Synthesis, Crystal Structure, DFT Calculations, Hirshfeld Surface Analysis and In Silico Drug-Target Profiling of (R)-2-(2-(1,3-Dioxoisoindolin-2-yl)propanamido)benzoic Acid Methyl Ester
Abstract
1. Introduction
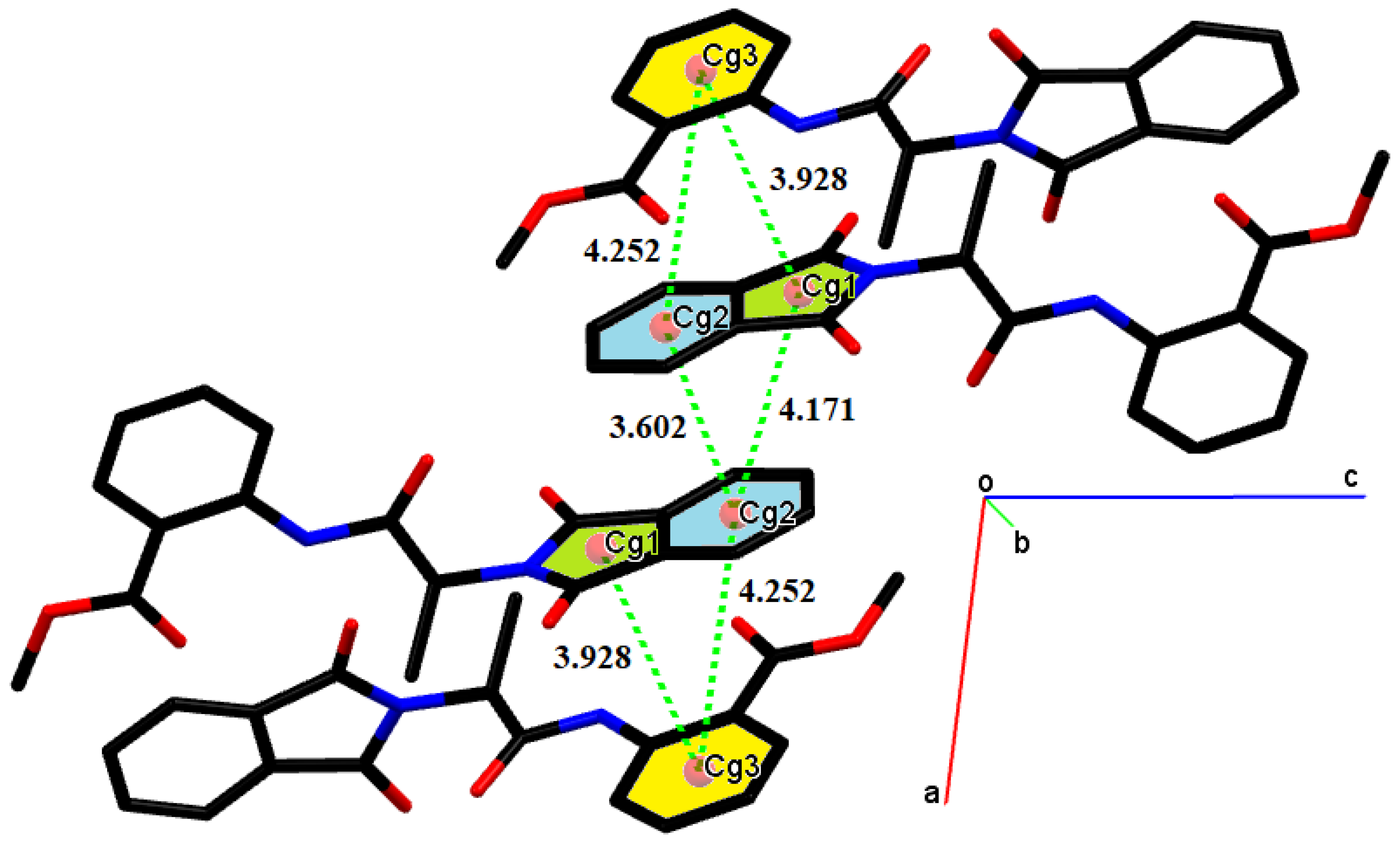
2. Results and Discussions
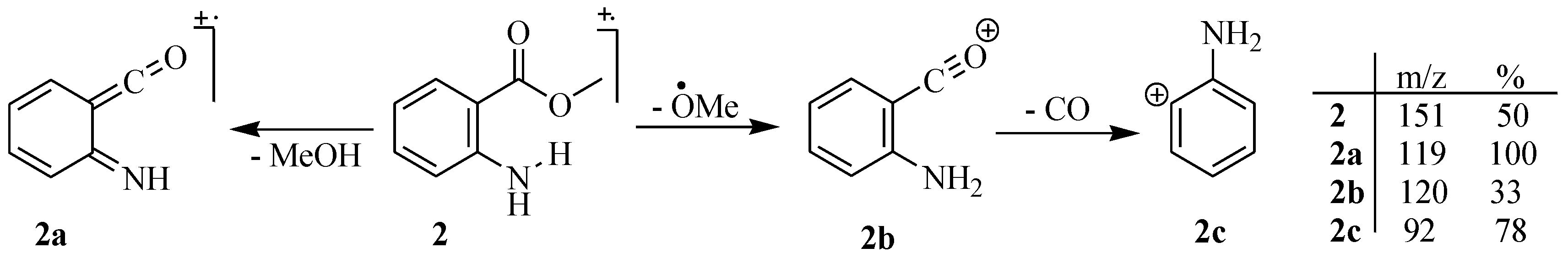
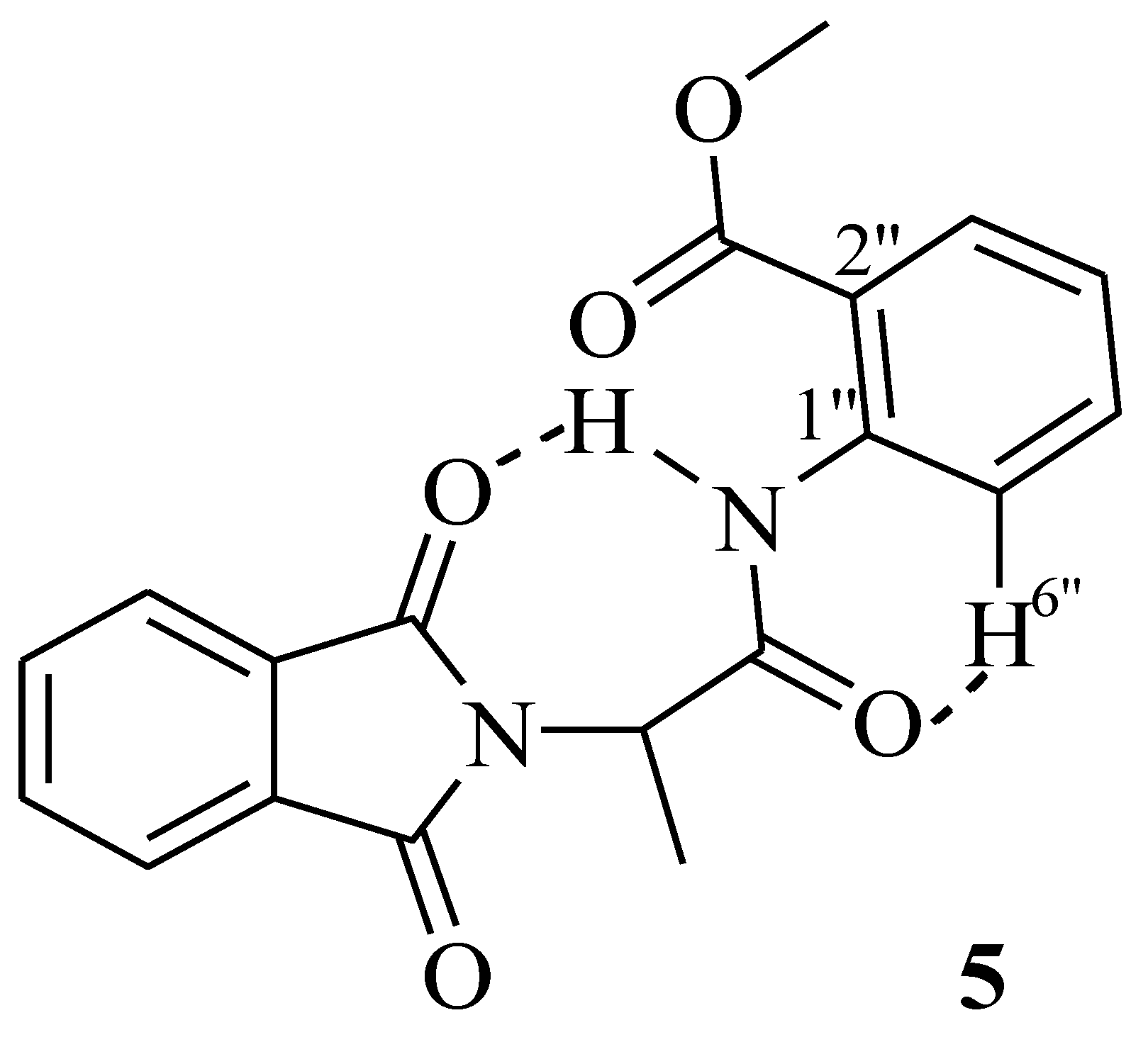
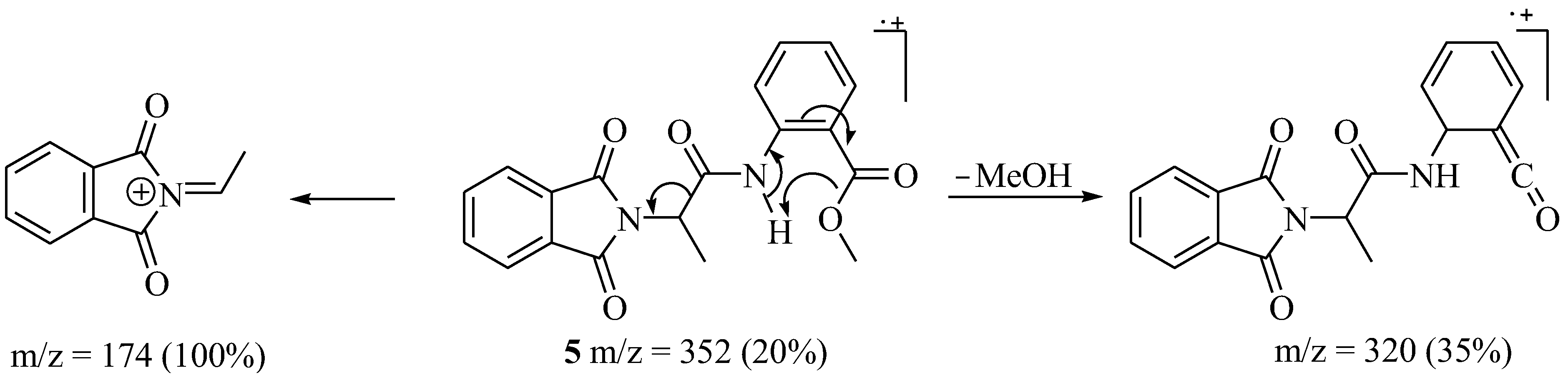
2.1. Characterization
2.2. DFT Exploration
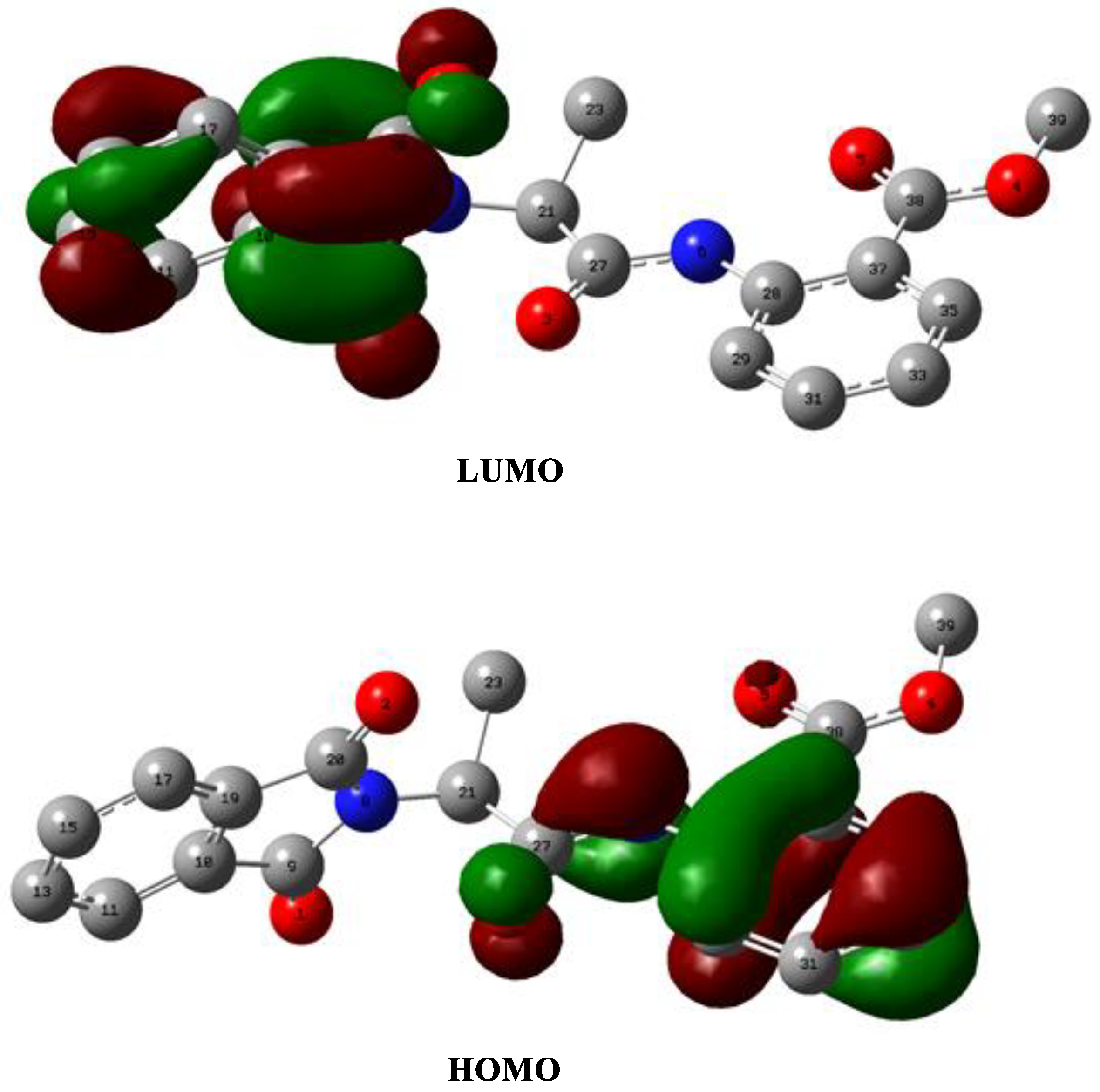
2.2.1. Frontier Molecular Orbital Analysis (FMO) and Polarizability
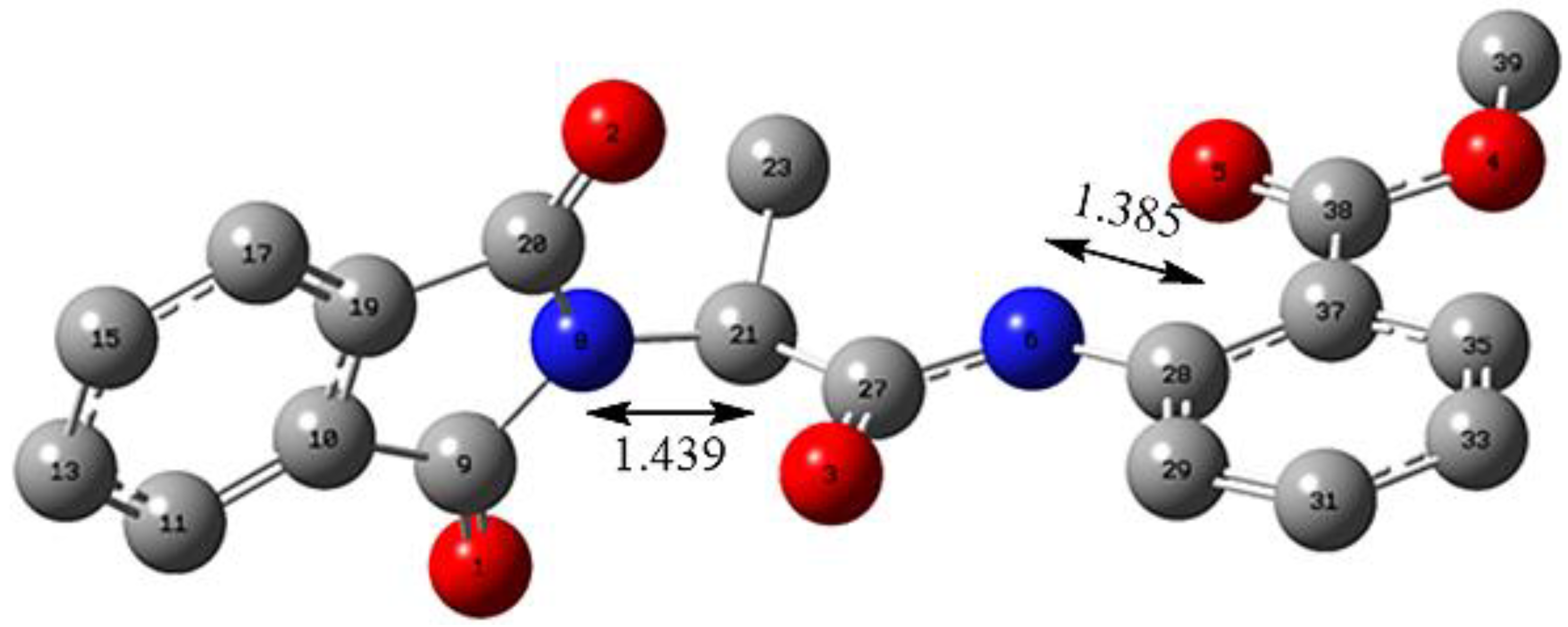
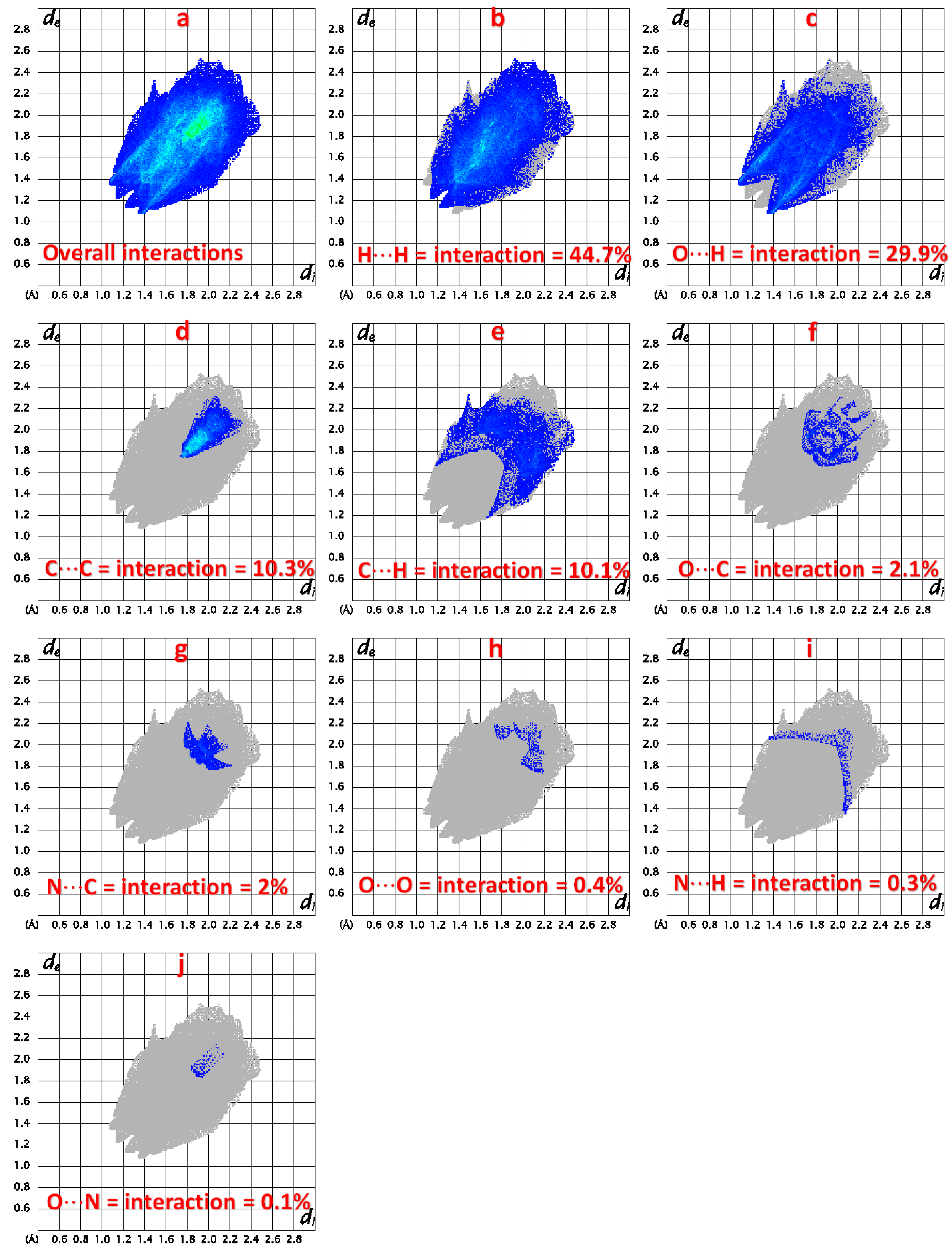
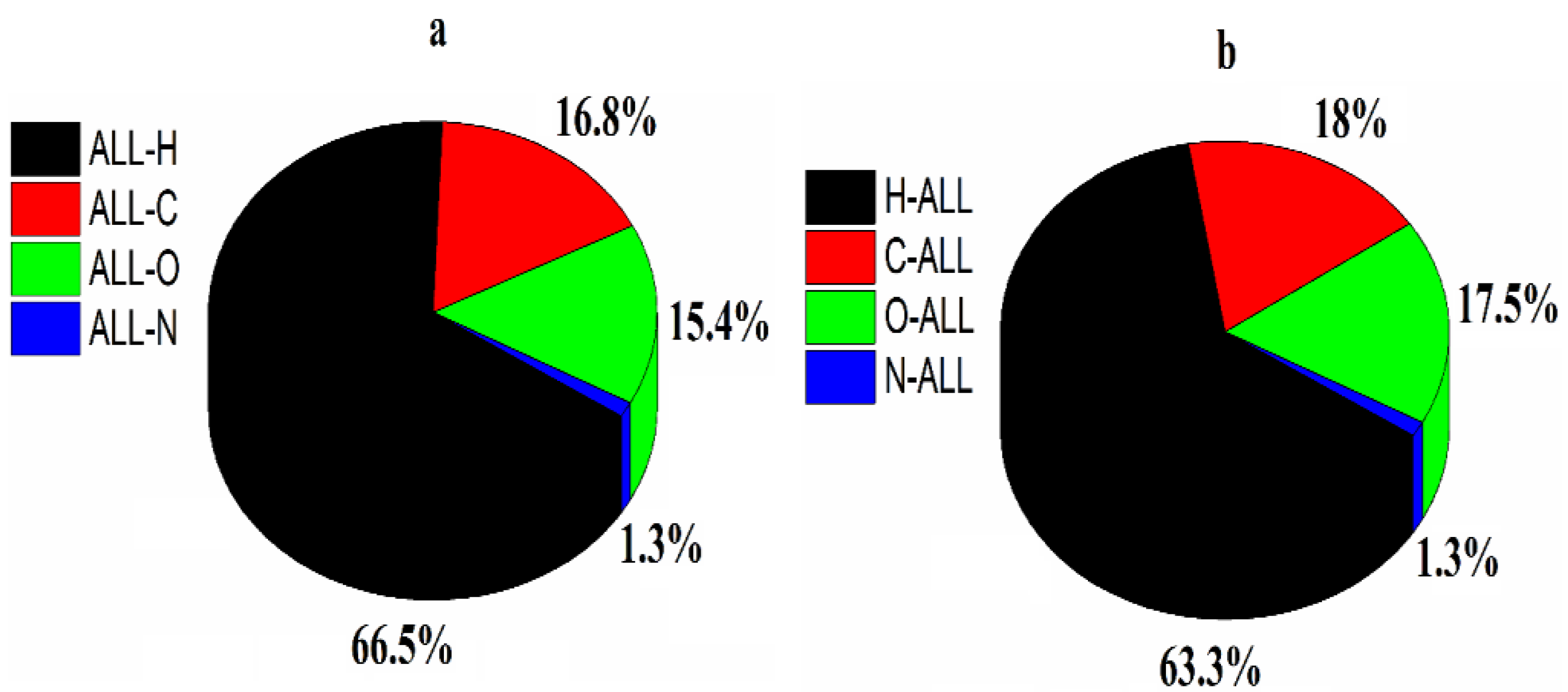
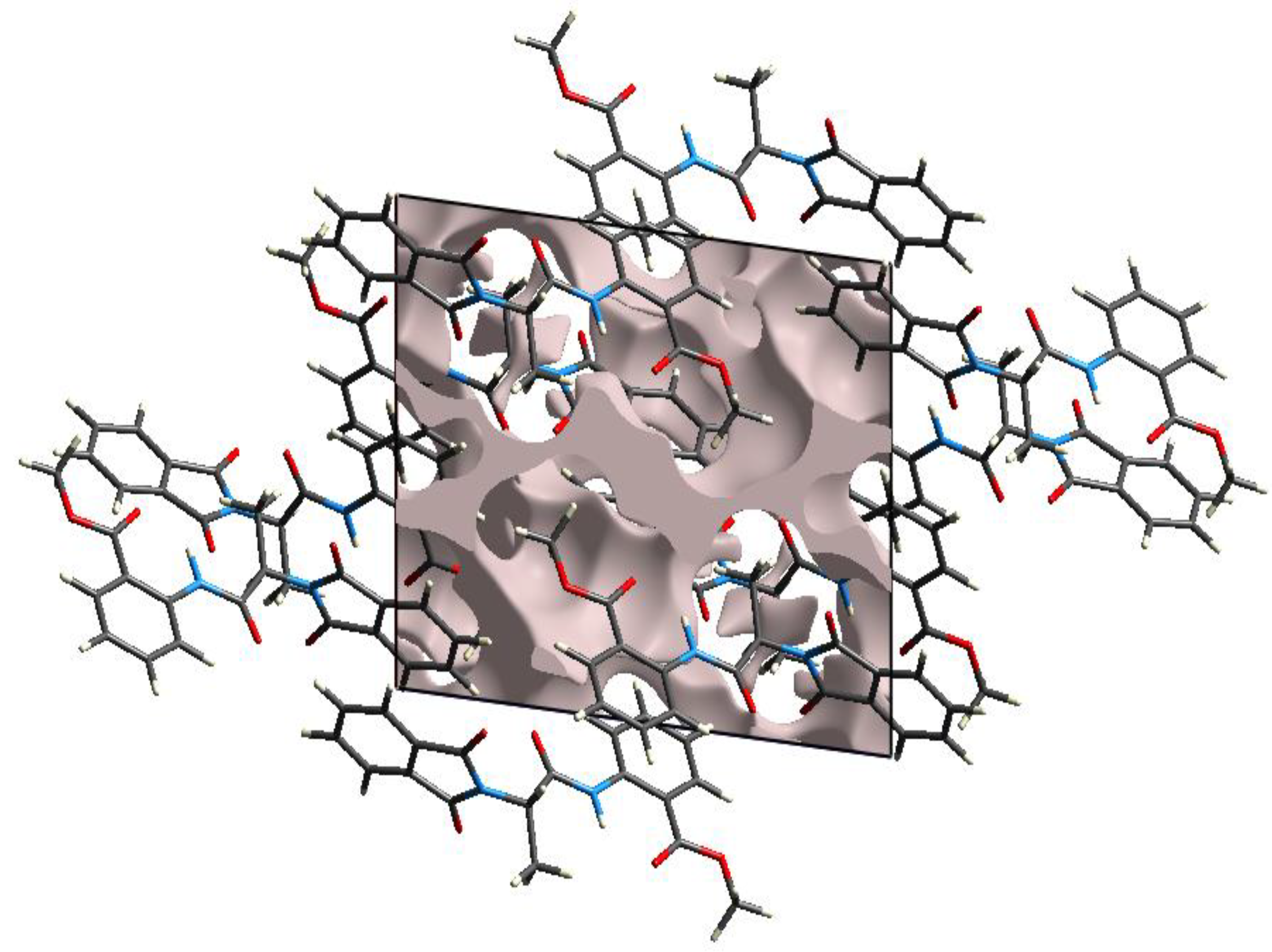
2.2.2. Hirshfeld Charge Analysis
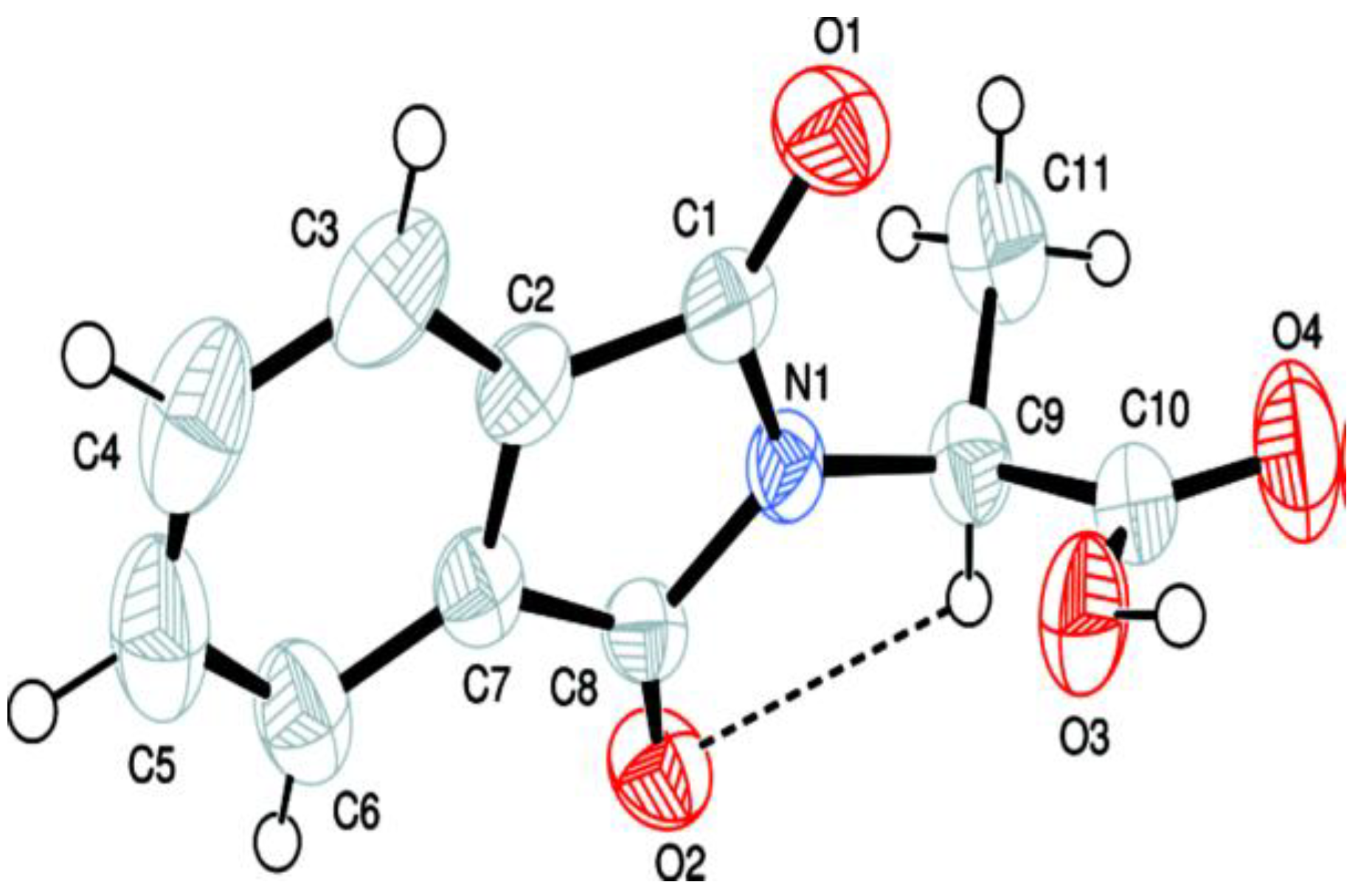
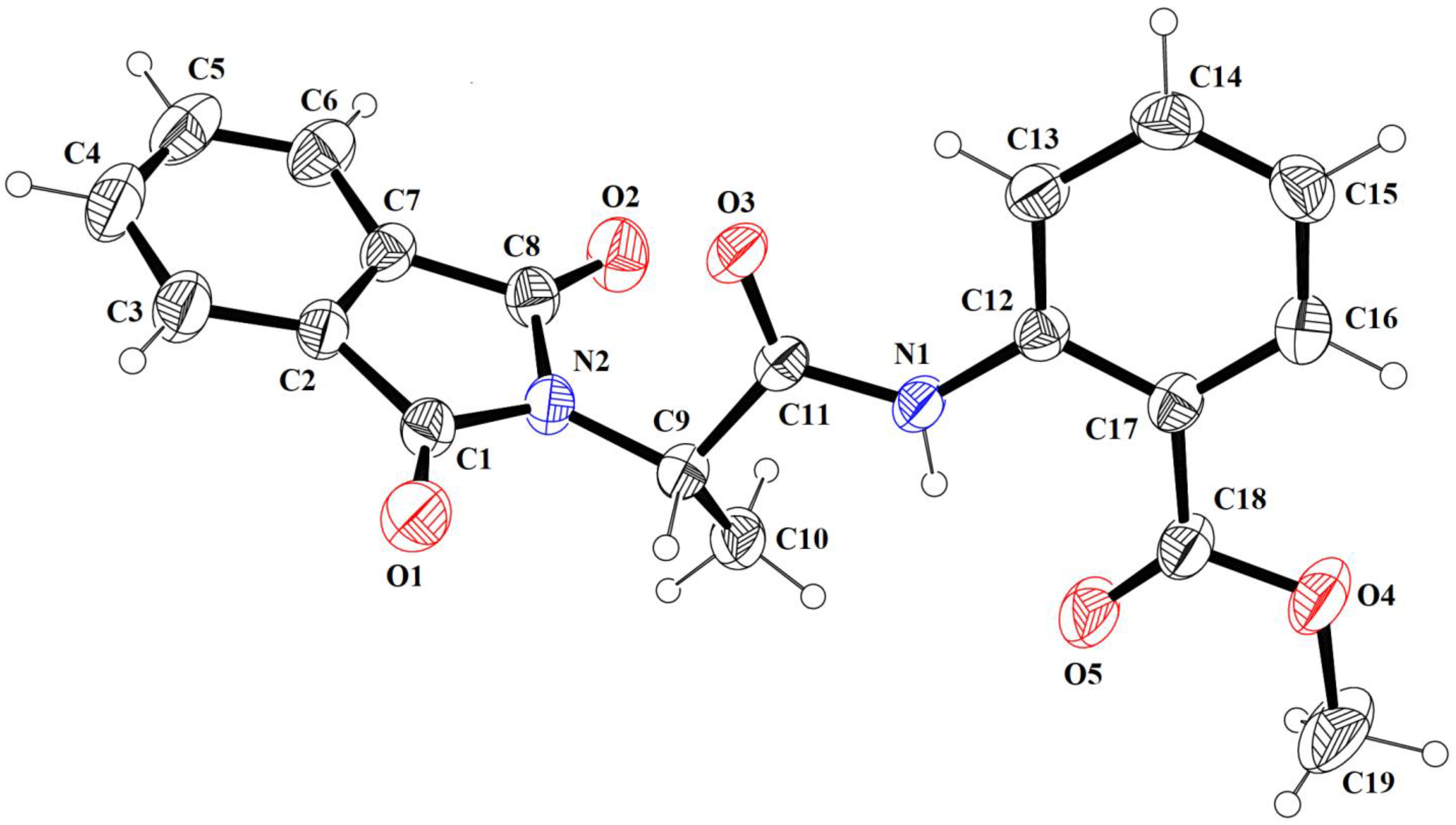
