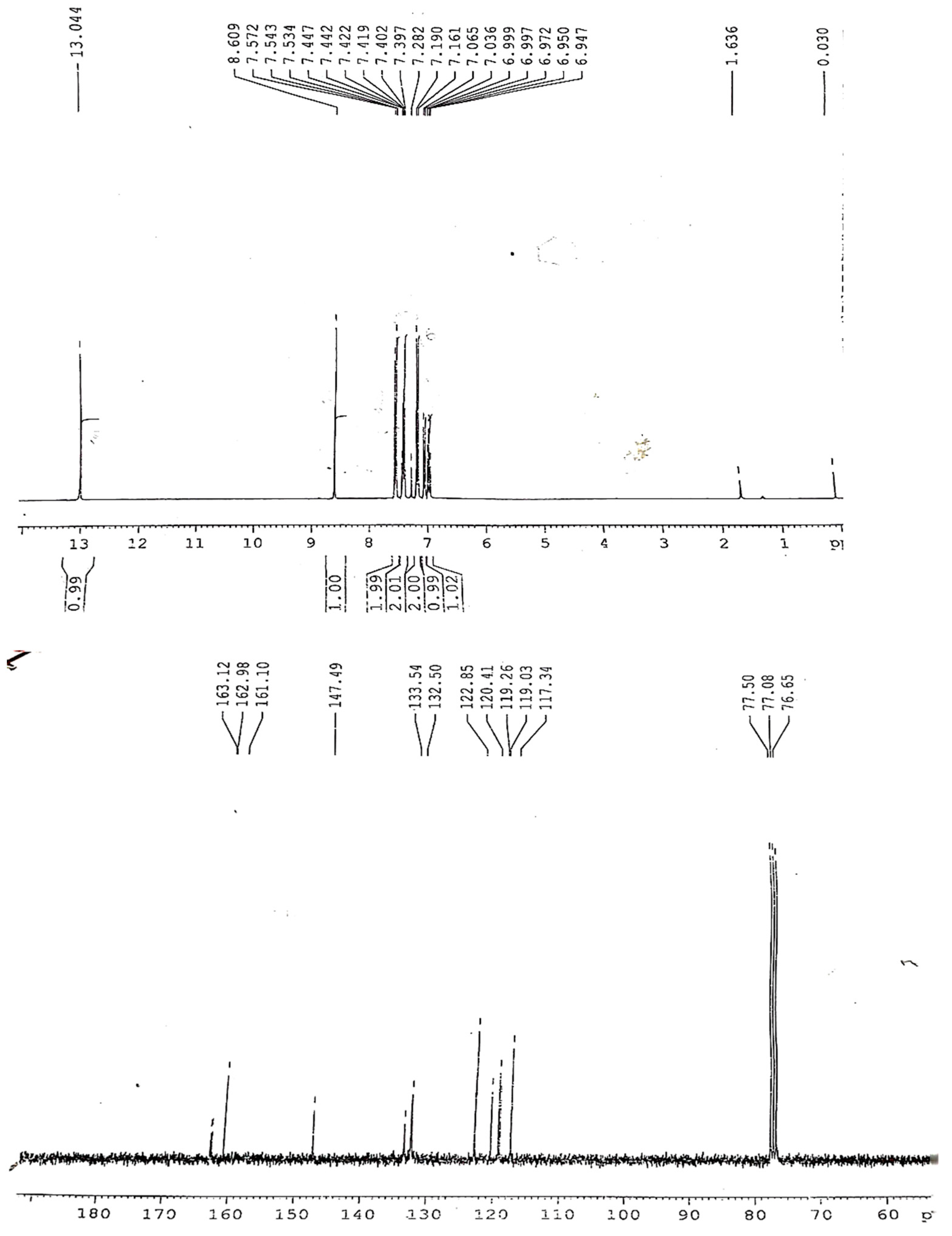
The broad peak for hydroxyl stretch is obscured by the NH band appearance. The azomethine is observed at the expected position. The formation of the ligand is further supported by the NMR spectra. The
1H-NMR of the ligand (H-pimp) revealed a singlet signal assigned for each azomethine moiety and the phenolic hydroxyl group. The rest of the spectrum shows peaks for the aromatic protons.
13C-NMR shows an imine peak at 162 ppm, and the hydroxyl peak at 148 ppm (
Figure 1). The Schiff base ligand reacted further with metal ions in a 2:1 molar ratio to produce metal derivatives of the anionic ligand with [M(pimp)
2] compositions as shown in
Scheme 1.
The IR spectra of the metal complexes share combine features of bonding irrespective of the metal center. The H-pimp ligand is behaving as a monoanionic ligand, offering coordination sites like O- and HC=N. Both the stretching bands were seen displaced from the position observed in the spectrum of the free ligand. The rest of the spectra show differences due to the complexation. The elemental compositions in metal complexes also revealed closeness for the [M(pimp)2] compositions in all the metal complexes. Therefore, it has been observed unambiguously that metal complexation occurs through the bonding of two molecules of the ligand with metal centers. The metal complexes were also observed to be non-electrolyte in nature.
The UV-visible spectra of the metal complexes were recorded within the region 320–1000 nm in a methanolic solution. The Co(pimp)2 complex showed a single and broadband at 860 nm assigned to the distorted square planar 2A2g →2B1g transition. The other transitions like 1A1g→1B1g and 1A1g→Eg were considered enfolded within the broad 2A2g →2B1g transition. Similarly, the visible spectrum of Ni(pimp)2 metal complex also revealed a single transition 1A1g → 1A2g for the distorted square planer geometry. The Cu(pimp)2 showed a weak transition at 660 nm assigned to the dz2 → dx2 − y2 in a distorted tetrahedral geometry. All the transitions in metal complexes were assigned charge transfer ligands to metal transitions where the imine group is involved in donating the electron density towards metal orbitals.
