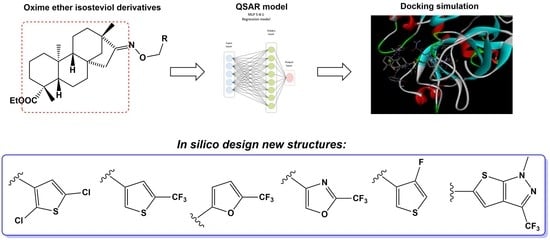
Novel Isosteviol-Based FXa Inhibitors: Molecular Modeling, In Silico Design and Docking Simulation
Abstract
1. Introduction
2. Results
2.1. Molecular Modeling
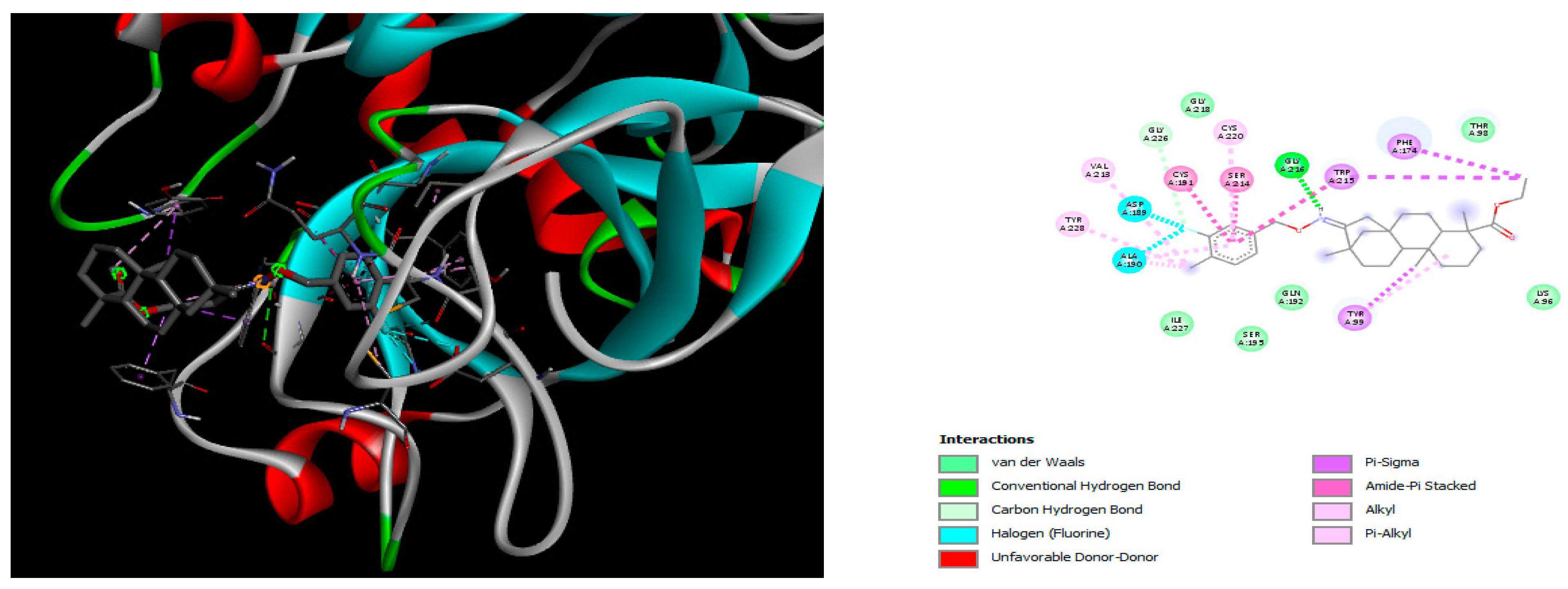
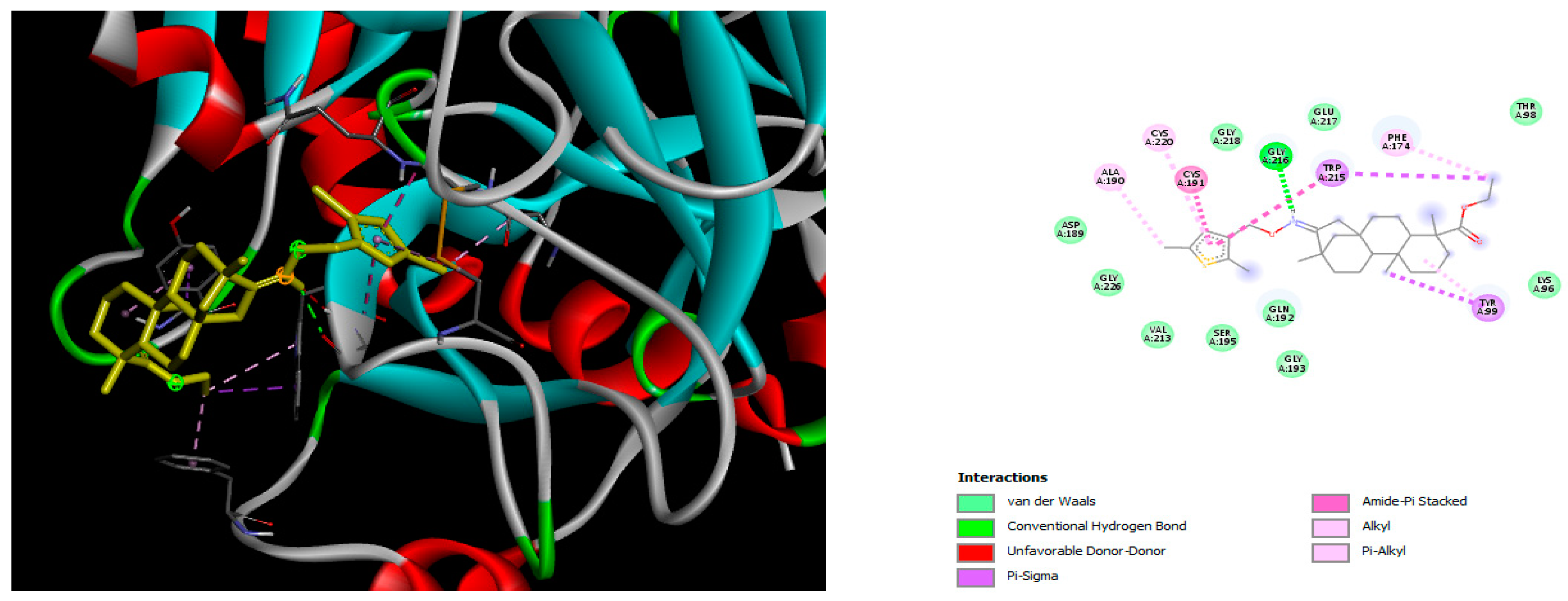
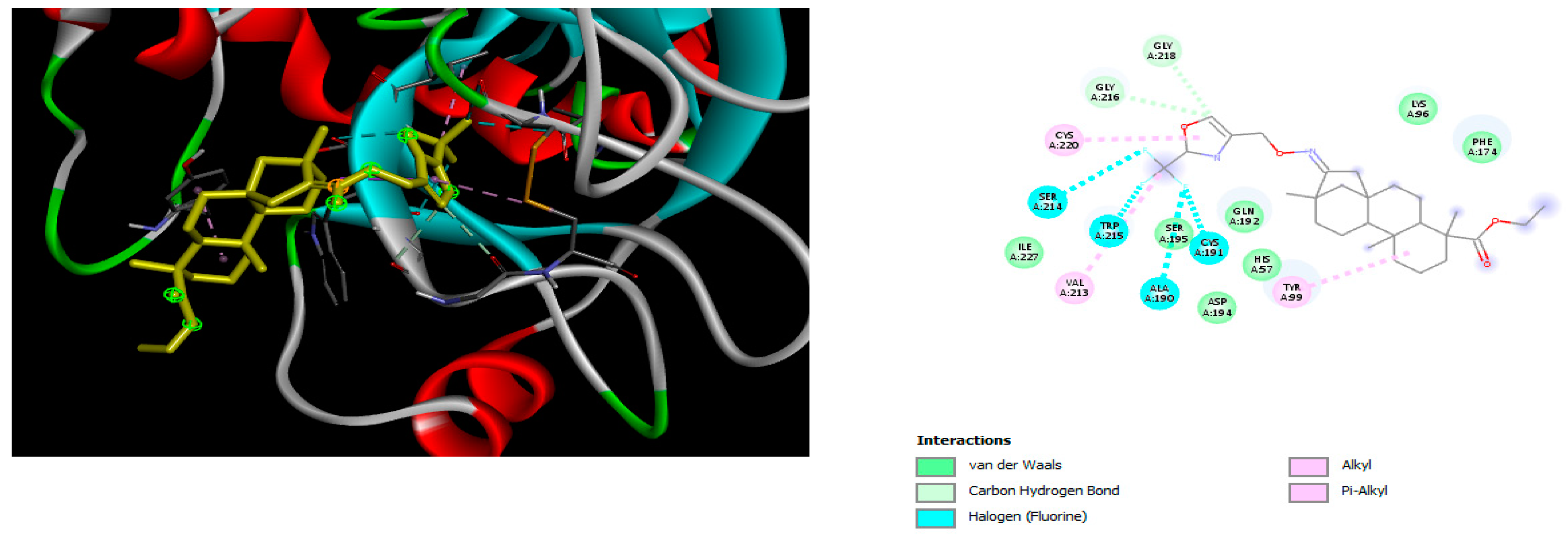
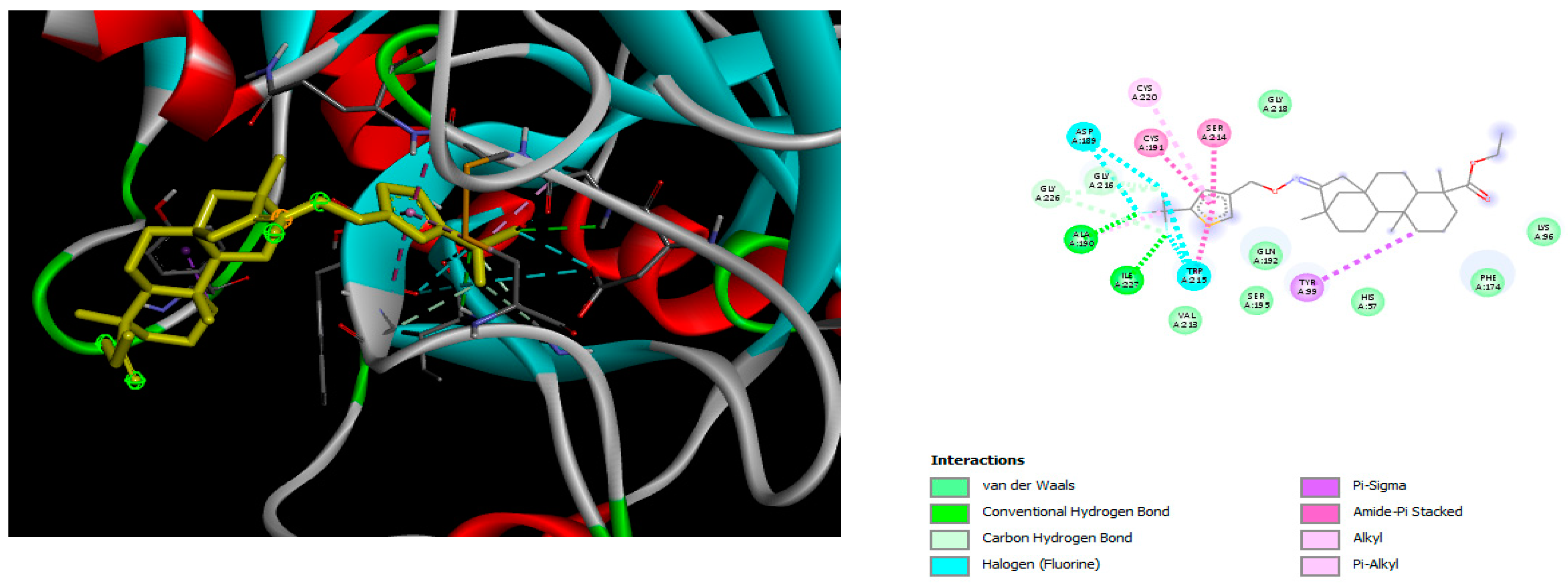
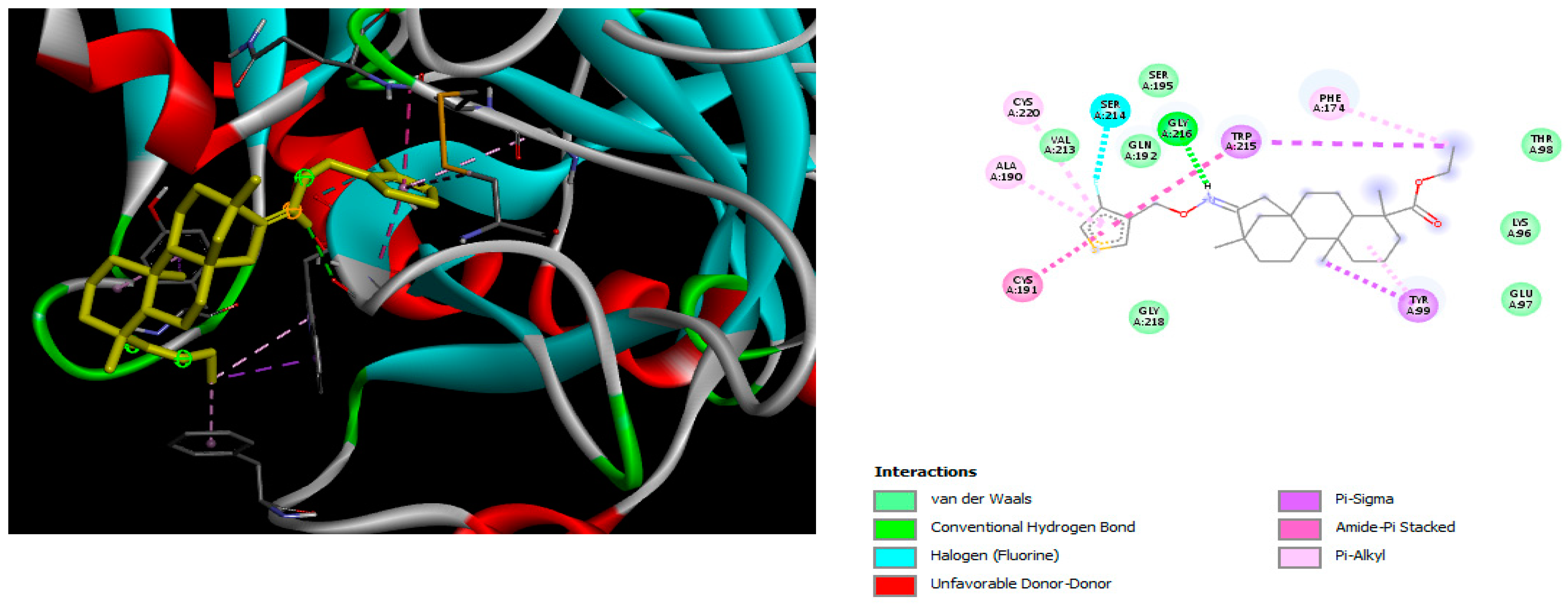
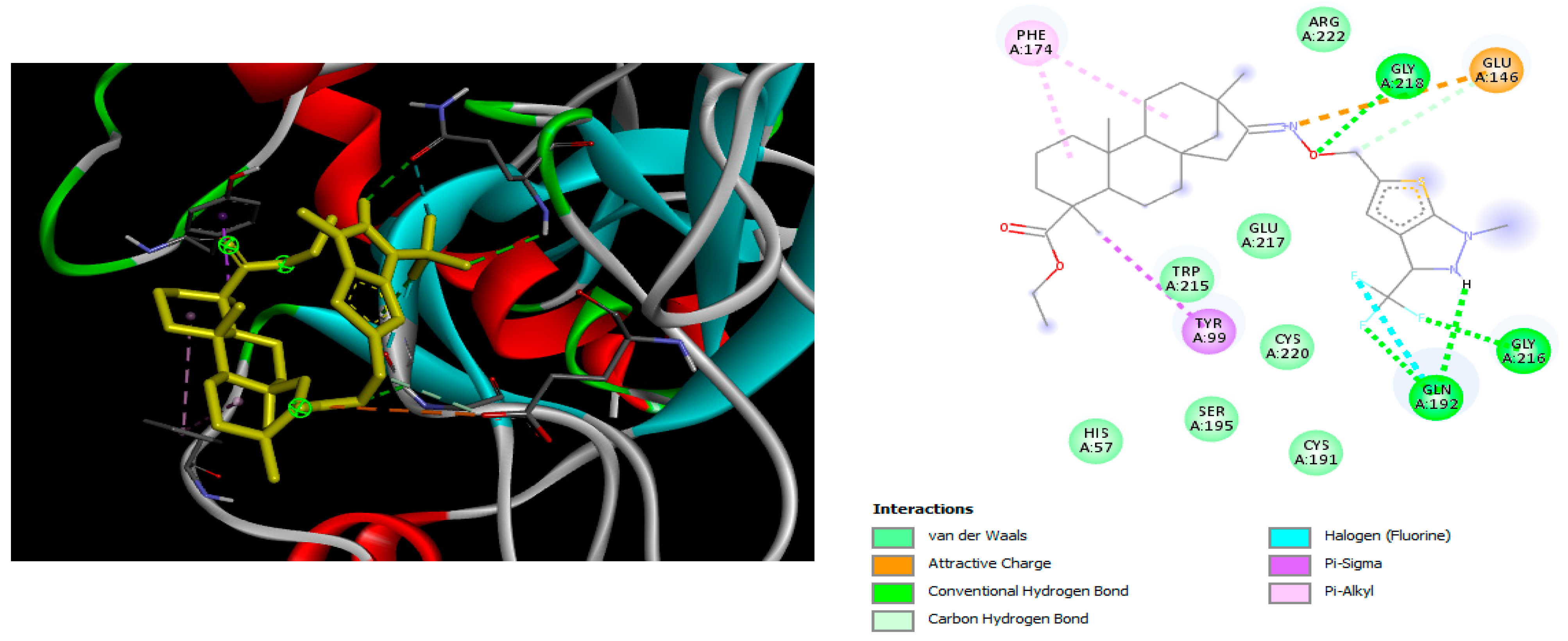
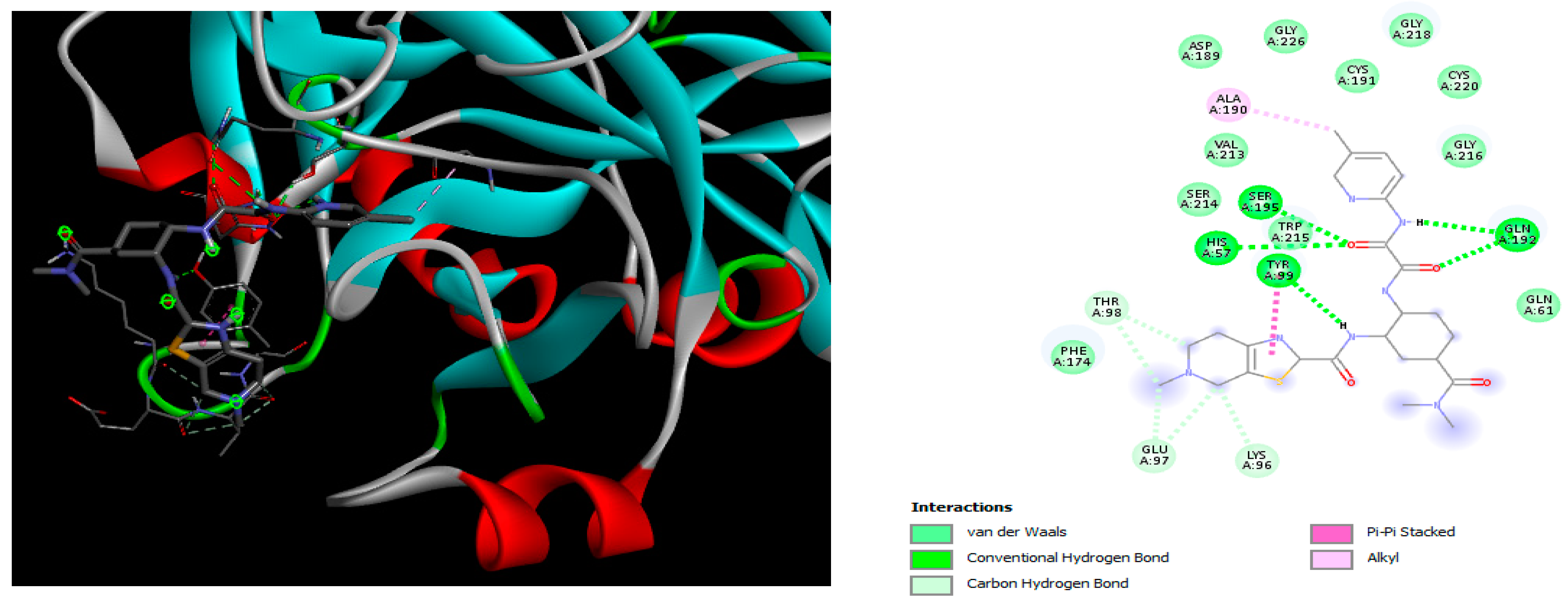
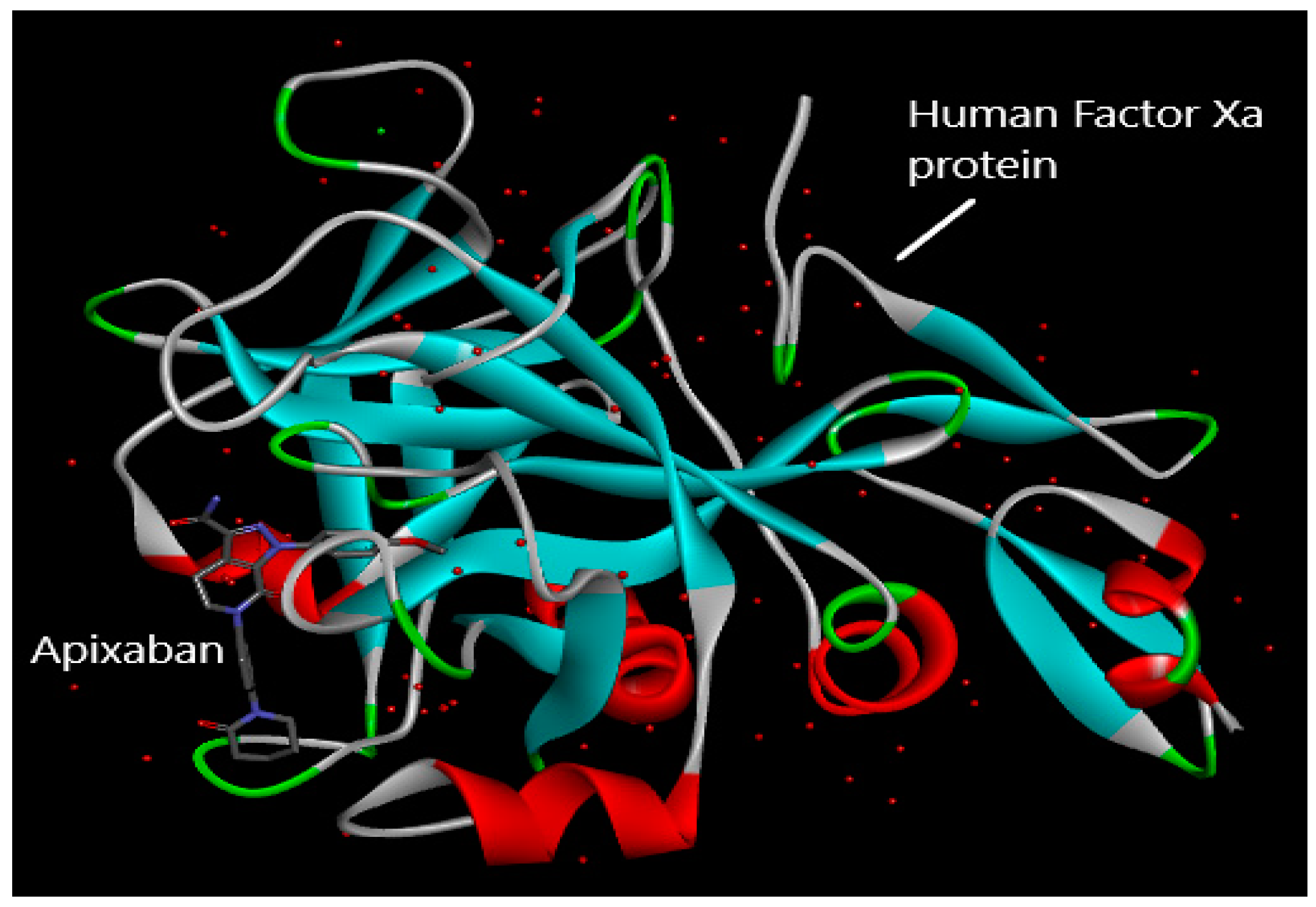
2.2. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Molecular Modeling
4.1.1. Energy Optimization of Isosteviol Analogues
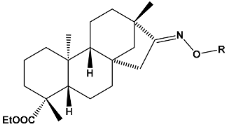 | ||||
| Compound | Sample | R * | Optimized Molecular Structure | Inhibition Constant (Ki), µM ** |
|---|---|---|---|---|
| a | train | 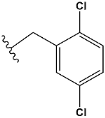 | 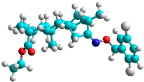 | 9.253 |
| b | train | 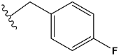 | 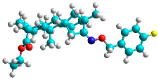 | 4.333 |
| d | train | 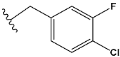 | 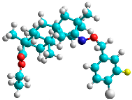 | 9.786 |
| e | train | 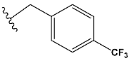 | 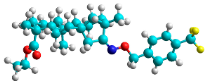 | 2.693 |
| f | train | 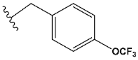 | 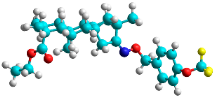 | 1.023 |
| g | train | 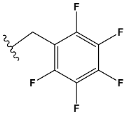 | 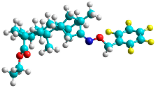 | 0.321 |
| h | test | 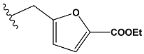 | 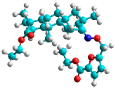 | 9.877 |
| i | train | 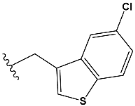 | 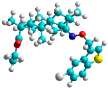 | 0.515 |
