Evaluation of Antimicrobial, Anticholinesterase Potential of Indole Derivatives and Unexpectedly Synthesized Novel Benzodiazine: Characterization, DFT and Hirshfeld Charge Analysis
Abstract
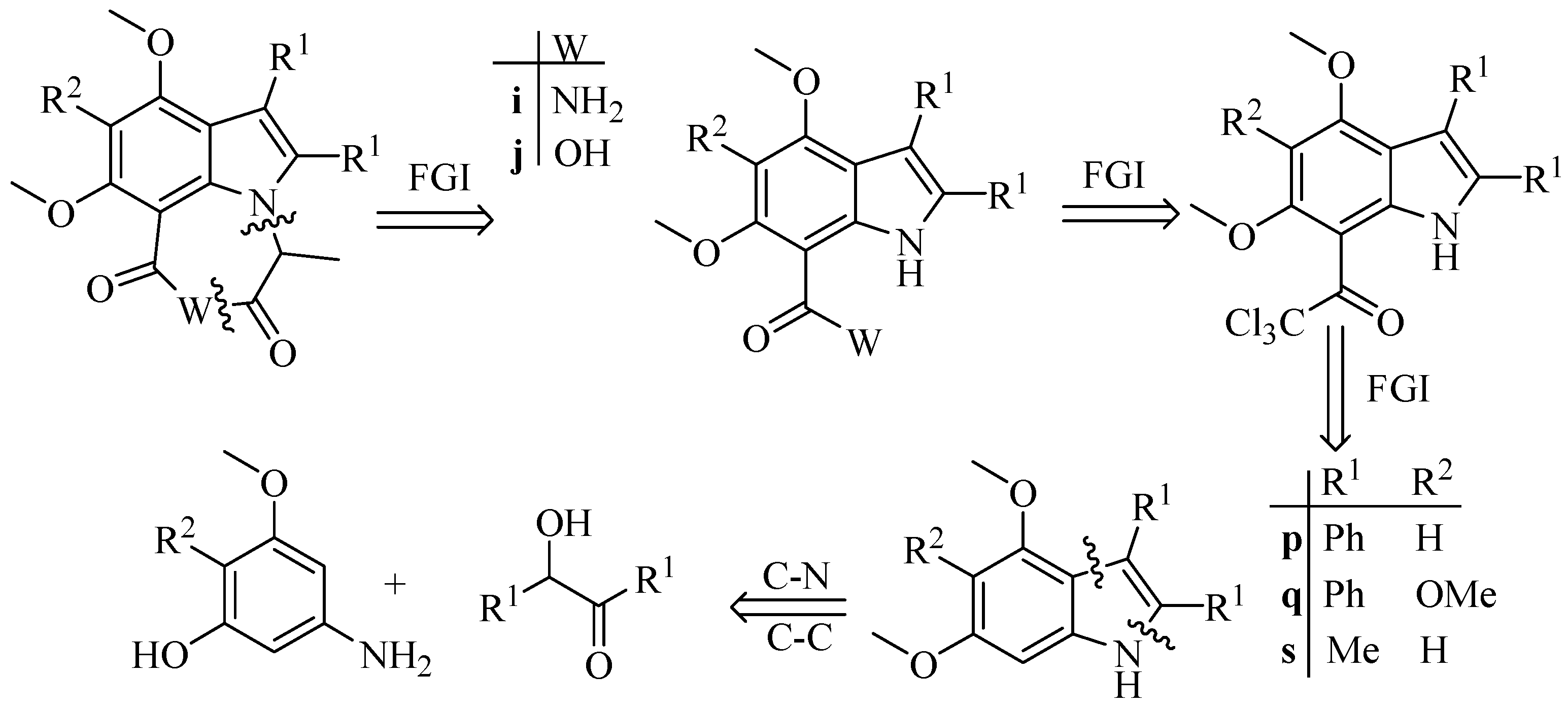
1. Introduction
2. Results and Discussions
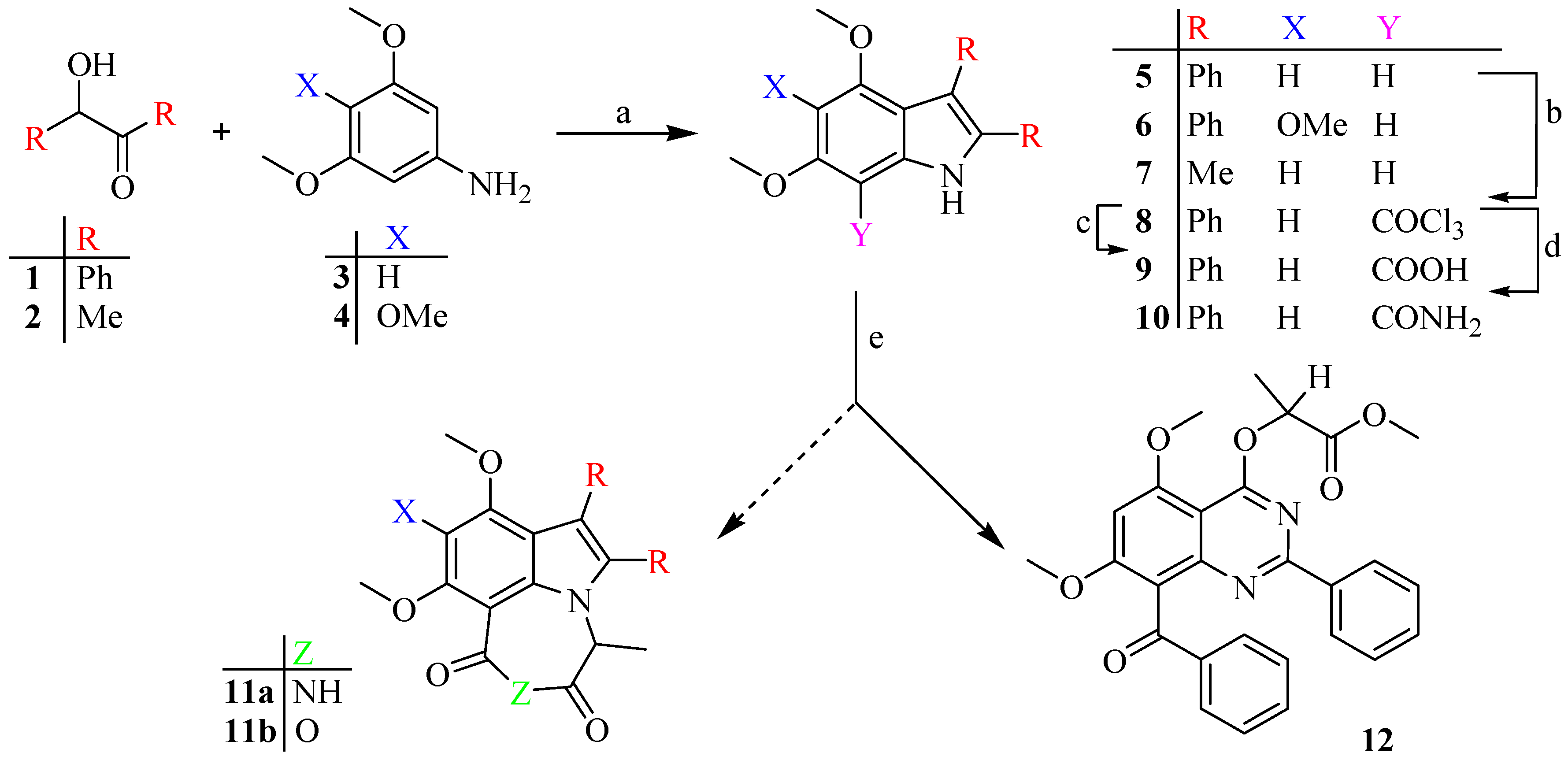
2.1. Synthesis
2.2. DFT Studies
Frontier Molecular Orbital (FMO) Analysis and Hyperpolarizability
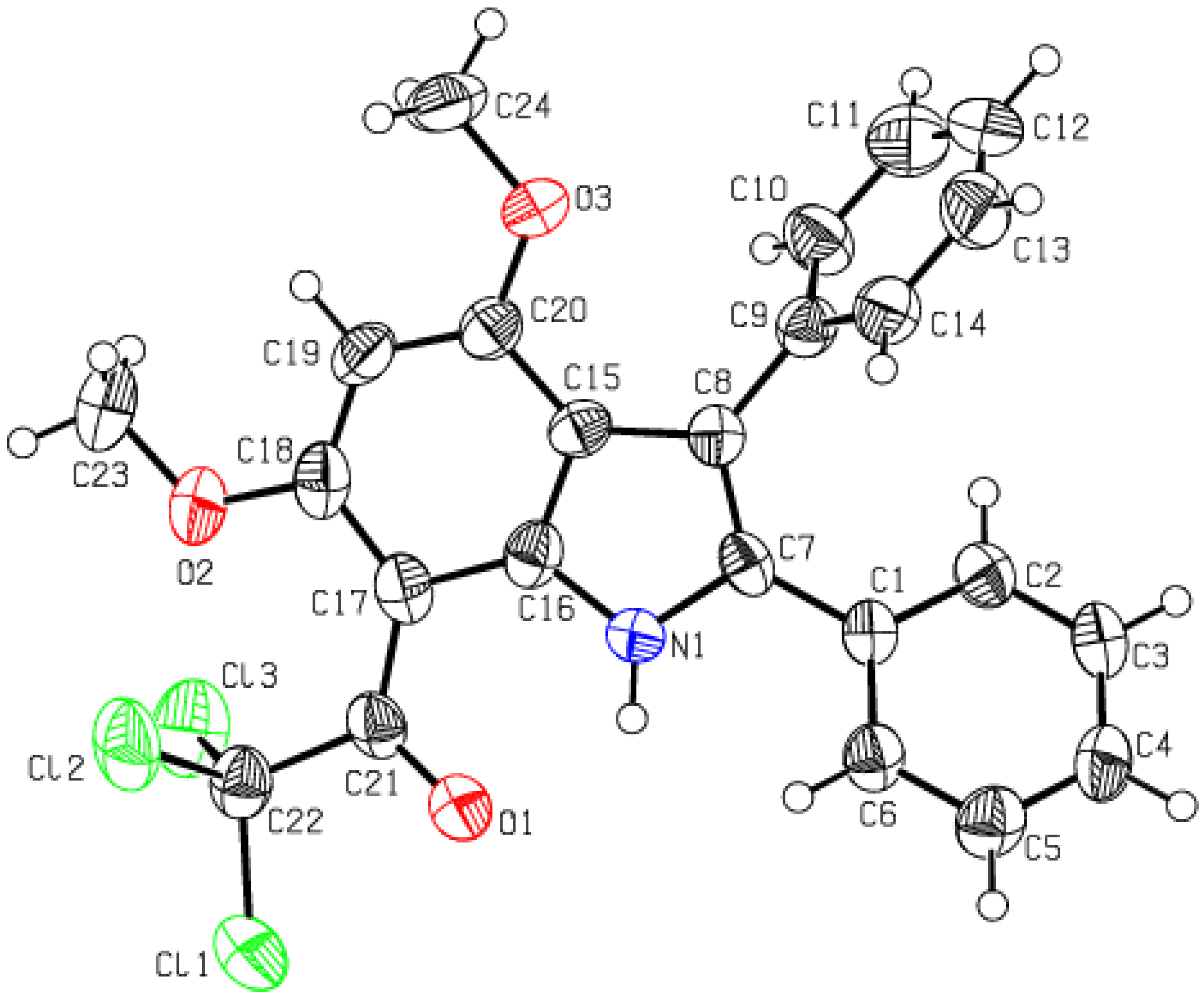
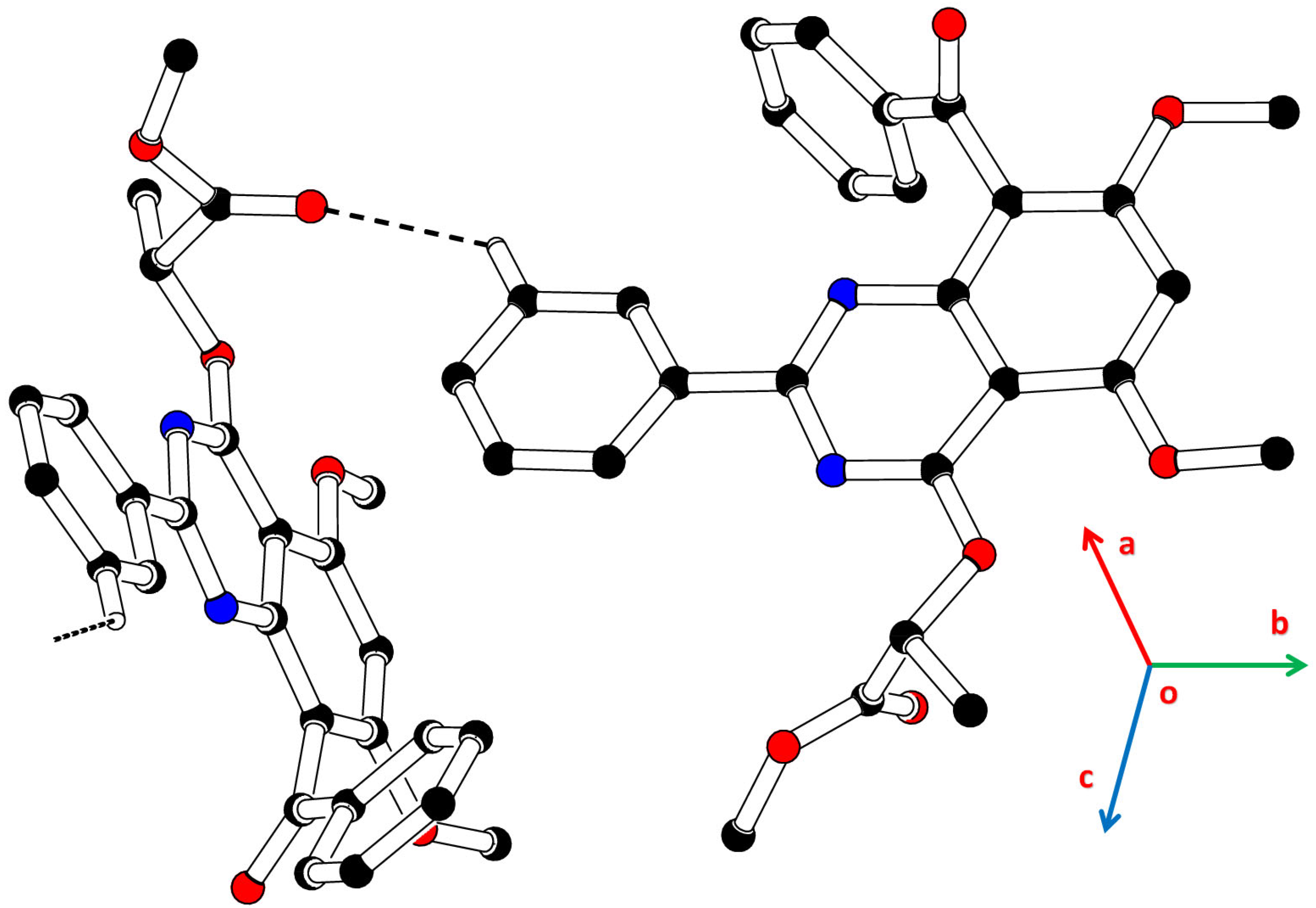
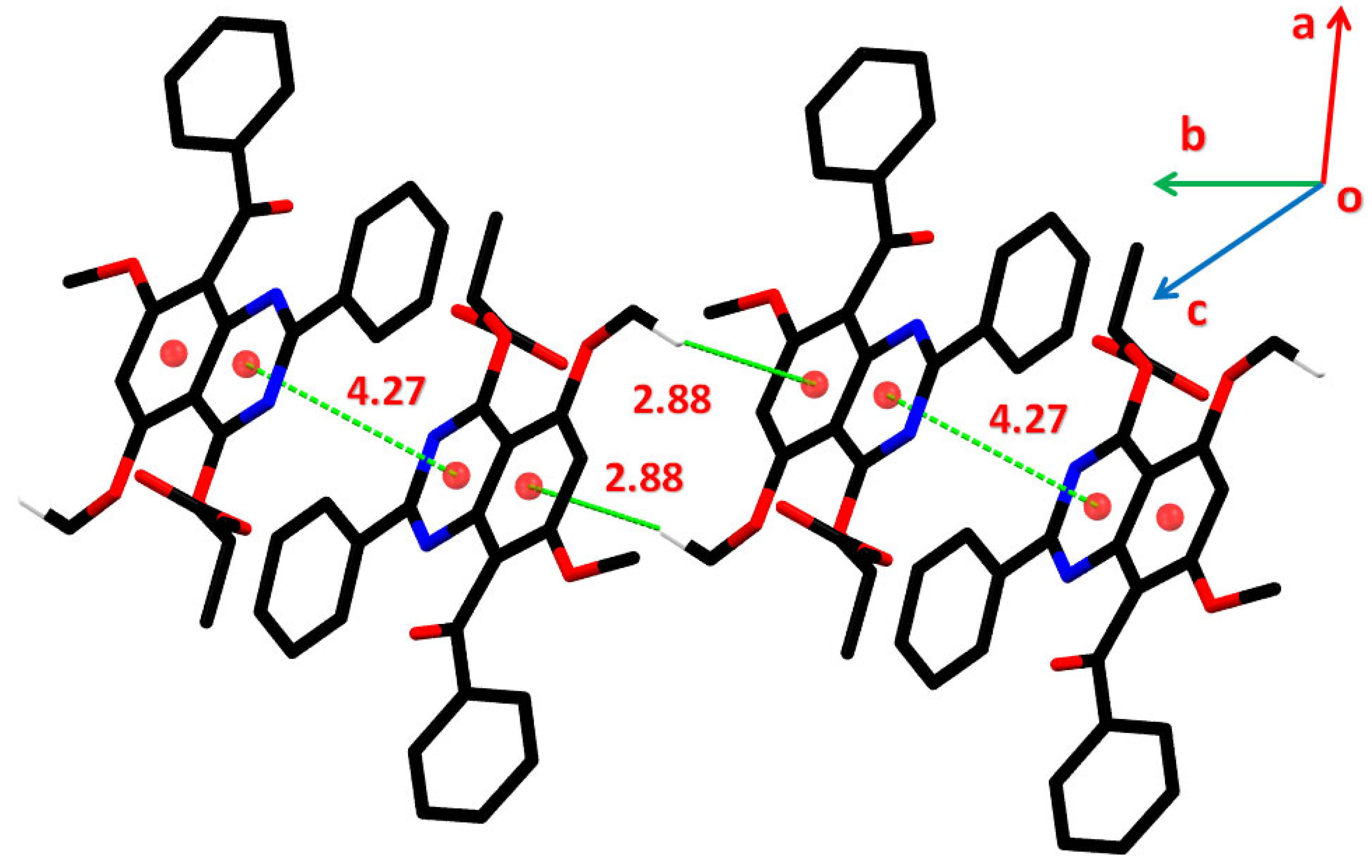
2.3. Crystal Data
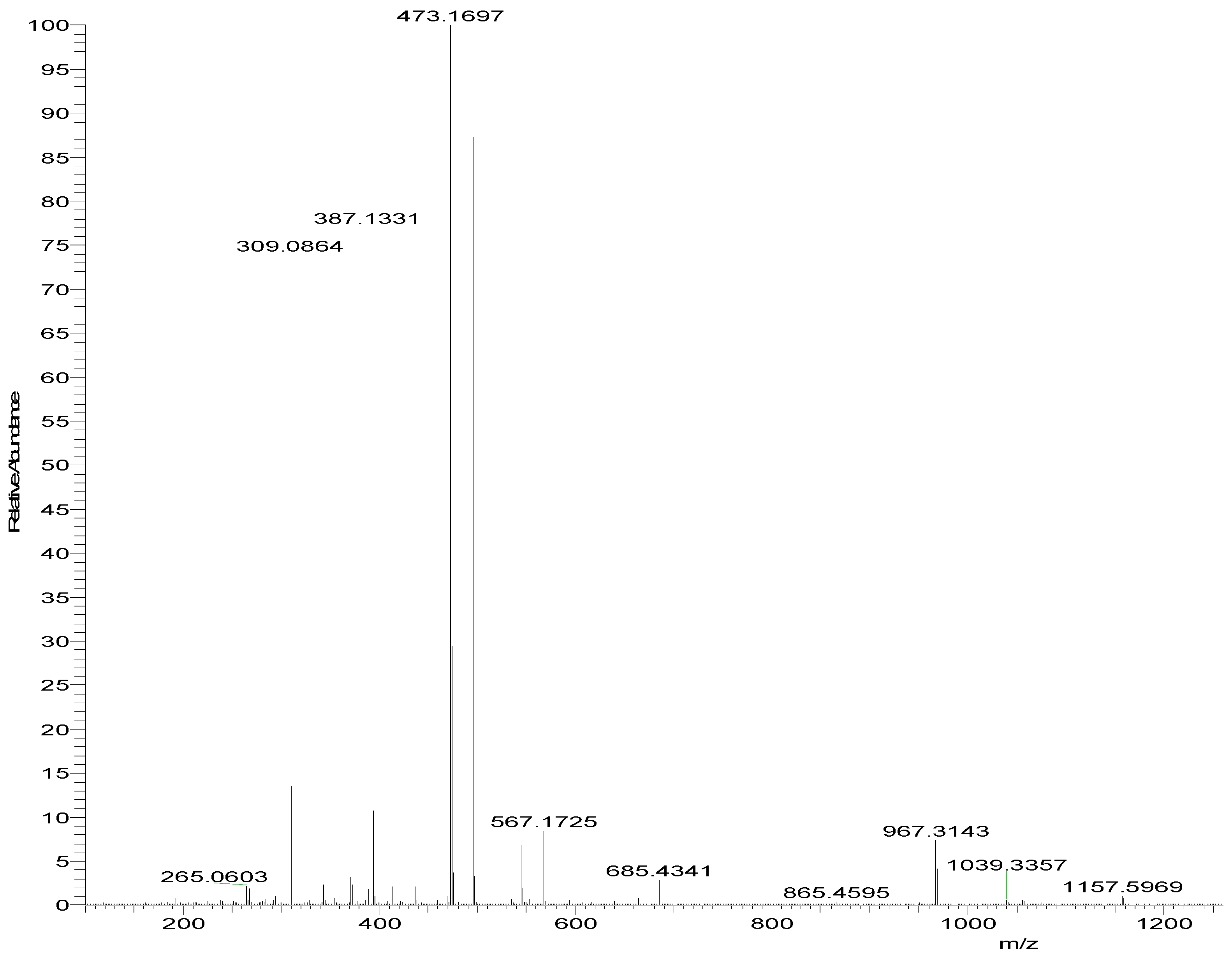
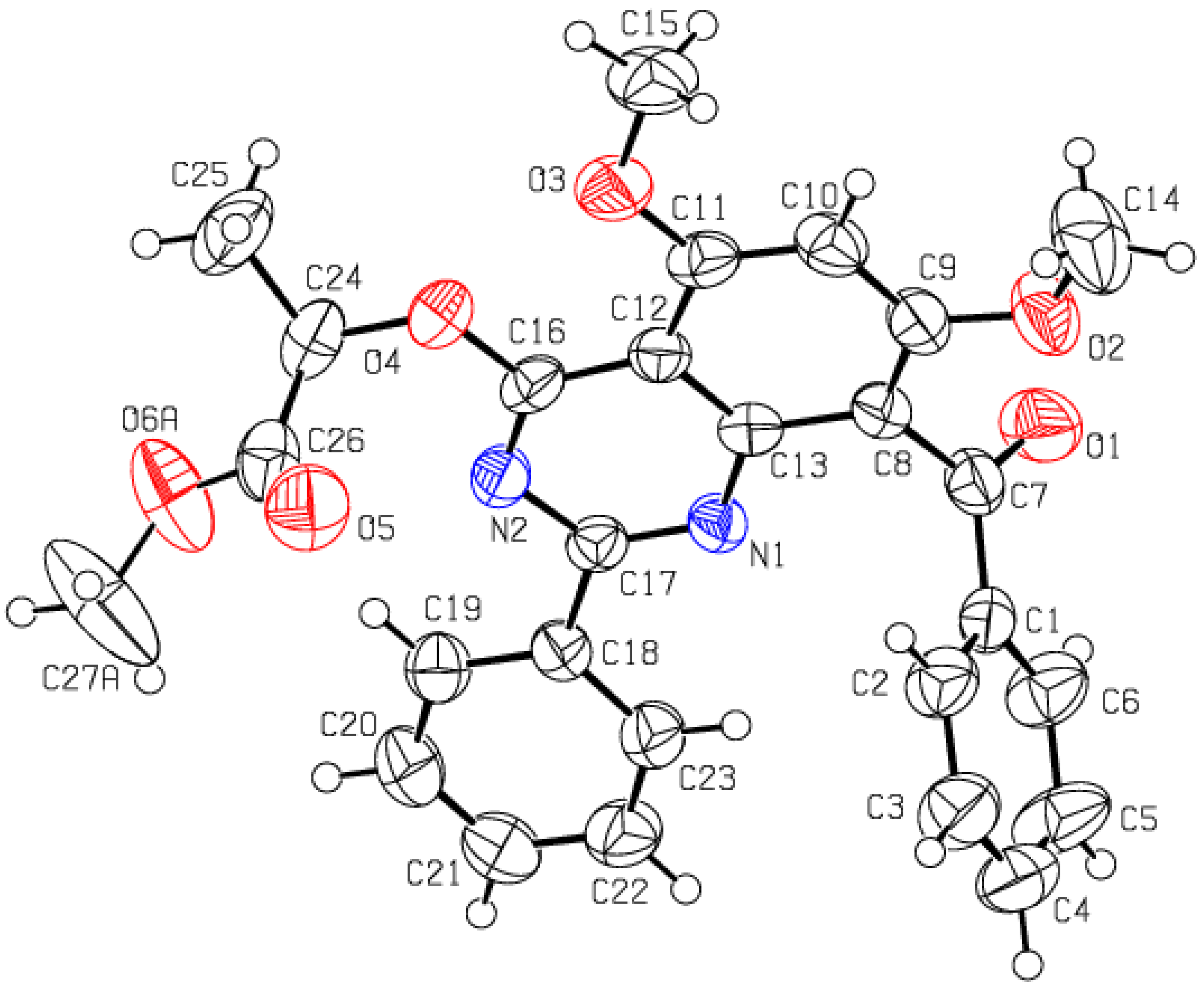
2.3.1. Single Crystal XRD Depiction of 8 and 12
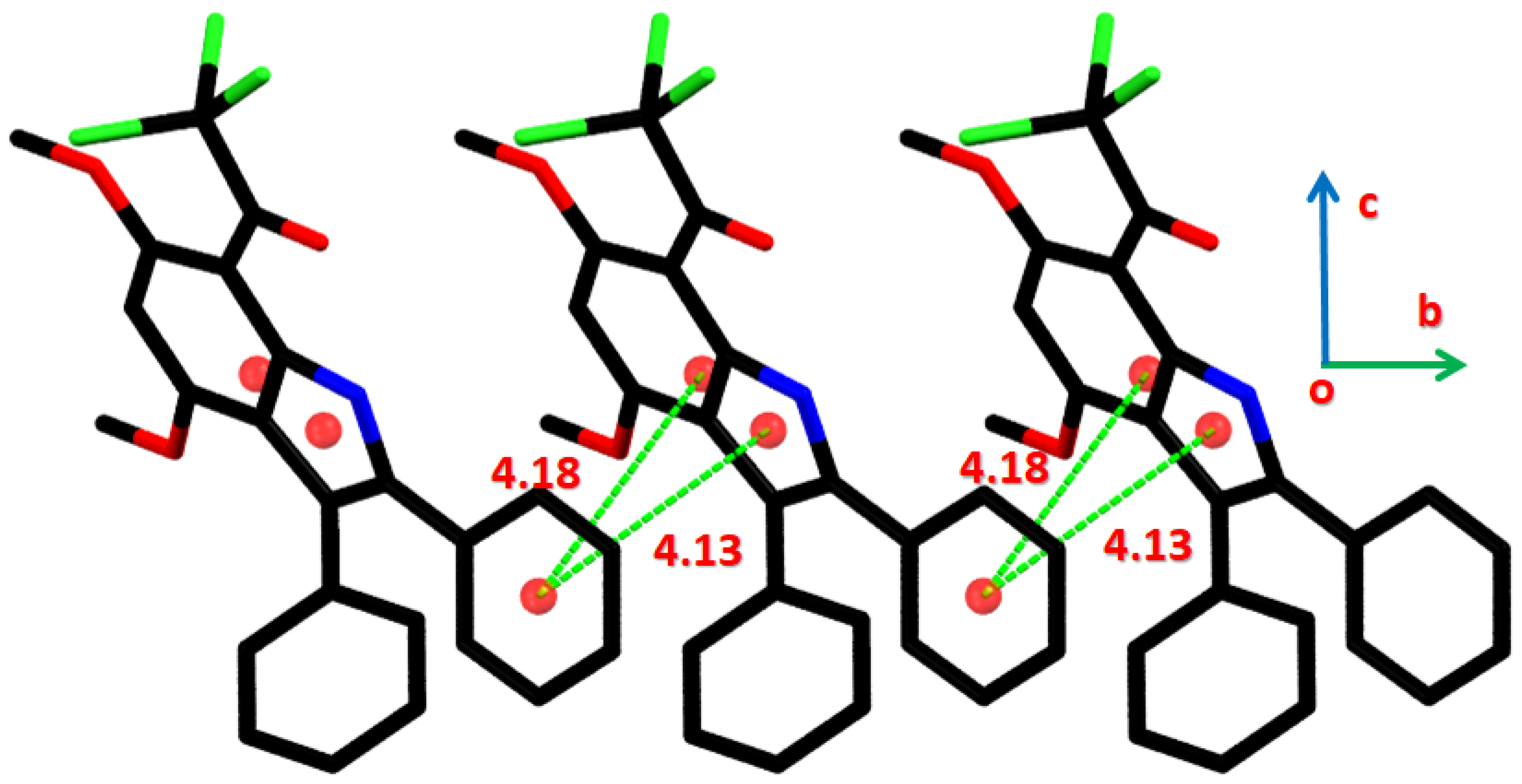
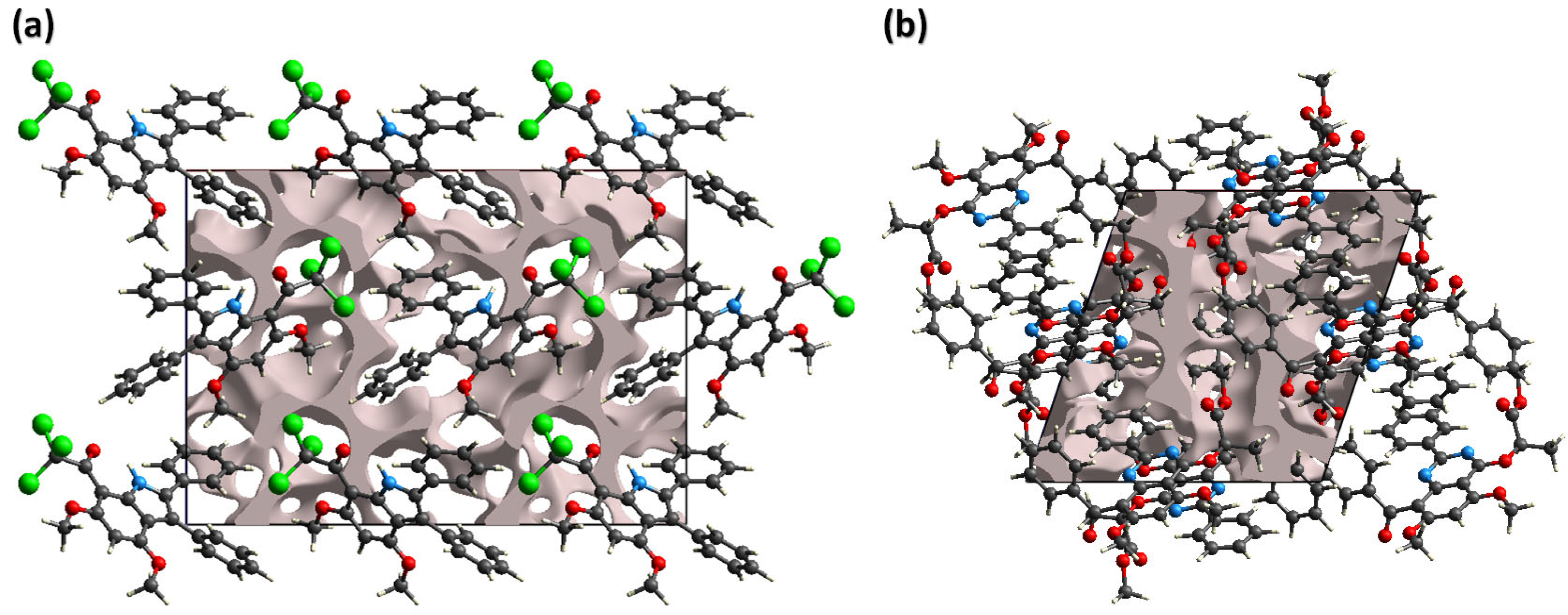
2.3.2. Hirshfeld Surface Analysis
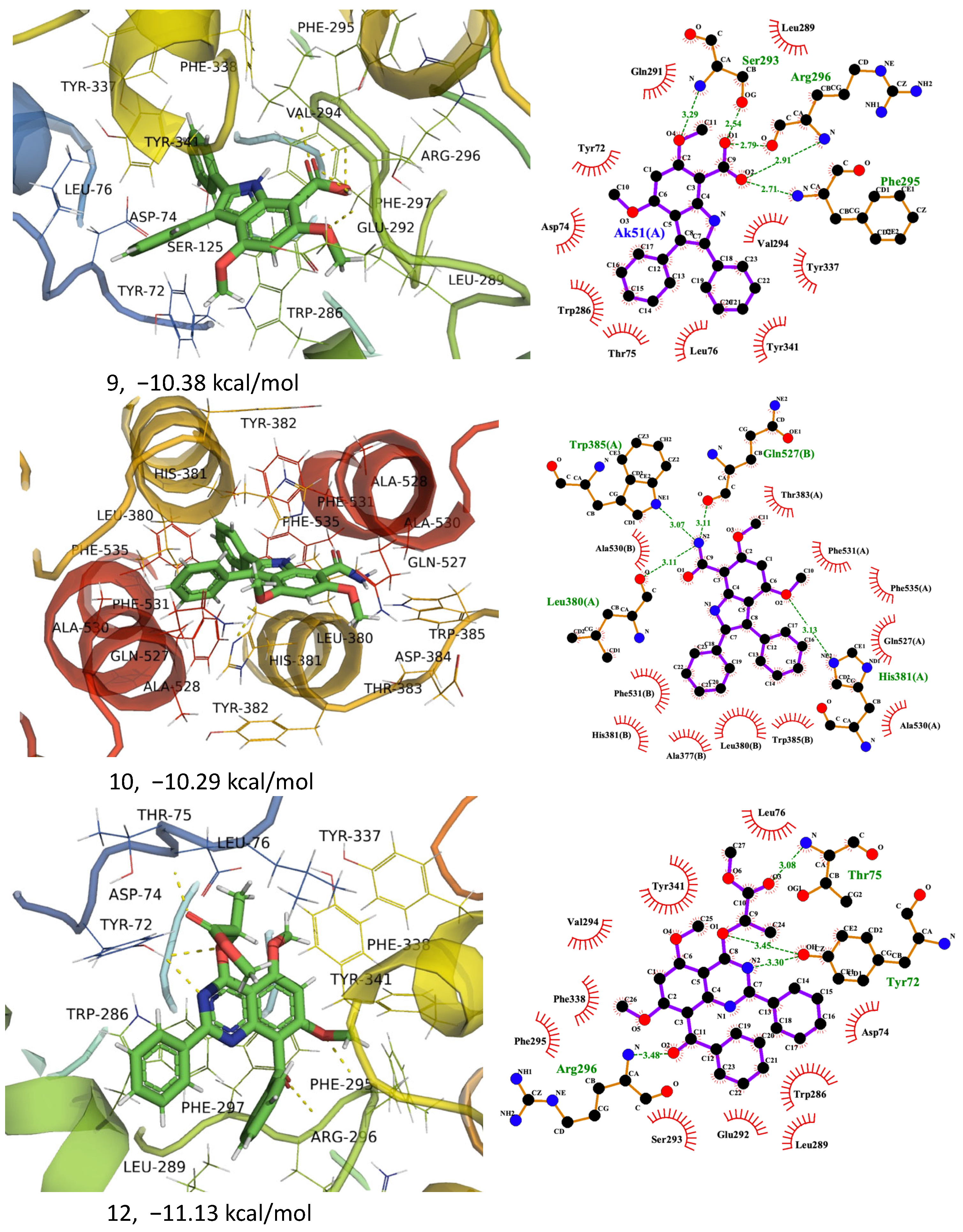
2.4. In Silico Anticholinesterase Activity
2.5. Anticholinesterase and Antibacterial Activities