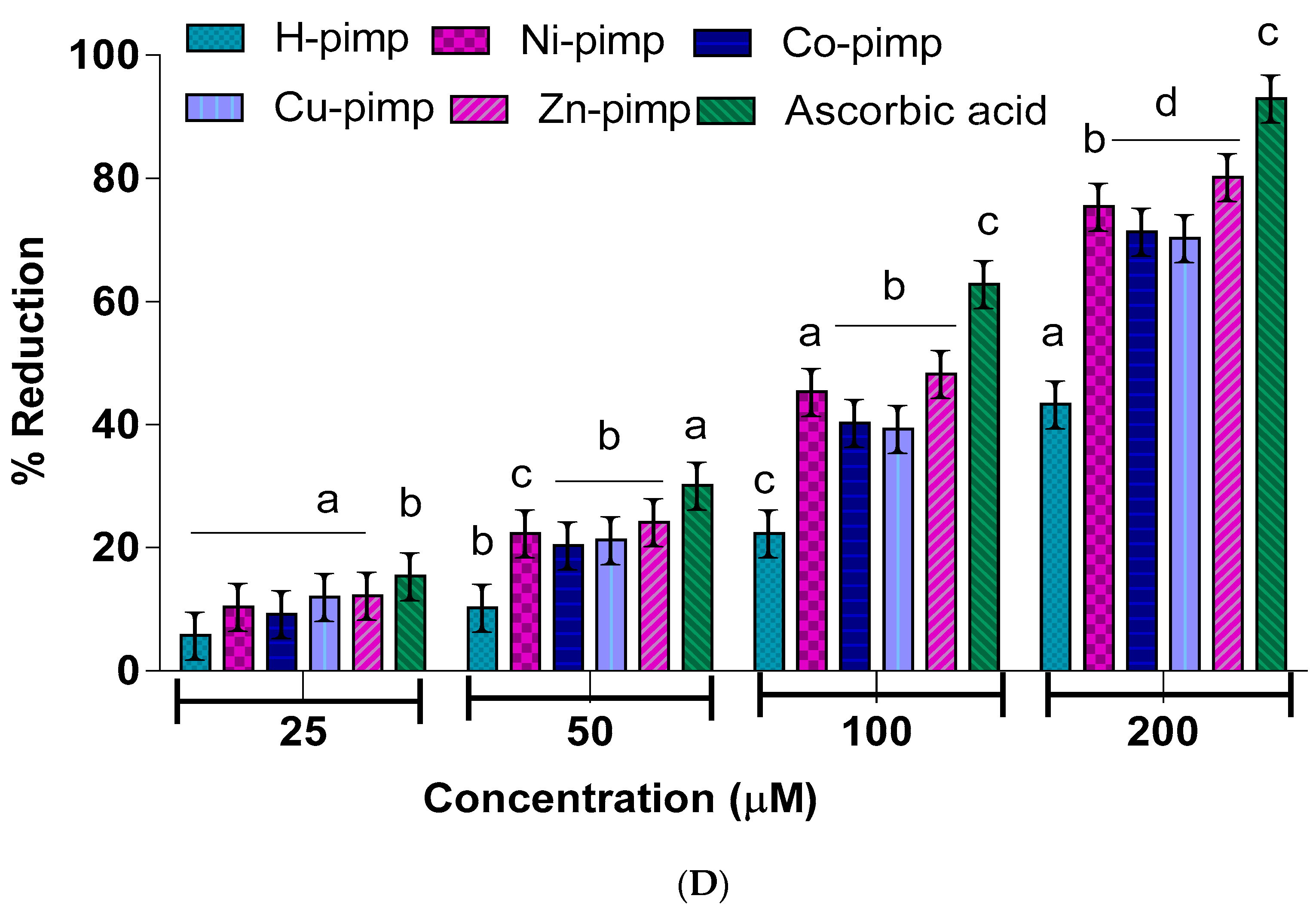
2.1. In Vitro Antioxidant Potential of the Synthesized Compounds
The antioxidant potential of the Schiff base ligand H-pimp and its synthesized metal complexes M(pimp)
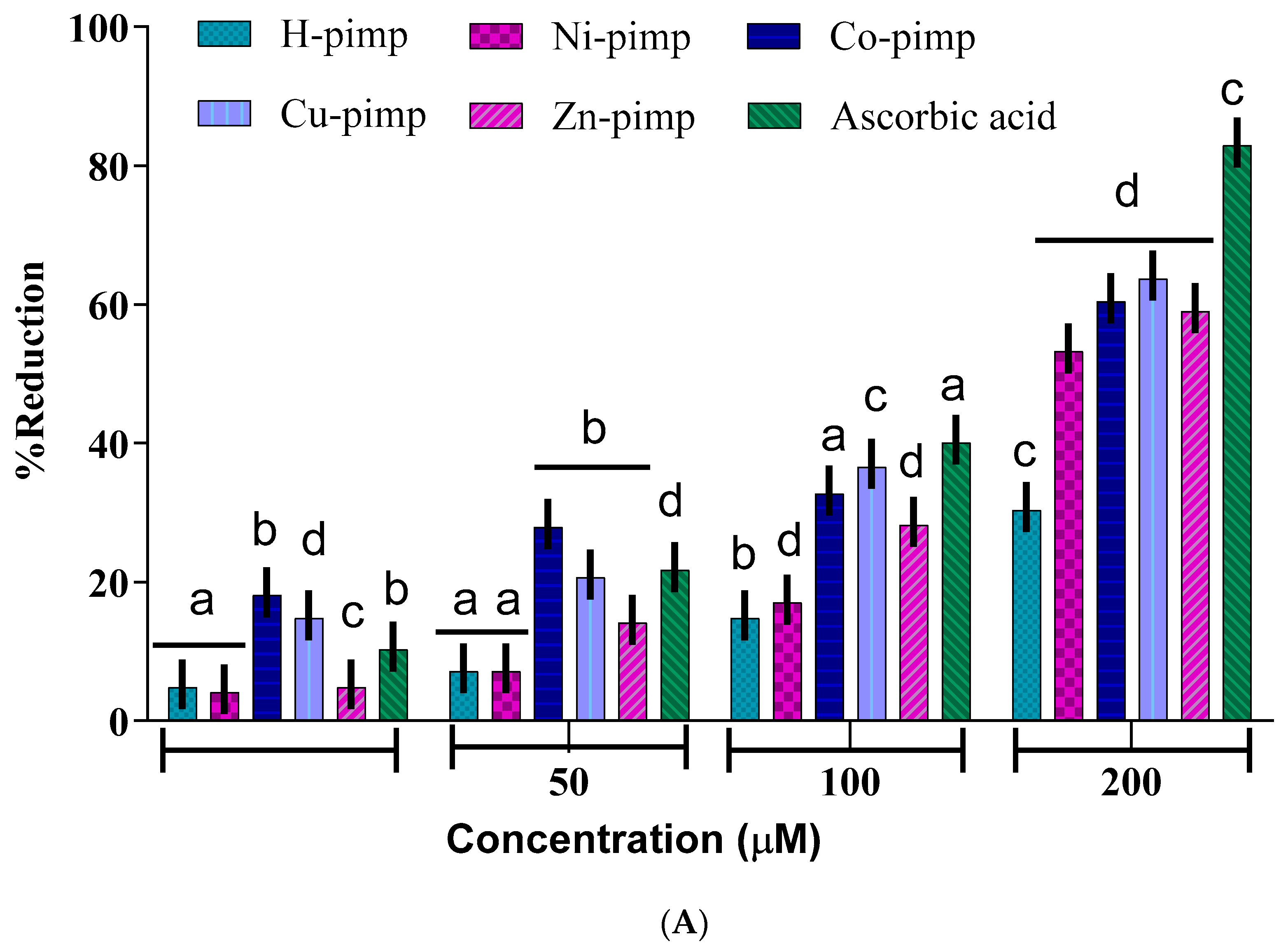
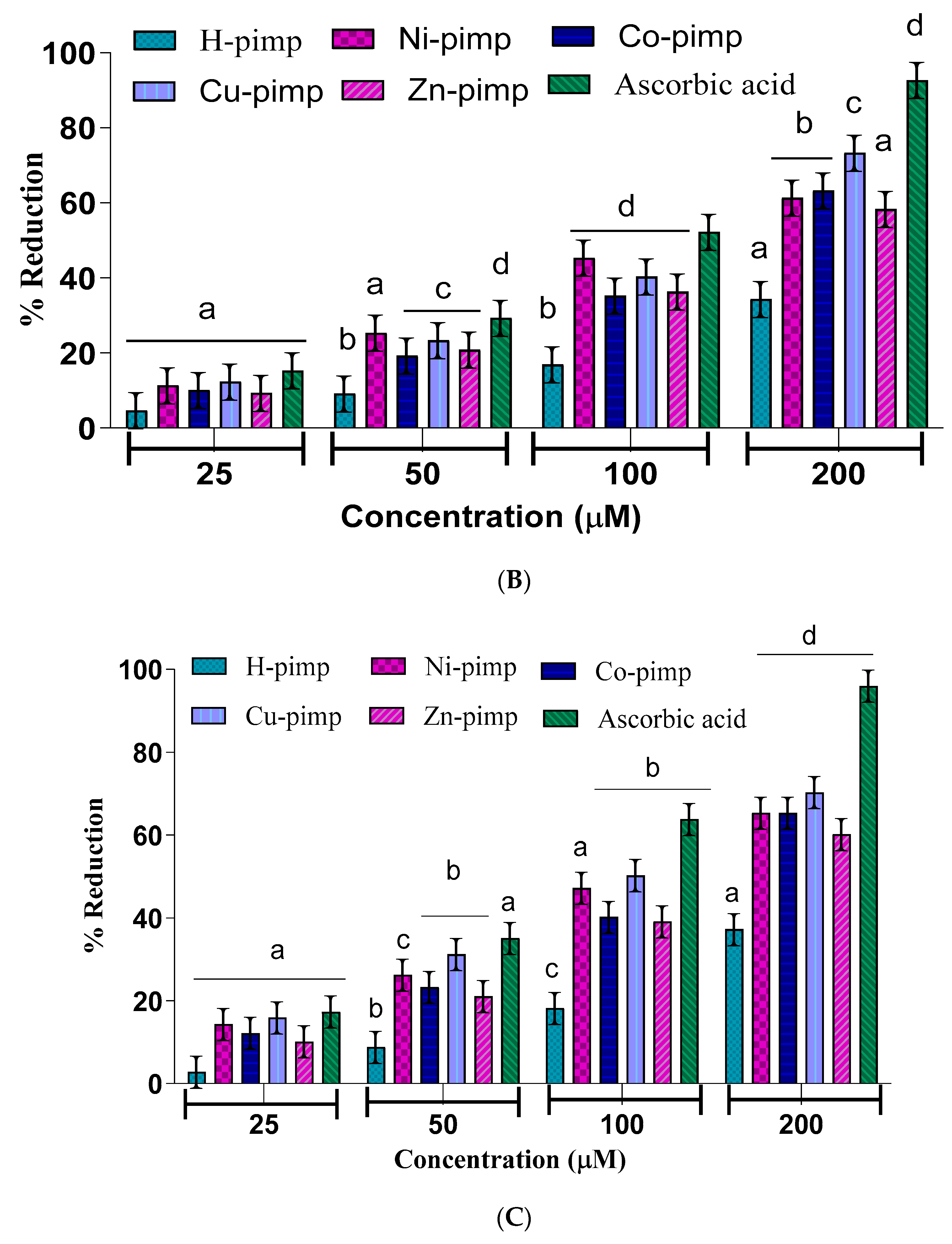
2 was checked by different assays, including a DPPH-free radical scavenging assay, a ferric ion reducing power assay (FRAP assay), total antioxidant activity (TAA assay) and hydroxyl radical scavenging activity (•OH assay) against ascorbic acid as a standard according to spectrophotometric measurements. The experimental findings presented in
Figure 2A–D have shown dose-dependent antioxidant potential for the synthesized compounds. The antioxidant potential data in terms of IC
50 values for the studied assays are given in
Table 1.
The
Figure 2A–D shows the concentration of the reducing agent required to produce 50% scavenging activity. A lower IC
50 value corresponds to a higher free radical scavenging or antioxidant activity.
The IC
50 value data shown in
Table 1 reflect the ligand H-pimp and its transition metal complexes M(pimp)
2, where M = Cu
2+, Ni
2+, Co
2+ and Zn
2+ show lesser antioxidant potential compared to the standard ascorbic acid in all assays. Another important point is the higher antioxidant potential of the synthesized metal complexes compared to the free ligand H-pimp.
Figure 2A–D shows that the higher antioxidant potential after complexation could be attributed to the presence of an electropositive metal center, resulting in the activation of π electronic cloud of the benzene ring of the ligand center for antioxidant activity. In addition, the chelate formation during complexation results in the redistribution of the electronic cloud over the whole chelate ring, which could enhance the electron-donating ability of the complex, hence its antioxidant potential. Among metal complexes, the highest antioxidant potential was shown by the copper complex Cu(pimp)
