Establishing the Role of Iridoids as Potential Kirsten Rat Sarcoma Viral Oncogene Homolog G12C Inhibitors Using Molecular Docking; Molecular Docking Simulation; Molecular Mechanics Poisson–Boltzmann Surface Area; Frontier Molecular Orbital Theory; Molecular Electrostatic Potential; and Absorption, Distribution, Metabolism, Excretion, and Toxicity Analysis
Abstract
1. Introduction
2. Results
2.1. Molecular Docking Interaction Data
2.2. MD Simulation Data
2.3. MM/PBSA Analysis Data
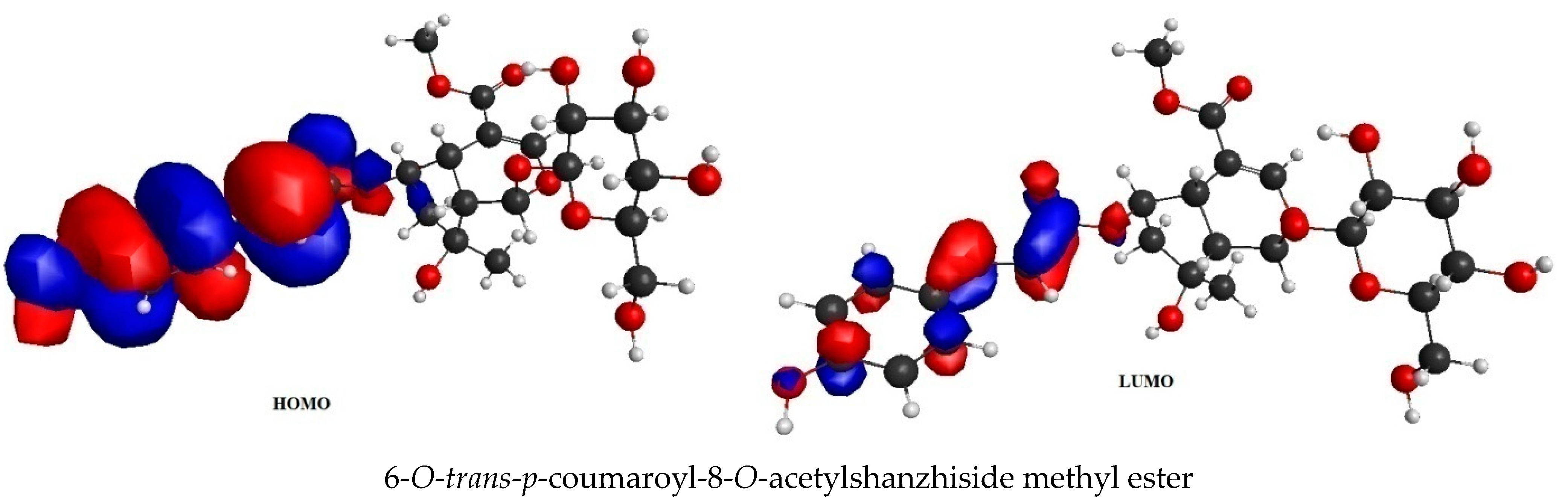
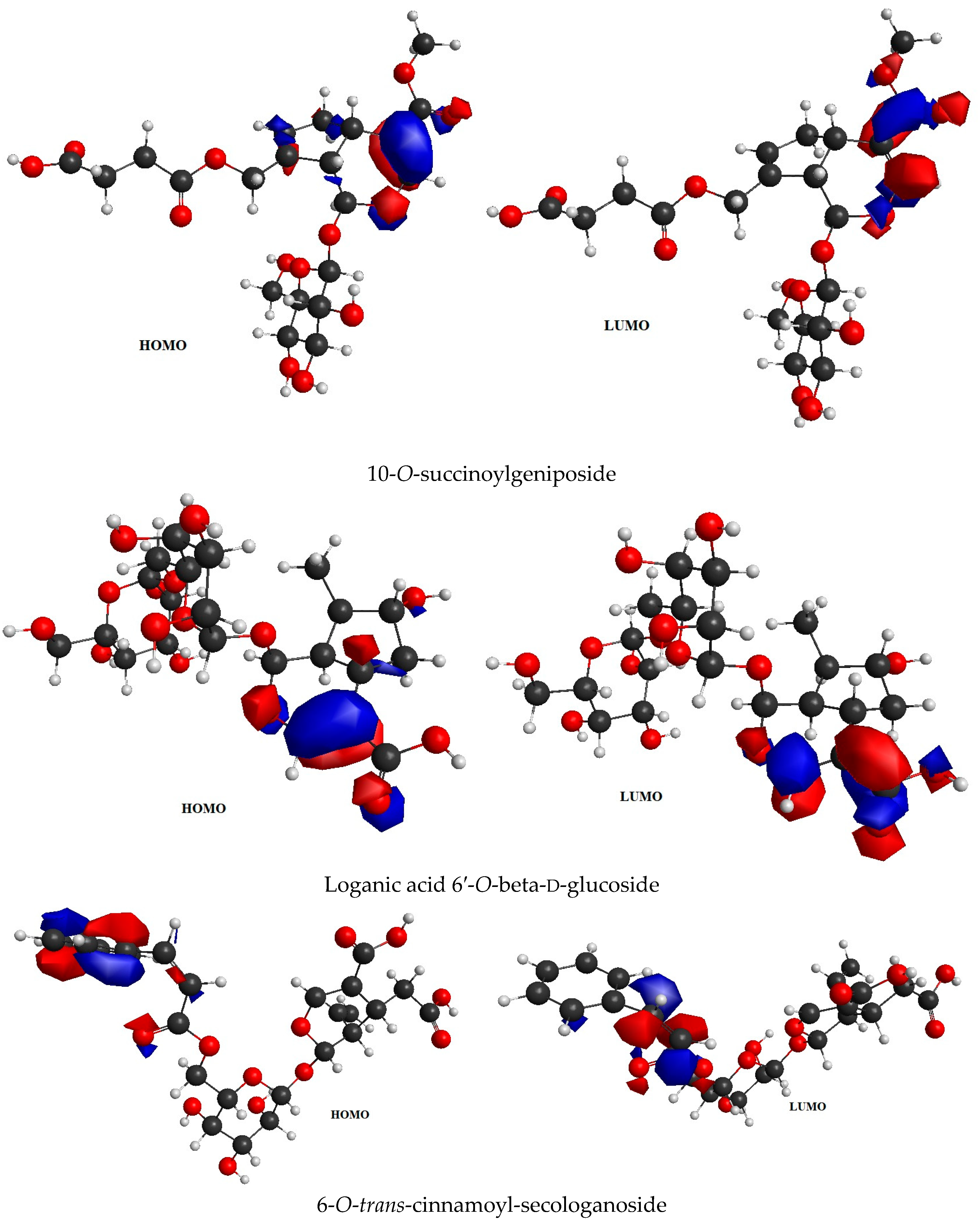
2.4. Frontier Molecular Orbital (FMO) Analysis Data
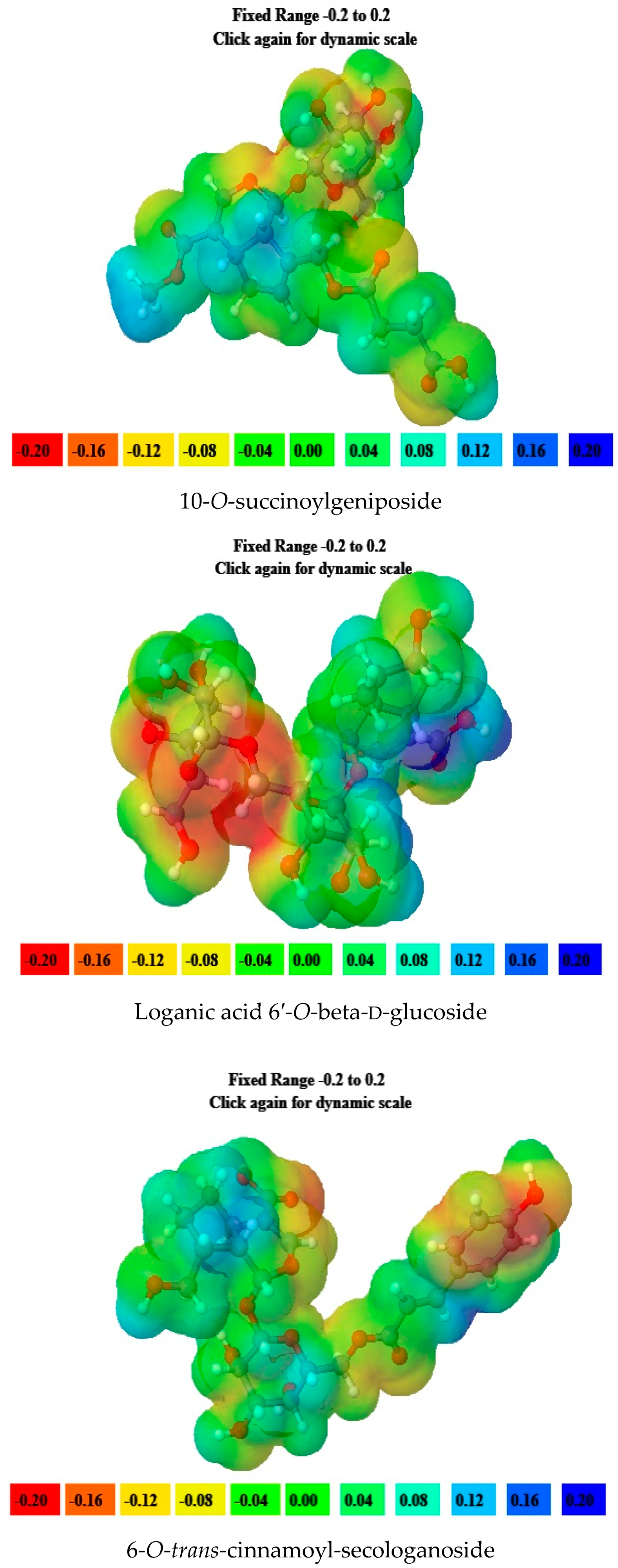
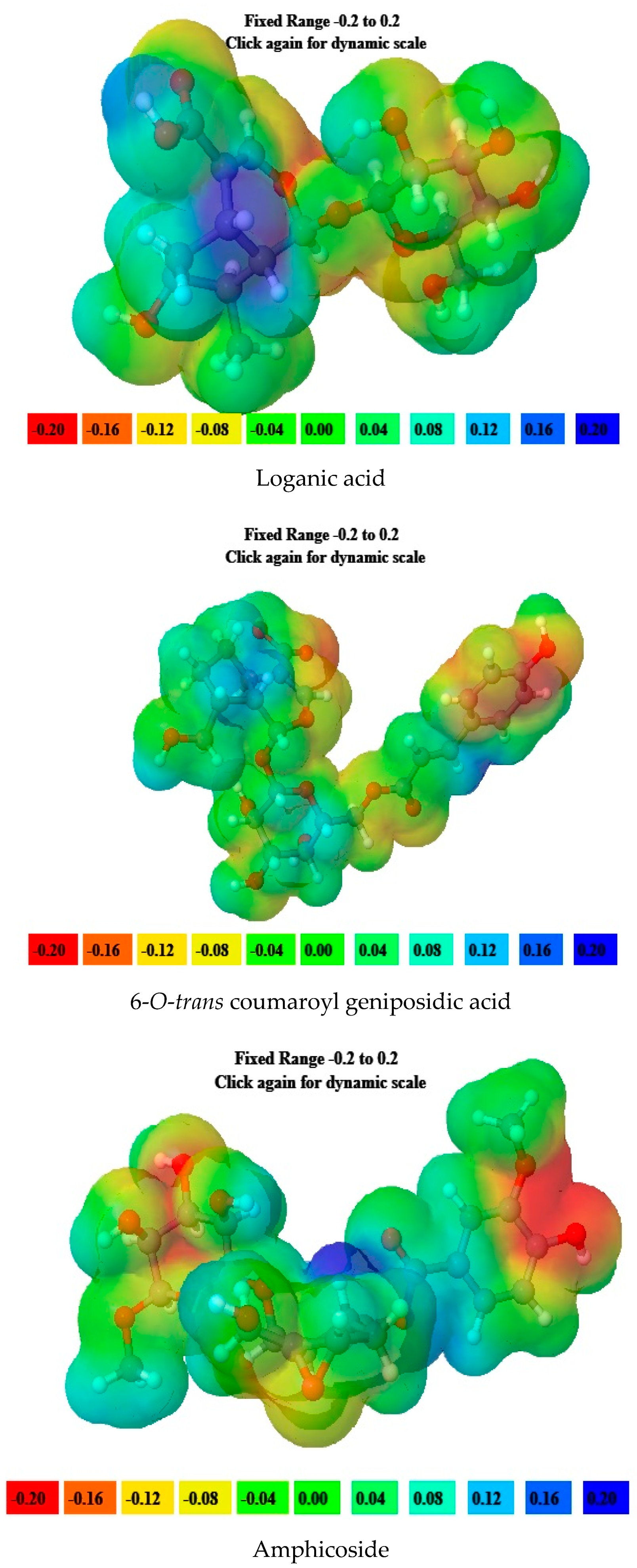
2.5. Molecular Electrostatic Potential (MEP) Data
2.6. ADMET (Absorption, Distribution, Metabolism, Excretion, Toxicity) Data
3. Discussion
4. Materials and Methods
4.1. Molecular Docking Studies
4.1.1. Selection and Preparation of the Receptor
4.1.2. Preparation of Ligand Molecules
4.1.3. Molecular Docking Parameters of the Molecules Interacting with the Receptors
4.2. MD Simulation Study
4.3. MM/PBSA: Calculation of Binding-Free Energy
4.4. FMO Analysis
4.5. MEP Investigation
4.6. ADMET Calculation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bar-Sagi, D.; Knelson, E.H.; Sequist, L.V. A bright future for KRAS inhibitors. Nat. Cancer 2020, 1, 25–27. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Guo, Z.; Wang, F.; Liwu, F. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef] [PubMed]
- Hai-Zhou, W.; Jia-Qi, X.; Song-Shu, X.; Yan, C. KRAS: A Promising Therapeutic Target for Cancer Treatment. Curr. Top. Med. Chem. 2019, 19, 2081–2097. [Google Scholar] [CrossRef]
- Liu, C.; Zheng, S.; Wang, Z.; Wang, S.; Wang, X.; Yang, L.; Xu, H.; Cao, Z.; Feng, X.; Xue, Q.; et al. KRAS-G12D mutation drives immune suppression and the primary resistance of anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer. Cancer Commun. 2022, 42, 828–847. [Google Scholar] [CrossRef] [PubMed]
- Uprety, D.; Adjei, A.A. KRAS: From undruggable to a druggable Cancer Target. Cancer Treat. Rev. 2020, 89, 102070. [Google Scholar] [CrossRef]
- Pantsar, T. The current understanding of KRAS protein structure and dynamics. Comput. Struct. Biotechnol. J. 2020, 18, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Alphonse, M.P.; Yan, L.; Qi, L. Targeting Mutant KRAS for Anticancer Therapy. Curr. Top. Med. Chem. 2019, 19, 2098–2113. [Google Scholar] [CrossRef]
- Christensen, J.G.; Olson, P.; Briere, T.; Wiel, C.; Bergo, M.O. Targeting Krasg12c -mutant cancer with a mutation-specific inhibitor. J. Intern. Med. 2020, 288, 183–191. [Google Scholar] [CrossRef]
- Zhongwei, M.; Hongying, X.; Panpan, S.; Yu, Y.; Jing, X.; Yunyun, Y.; Yanguo, S.; Lilan, Z.; Xin, L.; Yuying, Z.; et al. KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge. Cell Discov. 2022, 8, 5. [Google Scholar] [CrossRef]