3. Materials and Methods
3.1. Synthesis
3.1.1. General Procedure for the Synthesis Indoles, 5–6
3.1.2. 4,6-Dimethoxy-2,3-Diphenyl-1H-Indole, 5
3.1.3. 4,5,6-Trimethoxy-2,3-Diphenyl-1H-Indole, 6
3.1.4. 4,6-Dimethoxy-2,3-Dimethyl -1H-Indole, 7
3.1.5. 7-Trichloro-acetyl-4,6-Dimethoxy-2,3-Diphenyl-1H-Indole, 8
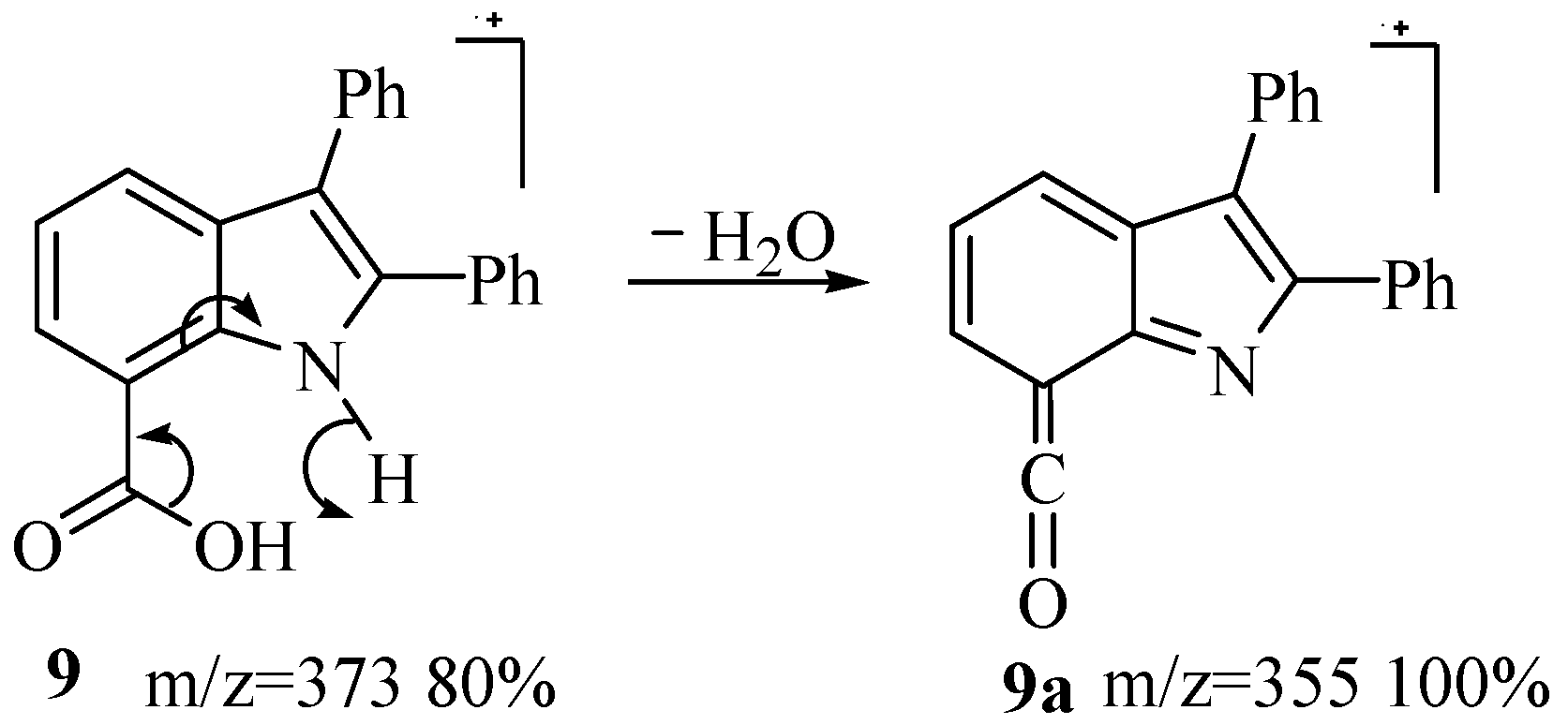
3.1.6. 4,6-Dimethoxy-2,3-Diphenylindole-7-Carboxylic Acid, 9
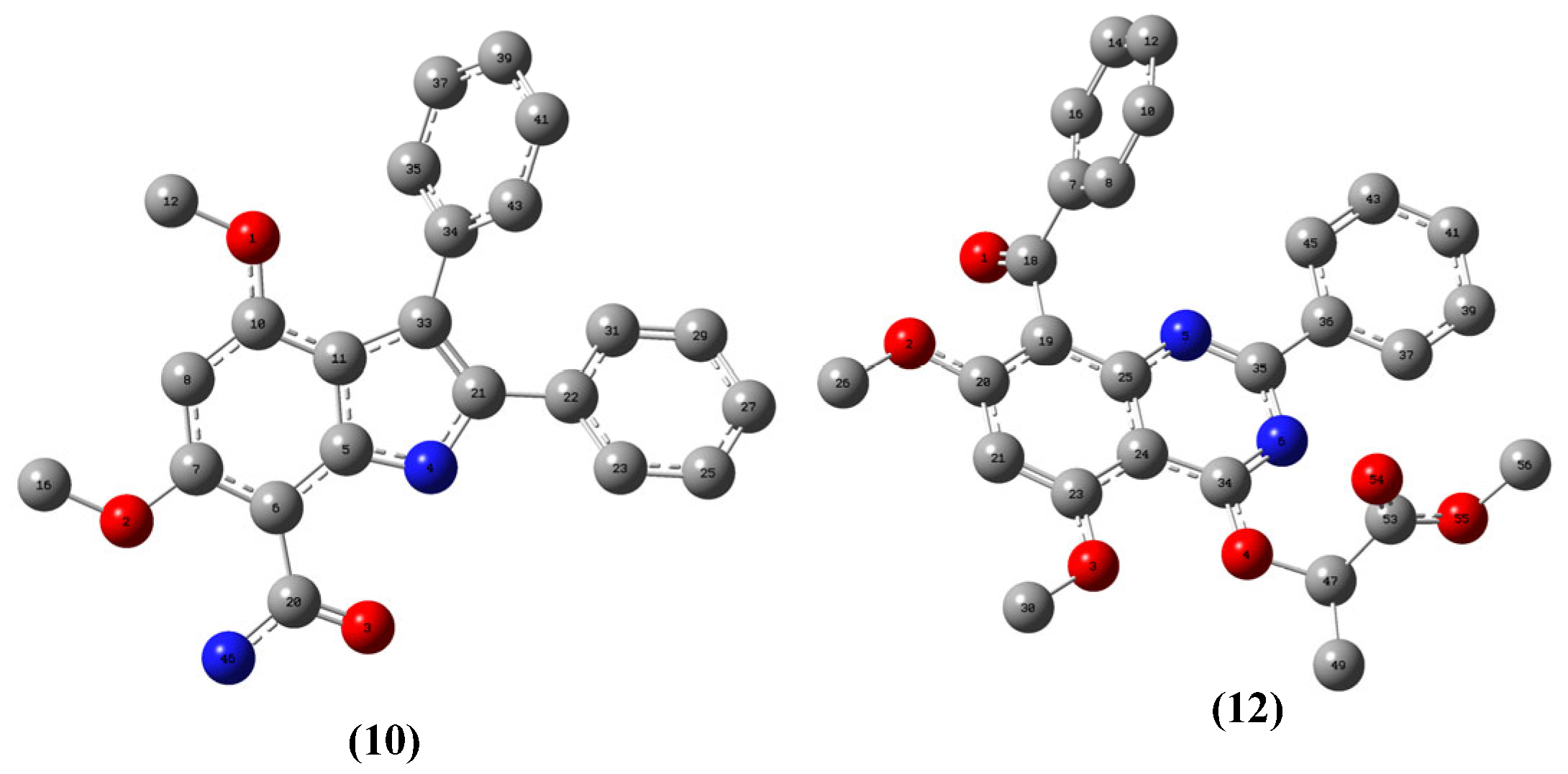
3.1.7. 4,6-Dimethoxy-2,3-Diphenyl-1H-Indole-7-Carboxamide, 10
3.1.8. 8-Benzoyl-5,7-Dimethoxy-4-(1‴-Methyoxycarbonylethoxy)-2-Phenyl-1,3-Benzodiazine, 12
3.2. Computational Methods
3.3. Antibacterial and Anticholinesterase Activities
3.4. Single Crystal XRD Details of 10 and 12
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Chen, Q.; Wu, C.; Zhu, J.; Li, E.; Xu, Z. Therapeutic potential of indole derivatives as anti-HIV agents: A mini-review. Curr. Top. Med. Chem. 2022, 22, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, H.; Mathur, J.; Guleria, A. Indole derivatives as potential anticancer agents: A review. J. Chil. Chem. Soc. 2020, 65, 4900–4907. [Google Scholar] [CrossRef]
- Devi, N.; Kaur, K.; Biharee, A.; Jaitak, V. Recent development in indole derivatives as anticancer agent: A mechanistic approach. Anti-Cancer Agents Med. Chem. 2021, 21, 1802–1824. [Google Scholar] [CrossRef]
- Sayed, M.; Kamal El-Dean, A.M.; Ahmed, M.; Hassanien, R. Design, synthesis, and characterization of novel pyrimidines bearing indole as antimicrobial agents. J. Chin. Chem. Soc. 2019, 66, 218–225. [Google Scholar] [CrossRef]
- Kherkhache, H.; Benabdelaziz, I.; Silva, A.M.; Lahrech, M.B.; Benalia, M.; Haba, H. A new indole alkaloid, antioxidant and antibacterial activities of crude extracts from Saccocalyx satureioides. Nat. Prod. Res. 2020, 34, 1528–1534. [Google Scholar] [CrossRef] [PubMed]
- Rubab, S.L.; Nisar, B.; Raza, A.R.; Saadia, M.; Tahir, M.N.; Sajjad, N.; Shajahan, S.; Sharmila, V.; Acevedo, R. Synthesis and antioxidant screening of Novel indole amines. J. Iran. Chem. Soc. 2022, 19, 2693–2704. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, N.; Chandra, R. The indole nucleus as a selective COX-2 inhibitor and anti-inflammatory agent (2011–2022). Org. Chem. Front. 2022, 9, 3624–3639. [Google Scholar]
- Gondal, H.Y.; Tariq, S.; Akhter, S.; Raza, A.R.; Ur Rehman, M.F.; Rubab, S.L.J.R.A. Synthesis, characterization, and in vitro anti-cholinesterase screening of novel indole amines. RSC Adv. 2023, 13, 1203–1215. [Google Scholar] [CrossRef]
- Kumar, R.R.; Kumar, V.; Kaur, D.; Nandi, N.K.; Dwivedi, A.R.; Kumar, V.; Kumar, B.J.C. Investigation of indole-3-piperazinyl derivatives as potential antidepressants: Design, synthesis, in-vitro, in-vivo and in-silico analysis. ChemistrySelect 2021, 6, 11276–11284. [Google Scholar] [CrossRef]