2 in all assays in
Figure 2A–D. The zinc complex Zn(pimp)
2 was found to be relatively lower in antioxidant potential, probably due to the redox inert closed stable 3d
10 electronic configuration for the Zn
2+ ion. The synthesized metal complexes have the same metal-to-ligand ratio of (1:2), so the difference in the activity could be attributed to the differences in their electronic configuration.
2.2. DNA Binding Activity
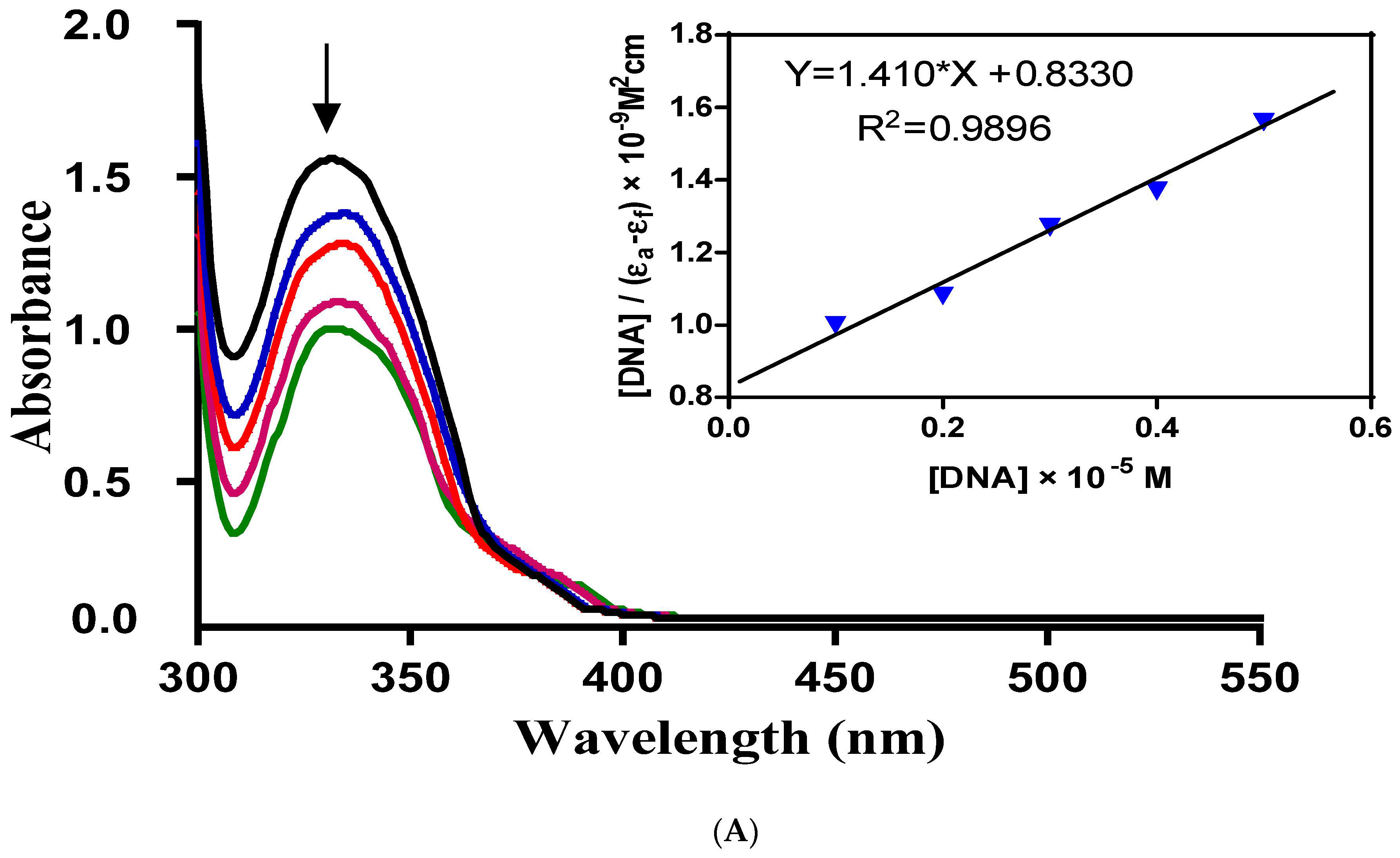
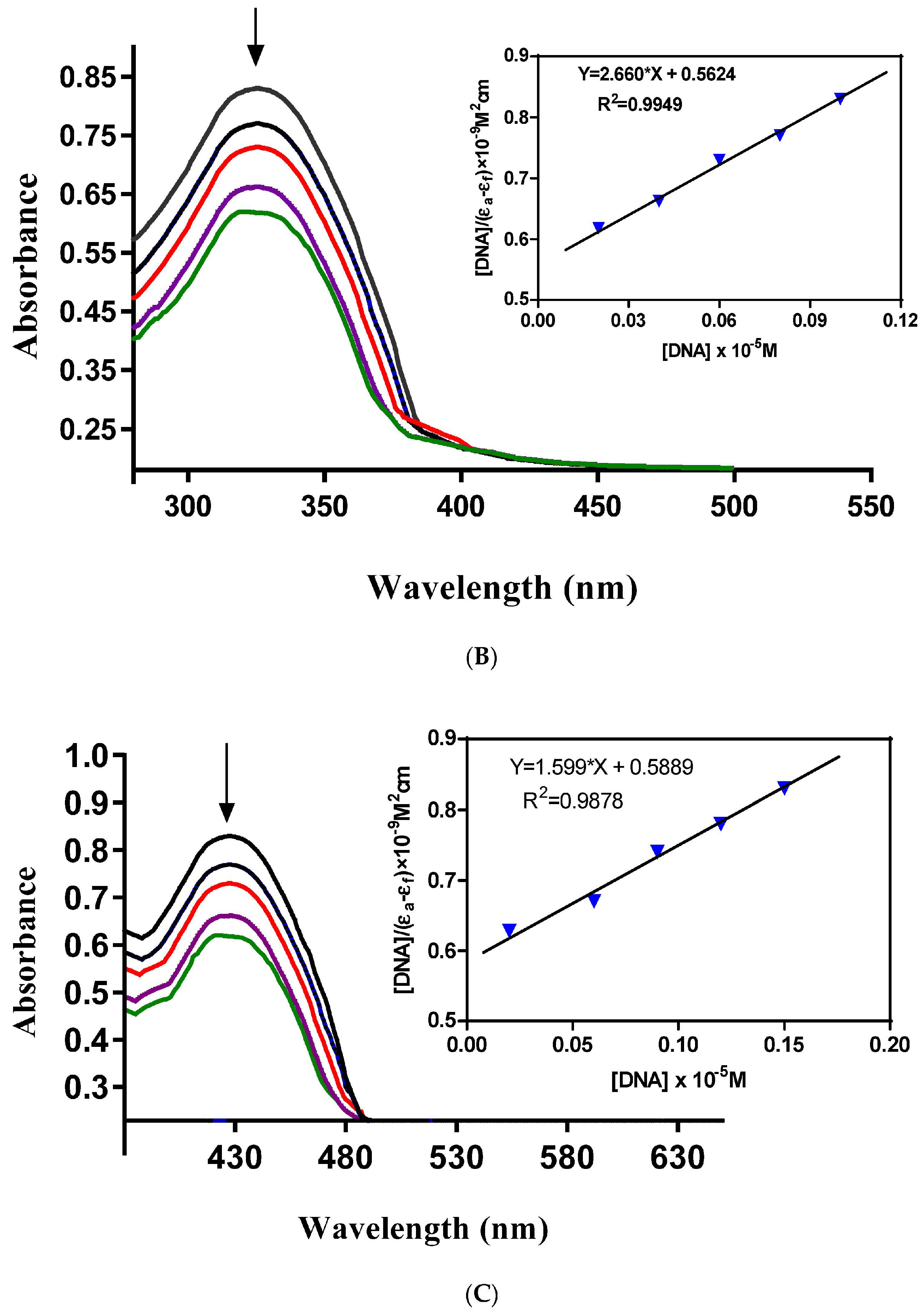
Metal-based complexes have offered an efficient solution for the treatment of cancer, a lethal disease caused by rapid DNA replication. Mostly, metal complexes directly interact with DNA, alter its shape and thus stop its rapid replication and, hence, cancer. This makes the study of complex-DNA interaction an important research area to overcome the life-threatening disease, cancer.
Electronic absorption spectroscopy can be used as a preliminary technique to study the metal complex-DNA interaction. Metal complex-DNA interaction alters UV-Vis spectra of the complexes in terms of λ
max shift (blue/red) or intensity changes (hypo/hyperchrpmisim). The extent of wavelength shift or change in absorption intensity shows the strength of interaction. The hypochromisim is associated with a red shift observed in the complex spectra. The increasing concentration of DNA during electronic absorption spectroscopic titrations indicates complex-DNA interaction in the form of intercalation [
11,
12]. This interaction is a consequence of stacking interactions between an aromatic chromophore of the complex and the DNA base pairs. After intercalation, the π* orbital of the complexes ligand couple with π orbitals of the base pairs of DNA. This coupling results in a decrease in energy required for π-π* transition and causes a red shift. If the coupling π* orbitals are partially filled by the electrons then transition probabilities also decrease causing hypochromisim [