- Broker, J.; Waterson, A.G.; Smethurst, C.; Kessler, D.; Bottcher, J.; Mayer, M.; Gmaschitz, G.; Phan, J.; Little, A.; Abbott, J.R.; et al. Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS G12C Inhibitor. J. Med. Chem. 2022, 65, 14614–14629. [Google Scholar] [CrossRef]
- Tanaka, N.; Bwalya, A.W.; Nontobeko, P.M.; Madan, S.P.; Phumzile, P.S.; Patrick, H.D.; Beverley, S.; Xavier, S. Iridoid Derivatives as Anticancer Agents: An Updated Review from 1970–2022. Cancers 2023, 15, 770. [Google Scholar] [CrossRef]
- Cho-Won, K.; Kyung-Chul, C. Potential Roles of Iridoid Glycosides and Their Underlying Mechanisms against Diverse Cancer Growth and Metastasis: Do They Have an Inhibitory Effect on Cancer Progression? Nutrients 2021, 13, 2974. [Google Scholar] [CrossRef]
- Hidayat, H.; Ivan, R.G.; Muhammad, S.; Muhammad, L.R.; Mamona, N. Therapeutic Potential of Iridoid Derivatives: Patent Review. Inventions 2019, 4, 29. [Google Scholar] [CrossRef]
- Hayato, Y.; Takahiro, M.; Takahiro, K.; Masaya, O.; Tomoe, O.; Tatsusada, Y.; Tetsushi, W. Anti-Proliferative Effects of Iridoids from Valeriana fauriei on Cancer Stem Cells. Int. J. Mol. Sci. 2022, 23, 14206. [Google Scholar] [CrossRef]
- María, L.C.; Tatiana, M.; Alarcón-de-la-Lastra, C.; Sánchez-Hidalgo, M. Potential Protective Role Exerted by Secoiridoids from Olea europaea L. in Cancer, Cardiovascular, Neurodegenerative, Aging-Related, and Immunoinflammatory Diseases. Antioxidants 2020, 9, 149. [Google Scholar] [CrossRef]
- Maria, K.; Olga, B.; Maria, K.; Merike, V. Anticancer Effect of the Iridoid Glycoside Fraction from Dipsacus fullonum L. Leaves. Nat. Prod. Commun. 2020, 15, 1934578X20951417. [Google Scholar] [CrossRef]
- Mohammad Rizki Fadhil, P.; Hadi, P.; Siswandono, S. Introducing a two-dimensional graph of docking score difference vs. similarity of ligand-receptor interactions. Indones. J. Biotechnol. 2021, 26, 54–60. [Google Scholar] [CrossRef]
- Mullaguri, S.C.; Akula, S.; Sahoo, P.S.; Ashireddygari, V.R.; Mupparapu, V.; Silveri, R.; Prasad Burra, V.L.S.; Kancha, R.K. Molecular docking analysis reveals differential binding affinities of multiple classes of selective inhibitors towards cancer-associated KRAS mutants. 3 Biotech 2022, 12, 343. [Google Scholar] [CrossRef]
- Kawsar, S.M.A.; Almalki, F.A.; Hadd, T.B.; Laaroussi, H.; Khan, M.A.R.; Hosen, M.A.; Mahmud, S.; Aounti, A.; Maideen, N.M.P.; Heidarizadeh, F.; et al. Potential antifungal activity of novel carbohydrate derivatives validated by POM, molecular docking and molecular dynamic simulations analyses. Mol. Simul. 2023, 49, 60–75. [Google Scholar] [CrossRef]
- Wang, C.; Nguyen, P.H.; Pham, K.; Huynh, D.; Le, T.-B.N.; Wang, H.; Ren, P.; Luo, R. Calculating protein-ligand binding affinities with MMPBSA: Method and error analysis. J. Comput. Chem. 2016, 37, 2436–2446. [Google Scholar] [CrossRef]