| j | test | 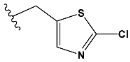 | 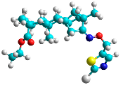 | 1.941 |
| k | train | 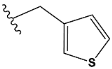 | 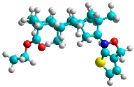 | 0.015 |
| l | train | 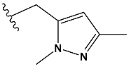 | 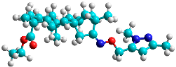 | 4.025 |
| m | train | 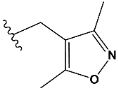 |  | 2.875 |
| n | train | 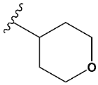 | 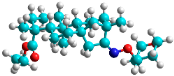 | 1.809 |
| o | train |  | 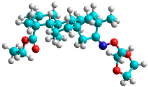 | 1.612 |
| p | validation | 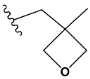 |  | 0.028 |
| q | train | 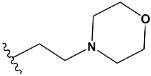 | 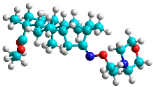 | 0.785 |
| r | validation | 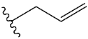 |  | 8.607 |
4.1.2. Molecular Descriptors
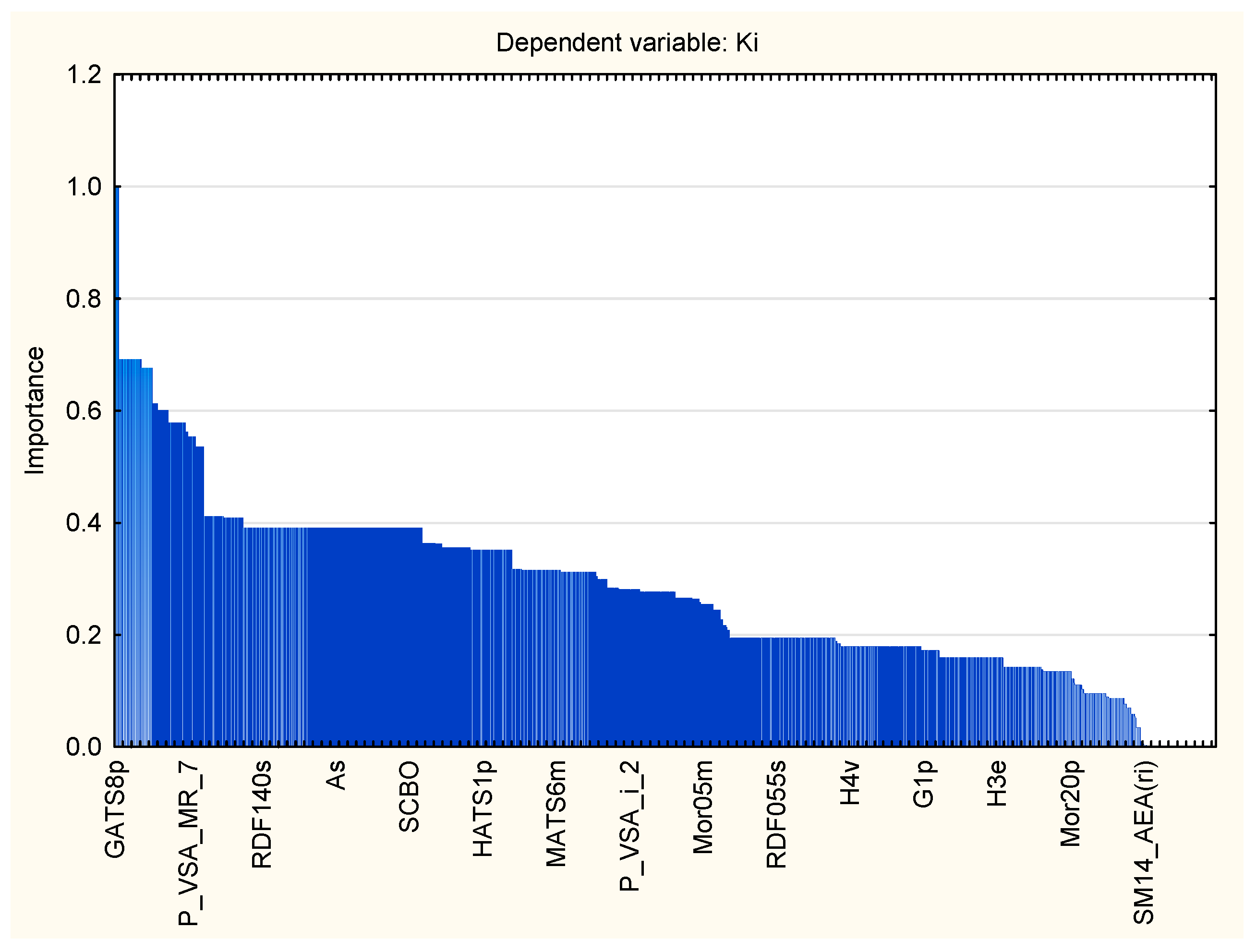
4.1.3. Regression Analysis
4.2. Molecular Docking Study
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kushnir, M.; Choi, Y.; Eisenberg, R.; Rao, D.; Tolu, S.; Gao, J.; Mowrey, W.; Billett, H.H. Efficacy and Safety of Direct Oral Factor Xa Inhibitors Compared with Warfarin in Patients with Morbid Obesity: A Single-Centre, Retrospective Analysis of Chart Data. Lancet Haematol. 2019, 6, e359–e365. [Google Scholar] [CrossRef]
- Mariño-Ocampo, N.; Rodríguez, D.F.; Guerra Díaz, D.; Zúñiga-Núñez, D.; Duarte, Y.; Fuentealba, D.; Zacconi, F.C. Direct Oral FXa Inhibitors Binding to Human Serum Albumin: Spectroscopic, Calorimetric, and Computational Studies. Int. J. Mol. Sci. 2023, 24, 4900. [Google Scholar] [CrossRef] [PubMed]