- Sravanthi, T.; Manju, S. Indoles—A promising scaffold for drug development. Eur. J. Pharm. Sci. 2016, 91, 1–10. [Google Scholar] [CrossRef]
- Borg, H.M.; Kabel, A.; Abdel-Kareem, M. Effect of metformin and indole-3-carbinol on a rat model of Parkinson’s disease induced by 6-hydroxydopamine. Bull. Egypt. Soc. Physiol. Sci. 2020, 40, 1–14. [Google Scholar] [CrossRef]
- Han, Y.; Dong, W.; Guo, Q.; Li, X.; Huang, L. The importance of indole and azaindole scaffold in the development of antitumor agents. Eur. J. Med. Chem. 2020, 203, 112506. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.-Y.; Li, X.; Zhu, H.-P.; Huang, W. Regulation of the ras-related signaling pathway by small molecules containing an indole core scaffold: A potential antitumor therapy. Front. Pharmacol. 2020, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Zhou, Z.; Jiang, Z.; Zhu, W.; Qiao, D.J.M. Indole-Based Tubulin Inhibitors: Binding Modes and SARs Investigations. Molecules 2022, 27, 1587. [Google Scholar] [CrossRef]
- Mohamed, F.A.M.; Alakilli, S.Y.M.; El Azab, E.F.; Baawad, F.A.M.; Shaaban, E.I.A.; Alrub, H.A.; Hendawy, O.; Gomaa, H.A.M.; Bakr, A.G.; Abdelrahman, M.H.; et al. Discovery of new 5-substituted-indole-2-carboxamides as dual epidermal growth factor receptor (EGFR)/cyclin dependent kinase-2 (CDK2) inhibitors with potent antiproliferative action. RSC Med. Chem. 2023, 14, 734–744. [Google Scholar] [CrossRef]
- Bhathiwal, A.S.; Bendi, A.; Tiwari, A. A study on synthesis of benzodiazepine scaffolds using biologically active chalcones as precursors. J. Mol. Struct. 2022, 1258, 132649. [Google Scholar] [CrossRef]
- Ahwazi, H.H.; Abdijadid, S. Chlordiazepoxide. 2019. Available online: https://europepmc.org/article/NBK/nbk547659 (accessed on 12 January 2023).
- Singh, R.; Abdijadid, S. Oxazepam. 2019. Available online: https://europepmc.org/article/NBK/nbk544349 (accessed on 12 January 2023).