- Kerru, N.; Gummidi, L.; Bhaskaruni, S.V.H.S.; Maddila, S.N.; Singh, P.; Jonnalagadda, S.B. A comparison between observed and DFT calculations on structure of 5-(4-chlorophenyl)-2-amino-1,3,4-thiadiazole. Sci. Rep. 2019, 9, 19280. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Álvarez, D.; Torres-Rojas, M.F.; Lara-Ramirez, E.E.; Marchat, L.A.; Rivera, G. Ligand-Based Virtual Screening, Molecular Docking, and Molecular Dynamic Simulations of New β-Estrogen Receptor Activators with Potential for Pharmacological Obesity Treatment. Molecules 2023, 28, 4389. [Google Scholar] [CrossRef] [PubMed]
- Devereux, C.; Smith, J.S.; Huddleston, K.K.; Barros, K.; Zubatyuk, R.; Isayev, O.; Roitberg, A.E. Extending the applicability of the ANI deep learning molecular potential to sulfur and halogens. J. Chem. Theory Comput. 2020, 16, 4192–4202. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.C.; Juyal, V.; Sah, A.N.; Saha, S. Computational Investigation of Geniposidic Acid as an Anticancer Agent Using Molecular Docking, Molecular Dynamic Simulation, DFT Calculation, and OSIRIS-Molinspiration Profiling. Phys. Chem. Res. 2023, 11, 801–823. [Google Scholar] [CrossRef]
- Amin, M.L. P-glycoprotein Inhibition for Optimal Drug Delivery. Drug Target Insights 2013, 7, 27–34. [Google Scholar] [CrossRef]
- Panse, N.; Gerk, P.M. The Caco-2 Model: Modifications and enhancements to improve efficiency and predictive performance. Int. J. Pharm. 2022, 624, 122004. [Google Scholar] [CrossRef]
- Takaki, T.; Shogo, N.; Yuta, Y.; Takeshi, I.; Sohei, I.; Vladimir, S.; Kaori, F.; Yuji, M.; Hiroaki, T.; Fuminori, M.; et al. Development of an Analysis Toolkit, AnalysisFMO, to Visualize Interaction Energies Generated by Fragment Molecular Orbital Calculations. J. Chem. Inf. Model. 2019, 59, 25–30. [Google Scholar] [CrossRef]
- Poli, G.; Granchi, C.; Rizzolio, F.; Tuccinardi, T. Application of MM-PBSA Methods in Virtual Screening. Molecules 2020, 25, 1971. [Google Scholar] [CrossRef]
- Krishnan, V.; Verma, P.; Saha, S.; Singh, B.; Vinutha, T.; Kumar, R.R.; Kulshreshta, A.; Singh, S.P.; Sathyavathi, T.; Sachdev, A.; et al. Polyphenol-enriched extract from pearl millet (Pennisetum glaucum) inhibits key enzymes involved in post prandial hyper glycemia (α-amylase, α-glucosidase) and regulates hepatic glucose uptake. Biocatal. Agric. Biotechnol. 2022, 43, 102411. [Google Scholar] [CrossRef]
- Saha, S.; Yeom, G.S.; Nimse, S.B.; Pal, D. Combination Therapy of Ledipasvir and Itraconazole in the Treatment of COVID-19 Patients Coinfected with Black Fungus: An In Silico Statement. BioMed Res. Int. 2022, 2022, 5904261. [Google Scholar] [CrossRef]
- Vishvakarma, V.K.; Singh, M.B.; Jain, P.; Kumari, K.; Singh, P. Hunting the main protease of SARS-CoV-2 by plitidepsin: Molecular docking and temperature-dependent molecular dynamics simulations. Amino Acids 2022, 54, 205–213. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R. Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa-A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Singh, A.K.; Bansal, T.; Yadav, A.; Prajapati, K.S.; Mohd, S.; Kumar, S. Identification of Natural Inhibitors Against SARS-CoV-2 Drugable Targets Using Molecular Docking, Molecular Dynamics Simulation, and MM-PBSA Approach. Front. Cell. Infect. Microbiol. 2021, 11, 730288. [Google Scholar] [CrossRef] [PubMed]