- Schreuder, M.; Reitsma, P.H.; Bos, M.H.A. Reversal Agents for the Direct Factor Xa Inhibitors: Biochemical Mechanisms of Current and Newly Emerging Therapies. Semin. Thromb. Hemost. 2020, 46, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, H.; Shimada, M.; Narita, M.; Narita, I.; Kimura, Y.; Tanaka, M.; Osanai, T.; Okumura, K.; Tomita, H. Rivaroxaban, a Direct Factor Xa Inhibitor, Ameliorates Hypertensive Renal Damage Through Inhibition of the Inflammatory Response Mediated by Protease-Activated Receptor Pathway. J. Am. Heart Assoc. 2019, 8, e012195. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, L. Top Companies and Drugs by Sales in 2021. Nat. Rev. Drug Discov. 2022, 21, 251. [Google Scholar] [CrossRef]
- Gosselin, R.C.; Adcock, D.M.; Douxfils, J. An Update on Laboratory Assessment for Direct Oral Anticoagulants (DOACs). Int. J. Lab. Hematol. 2019, 41, 33–39. [Google Scholar] [CrossRef]
- Wang, W.; Yuan, J.; Fu, X.; Meng, F.; Zhang, S.; Xu, W.; Xu, Y.; Huang, C. Novel Anthranilamide-Based FXa Inhibitors: Drug Design, Synthesis and Biological Evaluation. Molecules 2016, 21, 491. [Google Scholar] [CrossRef]
- Rodríguez, D.F.; Durán-Osorio, F.; Duarte, Y.; Olivares, P.; Moglie, Y.; Dua, K.; Zacconi, F.C. Green by Design: Convergent Synthesis, Computational Analyses, and Activity Evaluation of New FXa Inhibitors Bearing Peptide Triazole Linking Units. Pharmaceutics 2022, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Gackowski, M.; Golec, K.S.; Katarzyna, M.; Pluskota, R.; Koba, M. Quantitative Structure—Activity Relationship Analysis of Isosteviol—Related Compounds as Activated Coagulation Factor X (FXa) Inhibitors. Nutrients 2022, 14, 3521. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, D.; Li, M.; Wu, Q.; Lam, Y.P.Y.; Guo, Y.; Chen, C.; Bai, N.; Malhotra, S.; Li, W.; et al. Discovery of Novel, Potent, Isosteviol-Based Antithrombotic Agents. Eur. J. Med. Chem. 2019, 183, 111722. [Google Scholar] [CrossRef]
- Quan, M.L.; Ellis, C.D.; Liauw, A.Y.; Alexander, R.S.; Knabb, R.M.; Lam, G.; Wright, M.R.; Wong, P.C.; Wexler, R.R. Design and Synthesis of Isoxazoline Derivatives as Factor Xa Inhibitors. 2. J. Med. Chem. 1999, 42, 2760–2773. [Google Scholar] [CrossRef]
- Dudley, D.A.; Bunker, A.M.; Chi, L.; Cody, W.L.; Holland, D.R.; Ignasiak, D.P.; Janiczek-Dolphin, N.; McClanahan, T.B.; Mertz, T.E.; Narasimhan, L.S.; et al. Rational Design, Synthesis, and Biological Activity of Benzoxazinones as Novel Factor Xa Inhibitors. J. Med. Chem. 2000, 43, 4063–4070. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Wang, M.; Li, H.; Xu, F.; Gao, X.; Li, J.; Xu, S.; Zhang, D.; Wu, X.; Xu, J.; Hua, H.; et al. Diterpenoid Lead Stevioside and Its Hydrolysis Products Steviol and Isosteviol: Biological Activity and Structural Modification. Eur. J. Med. Chem. 2018, 156, 885–906. [Google Scholar] [CrossRef]
- Ullah, A.; Munir, S.; Mabkhot, Y. Bioactivity Profile of the Diterpene Isosteviol and Its Derivatives. Molecules 2019, 24, 678. [Google Scholar] [CrossRef]
- Ooms, F. Molecular Modeling and Computer Aided Drug Design. Examples of Their Applications in Medicinal Chemistry. Curr. Med. Chem. 2012, 7, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Aydın, S.G. Review on Molecular Modeling and Docking. SAR J. Sci. Res. 2022, 5, 206–210. [Google Scholar] [CrossRef]