10].
In order to assess the DNA-binding potential of the synthesized Schiff base ligand (H-pimp) and its Cu(pimp)
2, Ni(pimp)
2, Co(pimp)
2 and Zn(pimp)
2 complexes, electronic absorption spectral titrations were performed with fixed and variable concentrations of synthesized compounds and DNA, respectively. The results of the spectroscopic titrations are shown in
Figure 3A–E and for H-pimp, Ni(pimp)
2, Co(pimp)
2, Cu(pimp)
2 and Zn(pimp)
2, respectively, indicating hypochromism associated with a very small red shift with increasing concentrations of DNA solutions, thus indicating intercalation [
13].
In order to find the extent of this intercalative interaction between synthesized compounds and DNA, the intrinsic binding constants (
Kb) values were calculated according to the equation {[DNA]/(Ɛ
a − Ɛ
f) = DNA/(Ɛ
a − Ɛ
f) + 1/[
Kb(Ɛ
b − Ɛ
f)]} [
10]. In the equation, Ɛ
a, Ɛ
f, and Ɛ
b are the apparent, free and bound compound extinction coefficients, respectively. A plot of [DNA]/(Ɛ
b − Ɛ
f) versus [DNA], gave a slope of 1/(Ɛ
b − Ɛ
f) and a Y-intercept equal to [
Kb/(Ɛ − Ɛ
f)]
−1;
Kb is the ratio of the slope to the Y-intercept.
The intrinsic binding constants showing the binding strength of the compounds with DNA varied in the order: Zn(pimp)
2 {9.118 × 10
5 M
−1}> H-pimp {3.487 × 10
5 M
−1} > Co(pimp)
2 {3.090 × 10
5 M
−1} > Ni(pimp)
2 {1.858 × 10
5 M
−1} > Cu(pimp)
2 {1.367 × 10
5 M
−1}. The binding strength order seems to be related to the electronic configuration of the complexes. Zinc(pimp)
2 with a closed 3d
10 symmetric configuration shows the highest binding strength with DNA. The calculated
Kb values were found slightly higher than a classical intercalator, cisplatin (5.71 × 10
4 M
−1) [
9].