- Perri, M.J.; Weber, S.H. Web-Based Job Submission Interface for the GAMESS Computational Chemistry Program. J. Chem. Educ. 2014, 91, 2206–2208. [Google Scholar] [CrossRef]
- Hanson, R.M.; Prilusky, J.; Renjian, Z.; Nakane, T.; Sussman, J.L. JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem. 2013, 53, 207–216. [Google Scholar] [CrossRef]
- Giuseppe, M.J.B.; Colleen, B.; Laura, C.; Datta, D.; De Silva, N.; Emiliano, D.J.; Dmitri, G.F.; Jeffrey, R.G.; Anastasia, O.G.; Emilie, G.; et al. Recent developments in the general atomic and molecular electronic structure system. J. Chem. Phys. 2020, 152, 154102. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
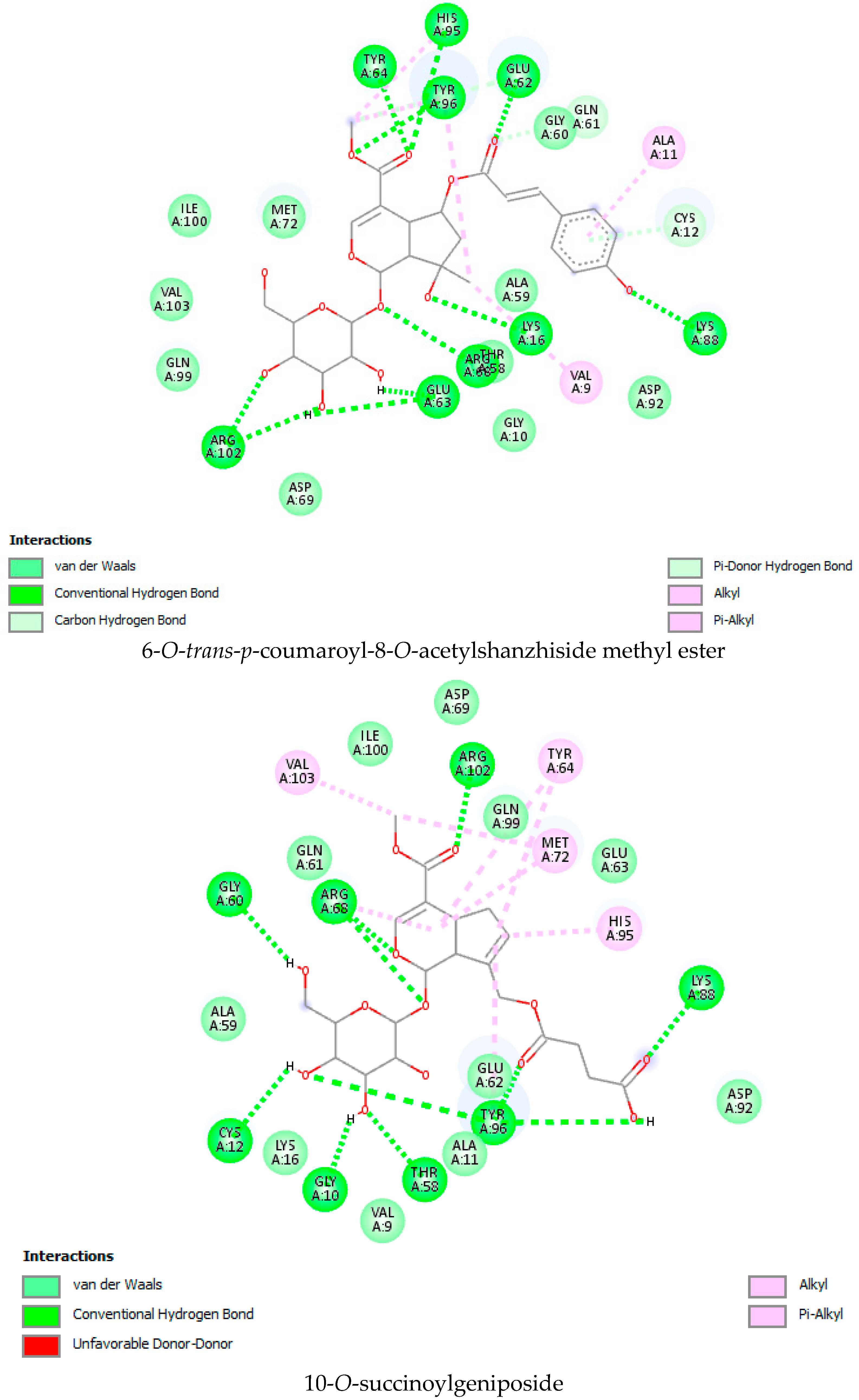
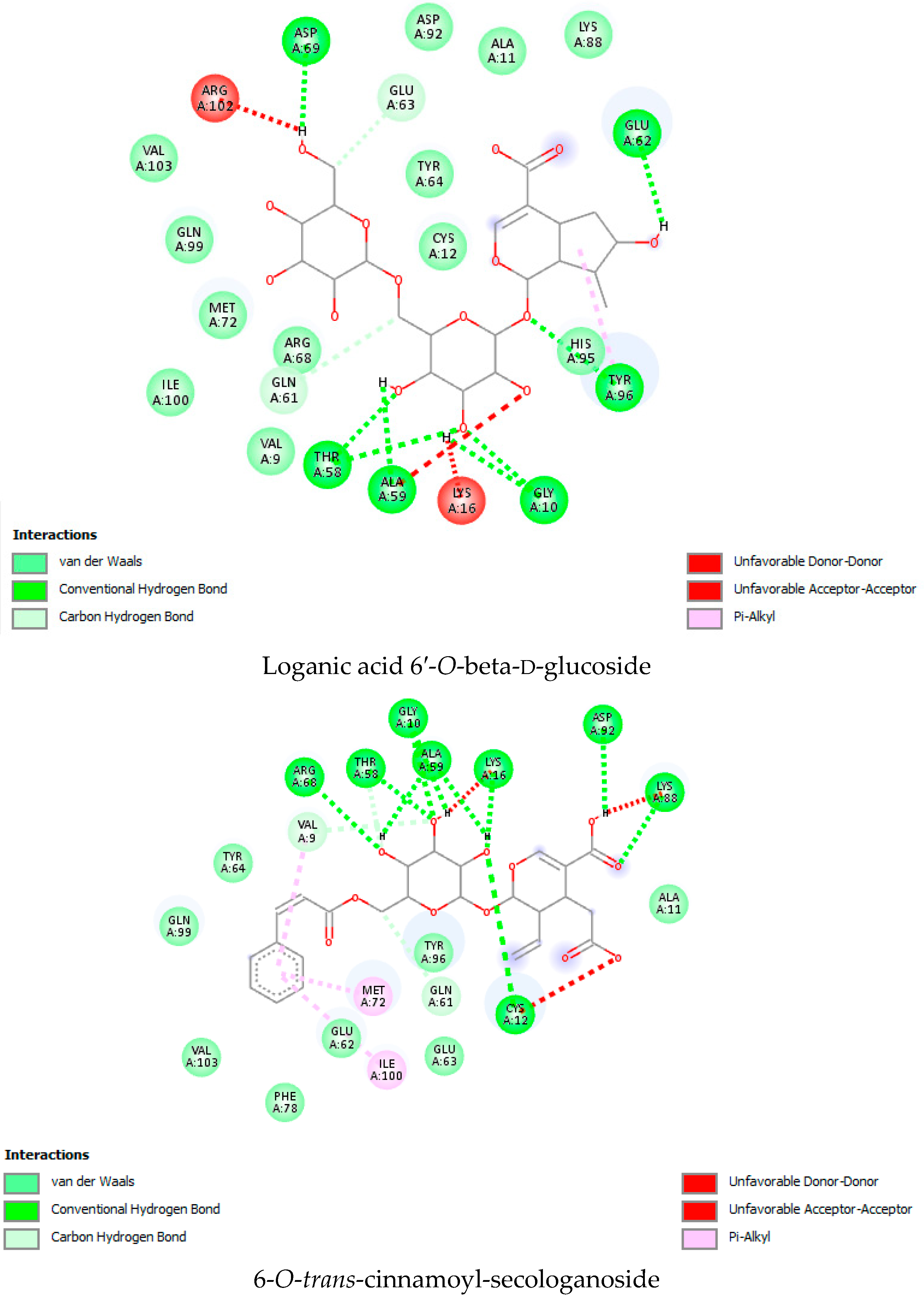
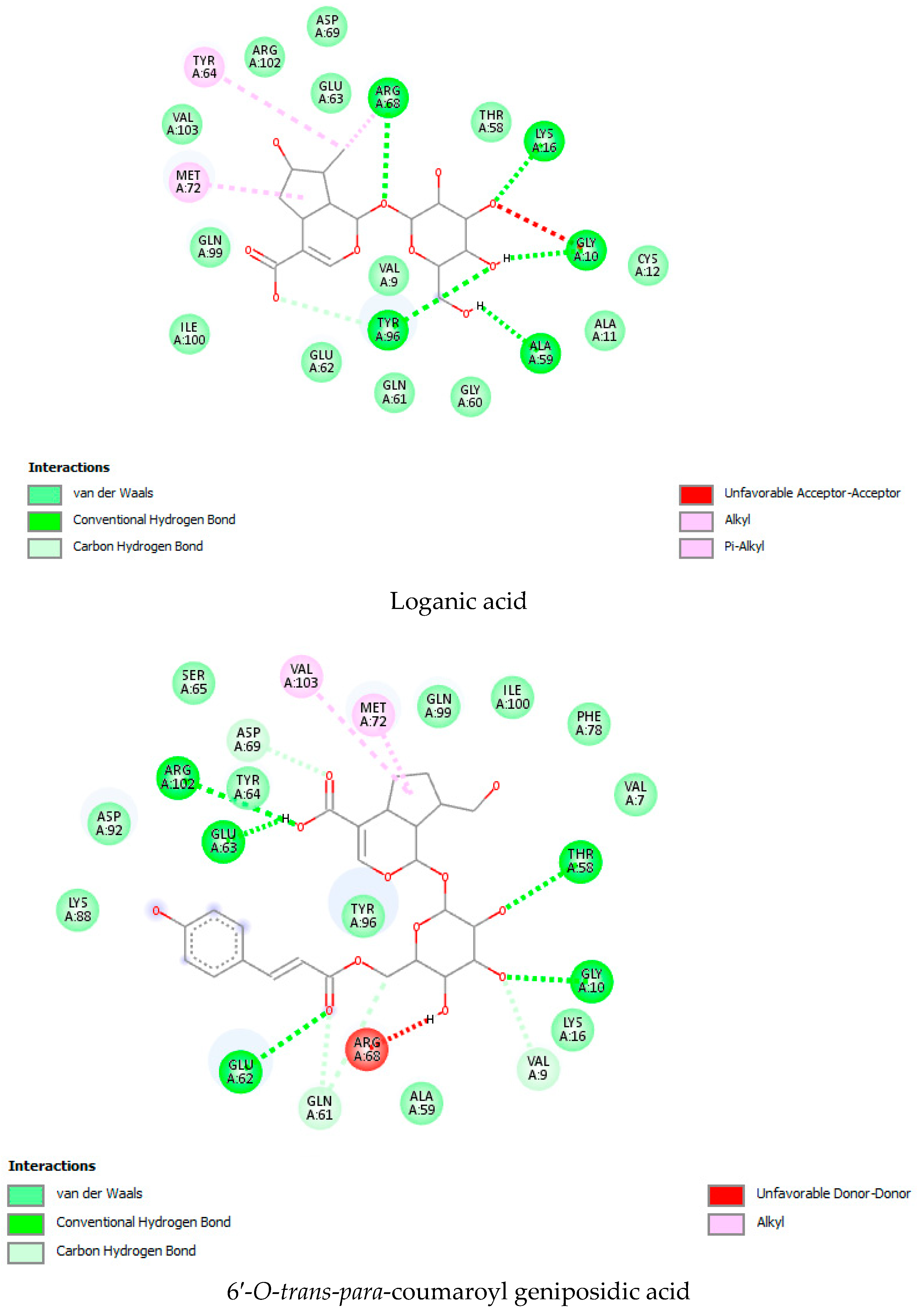
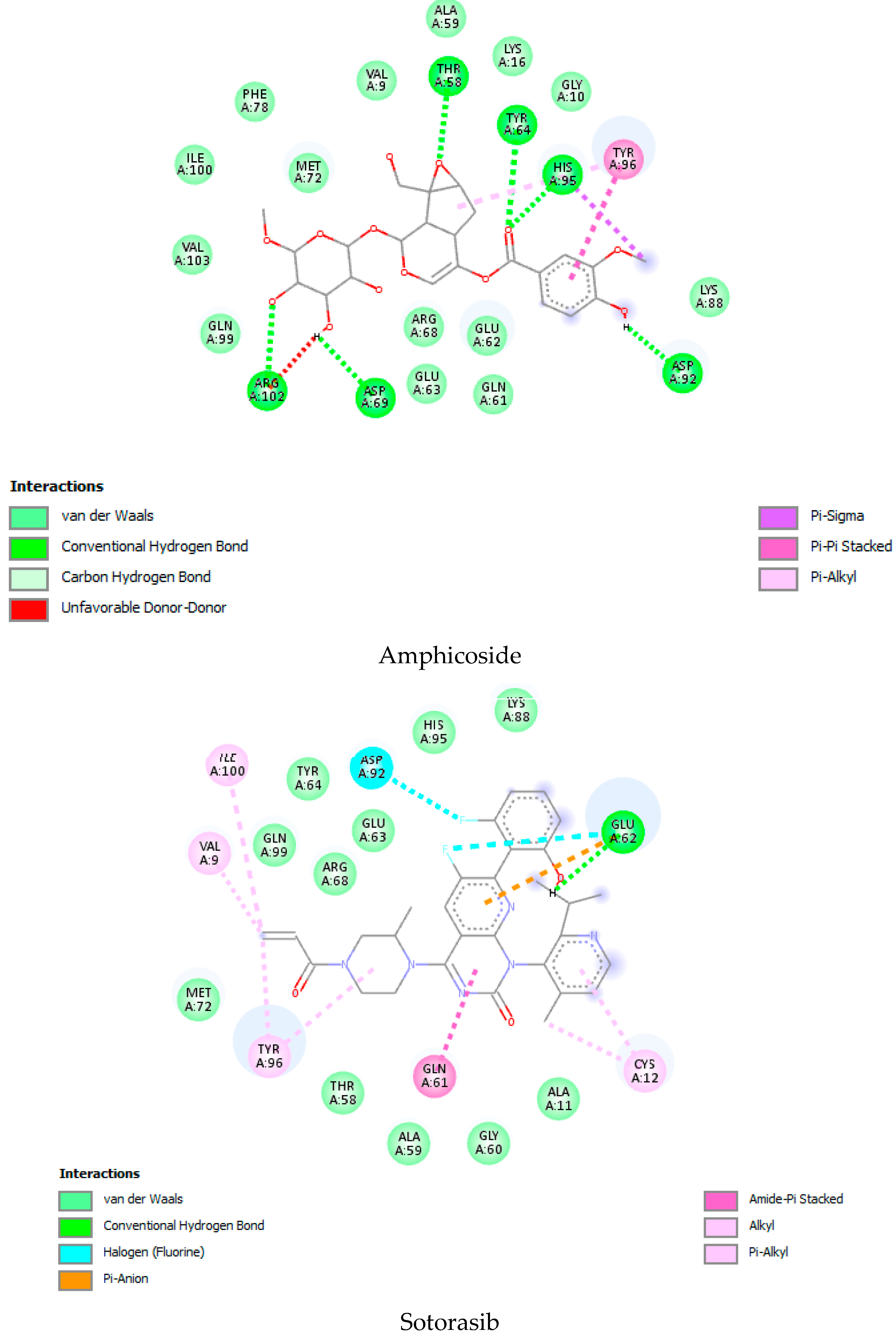
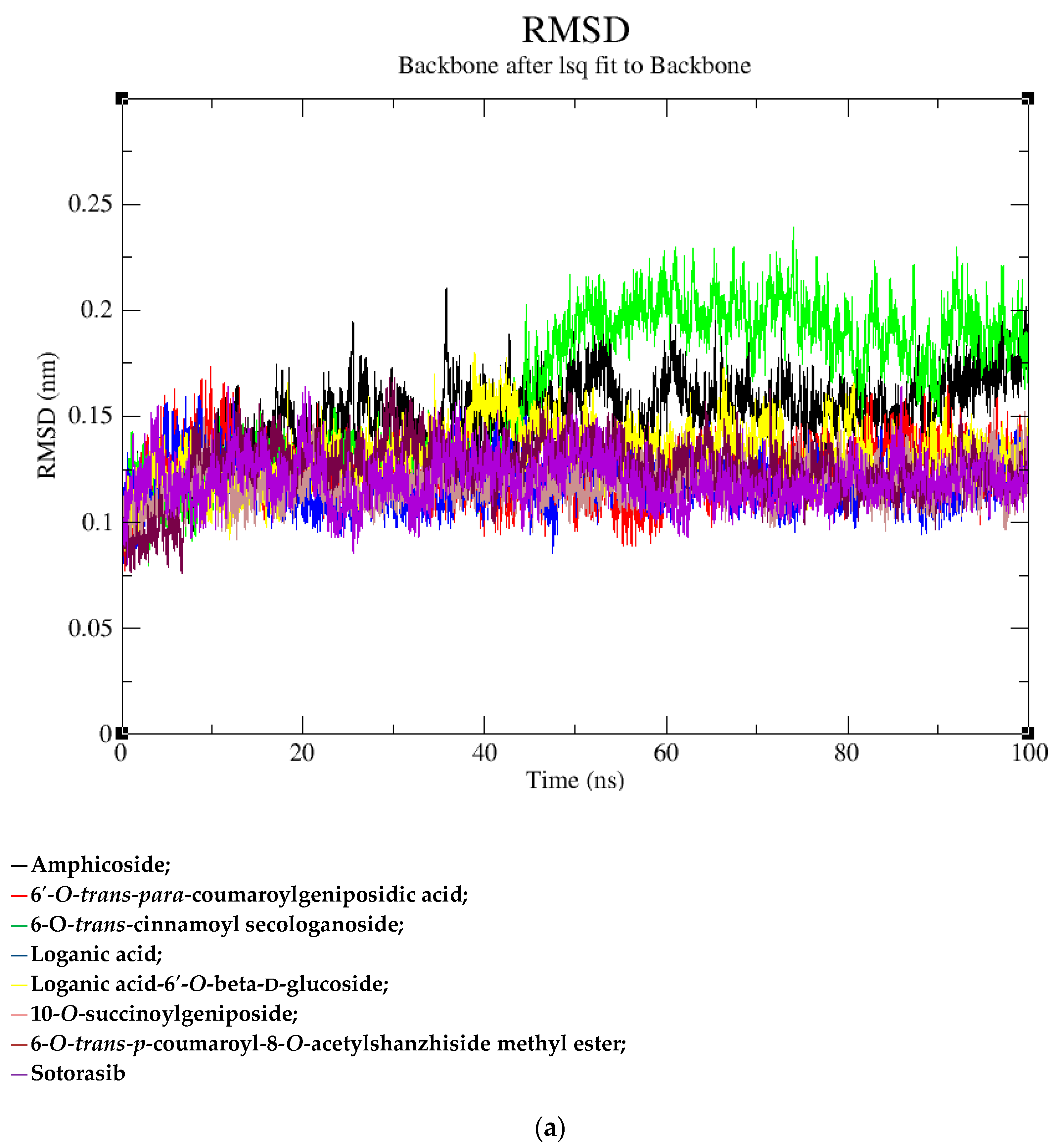
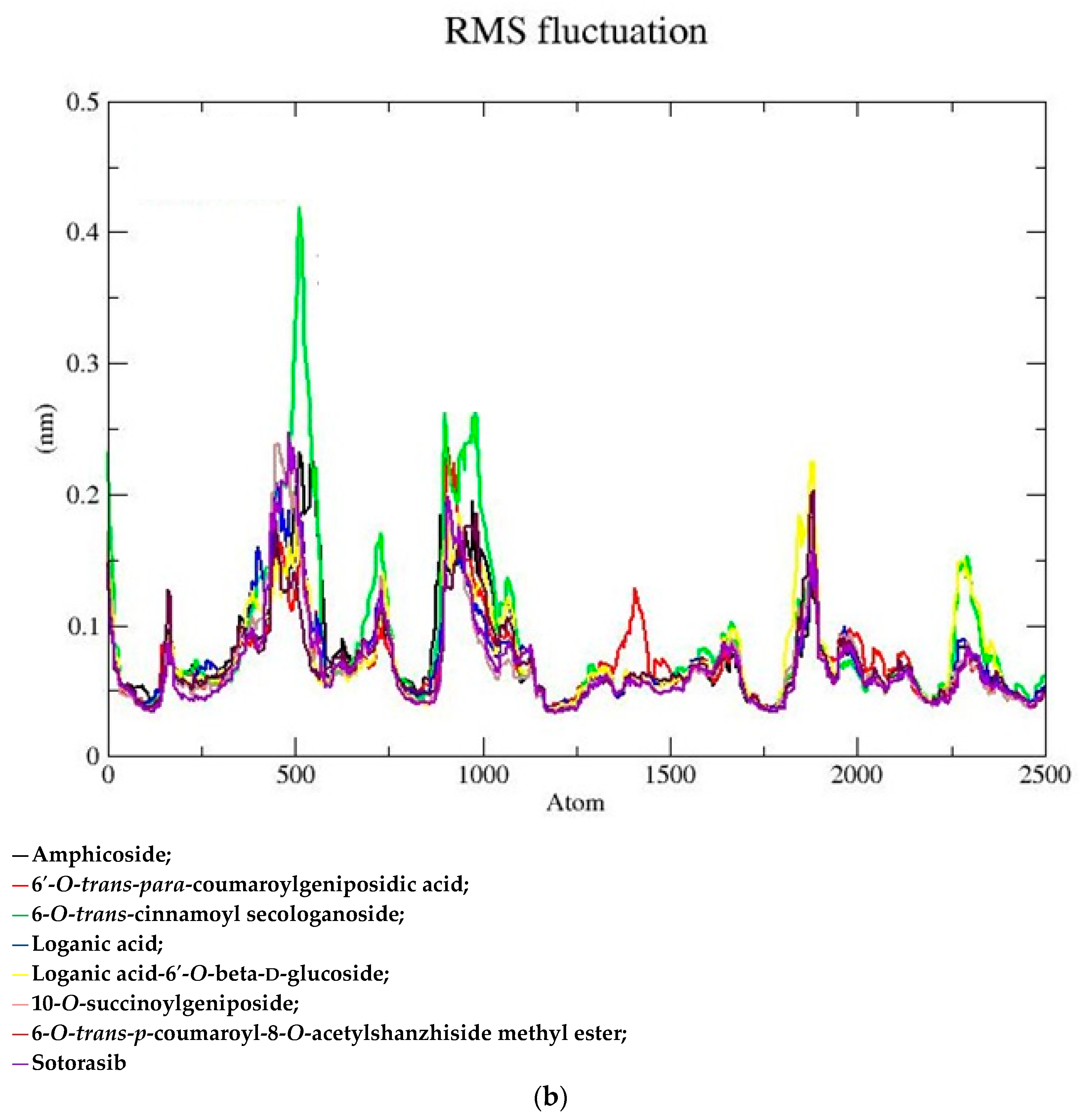

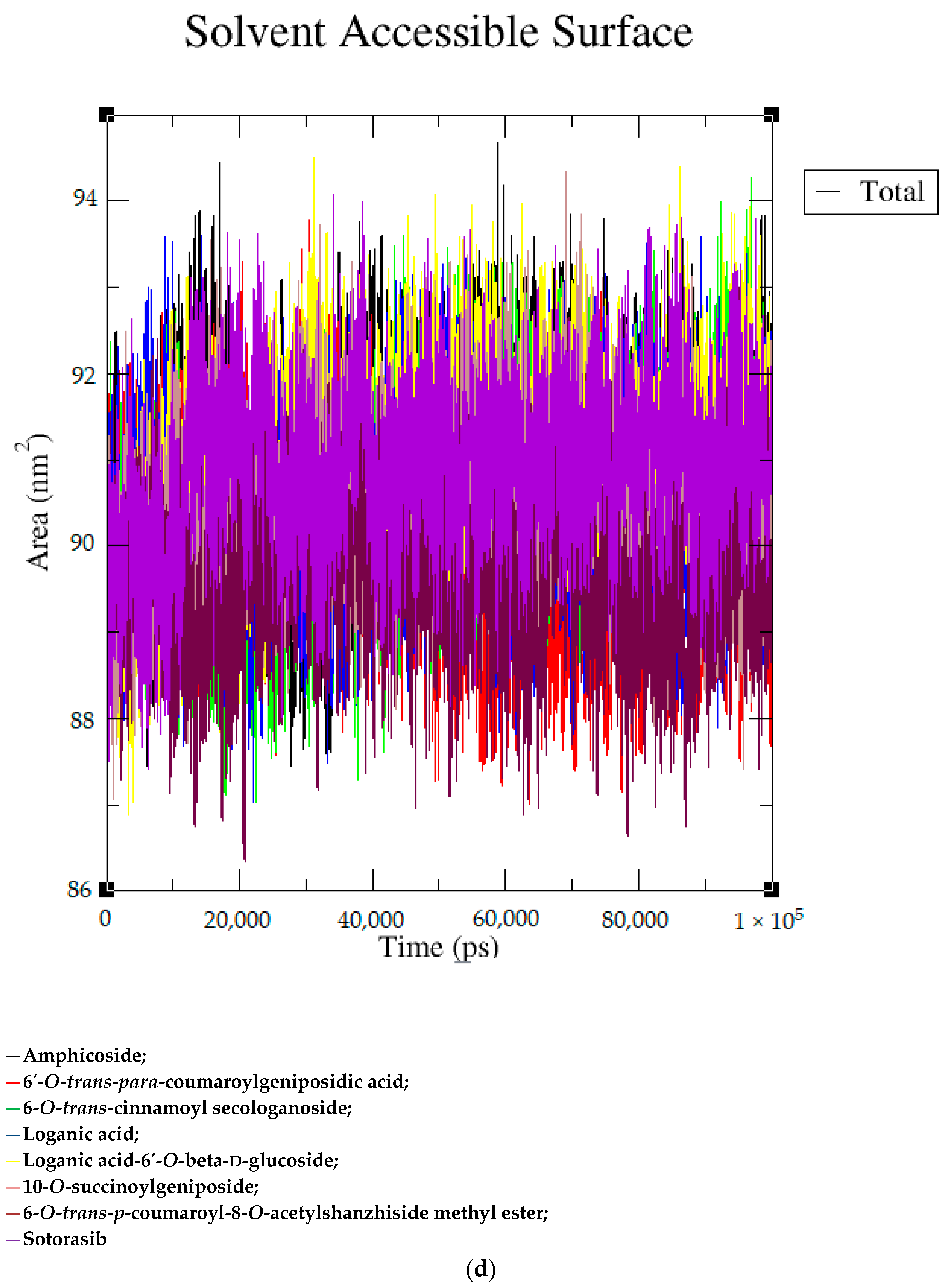
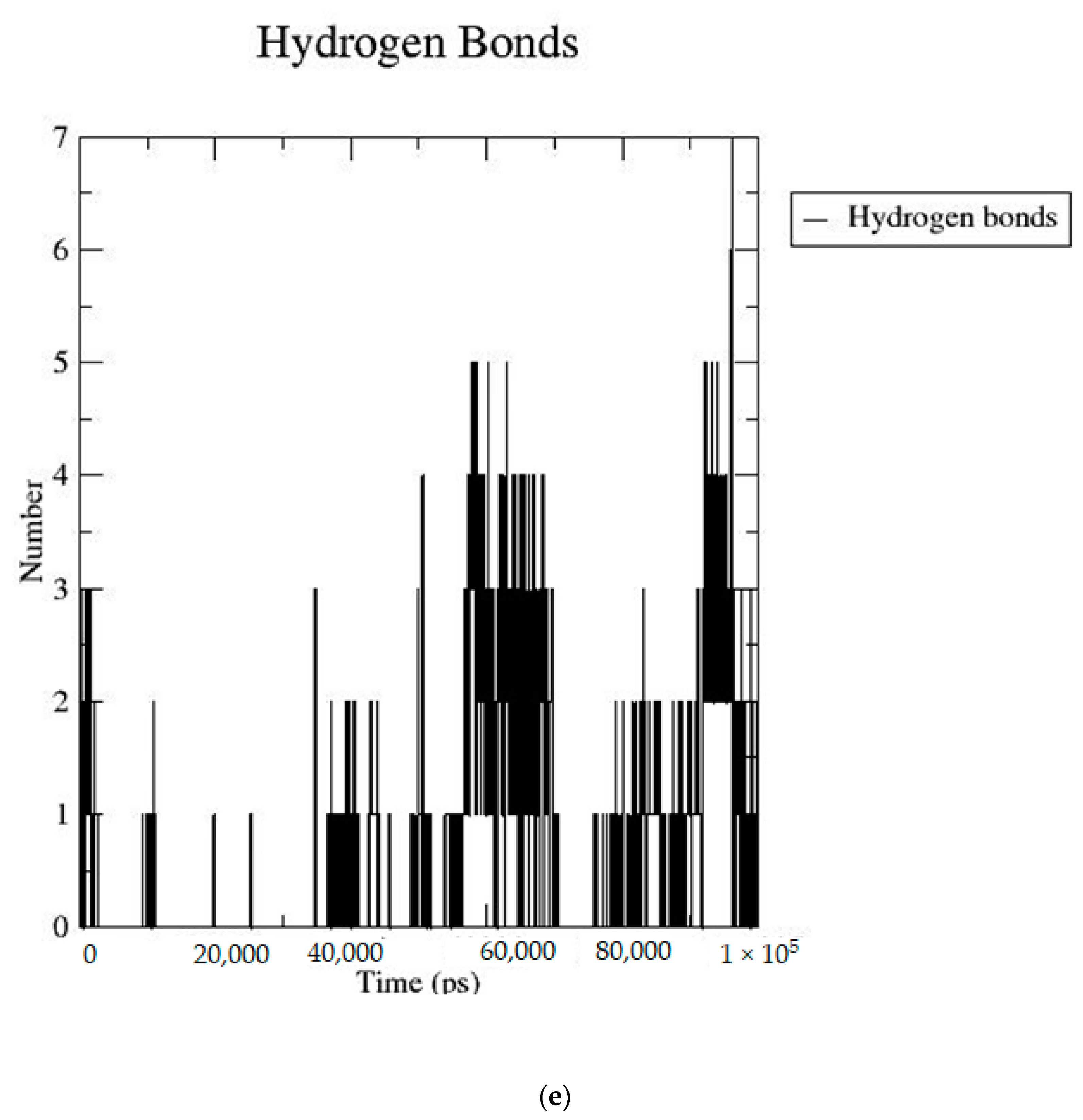






| Sr. No | Name of the Molecules | Docking Score (kcal/mol) | Interacting Amino Acid Residues |
|---|---|---|---|
| 1. | 6-O-alpha-d-galactopyranosylharpagoside | −7.8 | CYS12 b, LYS 88 b, ASP 92 b, GLY 60 b, TYR 96 b THR 35 a, GLN 61 c, GLU 62 c, ARG 68 c, GLU 63 c, TYR 64 c, MET 72 d, ALA 59 c, ALA 11 a |
| 2. | 6′-O-sinapoyl-geniposide | −9.0 | GLU 63 b, ARG 68 b, GLN 61 b, ALA 59 b, PHE 78, TYR 64 a, ASP 69 a, GLN 99 b, HIS 95 c, ASP 92 c, ALA 11 a, HIS 94 a, LYS 16 a, THR 58 a |
| 3. | 6-O-trans-cinnamoyl-secologanoside | −9.5 | ARG 68 b, GLY10 b, THR 58 b, ALA 59 b, LYS 88, CYS 12 d, MET 72 d, GLN 99 a, GLN 61, TYR 96 a, LYS 16 a, ASP 92 b, ALA 11 a |
| 4. | 6′-O-trans-para-coumaroylgeniposide | −9.0 | TYR 96 b, GLY 10 b, ALA 59 b, TYR 64 b, HIS 95 b, ASP 92 a, CYS 12 b, LYS 88 a, ALA 11 a,GLU 62 a, GLN 61 a, ARG 68 a, THR 58 a, GLN 99 a, PHE 78 a, MET 72 d. |
| 5. | 6′-O-trans-para-coumaroylgeniposidic acid | −9.6 | ASP 69 b, THR 58 b, GLY10 b, ALA59 b, LYS88 b, GLU 63 a, ARG 68 a, TYR 64 a, PHE78 a, GLN 61 a, ASP 92 a, MET 72 d, GLN 99 a. |
| 6. | 6-O-trans-p-coumaroyl-8-O-acetylshanzhiside methyl ester | −9.9 | LYS 88 b, ASP 92 b, TYR 96 b, ASP 69 b, GLU 63 b, ARG 68 b, ALA 11 a, CYS 12 d, THR 58 a, ALA 59 a, GLN 61 a, THR 64 a, MET 72 c. |
| 7. | 7-hydroxy eucommiol | −5.8 | ARG 68 b, GLU 63 b, TYR 96 b, TYR 64 a, GLU 62 a, HIS 95 a, GLN 99 a, MET 72 d, THR 58 c, VAL9 c. |
| 8. | 8-epideoxyloganic acid | −8.4 | TYR 96 b, ALA 59 b, CYS 12 d, GLU 62 c, GLN 61 c, GLU 63 a, TYR 64 a, ARG 68 a, MET 72 d, PHE 78 c, GLN99 a. |
| 9. | 8-p-coumaroylharpagide | −8.1 | LYS 88 b, CYS 12 b, ASP 92 b, GLY 60 b, TYR 96 b, ALA 11 a, GLU 62 a, ALA 59 a, GLN 61 c, VAL 9 c. |