2.3. Crystallographic Data
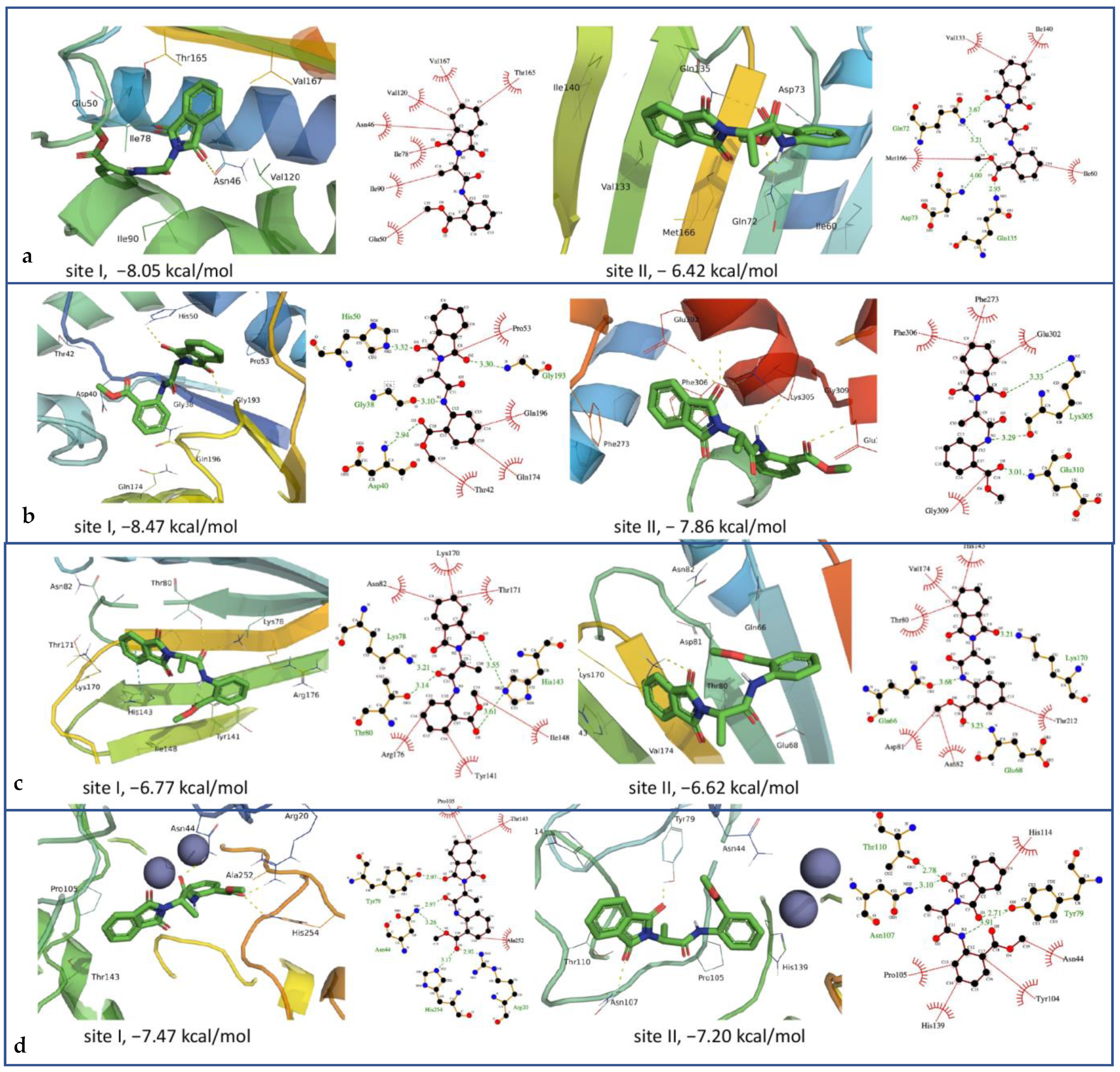
2.4. In Vitro and In Silico Antimicrobial Activities
3. Experimental Section
3.1. General Experimental Procedure
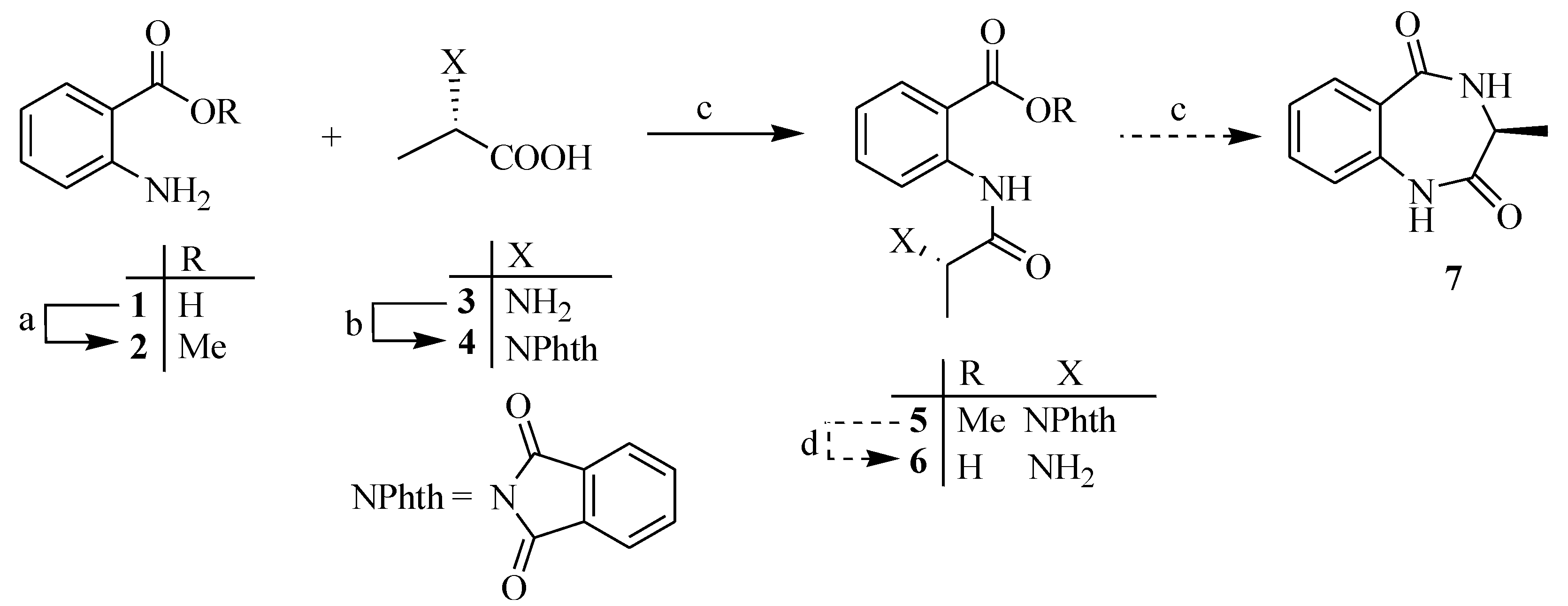
3.2. Synthesis
3.2.1. Methyl 2-Aminobenzoate (2)
3.2.2. (2S)-2-(1′,3′-Dioxoisoindolin-2-yl)propanoic Acid (4)
3.2.3. (R)-2-(2-(1,3-Dioxoisoindolin-2-yl)propanamido)benzoic Acid Methyl Ester (5)
3.3. In Vitro and In Silico Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Csende, F.; Porkoláb, A. Antiviral activity of isoindole derivatives. J. Med. Chem. Sci 2020, 3, 254–285. [Google Scholar]
- Kanamitsu, N.; Osaki, T.; Itsuji, Y.; Yoshimura, M.; Tsujimoto, H.; Soga, M. Novel water-soluble sedative-hypnotic agents: Isoindolin-1-one derivatives. Chem. Pharm. Bull. 2007, 55, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Kaur Bhatia, R. Isoindole derivatives: Propitious anticancer structural motifs. Curr. Top. Med. Chem. 2017, 17, 189–207. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.; Kizilkaya, S.; Noma, S.A.; Ates, B.; Kara, Y. Novel hybrid isoindole-1, 3 (2H)-dione compounds containing a 1H-tetrazole moiety: Synthesis, biological evaluation, and molecular docking studies. J. Biochem. Mol. Toxicol. 2022, 36, e23015. [Google Scholar] [CrossRef] [PubMed]
- Hirose, A.; Kato, T.; Ohno, Y.; Shimizu, H.; Tanaka, H.; Nakamura, M.; Katsube, J. Pharmacological actions of SM-9018, a new neuroleptic drug with both potent 5-hydroxytryptamine2 and dopamine2 antagonistic actions. Jpn. J. Pharmacol. 1990, 53, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, N.; Roy, P.; Sondhi, S.M. Synthesis, anti-inflammatory, and antiproliferative activity evaluation of isoindole, pyrrolopyrazine, benzimidazoisoindole, and benzimidazopyrrolopyrazine derivatives. Mol. Divers. 2013, 17, 753–766. [Google Scholar] [CrossRef]
- Csende, F.; Porkoláb, A. A review on antibacterial activity of some isoindole derivatives. Der Pharma Chem. 2018, 10, 43–50. [Google Scholar]
- El-Gohary, N.; Shaaban, M. Synthesis and biological evaluation of a new series of benzimidazole derivatives as antimicrobial, antiquorum-sensing and antitumor agents. Eur. J. Med. Chem. 2017, 131, 255–262. [Google Scholar] [CrossRef]
- May, H.; Kretzschmar, R.; Neumann, W.; Gries, J. Chemistry and biological properties of substituted 3-amino-1H-isoindoles (author’s transl). Arzneim.-Forsch. 1980, 30, 1487–1493. [Google Scholar]