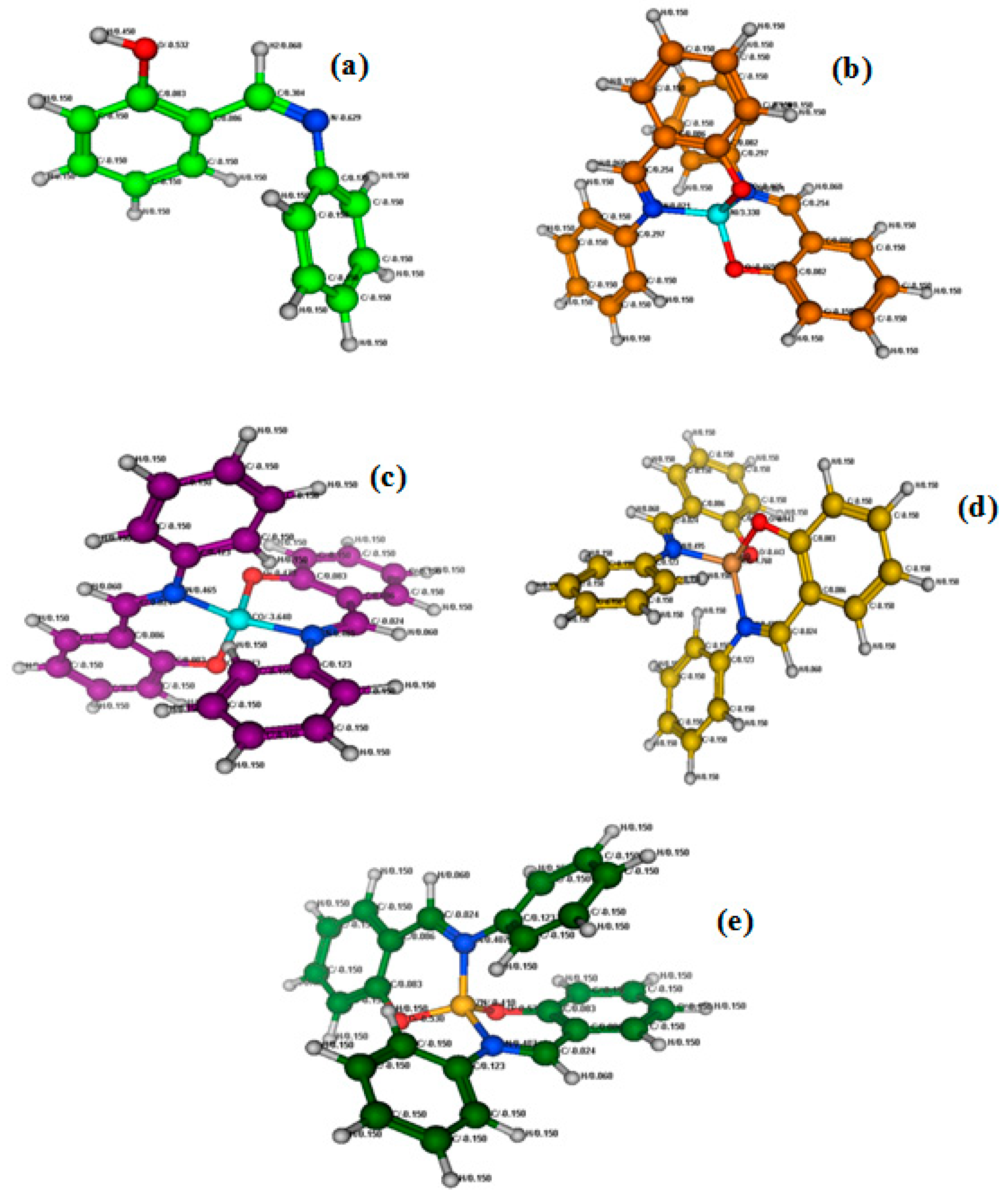
2.3. Structural Analysis and Molecular Docking
The structural analysis of 2-{(phenylimino) methyl}phenol ligandH-pimp and its metal complexes M(pimp)
2 was carried out using a semiempirical PM3 method (
Figure 4a–e) for their charge distribution and molecular docking with doubly stranded DNA (PDB:1D66). The molecular docking studies determined all possible configurations of computationally anticipated Schiff base ligand H-pimp, Ni(pimp)
2, Co(pimp)
2, Cu(pimp)
2, and Zn(pimp)
2 complexes with DNA to understand the molecular mechanism and physical mode of interaction. DNA is the primary pharmacological target of a number of anticancer compounds. Thus, the interaction between DNA and metal complexes is of vital prominence in understanding the mechanism. Transition metal complexes interact with DNA via both covalent and/or non-covalent interactions. Non-covalent DNA interactions include intercalative, electrostatic and groove binding of metal complexes with a DNA helix [
11]. A pose view analysis of H-pimp and its metal complexes was also performed to substantiate the mode of DNA binding, shown in
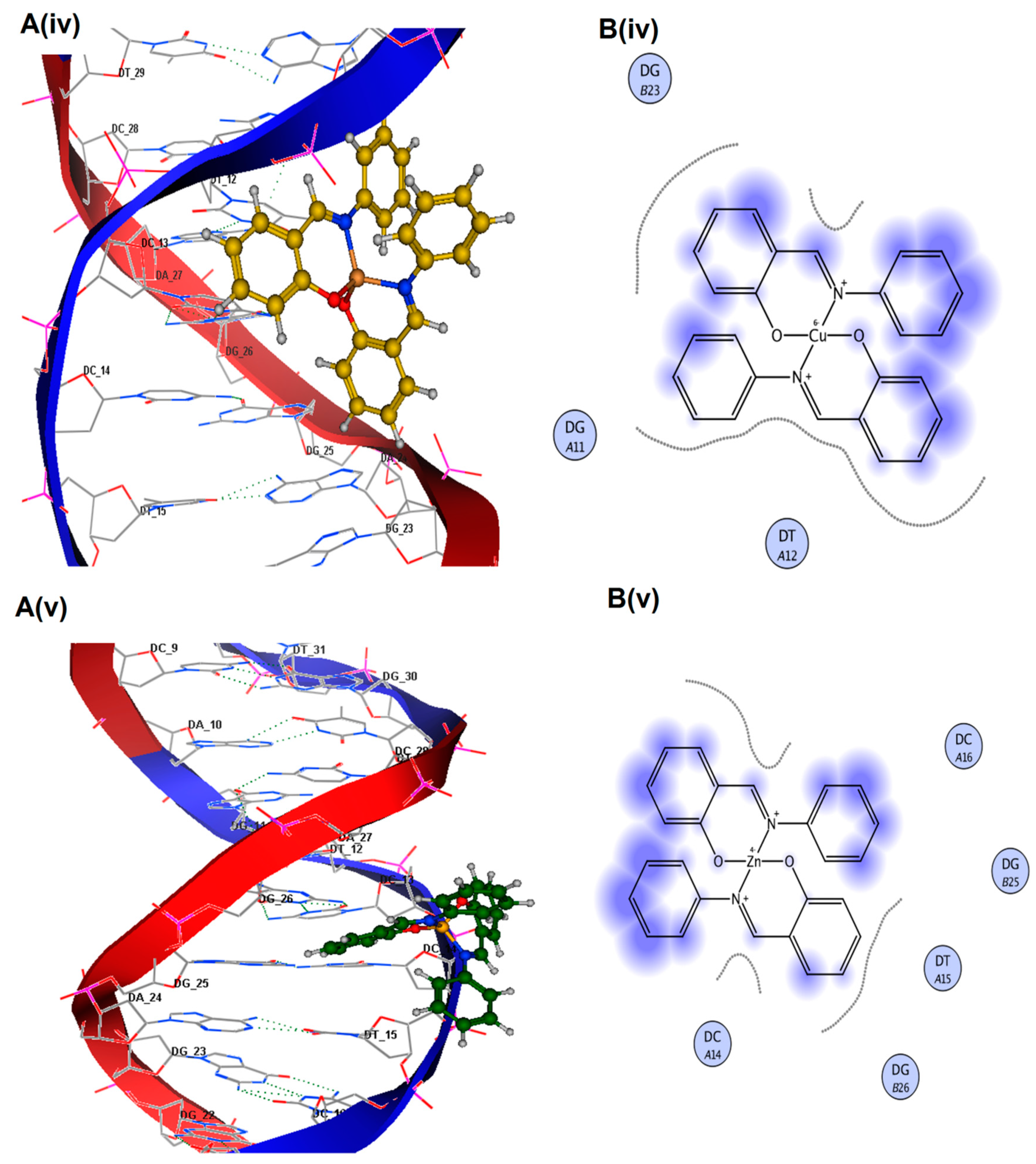
Figure 4a–e. Least energy confirmation pose of H-pimp revealed that the planar aromatic part of the ligand intercalates between stacked base pairs of DNA by π-stacking interactions. The 2D lig plot of the ligand showed that H of the H-O group develops H-bonding with adenine DA (A8) of DNA, whereas O of the H-O group builds H-bonding with H atom of Guanine DG(A7), as shown in
Figure 5A(i–v),B(i–v). H-bonding characteristics of ligands rendered significantly high binding affinity with DNA, as shown in
Table 2. Ni(II) complex of ligand exhibited mixed mode of intercalation and groove binding. It can be seen that the Ph-ring of the Zn(II) complex intercalates between DNA base pairs while the rest of the bulky structure latches onto the minor groove of DNA, establishing Vander Waal’s interactions with grooves, as visible in
Figure 5B(ii). It was observed that Co(II) and Cu(II) complexes of H-pimp fit well into the minor groove of DNA via hydrophobic and Vander Waal’s interactions, as shown in