| 10. | 10-Isovaleroyl-dihydropenstemide | −8.3 | TYR 96 b, ARG 102 b, ASP 69 b, LYS 88 a, LYS 16 a, GLY 10 a, ALA 59 a, THR 58 c, VAL 9 c, MET 72 d, TYR 64 a, GLN 99 a, ASP 92 a. |
| 11. | 10-O-acetylgeniposide | −8.7 | ARG 102 b, ARG 68 b, TYR 96 b, GLN 61 b, ALA59 b, HIS 95 a, GLU 62 a, TYR 64 a, MET 72 d, GLN 99 c, GLY 10 a, THR 58 c, LYS 16 a. |
| 12. | 10-O-succinoylgeniposide | −9.4 | ARG 102 b, ARG 68 b, THR 58 b, GLY 10 b, ALA 59 b,TYR 96 b, ASP 92 b, GLY60 b, MET 72 d, GLN99 c, LYS 16 a, TYR 64 a, GLU 62 a, HIS 95 a, CYS 12 d, ALA 11 a. |
| 13. | Acetylgeniposide | −6.6 | LYS 88 b, TYR 96 b, TYR 64 b, ARG 68 b, CYS 12 d, ALA 11 a, GLY60 a, ALA59 a, THR 58 a, HIS 95 a, GLN 99 a, GLU 62 c MET 72 d, GLY 10 a. |
| 14. | Acetylbarlerin | −7.7 | GLU 63 b, GLN 61 b, GLU 62 b, THR 96 b, CYS 12 b, THR 58 b, ARG 68 b, VAL9 a, ASP 92 a, ALA 11 a, MET 72 d. |
| 15. | Amphicoside | −9.2 | LYS 88 b, ASP 92 b, HIS 95 b, THR 58 b, ASP69 b, ARG 102 b, TYR64 b, GLU63 b, VAL 9 a, GLN 99 a, MET 72 d, ARG 68 c, TYR 96 c. |
| 16. | Asperuloside | −8.9 | ARG 68 b, ALA59 b, TYR96 b, TYR 64 a, GLN99 b, VAL9 b, GLN61 a, GLU62 a, HIS 95 c, MET 72 d. |
| 17. | Barlerin | −8.4 | ALA 59 b, GLU 62 b, ARG 68 b, ARG 102 b, TYR64 b, HIS 95 b, ASP 92 b, TYR 96 b, GLN 61 a, GLY 60 a, GLY 10 a, THR 58 a, ASP 69 c, MET 72 d, GLN99 c. |
| 18. | Brasoside | −8.6 | ALA 59 b, GLU 62 b, ARG 68 b, ARG 102 b, TYR 64 b, HIS 95 b, ASP 92 b, TYR 96 b, GLN 61 a, GLY 60 a, GLY 10 a, THR58 c, ASP69 c, MET 72 d, GLN 99 c. |
| 19. | Buddlejoside A9 | −7.5 | CYS 12 b, TYR 96 b, GLY 60 b, PRO34 a, ALA 11 a, GLU 62 a, GLY 10 c, TYR 64 a, THR 58 a, ARG 68 a, ASP 69 c, GLN 99 a, ALA 59 a, MET 72 d, HIS 95 c. |
| 20. | Cantleyoside | −8.1 | GLY 60 b, TYR 96 b, ARG 102 b, ASP 69 b, THR 58 b, GLY 10 b, PRO 34 b, THR 35 a, CYS 12 a, TYR 64 a, GLN99 c, ARG 68 c, MET 72 d, LYS 16 c, GLU 62 a, ALA 59 a. |
| 21. | Deacetyl asperuloside | −8.6 | TYR 96 b, ALA 59 b, ARG 68 b, GLU 62 a, GLU 63 a, GLN 99 c, MET 72 d, CYS 12 d. |
| 22. | Euphroside | −8.6 | GLY 10 b, THR 58 b, ASP64 b, ARG 68 b, ALA59 b, TYR 96 b, VAL9 a, GLN 99 a, MET 72 d, TYR 64 c, GLU 62 c, GLY 60 c, GLN 61 c. |
| 23. | Eurostoside | −8.6 | ALA 59 b, ARG 68 b, THR58 b, GLY 10 b, GLY 60 a, GLU62 a, TYR96 c, HIS 95 c, TYR64 a, GLN99 c, MET72 d, LYS 16 a, VAL 9 a, GLN61 c, CYS 12 d. |
| 24. | Garjasmine | −7.2 | TYR 64 b, ARG 68 b, GLU 62 a, TYR 96 a, GLN99 a, PHE 78 a, MET 72 d, VAL 9 c, GLN61 c, GLU 63 c. |
| 25. | Geniposidic Acid | −8.9 | CYS 12 b, ALA 59 b, THR58 b, ARG68 b, TYR64 b, ARG 102 b, HIS 95 b, ASP92 b, TYR96 b, GLY 10 b, MET 72 d, ASP69 c, GLN 99 a, GLU 62 a, LYS 16 a. |
| 26. | Gentiopicroside | −8.9 | TYR 96 b, GLY 10 b, THR 58 b, ARG 68 b, ALA 59 b, LYS16 a, VAL9 a, GLN99 c, ILE100 c, ASP69 a, MET72 d, TYR 64 a, GLU63 c, CYS12 d. |
| 27. | Isojaslanceoside B | −8.6 | LYS 88 b, CYS12 b, ARG 68 b, GLN61 b, ASP69 b, GLU63 b, TYR64 b, HIS 95 b, ASP92 b, GLU 62 a, ALA59 a, ARG102 a, GLN99 a, MET72 d, TYR 96 c. |