- Nadendla, R.R. Molecular Modeling: A Powerful Tool for Drug Design and Molecular Docking. Resonance 2004, 9, 51–60. [Google Scholar] [CrossRef]
- Talete SRL List of Molecular Descriptors Calculated by Dragon. Available online: http://www.talete.mi.it/products/dragon_molecular_descriptor_list.pdf (accessed on 18 April 2023).
- Consonni, V.; Todeschini, R.; Pavan, M.; Gramatica, P. Structure/Response Correlations and Similarity/Diversity Analysis by GETAWAY Descriptors. 2. Application of the Novel 3D Molecular Descriptors to QSAR/QSPR Studies. J. Chem. Inf. Comput. Sci. 2002, 42, 693–705. [Google Scholar] [CrossRef]
- Wong, K.Y.; Mercader, A.G.; Saavedra, L.M.; Honarparvar, B.; Romanelli, G.P.; Duchowicz, P.R. QSAR Analysis on Tacrine-Related Acetylcholinesterase Inhibitors. J. Biomed. Sci. 2014, 21, 84. [Google Scholar] [CrossRef]
- Fatemi, M.H.; Gharaghani, S.; Mohammadkhani, S.; Rezaie, Z. Prediction of Selectivity Coefficients of Univalent Anions for Anion-Selective Electrode Using Support Vector Machine. Electrochim. Acta 2008, 53, 4276–4282. [Google Scholar] [CrossRef]
- Abreu, R.M.V.; Ferreira, I.C.F.R.; Queiroz, M.J.R.P. QSAR Model for Predicting Radical Scavenging Activity of Di(Hetero)Arylamines Derivatives of Benzo[b]Thiophenes. Eur. J. Med. Chem. 2009, 44, 1952–1958. [Google Scholar] [CrossRef] [PubMed]
- Khan, P.M.; Rasulev, B.; Roy, K. QSPR Modeling of the Refractive Index for Diverse Polymers Using 2D Descriptors. ACS Omega 2018, 3, 13374–13386. [Google Scholar] [CrossRef]
- Consonni, V.; Todeschini, R. Multivariate Analysis of Molecular Descriptors. In Statistical Modelling of Molecular Descriptors in QSAR/QSPR; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2012; Volume 2, pp. 111–147. ISBN 9783527324347. [Google Scholar]
