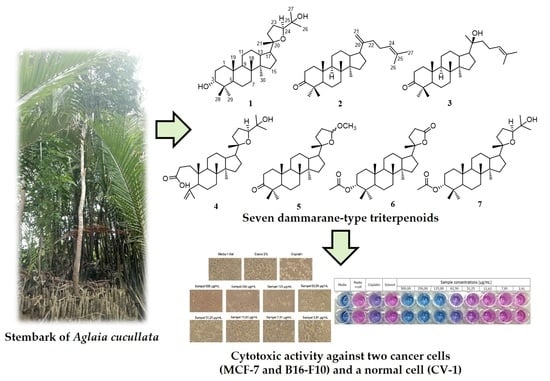
The Cytotoxic Activity of Dammarane-Type Triterpenoids Isolated from the Stem Bark of Aglaia cucullata (Meliaceae)
Abstract
1. Introduction
2. Results and Discussions
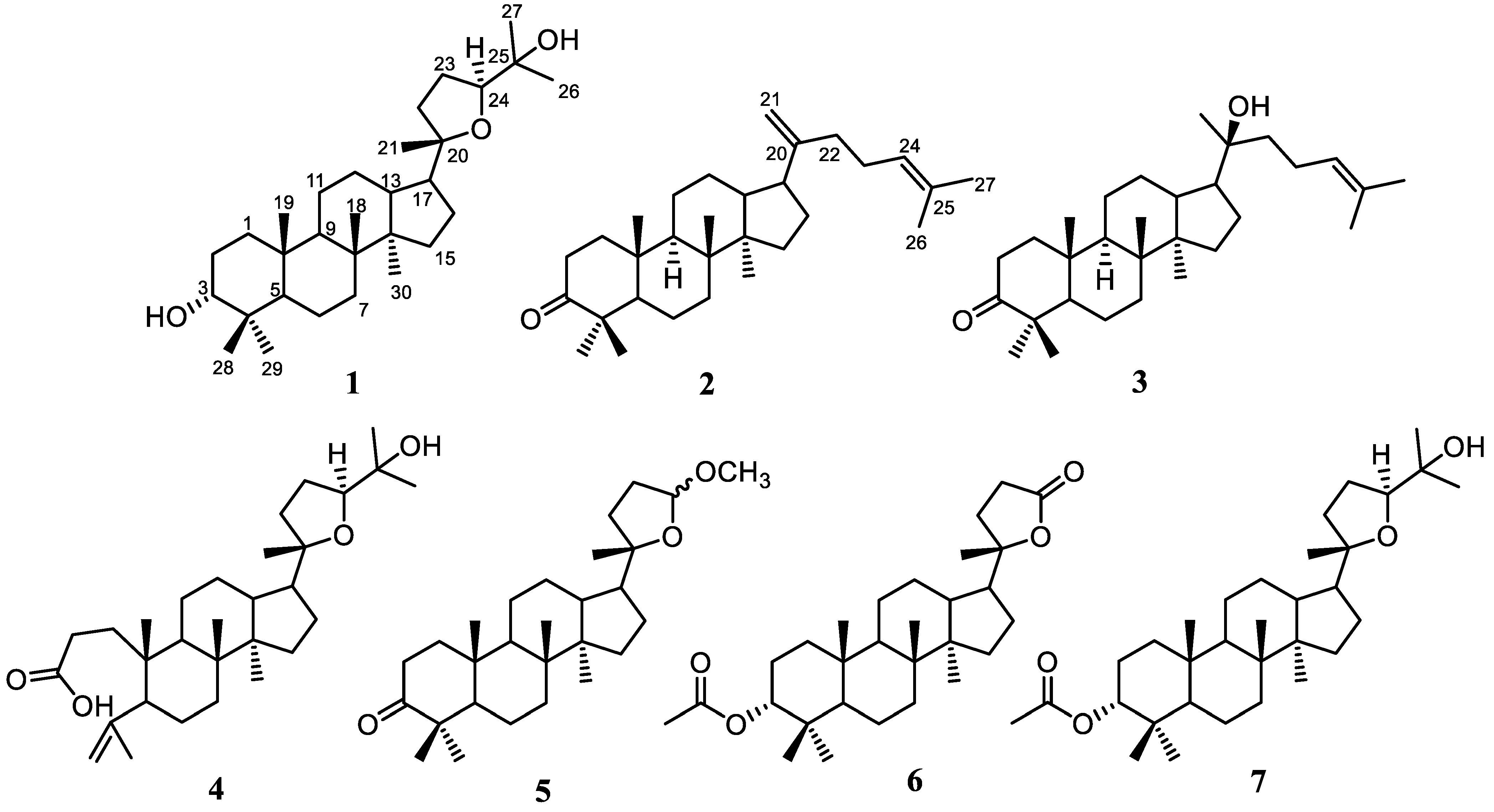
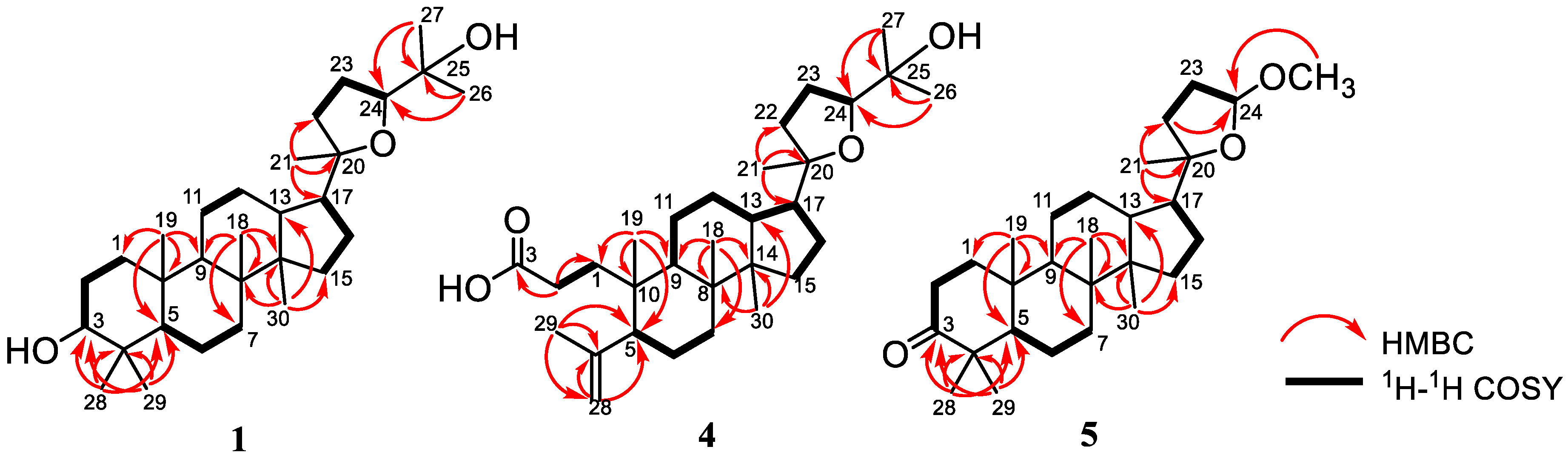
2.1. Structural Identification of the Isolated Compounds
2.2. Cytotoxic Activity Compounds 1–7
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Determination of Cytotoxic Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Heywood, V.H.; Brummitt, R.K.; Culham, A.; Seberg, O. Flowering Plant Families of the World; Firefly Books: Richmond Hill, ON, Canada, 2007; p. 207. [Google Scholar]
- Harneti, D.; Supratman, U. Phytochemistry and biological activities of Aglaia species. Phytochemistry 2021, 181, 112540. [Google Scholar] [PubMed]
- Pannell, C.M. Taxonnomic Monograph of the Genus Aglaia lour. (Meliaceae); Kew Bulletin Additional Series XVI; HMSO: Kew, UK, 1992. [Google Scholar]