| 28. | Kutkin | −1.8 | SER 17 b, ILE 36 b, THR58 b, MG202 b, GDP 201 b, ASP57 a, ALA59 a. |
| 29. | Laciniatoside I | −8.6 | ASP 92 b, CYS12 b, TYR 96 b, GLU62 b, ALA11 a, GLN99 a, ARG 68 a, ILE100 c, VAL103 c, MET 72 d, GLN 61 c, VAL9 a, GLU 63 c, HIS 95 a. |
| 30. | LaciniatosideII | −8.7 | ASP92 b, HIS95 b, TYR64 b, ASP69 b, GLU63 b, CYS12 a, GLU62 a, GLY10 a, ALA59 a, ARG68 c, LYS16 a, MET 72 d, VAL103 a, GLN99 a, ARG 102 c. |
| 31. | Loganic acid | −9.4 | TYR 96 b, GLY10 b, THR 58 b, ARG68 b, ALA 59 b, LYS 16 a, ILE 100 a, GLN99 a, VAL 9 a, TYR64 c, MET72 c, GLU 63 a, VAL 103 a, ASP69 a, GLY60 a. |
| 32. | Loganic acid 6′-O-beta-d-glucoside | −9.5 | TYR 96 b, ASP92 b, ASP69 b, GLY10 b, THR58 b, ALA59 b, HIS95 a, GLU62 a, ALA11 a, GLN99 a, TYR64 a, GLU63 a, MET72 a, ARG68 a, GLN61 a, CYS12 a. |
| 33. | Minecoside | −8.9 | TYR 96 b, ARG102 b, ARG68 b, GLN99 a, TYR64 a, GLN61 a, GLU62 c, HIS95 c, ASP92 c, THR 58 a, ALA59 a, VAL 9 a, MET 72 a, GLU 63 a, ILE 100 a. |
| 34. | Mussaenoside | −8.2 | TYR 96 a, TYR64 a, HIS 95 a, ARG 102 b, ASP 92 a, ARG 68 b, GLY 60 b, ALA 59 a, GLN61 a, VAL 103 a, GLN99 a, MET 72 c, VAL 9 c, GLU 62 a. |
| 35. | Ninpogenin | −6.0 | GLN61 a, GLU63 b, ARG68 b, ASP92 a, HIS95 b, TYR64 a, GLU62 a, MET 72 c, GLN 99 a, TYR 96 a. |
| 36. | Nuezhenelenoliciside | −8.7 | ASP 92 b, GLU 63 b, ASP69 b, ARG69 b, ALA 59 b, GLY60 b, HIS 95 b, TYR 64 b, LYS 88 a, TYR 96 a, MET 72 c, GLY 10 a, VAL 9 a, THR 58 a, LYS 16 a, GLN 61 a, GLU 62 a. |
| 37. | Nuezhenide | −8.9 | GLU62 b, LYS 88 b, CYS12 b, TYR96 b, GLY10 b, ARG 68 b, HIS 95 a, ASP 92 a, ALA 11 a, LYS 16 a, ALA59 a, VAL9 a, GLN 99 a, ILE 100 a, VAL 103 a, MET 72 c. |
| 38. | Oleoside dimethyl ester | −7.1 | GLU 62 b, LYS 88 b, ASP 92 b, HIS 95 b, ARG 68 b, GLY 60 b, CYS 12 a, ALA 11 c, TYR 96 a, TYR 64 a, MET 72 a, ALA 59 a, GLN 99 a, GLU 63 a, VAL 9 a, VAL 103 a, GLN 61 a, THR 35 a, PRO 34 a. |
| 39. | Oleuropein | −8.4 | LYS 88 b, ASP 92 b, HIS 95 b, TYR 64 b, ARG 68 b, TYR 96 c, ALA 11 a, VAL9 a, GLN 99 a, VAL103 c, ILE 100 a, ARG 102 a, ASP 69 a, ALA 59 a, GLU 62 a. |
| 40. | Patrinalloside A | −8.9 | GLU 62 b, LYS 88 b, ASP 92 b, TYR 96 b, THR 58 b, GLY 10 b, VAL 9 c, GLN99 a, ILE 100 c, VAL 103 c, MET 72 c, LYS 16 a, ALA 59 a, HIS 95 a. |
| 41. | Picroside-II | −9.0 | CYS 12, ASP 92, GLY 10, TYR 96, ARG 102, ARG 68; by H- bond interactions, ALA 11, GLN 61, GLU 62, GLU 63, TYR 64, MET 72, ALA 59, VAL 9, THR 58 by hydrophobic interactions. |
| 42. | Picroside-III | −8.9 | LYS 88 b, HIS 95 b, GLU 62 b, TYR 64 b, GLN99 b, THR58 b, GLY 10 b, TYR 96 b, ASP92 b, VAL 103 a, GLU 63 a, ARG 68 a, ASP 69 a, VAL 9 a, GLN 61 a, ALA 11 a, CYS 12 c. |
| 43. | Pinnatoside | −7.7 | ALA 59 b, THR 58 b, ARG 68 b, GLU 63, GLN 61 b, GLU 62, LYS 16, TYR 96, GLN 99 b,ARG 102 b, ILE 100 a, VAL 103 a, MET 72 a. |
| 44. | Plantarenaloside | −8.6 | TYR 96 c, HIS 95 b, ASP 92 b, TYR 64 b, ARG 68 b, GLY 60 b, ALA 59 b, GLU 62 a, GLN 99 a, MET 72 c, VAL 9 c, THR 58 a, GLN 61 a. |