- Kamiński, K.; Obniska, J. Design, synthesis, and anticonvulsant activity of N-phenylamino derivatives of 3, 3-dialkyl-pyrrolidine-2, 5-diones and hexahydro-isoindole-1, 3-diones. Bioorg. Med. Chem. 2008, 16, 4921–4931. [Google Scholar] [CrossRef]
- Rubab, S.L.; Nisar, B.; Raza, A.R.; Ullah, N.; Tahir, M.N. Asymmetric Synthesis of 4,1-Benzoxazepine-2,5-Diones—Effect of the Halogen of (2S)-α-Haloacids. Molecules 2014, 19, 139–148. [Google Scholar] [CrossRef]
- Yadav, G.; Krishnan, M. An ecofriendly catalytic route for the preparation of perfumery grade methyl anthranilate from anthranilic acid and methanol. Org. Process Res. Dev. 1998, 2, 86–95. [Google Scholar] [CrossRef]
- Zeng, Q.; Wang, H.; Liu, Z.; Li, B.; Zhao, Y. Facile synthesis of optically pure (S)-3-p-hydroxyphenyllactic acid derivatives. Amino Acids 2007, 33, 537–541. [Google Scholar] [CrossRef]
- Taylor, R. Which intermolecular interactions have a significant influence on crystal packing? CrystEngComm 2014, 16, 6852–6865. [Google Scholar] [CrossRef]
- Israr, H.; Rasool, N.; Rizwan, K.; Hashmi, M.A.; Mahmood, T.; Rashid, U.; Hussein, M.Z.; Akhtar, M.N. Synthesis and reactivities of triphenyl acetamide analogs for potential nonlinear optical material uses. Symmetry 2019, 11, 622. [Google Scholar] [CrossRef]
- Wolff, S.; Grimwood, D.; McKinnon, J.; Turner, M.; Jayatilaka, D.; Spackman, M. Crystal Explorer; University of Western Australia: Perth, WA, USA, 2012. [Google Scholar]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. Gaussian 09; Revision D. 01; Gaussian, Inc.: Wallingford, CT, USA, 2009; Available online: http://www.gaussian.com (accessed on 11 October 2020).