- Boddu, S.H.; Kumari, S.J.P. A short review on the intranasal delivery of diazepam for treating acute repetitive seizures. Pharmaceutics 2020, 12, 1167. [Google Scholar] [CrossRef]
- Mathew, T.; Papp, A.Á.; Paknia, F.; Fustero, S.; Prakash, G.S. Benzodiazines: Recent synthetic advances. Chem. Soc. Rev. 2017, 46, 3060–3094. [Google Scholar] [CrossRef]
- Liu, F.; Huang, Y. Physiology. Antifungal bioactivity of 6-bromo-4-ethoxyethylthio quinazoline. Pestic. Biochem. Physiol. 2011, 101, 248–255. [Google Scholar] [CrossRef]
- Kuneš, J.; Bažant, J.; Pour, M.; Waisser, K.; Šlosárek, M.; Janota, J. Quinazoline derivatives with antitubercular activity. Il Farm. 2000, 55, 725–729. [Google Scholar] [CrossRef]
- Marzaro, G.; Guiotto, A.; Chilin, A. Quinazoline derivatives as potential anticancer agents: A patent review (2007–2010). Expert Opin. Ther. Patents 2012, 22, 223–252. [Google Scholar] [CrossRef] [PubMed]
- Alafeefy, A.M.; Kadi, A.A.; Al-Deeb, O.A.; El-Tahir, K.E.; Al-Jaber, N.A. Synthesis, analgesic and anti-inflammatory evaluation of some novel quinazoline derivatives. Eur. J. Med. Chem. 2010, 45, 4947–4952. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Li, X.; Zhang, C.; Lv, L.; Gao, B.; Li, M. Research progress on antibacterial activities and mechanisms of natural alkaloids: A review. Antibiotics 2021, 10, 318. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.-L.; Liu, J.; Fang, W.-Y.; Ravindar, L.; Rakesh, K. Indole-based derivatives as potential antibacterial activity against methicillin-resistance Staphylococcus aureus (MRSA). Eur. J. Med. Chem. 2020, 194, 112245. [Google Scholar] [CrossRef]
- Song, F.; Li, Z.; Bian, Y.; Huo, X.; Fang, J.; Shao, L.; Zhou, M. Indole/isatin-containing hybrids as potential antibacterial agents. Arch. Pharm. 2020, 353, 2000143. [Google Scholar] [CrossRef]
- Ciulla, M.G.; Kumar, K. The natural and synthetic indole weaponry against bacteria. Tetrahedron Lett. 2018, 59, 3223–3233. [Google Scholar] [CrossRef]
- Phuntsho, N. The Electrochemistry and Fluorescence Studies of 2, 3-Diphenyl Indole Derivatives. Ph.D. Thesis, University of New South Wales, Kensington, Australia, 2014. [Google Scholar]
- Wood, K.; Black, D.S.; Kumar, N.J.T. Ring closing metathesis strategies towards functionalised 1, 7-annulated 4, 6-dimethoxyindoles. Tetrahedron 2011, 67, 4093–4102. [Google Scholar] [CrossRef]
- Arshad, M.N.; Bibi, A.; Mahmood, T.; Asiri, A.M.; Ayub, K. Synthesis, crystal structures and spectroscopic properties of triazine-based hydrazone derivatives; a comparative experimental-theoretical study. Molecules 2015, 20, 5851–5874. [Google Scholar] [CrossRef]
- Beyer, R.L.; Kandemir, H.; Bhadbhade, M.; Sengul, I.F.; Leu, C.-w.; Wenholz, D.; Kumar, N.; Black, D.S. Synthesis of dipyrrolo [2, 3-a: 1′, 2′, 3′-fg] acridin-12 (1H)-ones. Tetrahedron Lett. 2018, 59, 4483–4486. [Google Scholar] [CrossRef]
- Corey, E.; Link, J.O.; Sarshar, S.; Shao, Y. X-ray diffraction studies of crystalline trihalomethyl ketones (RCOCX3) reveal an unusual structural deformation about the carbonyl group. Tetrahedron Lett. 1992, 33, 7103–7106. [Google Scholar] [CrossRef]
- Wang, X.; Lerchen, A.; Glorius, F. A comparative investigation: Group 9 Cp* M (III)-catalyzed formal [4 + 2] cycloaddition as an atom-economic approach to quinazolines. Org. Lett. 2016, 18, 2090–2093. [Google Scholar] [CrossRef]
- Wang, J.; Zha, S.; Chen, K.; Zhang, F.; Song, C.; Zhu, J. Quinazoline synthesis via Rh (III)-catalyzed intermolecular C–H functionalization of benzimidates with dioxazolones. Org. Lett. 2016, 18, 2062–2065. [Google Scholar] [CrossRef]
- Lingayya, R.; Vellakkaran, M.; Nagaiah, K.; Tadikamalla, P.R.; Nanubolu, J.B. Palladium (II)-catalyzed direct O-alkenylation of 2-arylquinazolinones with N-tosylhydrazones: An efficient route to O-alkenylquinazolines. Chem. Commun. 2017, 53, 1672–1675. [Google Scholar] [CrossRef] [PubMed]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M. A32. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Spackman, M.A.; Jayatilaka, D.J.C. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Visualisation and characterisation of voids in crystalline materials. CrystEngComm 2011, 13, 1804–1813. [Google Scholar] [CrossRef]
- Bingul, M.; Ercan, S.; Boga, M.; Bingul, A.A. Antioxidant and Anticholinesterase Potentials of Novel 4,6-Dimethoxyindole based Unsymmetrical Azines: Synthesis, Molecular Modeling, In Silico ADME Prediction and Biological Evaluations. Polycycl. Aromat. Compd. 2023, 1–22. [Google Scholar] [CrossRef]
- Botros, S.; Naguib, B.; Osman, A. Synthesis and Tranquillizing Effect of New 1, 4-Benzoxazepines. Egypt. J. Chem. 2011, 54, 565–578. [Google Scholar]
- Odame, F.; Neglo, D.; Sedohia, D.; Arthur, R. Antifungal synergistic effects and anti-biofilm formation activities of some bioactive 2,3-dihydro-1,5-benzoxazepine derivatives. Arch. Microbiol. 2022, 205, 39. [Google Scholar] [CrossRef]
- Stefaniak, M.; Olszewska, B. 1,5-Benzoxazepines as a unique and potent scaffold for activity drugs: A review. Archiv. Der Pharm. 2021, 354, 2100224. [Google Scholar] [CrossRef]
- Chouiter, A.D.; Mousser, M.O.; Mousser, H.B.; Krid, A.; Belkhiri, L.; Fleutot, S.; François, M. Synthesis, spectra, crystal, DFT, molecular docking and in vitro cholinesterase inhibition evaluation on two novel symmetrical Azine Schiff bases. J. Mol. Struct. 2023, 1281, 135171. [Google Scholar] [CrossRef]
- Ayyash, A.N. Design, Synthesis, and Antimicrobial Studies of Novel Derivatives: Benzoxazepine-4, 7-dione and Benzodiazepine-4, 7-dione. Indian J. Heterocycl. Chem. 2019, 29, 255–259. [Google Scholar]
- Ponduri, R.; Kumar, P.; Vadali, L.R. Synthesis and Antimicrobial Activity of New Isoxazolyl Benzo[1, 4] diazepine-5-one Derivatives. Chem. Sel. 2018, 3, 10108–10112. [Google Scholar] [CrossRef]
- Kendre, B.V.; Landge, M.G.; Bhusare, S.R. Synthesis and biological evaluation of some novel pyrazole, isoxazole, benzoxazepine, benzothiazepine and benzodiazepine derivatives bearing an aryl sulfonate moiety as antimicrobial and anti-inflammatory agents. Arab. J. Chem. 2019, 12, 2091–2097. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D. 01; Gaussian Inc.: Wallingford, UK, 2010.