| 45. | Polystachyn A | −8.5 | ARG 68 b, TYR 96 b, GLN 61 a, ALA59 a, TYR 64 c, GLU 62 a, MET 72 c, VAL 9 a, GLU 63 a, ASP 69 a, GLN 99 a, ARG 102 a, VAL 103 a, PHE 78 a. |
| 46. | Shanzhiside methyl ester | −8.4 | GLU 62 b, HIS 95 b, ASP 92 b, TYR 64 b, ARG 68 b, GLU 63 b, TYR 96 b, VAL 9 c, GLN 99 a, MET 72 c, ILE 100 c, ALA 11 a, CYS 12 a. |
| 47. | Specioside | −8.6 | LYS 88 b, ASP92 b, ARG 68 b, GLU 63 b, TYR 96 b, TYR 64 a, HIS 95 a, VAL 9 a, VAL103 c, MET 72 c, ILE 100 c, PHE 78 a, ALA 59 a, CYS 12 a, GLU 62 a. |
| 48. | Sylvestroside I | −8.0 | GLU 63 b, ASP 92 b, HIS 95 b, HIS 94 b, GLU 98 b, ARG 68 b, GLN 61 a, TYR 64 a, GLU 62 a, TYR 96 a, VAL 9 c. |
| 49. | Sylvestroside III | −8.6 | LYS 88 b, ASP92 b, TYR 64 b, ASP 69 b, HIS 95 b, GLN 99 b, ARG 102 b, TYR 96 b, ARG 68 b, PHE 78 a, MET 72 a, VAL 103 a, ILE 100 a, GLU62 a, GLU 63 a, ALA 59 a, GLN 61 a. |
| 50. | Sylvestroside III dimethyl acetal | −8.3 | LYS 88 b, CYS 12 b, THR 58 b, ASP 69 b, TYR 64 b, GLU 63 b, TYR 96 b, ALA 11 b, ALA 59 b, VAL 9 a, ILE 100 a, VAL 103 a, HIS 95 a, GLN 61 a, GLU 62 a. |
| 51. | Sylvestroside IV | −8.8 | CYS 12 b, ASP 92 b, TYR 96 b, ARG 68 b, ARG 102 b, HIS 95 a, GLY 10 a, ALA 59 a, THR 58 a, LYS 16 a, VAL 9 c, MET 72 c, GLU 63 a, ASP 69 a, GLN 99 a, VAL 103 a. |
| 52. | Verminoside | −8.9 | LYS 88 b, ASP92 b, HIS 95 b, ARG 102 b, ASP69 b, TYR 64 b, ALA 11 c, CYS 12 d, GLN 61 a, GLU 62 a, ALA 59 a, ARG 68 a, THR 58 a, VAL 9 c, MET 72 a, GLN 99 a, PHE 78 a, ILE 100 a, VAL 103 a, TYR 96 a. |
| 53. | Sotorasib | −9.1 | MET 72 a, ILE 100 a, GLN 99 a, ASP92 a, LYS 88 a, GLU 62 b, CYS 12 a, GLN 61 c, TYR 96 c, ARG 68 a. |
| 54. | LXD | −11.7 | ASP 92 b, HIS 95 b, TYR 64 b, ASP 69 b, GLU 63 b, CYS 12 a, GLU 62 a, ALA 11 a, LYS 88 a, TYR 96 a, VAL 9 c, GLN 99 a, VAL 103 c, MET 72 d, ARG 68 c. |
| Sr. No. | Name of the Molecules | Van der Waal Energy (kJ/mol) | Electrostatic Energy (kJ/mol) | Polar Solvation Energy (kJ/mol) | SASA Energy (kJ/mol) | Binding Energy (kJ/mol) |
|---|---|---|---|---|---|---|
| 1. | 6-O-trans-p-coumaroyl-8-O-acetylshanzhiside methyl ester | −5.656 | −2.468 | 1.643 | −0.828 | −7.309 |
| 2. | 10-O-succinoylgeniposide | −16.228 | −6.363 | 24.886 | −2.391 | −0.096 |
| 3. | Loganic acid 6′-O-beta-d-glucoside | −21.887 | −10.770 | 33.348 | −2.841 | −2.150 |
| 4. | 6-O-trans-cinnamoyl-secologanoside | −9.640 | −11.466 | 27.814 | −1.737 | −4.971 |
| 5. | Loganic acid | −15.230 | −6.953 | 25.109 | −2.009 | −0.917 |
| 6. | 6′-O-trans-para-coumaroyl geniposidic Acid | −25.501 | −5.342 | 23.845 | −3.154 | −6.153 |
| 7. | Amphicoside | −11.486 | −3.441 | 12.217 | −1.416 | −4.216 |
| 8. | Sotorasib | −10.429 | −1.621 | 9.543 | −1.044 | −3.551 |
| Sr. No. | Name of the Molecules | EHOMO (eV) | ELUMO (eV) | ΔE gap (eV) | I | A | η | ζ | μ | Ψ |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | 6-O-trans-p-coumaroyl-8-O-acetylshanzhiside Methyl Ester | −6.44 | −2.14 | 4.3 | 6.44 | 2.14 | 2.15 | 0.23 | 4.29 | 4.28 |