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Aslam, M.; Shahid, M.; ur Rehman, F.; Murtaza, M.A.; Sharif, S.; Ata, A.; Noor, S. Production optimization and characterization of a low molecular weight bacteriocin from Lactococcus lactis subsp. lactis. Afr. J. Microbiol. Res. 2012, 6, 5924–5933. [Google Scholar]
- Rehman, M.F.u.; Akhter, S.; Batool, A.I.; Selamoglu, Z.; Sevindik, M.; Eman, R.; Mustaqeem, M.; Akram, M.S.; Kanwal, F.; Lu, C. Effectiveness of natural antioxidants against SARS-CoV-2? Insights from the In-Silico World. Antibiotics 2021, 10, 1011. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L.; Bromberg, S. PyMOL User’s Guide; DeLano Scientific LLC: San Carlos, CA, USA, 2004; Volume 629. [Google Scholar]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
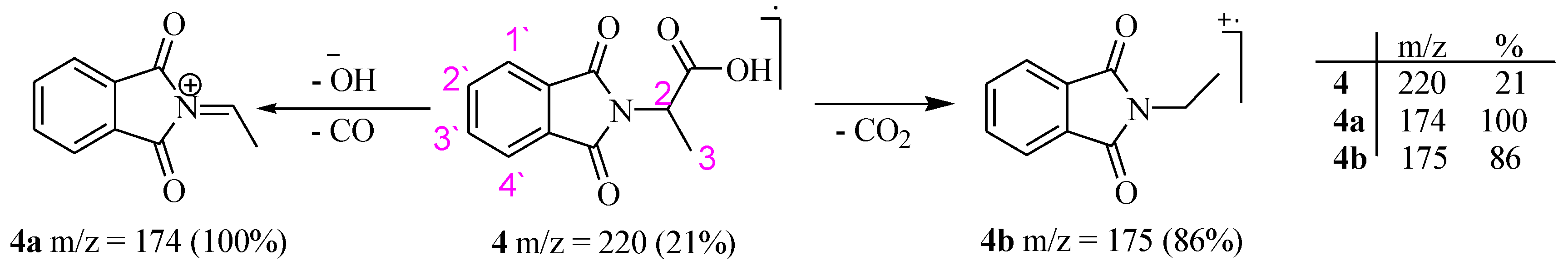

| ύ (cm−1) | λmax (nm) | log ε (L.cm−1.M−1) | |
|---|---|---|---|
| O–H | C=O | ||
| 3480 | 1720 | 295.5 | 3.2003 |
| δ ppm (J = Hz) | ||
|---|---|---|
| H2 | H3 | H1′–4′ |
| 5.00 (q, J = 7.2 Hz) | 1.65 (d, J = 7.2 Hz) | 7.85 (m) |
| Atom ^ | * H.C. | Atom | * H.C. | Atom | * H.C. |
|---|---|---|---|---|---|
| O1 | −0.27 | O5 | −0.26 | C20 | 0.19 |
| O2 | −0.27 | N6 | 0.00 | C28 | 0.07 |
| O3 | −0.28 | N8 | −0.06 | C38 | 0.22 |
| O4 | −0.11 | C9 | 0.19 | C39 | 0.15 |
| Crystallographic Data | |
|---|---|
| Chemical Formula | C19H16N2O5 |
| Mr Temperature (K) | 352.34 296 (2) |
| Crystal system, space group | Monoclinic, P21/n |
| a, b, c (Å) | 12.0628 (10), 9.7196 (8), 14.8898 (10) |
| α, β, γ° Z | 90, 96.520 (4), 90 4 |
| V (Å3) | 1734.5 (2) |
| F(000) Wavelength (λ) | 736 0.71073 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.099 |
| Crystal size (mm) Density (calculated)g/cm−3 | 0.30 × 0.25 × 0.23 1.349 |
| Data Collection | |
| Absorption correction Diffractometer | Multi-scan (SADABS; Bruker, 2007) Bruker APEXII CCD diffractometer |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 14529, 4154, 2311 |
| Rint | 0.032 |
| Theta range for data collection (°) | 2.062 to 27.944 |
| Index ranges | −15 ≤ h ≤ 15, −10 ≤ k ≤ 12, −19 ≤ l ≤ 19 |
| (sin θ/λ)max (Å−1) | 0.659 |
| Data Refinement | |
| No. of parameters No. of reflections R[F2 >2σ(F2)], wR(F2), S | 237 4154 0.051, 0.135, 1.01 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.16, −0.17 |
| Bond Lengths | Bond Angles | ||
|---|---|---|---|
| O1-C1 | 1.206 (2) | O1-C1-N2 | 124.3 (2) |
| N2-C1 | 1.387 (2) | C1-N2-C8 | 111.4 (1) |
| N2-C8 | 1.389 (2) | N2-C8-O2 | 125.1 (2) |
| O2-C8 | 1.201 (2) | C1-N2-C9 | 122.9 (2) |
| N2-C9 | 1.460 (2) | C8-N2-C9 | 124.9 (2) |
| O3-C11 | 1.209 (2) | O3-C11-N1 | 124.7 (2) |
| N1-C11 | 1.348 (2) | C11-N1-C12 | 129.7 (2) |
| N1-C12 | 1.396 (2) | O4-C18-O5 | 121.5(2) |
| O4-C18 | 1.331 (2) | C17-C18-O4 | 112.5 (2) |
| O5-C18 | 1.202 (3) | C17-C18-O5 | 125.9 (2) |
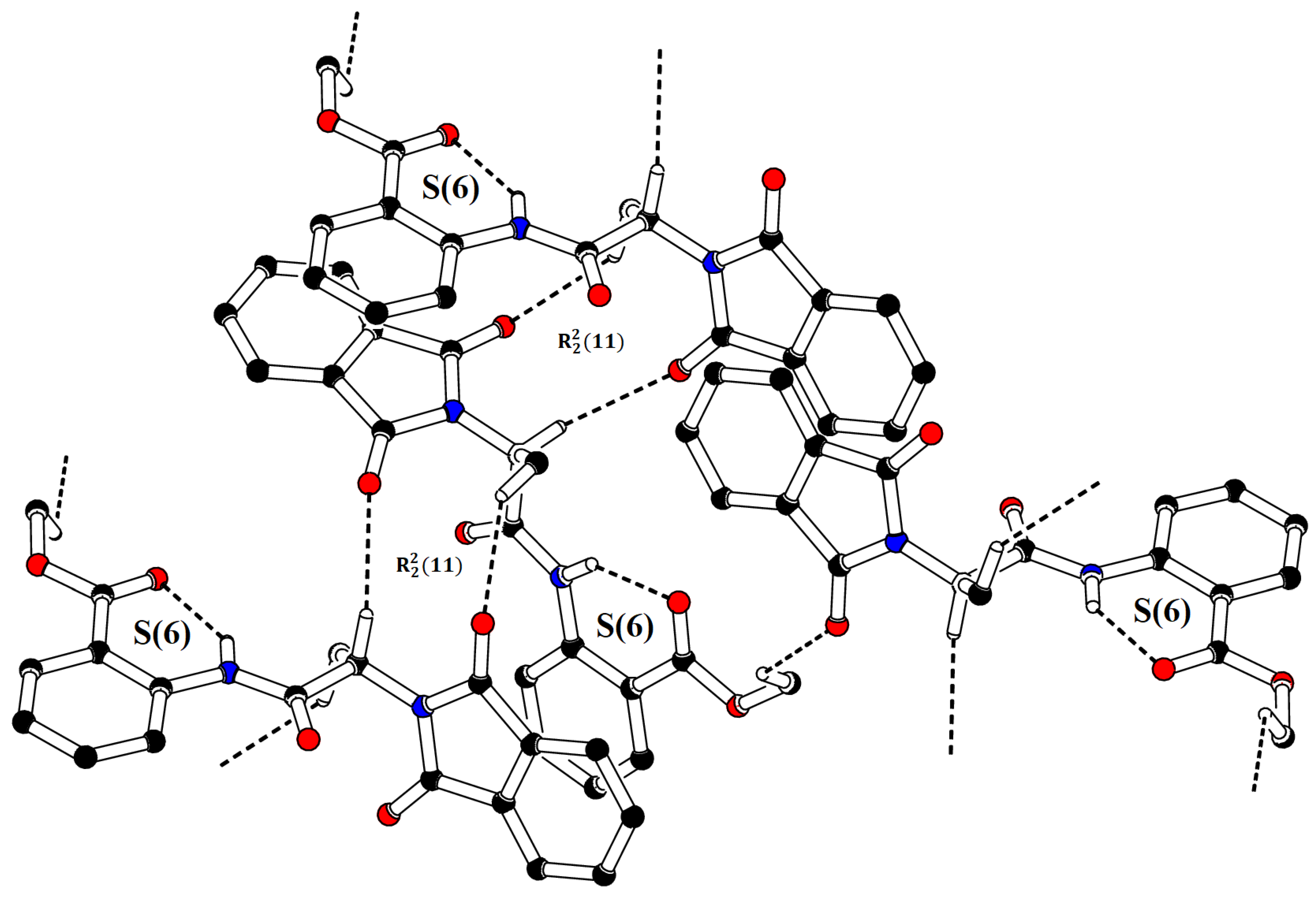
| D—H⋯A | H⋯A | D—H | D⋯A | <(D—H⋯A)° |
|---|---|---|---|---|
| C9—H9⋯O2i | 2.54 | 0.98 | 3.337 (2) | 139 |
| C10—H10A⋯O1ii | 2.57 | 0.96 | 3.402 (3) | 145 |
| C19—H19A⋯O1iii | 2.48 | 0.96 | 3.025 (3) | 116 |
| C10—H10A⋯O2 | 2.63 | 0.96 | 3.139 (2) | 113 |