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef]
- Perdew, J.P. Generalized gradient approximations for exchange and correlation: A look backward and forward. Phys. B Condens. Matter 1991, 172, 1–6. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104–154122. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.F.U.; Akhter, S.; Batool, A.I.; Selamoglu, Z.; Sevindik, M.; Eman, R.; Mustaqeem, M.; Akram, M.S.; Kanwal, F.; Lu, C. Effectiveness of natural antioxidants against SARS-CoV-2? Insights from the In-Silico World. Antibiotics 2021, 10, 1011. [Google Scholar] [CrossRef] [PubMed]
- Bilal, S.; Sami, A.J.; Hayat, A.; ur Rehman, M.F. Assessment of pesticide induced inhibition of Apis mellifera (honeybee) acetylcholinesterase by means of N-doped carbon dots/BSA nanocomposite modified electrochemical biosensor. Bioelectrochemistry 2022, 144, 107999. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr 2002, 40, 82–92. [Google Scholar]
- Naveed, M.; Ain, N.u.; Aziz, T.; Javed, K.; Shabbir, M.A.; Alharbi, M.; Alsahammari, A.; Alasmari, A.F. Artificial Intelligence Assisted Pharmacophore Design for Philadelphia Chromosome-Positive Leukemia with Gamma-Tocotrienol: A Toxicity Comparison Approach with Asciminib. Biomedicines 2023, 11, 1041. [Google Scholar] [CrossRef]
- Naveed, M.; Shabbir, M.A.; Ain, N.-u.; Javed, K.; Mahmood, S.; Aziz, T.; Khan, A.A.; Nabi, G.; Shahzad, M.; Alharbi, M.E.; et al. Chain-Engineering-Based De Novo Drug Design against MPXVgp169 Virulent Protein of Monkeypox Virus: A Molecular Modification Approach. Bioengineering 2023, 10, 11. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Aslam, M.; Shahid, M.; Ur Rehman, F.; Murtaza, M.A.; Sharif, S.; Ata, A.; Noor, S. Production optimization and characterization of a low molecular weight bacteriocin from Lactococcus lactis subsp. lactis. Afr. J. Microbiol. Res 2012, 6, 5924–5933. [Google Scholar]
- Sheldrick, G.M. SHELXT–Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.J. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed]




| CCDC | ||
|---|---|---|
| Chemical formula | C24H18Cl3NO3 | C27H24N2O6 |
| Mr | 474.74 | 472.48 |
| Crystal system, space group | Orthorhombic, Pca21 | Monoclinic, P21/n |
| a, b, c (Å) | 22.879 (5), 6.0315 (14), 16.084 (4) | 14.3363 (16), 12.0884 (13), 14.8394 (17) |
| Temperature (K) | 296 | 296 |
| V (Å3) | 2219.5 (9) | 2430.8 (5) |
| α, β, γ (°) | 90, 90, 90 | 90, 109.053 (6), 90 |
| Z | 4 | 4 |
| µ (mm−1) | 0.44 | 0.09 |
| Radiation type | Mo Kα | Mo Kα |
| Crystal size (mm) | 0.36 × 0.18 × 0.15 | 0.35 × 0.30 × 0.24 |
| Data Collection | ||
| Absorption correction | Multi-scan (SADABS; Bruker, 2007) | Multi-scan (SADABS; Bruker, 2007) |
| No. of observed, independent and measured [I > 2σ (I)] reflections | 1588, 3218, 9325 | 1588, 3218, 9325 |
| (sin θ/λ)max (Å−1) | 0.606 | 0.617 |
| Diffractometer | Bruker Kappa APEXII CCD | Bruker Kappa APEXII CCD |
| Rint | 0.095 | 0.083 |
| Refinement | ||
| No. of reflections | 3218 | 4764 |
| No. of restraints | 1 | 26 |
| No. of parameters | 270 | 341 |
| R [F2 > 2σ(F2)], wR(F2), S | 0.045, 0.091, 0.97 | 0.058, 0.144, 0.99 |
| Δρmax, Δρmin (e Å−3) | 0.20, −0.24 | 0.16, −0.16 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Compounds | Binding Energy (kcal/mol) | Dissociation Constants (nM) | Binding Sites |
|---|---|---|---|
| 5 | −10.48 | 20.74 | TYR 72, THR 75, ASP 74, LEU 76, LEU 289, TRP 286, GLN 291, SER 293, GLU 292, VAL 294, ARG 296, PHE 295, PHE 297, PHE 338, TYR 337, TYR 341 |
| 6 | −11.25 | 5.67 | ALA 377 (Chain A), LEU 380 (Chain A), HIS 381 (Chain A), THR 383 (Chain A), ASP 384 (Chain A), TRP 385, (Chain A), GLN 527 (Chain A), ALA 530 (Chain A), PHE 531 (Chain A), PHE 535 (Chain B), ALA 377 (Chain B), VAL 378 (Chain B), LEU 380 (Chain B), HIS 381 (Chain B), THR 383 (Chain B), TRP 385 (Chain B), GLN 527 (Chain B), ALA 530 (Chain B). PHE 531 (Chain B), PHE 535 |
| −9.79 | 66.97 | TYR 72, THR 75, ASP 74, LEU 76, GLY 121, SER 125, GLY 122, TRP 286, PHE 295, ARG 296, VAL 294, TYR 337, PHE 297, TYR 341, PHE 338, HIS 447 | |
| 7 | −10.50 | 20.13 | TYR 72, ASP 74, THR 75, LEU 76, GLY 121, GLY 122, SER 125, SER 203, TRP 286, LEU 289, SER 293, VAL 294, PHE 295, ARG 296, PHE 297, TYR 337, PHE 338, TYR 341, HIS 447 |
| 8 | −10.46 | 21.69 | TYR 72, THR 75, ASP 74, LEU 76, LEU 289, TRP 286, GLN 291, SER 293, GLU 292, VAL 294, ARG 296, PHE 295, PHE 297, PHE 338, TYR 337, TYR 341 |
| 9 | −10.38 | 24.58 | TYR 72, TYR 341, ASP 74, LEU 76, THR 75, TRP 286, GLN 291, LEU 289, SER 293, GLU 292, PHE 295, VAL 294, ARG 296, TYR 337, PHE 297, PHE 338, |
| 10 | −10.46 | 21.49 | ALA 377 (Chain A), LEU 380 (Chain A), HIS 381 (Chain A), THR 383 (Chain A), ASP 384 (Chain A), TRP 385, (Chain A), GLN 527 (Chain A), ALA 530 (Chain A), PHE 531 (Chain A), PHE 535 (Chain B), ALA 377 (Chain B), VAL 378 (Chain B), LEU 380 (Chain B), HIS 381 (Chain B), THR 383 (Chain B), TRP 385 (Chain B), GLN 527 (Chain B), ALA 530 (Chain B). PHE 531 (Chain B), PHE 535 |
| −10.29 | 28.76 | TYR 341, PHE 338, TYR 337, PHE 297, ARG 296, PHE 295, VAL 294, SER 293, GLU 292, GLN 291, LEU 289, LEU 76, ASP 74, THR 75, TYR 72 | |
| 12 | −11.13 | 6.97 | TYR 72, ASP 74, THR 75, TRP 286, THR 83, LEU 76, HIS 287, TRP 286, LEU 289, GLU 292, GLN 291, VAL 294, PHE 295, SER 293, ARG 296, TYR 337, PHE 297, PHE 338, GLY 342, TYR 341 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raza, A.R.; Rubab, S.L.; Ashfaq, M.; Altaf, Y.; Tahir, M.N.; Rehman, M.F.u.; Aziz, T.; Alharbi, M.; Alasmari, A.F. Evaluation of Antimicrobial, Anticholinesterase Potential of Indole Derivatives and Unexpectedly Synthesized Novel Benzodiazine: Characterization, DFT and Hirshfeld Charge Analysis. Molecules 2023, 28, 5024. https://doi.org/10.3390/molecules28135024
Raza AR, Rubab SL, Ashfaq M, Altaf Y, Tahir MN, Rehman MFu, Aziz T, Alharbi M, Alasmari AF. Evaluation of Antimicrobial, Anticholinesterase Potential of Indole Derivatives and Unexpectedly Synthesized Novel Benzodiazine: Characterization, DFT and Hirshfeld Charge Analysis. Molecules. 2023; 28(13):5024. https://doi.org/10.3390/molecules28135024
Chicago/Turabian StyleRaza, Abdul Rauf, Syeda Laila Rubab, Muhammad Ashfaq, Yasir Altaf, Muhammad Nawaz Tahir, Muhammad Fayyaz ur Rehman, Tariq Aziz, Metab Alharbi, and Abdullah F. Alasmari. 2023. "Evaluation of Antimicrobial, Anticholinesterase Potential of Indole Derivatives and Unexpectedly Synthesized Novel Benzodiazine: Characterization, DFT and Hirshfeld Charge Analysis" Molecules 28, no. 13: 5024. https://doi.org/10.3390/molecules28135024
APA StyleRaza, A. R., Rubab, S. L., Ashfaq, M., Altaf, Y., Tahir, M. N., Rehman, M. F. u., Aziz, T., Alharbi, M., & Alasmari, A. F. (2023). Evaluation of Antimicrobial, Anticholinesterase Potential of Indole Derivatives and Unexpectedly Synthesized Novel Benzodiazine: Characterization, DFT and Hirshfeld Charge Analysis. Molecules, 28(13), 5024. https://doi.org/10.3390/molecules28135024