Figure 5B(iii,iv). The interactions of the Zn(II) complex with DNA are attributed to the intercalation of the aromatic planar ring with flanking base pairs and, more importantly, with DC(A14), DG(B26) and DT(A15) of DNA, as can be seen in
Figure 5B(v). A substantial observation was that the Zn(II) complex showed enhanced interactions and binding affinity in terms of the highest
Kb value as compared to free ligands and other complexes, as shown in
Table 2. The negative values of free energy depicted that the physical interaction mechanism of compounds with DNA has been proven to be spontaneous.
For the comprehension of microscopic interactions between DNA and H-pimp ligand as well as its complexes, a number of physicochemical descriptors were calculated, listed as electronic descriptors and steric descriptors as shown in
Table 3 and
Table 4, respectively.
Electronic parameters of the ligand and complexes were calculated; however, semi-empirical calculations of transition metal compounds were complicated due to partially filled d-orbitals of the metal ions that are responsible for the multifarious structures with a large variety of possible coordination numbers and geometries [
14,
15]. Hence, electronic parameters of only ligand and Zn complex (completely filled d-orbital) were determined. E
HOMO and E
LUMO values gave an estimate of the electron-donating or electron-accepting character of a given compound. Consequently, a compound is considered more electron-donating as its E
HOMO value increases and more electron-accepting as its E
LUMO value decreases.
Table 3 depicts that Zn(II) complex is more electron donating as compared to due to the higher value of its E
HOMO.
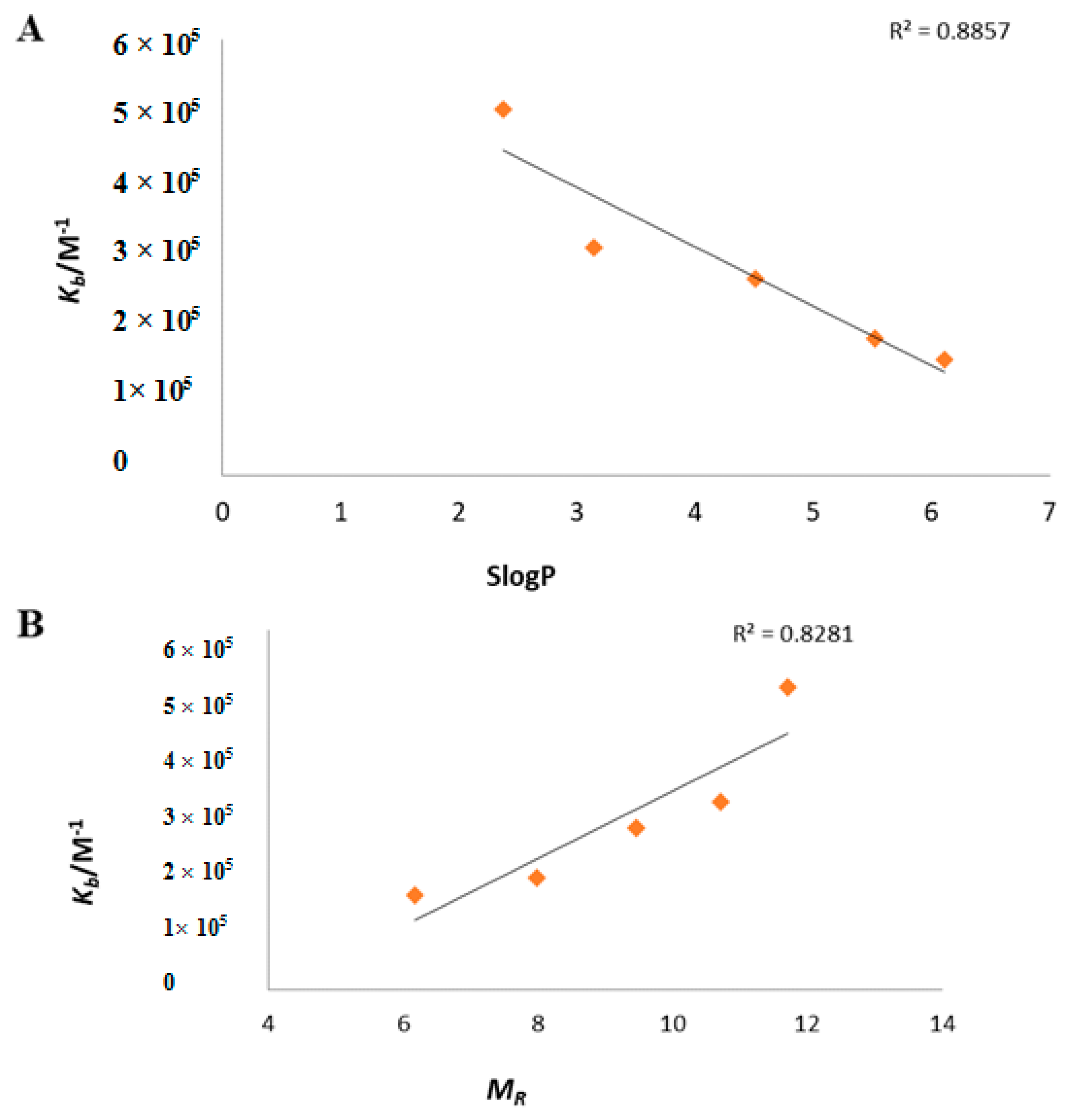
An excellent correlation of two steric parameters, i.e. partition coefficient (log
p) and Molar refractivity (M
R) with binding constant (
Kb), has been perceived. The partition coefficient (log
p) is illustrative of the hydrophobicity of the molecule. In this work,
slogP revealed a reasonable inverse correlation with the
Kb of ligand and its metal complexes (R
2 = 0.885), indicating that the compounds with a lower
slogP are anticipated to constitute stronger complexes, as shown in
Figure 6A.