| C13—H13⋯O3 | 2.24 | 0.93 | 2.857 (2) | 123 |
| N1—H1⋯O5 | 1.95 | 0.86 | 2.6562 (19) | 139 |
| Enzyme | PDB-ID | Binding Energy | Dissociation Constant (µM) | Active Site Residues |
|---|---|---|---|---|
| Dihydroorotase (E. coli) | 2KZN | −8.05 | 1.25 | Gly38, Ala39, Thr42, Asp40, Gly49, His50, Phe54, Pro53, Gly79, Asp80, Arg88, Lys84, Tyr170, Gln174, Gly193, Gly192, Ser194, Asp195, Asn199, Gln196, Pro222, Val224, Leu223 |
| −6.42 | 19.65 | Lys226, Asp228, Lys230, Lys231, Gly233, Lys234, Ser235, Val240, Thr247, Glu251, Gln254, Phe255, Asn258 | ||
| DNA gyrase (E. coli) | 2EG7 | −7.47 | 3.40 | His18, Arg20, Kcx102, Asn44, Tyr79, Tyr104, Pro105, Thr110, Asn107, His139, Thr143, His177, Leu222, Asp250, His254, Ala252, Ala266 |
| −7.20 | 6.31 | Asn44, Tyr79, Pro105, Tyr104, Asn107, Asn111, Thr110, His114, Gly115, Val116, Leu222, His139 | ||
| Tyrosyl-tRNA synthetase (S. aureus) | 1JIJ | −8.47 | 0.615 | Gly38, Ala39, Thr42, Asp40, Gly49, Pro53, His50, Phe54, Gly79, Asp80, Lys84, Gln174, Arg88, Tyr170, Gly193, Ser194, Gly192, Asp195 |
| −7.86 | 1.72 | Arg58, Asn109, Phe271, Thr272, Phe273, Glu302, Val303, Lys305, Phe306, Gly309, Glu310, Asp311 | ||
| DNA gyrase (S. aureus) | 3G7B | −6.77 | 10.79 | Lys78, Thr80, Asp81, Asn82, Tyr141, His143, Ile148, Lys170, Thr171, Gly172, Val174, Arg176 |
| −6.62 | 14.11 | Gln66, Ile67, Glu68, Thr80, Asp81, Asn82, His143, Lys170, Thr171, Gly172, Val174, Gln210, Thr212, Ser226 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubab, S.L.; Raza, A.R.; Nisar, B.; Ashfaq, M.; Altaf, Y.; Hussain, R.; Sajjad, N.; Akram, M.S.; Tahir, M.N.; Shaheen, M.A.; et al. Synthesis, Crystal Structure, DFT Calculations, Hirshfeld Surface Analysis and In Silico Drug-Target Profiling of (R)-2-(2-(1,3-Dioxoisoindolin-2-yl)propanamido)benzoic Acid Methyl Ester. Molecules 2023, 28, 4375. https://doi.org/10.3390/molecules28114375
Rubab SL, Raza AR, Nisar B, Ashfaq M, Altaf Y, Hussain R, Sajjad N, Akram MS, Tahir MN, Shaheen MA, et al. Synthesis, Crystal Structure, DFT Calculations, Hirshfeld Surface Analysis and In Silico Drug-Target Profiling of (R)-2-(2-(1,3-Dioxoisoindolin-2-yl)propanamido)benzoic Acid Methyl Ester. Molecules. 2023; 28(11):4375. https://doi.org/10.3390/molecules28114375
Chicago/Turabian StyleRubab, Syeda Laila, Abdul Rauf Raza, Bushra Nisar, Muhammad Ashfaq, Yasir Altaf, Riaz Hussain, Noreen Sajjad, Muhammad Safwan Akram, Muhammad Nawaz Tahir, Muhammad Ashraf Shaheen, and et al. 2023. "Synthesis, Crystal Structure, DFT Calculations, Hirshfeld Surface Analysis and In Silico Drug-Target Profiling of (R)-2-(2-(1,3-Dioxoisoindolin-2-yl)propanamido)benzoic Acid Methyl Ester" Molecules 28, no. 11: 4375. https://doi.org/10.3390/molecules28114375
APA StyleRubab, S. L., Raza, A. R., Nisar, B., Ashfaq, M., Altaf, Y., Hussain, R., Sajjad, N., Akram, M. S., Tahir, M. N., Shaheen, M. A., Rehman, M. F. u., & Ali, H. M. (2023). Synthesis, Crystal Structure, DFT Calculations, Hirshfeld Surface Analysis and In Silico Drug-Target Profiling of (R)-2-(2-(1,3-Dioxoisoindolin-2-yl)propanamido)benzoic Acid Methyl Ester. Molecules, 28(11), 4375. https://doi.org/10.3390/molecules28114375