- da Silva Rocha, S.F.L.; Olanda, C.G.; Fokoue, H.H.; Sant’Anna, C.M.R. Virtual Screening Techniques in Drug Discovery: Review and Recent Applications. Curr. Top. Med. Chem. 2019, 19, 1751–1767. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Mehrotra, R. An Overview of Molecular Simulation. JSM Chem. 2016, 4, 1024–1028. [Google Scholar]
- Shi, Y.; Pan, B.W.; Li, W.C.; Wang, Q.; Wu, Q.; Pan, M.; Fu, H.Z. Synthesis and Biological Evaluation of Isosteviol Derivatives as FXa Inhibitors. Bioorganic Med. Chem. Lett. 2020, 30, 126585. [Google Scholar] [CrossRef]
- Dobchev, D.; Karelson, M. Have Artificial Neural Networks Met Expectations in Drug Discovery as Implemented in QSAR Framework? Expert Opin. Drug Discov. 2016, 11, 627–639. [Google Scholar] [CrossRef]
- TIBCO Statistica® User’s Guide Statistica Automated Neural Networks (SANN)—Neural Networks Overview. Available online: https://docs.tibco.com/pub/stat/14.0.0/doc/html/UsersGuide/GUID-F60C241F-CD88-4714-A8C8-1F28473C52EE.html (accessed on 15 January 2023).
- Pappachen, L.K.; Zachariah, S.M.; Chandran, D. In Silico Design, Synthesis and Characterization of Some Novel Benzothiazole Derivatives as Anticancer Agents. Asian J. Pharm. Clin. Res. 2017, 10, 150–155. [Google Scholar] [CrossRef]
- RCSB Protein Data Bank 2P16: Factor Xa in Complex with the Inhibitor Apixaban (BMS-562247) AKA 1-(4-Methoxyphenyl)-7-OXO-6-(4-(2-OXO-1-Piperidinyl)Phenyl)-4,5,6,7-Tetrahydro-1H-Pyrazolo[3, 4-C]Pyridine-3-Carboxamide. Available online: https://www.rcsb.org/structure/2P16 (accessed on 14 March 2023).