Another imperative steric descriptor calculated in the present work was Molar refractivity (
MR), a measure of the total polarizability of a mole of a substance and its dependence on the temperature, the index of refraction and the pressure. A direct correlation of the
MR (R
2 = 0.8281) with
Kb was indicative of the fact that compounds with higher
MR have more binding affinity with DNA (
Figure 6B).
3. Materials and Methods
3.1. Chemicals and Reagents
Analytical grade salicylaldehyde, aniline, 2,2-diphenyl-1-picrylhydrazyl radical (DPPH), ascorbic acid, tris-HCl buffer, ferric chloride, O-phenenthroline, sulfuric acid, ammonium molybdate, potassium phosphate (monophosphate and di-phosphate), hydrogen peroxide, ethanol and salmon sperm DNA (SSDNA) were purchased from commercial suppliers and were used without any further purification.
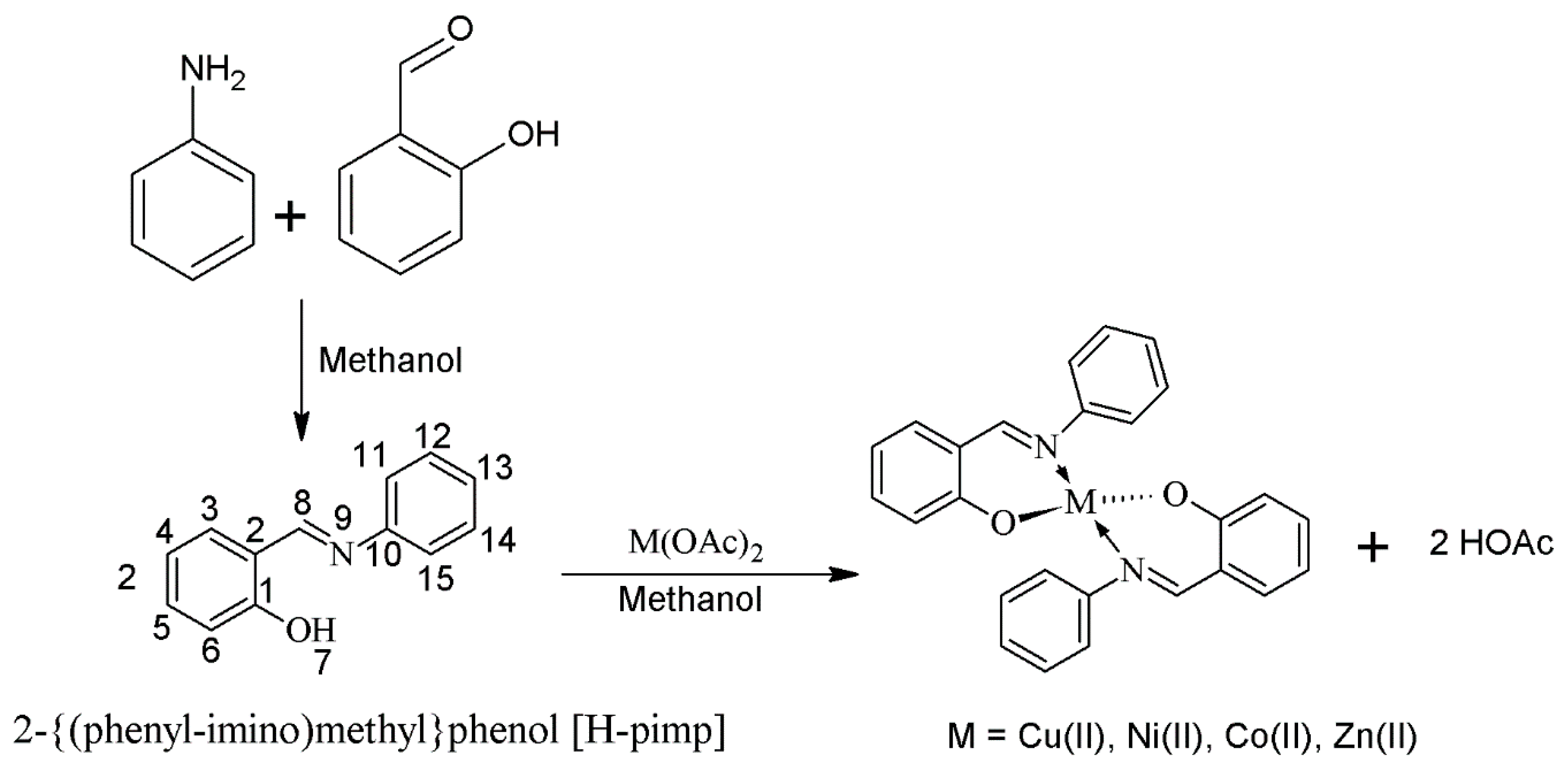
3.2. Synthesis of 2-[(Phenylimino)methyl]phenol{H-pimp}
The Schiff base ligand (H-pimp) was synthesized by reacting equimolar (5 mmol) methanolic solutions of aniline and salicyladehyde as shown in
Scheme 1. A yellowish product was produced instantaneously upon mixing which was filtered and recrystallized from a concentrated metahanolic solution.
Yield: 63%, M.pt: 48 °C, Elemental analysis (C13H11NO) Calc. (Exp.): C, 79.16 (80.63); H, 5.62 (5.12); N, 7.10 (7.01). IR (cm−1): 3200 (w), 1569 (s), 1483 (s), 1407 (s), 1363 (s), 1282 (s), 1156 (s), 1110 (s), 987 (s), 912 (s), 857 (s), 756 (s), 688 (s), 1H-NMR (300.13 MHz, CDCl3, 303K): δ = 6.94 (d, 3JHH = 7.03 Hz, 1H, H13), 6.97 (d, 3JHH = 7.03 Hz, 2H, H12 & H14), 6.99 (d, 3JHH = 7.01 Hz, 2H, H4 & H5), 7.02 (d, 3JHH = 7.2 Hz, 1H, H3), 7.16 (d, 3JHH = 7.31 Hz, 2H, H11 & H15), 7.28 (d, 3JHH = 6.9 Hz, 1H, H6), 8.60 (s, Ar HC=N), 13.0 (s, OH), 13C{1H}-NMR (75.47 MHz, CDCl3, 303k), 117 (CH, C13), 119 (CH, C12 & C14), 119.2 (CH, C4 & C5), 120 (CH, C3), 122 (CH, C11 & C15), 132 (CH, C6), 133 (C, C2), 148 (C, C1), 161 (C, C10), 163 (CH, Ar HC-N).
3.3. Synthesis of Metal Complexes {M(pimp)2}
The transition metal complexes of the Schiff base ligand (H-pimp) were synthesized (
Scheme 2) by a reported procedure [
11]. The metal salts M(OAc)
2 were initially dehydrated by keeping them in an oven for 3–4 h at 110 °C. A methanolic solution of ligand H-pimp (5 mmol) was added to the methanolic solution of metal acetate (2.5 mmol) and stirred for 3 h. The product was obtained either instantly or upon concentrating the solution through a rotary evaporator.