| 2. | 10-O-succinoylgeniposide | −6.63 | −1.06 | 5.57 | 6.63 | 1.06 | 2.78 | 0.17 | 3.85 | 2.66 |
| 3. | Loganic acid 6′-O-beta-d-glucoside | −6.83 | −1.41 | 5.42 | 6.83 | 1.41 | 2.71 | 0.18 | 4.12 | 3.13 |
| 4. | 6-O-trans-cinnamoyl-secologanoside | −6.55 | −1.57 | 4.98 | 6.55 | 1.57 | 2.49 | 0.20 | 4.06 | 3.30 |
| 5. | Loganic acid | −6.80 | −1.25 | 5.55 | 6.80 | 1.25 | 2.77 | 0.18 | 4.02 | 2.91 |
| 6. | 6′-O-trans-para-coumaroyl geniposidic acid | −6.34 | −1.95 | 4.39 | 6.34 | 1.95 | 2.19 | 0.22 | 4.14 | 3.91 |
| 7. | Amphicoside | −6.28 | −1.49 | 4.79 | 6.28 | 1.49 | 2.39 | 0.20 | 3.88 | 3.14 |
| 8. | Sotorasib | −6.34 | −2.74 | 3.6 | 6.34 | 2.74 | 1.8 | 0.27 | 4.54 | 5.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alamri, M.A.; Alawam, A.S.; Alshahrani, M.M.; Kawsar, S.M.A.; Prinsa; Saha, S. Establishing the Role of Iridoids as Potential Kirsten Rat Sarcoma Viral Oncogene Homolog G12C Inhibitors Using Molecular Docking; Molecular Docking Simulation; Molecular Mechanics Poisson–Boltzmann Surface Area; Frontier Molecular Orbital Theory; Molecular Electrostatic Potential; and Absorption, Distribution, Metabolism, Excretion, and Toxicity Analysis. Molecules 2023, 28, 5050. https://doi.org/10.3390/molecules28135050
Alamri MA, Alawam AS, Alshahrani MM, Kawsar SMA, Prinsa, Saha S. Establishing the Role of Iridoids as Potential Kirsten Rat Sarcoma Viral Oncogene Homolog G12C Inhibitors Using Molecular Docking; Molecular Docking Simulation; Molecular Mechanics Poisson–Boltzmann Surface Area; Frontier Molecular Orbital Theory; Molecular Electrostatic Potential; and Absorption, Distribution, Metabolism, Excretion, and Toxicity Analysis. Molecules. 2023; 28(13):5050. https://doi.org/10.3390/molecules28135050
Chicago/Turabian StyleAlamri, Mubarak A., Abdullah S. Alawam, Mohammed Merae Alshahrani, Sarkar M. A. Kawsar, Prinsa, and Supriyo Saha. 2023. "Establishing the Role of Iridoids as Potential Kirsten Rat Sarcoma Viral Oncogene Homolog G12C Inhibitors Using Molecular Docking; Molecular Docking Simulation; Molecular Mechanics Poisson–Boltzmann Surface Area; Frontier Molecular Orbital Theory; Molecular Electrostatic Potential; and Absorption, Distribution, Metabolism, Excretion, and Toxicity Analysis" Molecules 28, no. 13: 5050. https://doi.org/10.3390/molecules28135050
APA StyleAlamri, M. A., Alawam, A. S., Alshahrani, M. M., Kawsar, S. M. A., Prinsa, & Saha, S. (2023). Establishing the Role of Iridoids as Potential Kirsten Rat Sarcoma Viral Oncogene Homolog G12C Inhibitors Using Molecular Docking; Molecular Docking Simulation; Molecular Mechanics Poisson–Boltzmann Surface Area; Frontier Molecular Orbital Theory; Molecular Electrostatic Potential; and Absorption, Distribution, Metabolism, Excretion, and Toxicity Analysis. Molecules, 28(13), 5050. https://doi.org/10.3390/molecules28135050