- Morsy, M.A.; Ali, E.M.; Kandeel, M.; Venugopala, K.N.; Nair, A.B.; Greish, K.; El-Daly, M. Screening and Molecular Docking of Novel Benzothiazole Derivatives as Potential Antimicrobial Agents. Antibiotics 2020, 9, 221. [Google Scholar] [CrossRef]
- Ren, L.; You, T.; Li, Q.; Chen, G.; Liu, Z.; Zhao, X.; Wang, Y.; Wang, L.; Wu, Y.; Tang, C.; et al. Molecular Docking-Assisted Screening Reveals Tannic Acid as a Natural Protein Disulphide Isomerase Inhibitor with Antiplatelet and Antithrombotic Activities. J. Cell. Mol. Med. 2020, 24, 14257–14269. [Google Scholar] [CrossRef]

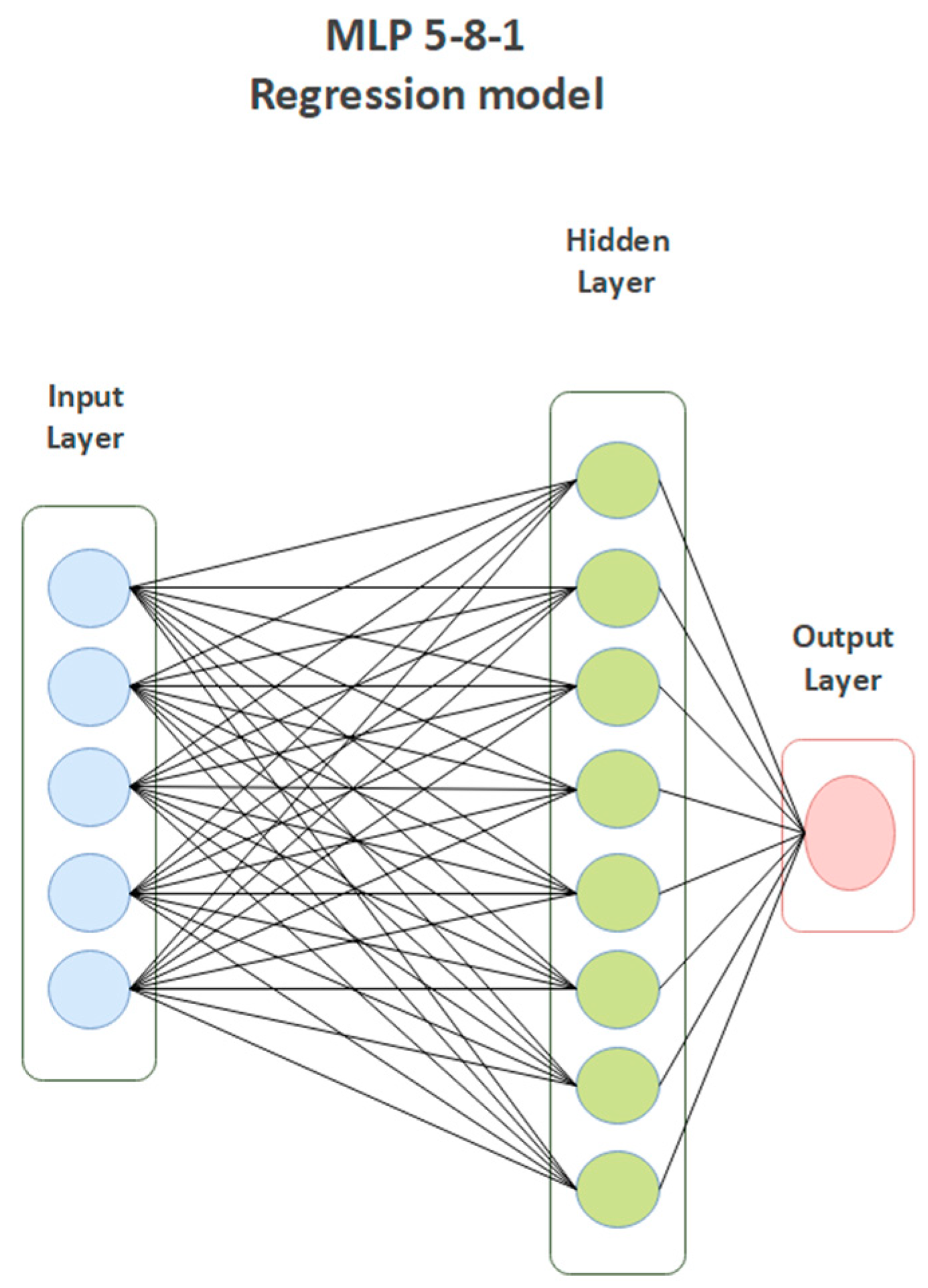
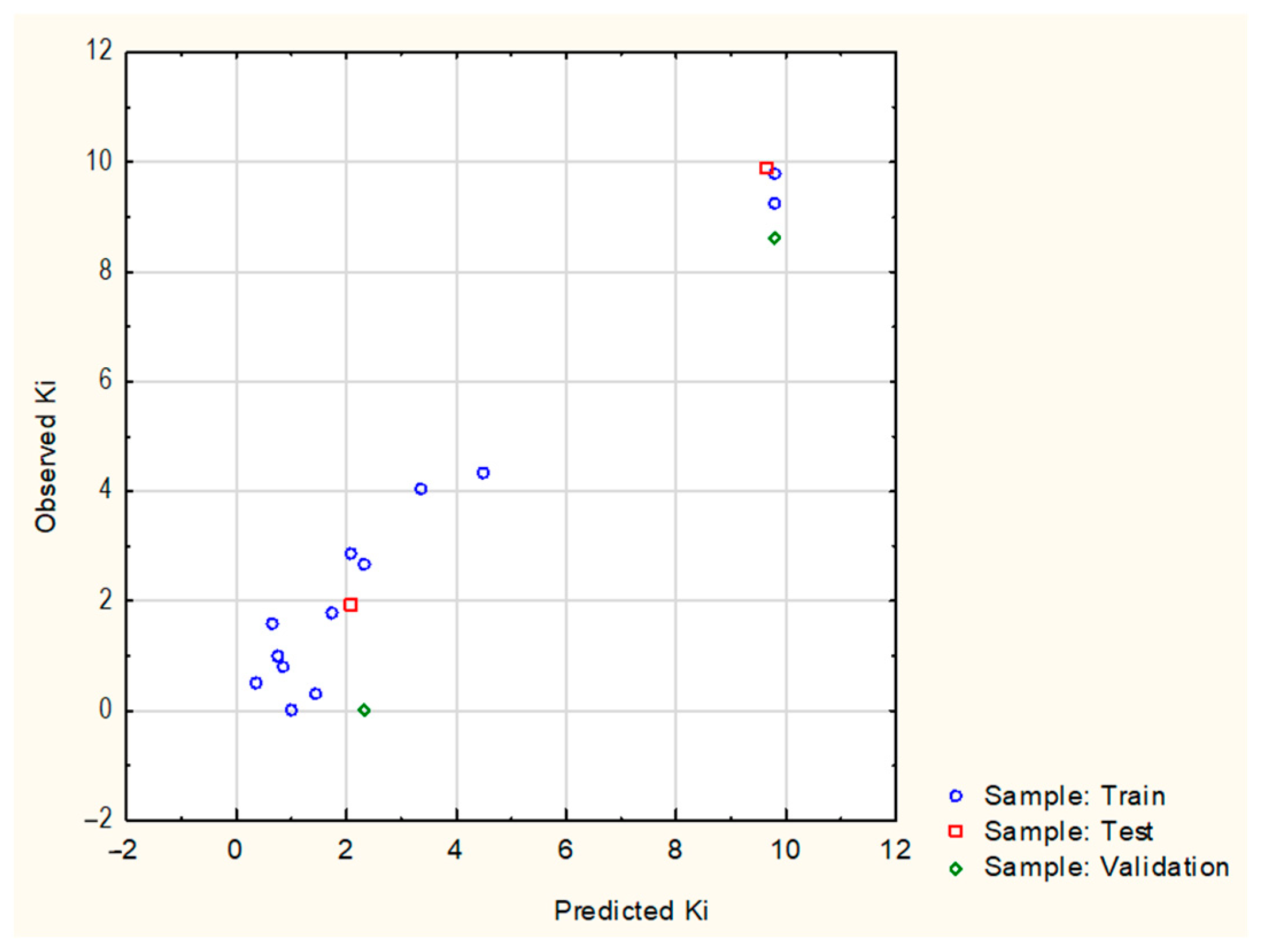

| MLP 5-8-1 Model | |
|---|---|
| Learning algorithm | BFGS 31 |
| Activation function (Hidden layer) | Logistic |
| Activation function (Output layer) | Tanh |
| Name | Definition | Category | Dimensionality | Error MLP 5-8-1 | Rank |
|---|---|---|---|---|---|
| GATS8p | Geary autocorrelation of lag 8 weighted by polarizability [19] | 2D autocorrelations | 2D | 16.75531 | 1 |
| HATS2i | leverage-weighted autocorrelation of lag 2/weighted by ionization potential [19] | GETAWAY descriptors | 3D | 14.49011 | 2 |
| R5e | R autocorrelation of lag 5/weighted by Sanderson electronegativity [19] | GETAWAY descriptors | 3D | 13.33332 | 3 |
| HATS2e | leverage-weighted autocorrelation of lag 2/weighted by Sanderson electronegativity [19] | GETAWAY descriptors | 3D | 8.866242 | 4 |
| SpMAD_B(v) | spectral mean absolute deviation from Burden matrix weighted by van der Waals volume [19] | 2D-matrix-based descriptors | 2D | 8.699860 | 5 |
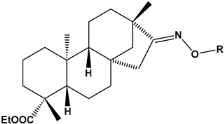 | ||
| Compound | R * | Predicted Inhibition Activity Against FXa: Inhibition Constant (Ki), [µM] ** |
|---|---|---|
| e1 | 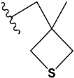 | 9.785990 |
| e2 | 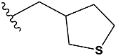 | 1.118201 |
| e3 | 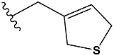 | 0.645249 |
| e4 | 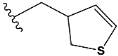 | 9.282278 |
| e5 | 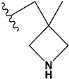 | 3.984843 |
| e6 | 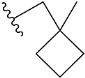 | 6.454281 |
| e7 | 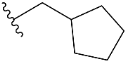 | 2.640193 |
| e8 | 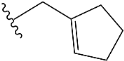 | 1.421969 |
| e9 | 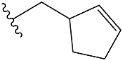 | 4.412301 |
| e10 | 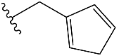 | 1.569841 |
| e11 |  | 1.133094 |
| e12 | 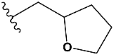 | 0.936761 |
| e13 |  | 0.372101 |
| e14 | 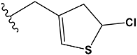 | 0.544725 |
| e15 | 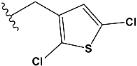 | 0.906731 |
| e16 | 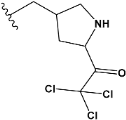 | 0.947983 |
| e17 | 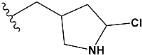 | 0.936441 |