3.4. Bis-(2-[(phenylimino)methyl]phenolate)cobalt(II) {(Co-pimp)2}
Yield: 43%. Elemental analysis (C26H20CoN2O2) Calc. (Exp.): C, 69.18 (69.88); H, 4.47 (5.11); Co, 13.06 (13.02); N, 6.21 (6.12)%. IR (cm−1): 1536 (s), 1482 (s), 1465 (s), 1450 (s), 1328 (s), 1180 (s), 1148 (s), 1122 (s), 1086 (s), 1010 (s), 976 (s), 929 (s), 858 (s), 837 (s), 758 (s), 698 (s), λmax = 860 nm (ε = 17.6 M−1 cm−1, 2A2g → 2B1g).
3.5. Bis-(2-[(phenylimino)methyl]phenolate)nickel(II) {(Ni-pimp)2}
Yield: 55%. Elemental analysis (C26H20N2NiO2) Calc. (Exp.): C, 69.22 (69.28%); H, 4.47 (4.90%); N, 6.21 (6.29%); Ni, 13.01 (12.11%). IR (cm−1): 1533 (s), 1464 (s), 1443 (s), 1416 (s), 1343 (s), 1260 (s), 1224 (s), 1181 (s), 1147 (s), 1123 (s), 1033 (w), 981(s), 946 (s), 871 (w), 821 (s), 810 (s), 761 (s), 751 (s), 728 (s), 689 (s), 671 (s), λmax = 690 nm (ε = 28.3 M−1 cm−1, 1A1g → 1A2g).
3.6. Bis-(2-[(phenylimino)methyl]phenolate)copper(II) {(Cu-pimp)2}
Yield: 68%. Elemental analysis (C26H20CuN2O2) Calc. (Exp.): C, 68.48 (69.33%); H, 4.42 (4.92%); Cu, 13.94 (13.01%); N, 6.14 (7.44%)%. IR (cm−1): 1555 (s), 1523 (s), 1478 (s), 1441 (s), 1388 (s), 1351 (s), 1323 (s), 1255 (s), 1205 (w), 1175 (s), 1151 (s), 1133 (s), 1098 (s), 1031 (s), 1009 (s), 987 (s), 937 (s), 834 (s), 767 (s), 699 (s), 623 (s), λmax = 660 nm (ε = 188.6 M−1 cm−1, dz2 → dx2 − y2 ).
3.7. Bis-(2-[(phenylimino)methyl]phenolate)zinc(II) {(Zn-pimp)2}
Yield: 55%. Elemental analysis (C26H20N2O2Zn) Calc. (Exp.): C, 68.20 (70.01%); H, 4.40 (4.78%); N 6.12 (6.89%); Zn, 14.29 (14.95%). IR (cm−1): 1581 (s), 1531 (s), 1459 (s), 1441 (s), 1389 (s), 1351 (s), 1326 (s), 1253 (s), 1169 (s), 1151 (s), 1096 (s), 1032 (s), 1008 (s), 927 (s), 829 (s), 789 (s), 762 (s), 688 (s), 596 (s).
3.8. In Vitro Antioxidant Studies
The synthesized compounds were studied for their in vtiro antioxidant potential by different assays using UV-Visible Spectrophotometer. In DPPH radical scavenging assay, different concentrations (25, 50, 100 and 200 µM) of compounds were mixed with an ethanolic solution containing 85 µM DPPH radical. The decrease in absorbance was measured at 517 nm. Ascorbic acid was used as a positive control to determine the maximal decrease in DPPH absorbance. The values are expressed in the percentage of inhibition of DPPH absorbance in relation to the control values without the compounds (ascorbic acid maximal inhibition was considered 100% of inhibition).
The ferric ion-reducing power of the newly synthesized compounds was determined according to the reported procedure [
12,
13]. Varying concentrations (25, 50, 100 and 200 µM) of compounds, 0.2 mL of 3.6 mM ferric chloride, 0.3 mL of 100 mM tris buffer (pH = 7.4), 0.1 mL of 9 mM O-phenanthroline were mixed and diluted up to 3.0 mL with ultra-pure distal water. Solutions were shaken for 10 min vigorously and left to stand at room temperature. The increase in absorbance of the sample solution was measured at 595 nm. Ascorbic acid at the same concentrations was utilized as a reference standard and without a compound sample mixture as a control.
The total antioxidant capacity of the compounds was evaluated using a phosphomolybdenum assay [
10] based on the conversion of Mo(VI) into Mo(V), with the test compound acting as a reducing agent in comparison with ascorbic acid used as a standard.
The reagent solution containing varying concentrations (25, 50, 100 and 200 µM) of synthesized compounds aliquot in ethanol, 0.7 mL of 0.6 M sulphuric acid, 1.0 mM ammonium molybdate, 1.0 mL of 28 Mm pottasium pasphate and ultra-pure distal water were incubated at 95 °C for 90 min. After cooling, at room temperature, the increase in absorbance of the mixture was measured at 695 nm. Ascorbic acid was utilized as the reference standard and without compounds sample mixture as control.
The scavenging activity of the synthesized compounds for hydroxyl radicals was measured with the Fenton reaction [
9]. Varying concentrations (25, 50, 100 and 200 µM) of synthesized compounds, 0.1 mL of 7.5 mM O-phenanthroline, 0.5 mL of 0.2 M phosphate buffer ( pH 6.6), 0.1 mL of 7.5 mM ferrous sulfate and 0.1 mL of H
2O
2 (0.1%) were mixed and diluted up to 3 mL with distilled water. The reaction mixture was incubated at room temperature for 30 min and the absorbance was measured at 510 nm. The reaction mixtures without synthesized compounds have been used as control and without compounds and H
2O
2 as a blank.
3.9. DNA Absorption Spectroscopic Studies
The electronic absorption spectroscopy was used to study the interaction between synthesized compounds and salmon sperm DNA (SSDNA). The solution of SSDNA in the buffer 50 mM NaCl/5 mM Tris–HCl (pH 7.2) in water gave an absorbance ratio of 1.9 at 260/280 nm, indicating that the DNA was sufficiently free from protein [
11]. The concentration of DNA was measured using its extinction coefficient at 260 nm (6600 M
−1 cm
−1) after 1:100 dilution. Concentrated stock solutions of the synthesized compounds were prepared by dissolving the compounds in ethanol and diluting them suitably with the corresponding buffer to the required concentration for all of the experiments.
3.10. Statistical Analysis
Linear regression analysis was used to calculate IC50 ± SEM values from data and graphs by using Graph pad prism 6®. Significant differences among the means of data were tested by the one-way ANOVA followed by the student’s t-test with significance level (p < 0.05). All the tests were conducted in triplicate.
3.11. Molecular Docking Methodology
The chemical structures of the Schiff base ligand and its complexes with Co(II), Ni(II), Cu(II) and Zn(II) were sketched and optimized on the MOE2017 window using MOE molecular builder and were entered into the MOE database. The 3-D structure of DNA with PDB ID-1D66, resolution of 2.7, and sequence of CCGGAGGACTGTCCTCCGG was obtained from the RCSB protein Data Bank [
16]. For the purpose of docking simulation, PDB internal coordinates of DNA were optimized using a molecular dynamic AMBER force field and semi-empirical PM3 approaches to attain minimum energy and stable conformation. Water molecules were removed from DNA structures using a sequence editor of MOE to exclude the effect of water on the interaction of DNA with the ligand and its metal complexes. Structures were protonated with their standard geometry followed by their energy optimization tool using MOPAC 7.0. The resulting structures were subjected to a systematic conformational search at default parameters with an RMS gradient of 0.01 kcal/mol using Site Finder to find out active sites of DNA molecules. Finally, a few docking runs were carried out to get a final binding pose from the scoring function. The best conformation was selected based on energetic ground and the minimum Final Docking Energy (∆G) [
17].