| e18 |  | 0.650819 |
| e19 | 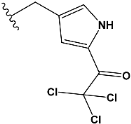 | 0.493827 |
| e20 | 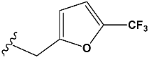 | 0.673789 |
| e21 | 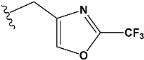 | 0.656182 |
| e22 | 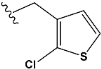 | 0.749590 |
| e23 | 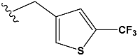 | 0.554588 |
| e24 | 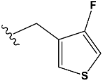 | 0.642575 |
| e25 | 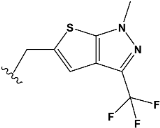 | 0.687738 |
| e26 | 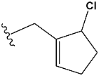 | 0.854700 |
| Compound | Binding-Free Energy (kcal/mol) |
|---|---|
| a | −8.7 |
| b | −8.3 |
| d | −9.3 |
| e | −8.1 |
| f | −8.8 |
| g | −8.3 |
| h | −8.4 |
| i | −7.4 |
| j | −8.0 |
| k | −8.1 |
| l | −8.7 |
| m | −7.8 |
| n | −6.9 |
| o | −7.7 |
| p | −7.0 |
| q | −8.0 |
| r | −6.8 |
| e1 | −6.9 |
| e2 | −7.7 |
| e3 | −8.0 |
| e4 | −7.4 |
| e5 | −7.2 |
| e6 | −7.1 |
| e7 | −7.4 |
| e8 | −7.4 |
| e9 | −7.5 |
| e10 | −7.6 |
| e11 | −7.3 |
| e12 | −7.7 |
| e13 | −7.7 |
| e14 | −7.2 |
| e15 | −8.3 |
| e16 | −6.9 |
| e17 | −7.7 |
| e18 | −7.4 |
| e19 | −6.9 |
| e20 | −8.1 |
| e21 | −8.3 |
| e22 | −7.0 |
| e23 | −8.1 |
| e24 | −8.2 |
| e25 | −8.2 |
| e26 | −7.0 |
| apixaban | −10.3 |
| edoxaban | −8.8 |
| rivaroxaban | −9.4 |
| Action | Reason | Number |
|---|---|---|
| deleted | constant | 1856 |
| near constant | 95 | |
| all missing | 2 | |
| one missing | 2 | |
| highly correlated (|r| > 0.95) | 1658 | |
| standard deviation < 0.0001 | 1856 | |
| retained | suitable for model-building | 1274 |
| Symbol | Variable Rank | Importance |
|---|---|---|
| GATS8p | 100 | 1 |
| R5e | 100 | 1 |
| HATS2i | 100 | 1 |
| HATS2e | 100 | 1 |
| SpMAD_B(v) | 100 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gackowski, M.; Madriwala, B.; Studzińska, R.; Koba, M. Novel Isosteviol-Based FXa Inhibitors: Molecular Modeling, In Silico Design and Docking Simulation. Molecules 2023, 28, 4977. https://doi.org/10.3390/molecules28134977
Gackowski M, Madriwala B, Studzińska R, Koba M. Novel Isosteviol-Based FXa Inhibitors: Molecular Modeling, In Silico Design and Docking Simulation. Molecules. 2023; 28(13):4977. https://doi.org/10.3390/molecules28134977
Chicago/Turabian StyleGackowski, Marcin, Burhanuddin Madriwala, Renata Studzińska, and Marcin Koba. 2023. "Novel Isosteviol-Based FXa Inhibitors: Molecular Modeling, In Silico Design and Docking Simulation" Molecules 28, no. 13: 4977. https://doi.org/10.3390/molecules28134977
APA StyleGackowski, M., Madriwala, B., Studzińska, R., & Koba, M. (2023). Novel Isosteviol-Based FXa Inhibitors: Molecular Modeling, In Silico Design and Docking Simulation. Molecules, 28(13), 4977. https://doi.org/10.3390/molecules28134977