- Heyne, K. The Useful Indonesian Plants, Research and Development Agency; Ministry of Forestry: Jakarta, Indonesia, 1982; pp. 1029–1031. [Google Scholar]
- Cai, X.; Wang, Y.; Zhao, P.; Li, Y.; Luo, X. Dolabellane diterpenoids from Aglaia odorata. Phytochemistry 2010, 71, 1020–1024. [Google Scholar] [PubMed]
- Yodsaoue, O.; Sonprasit, J.; Karalai, C.; Ponglimanont, C.; Tewtrakul, S.; Chantrapromma, S. Diterpenoids and triterpenoids with potential anti-inflammatory activity from the leaves of Aglaia odorata. Phytochemistry 2012, 76, 83–91. [Google Scholar] [PubMed]
- Othman, N.; Pan, L.; Mejin, M.; Voong, J.C.; Chai, H.B.; Pannell, C.M.; Kinghorn, A.D.; Yeo, T.C. Cyclopenta[b]benzofuran and Secodammarane Derivatives from the Stems of Aglaia stellatopilosa. J. Nat. Prod. 2016, 79, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Chai, H.; Mi, Q.; Riswan, S.; Kardono, L.B.S.; Afriastini, J.J.; Santarsiero, B.D.; Mesecar, A.D.; Fransworth, N.R.; Cordell, G.A.; et al. Activity-guided isolation of cytotoxic constituents from the bark of Aglaia crassinervia collected in Indonesia. J. Bioorg. Med. Chem. 2006, 14, 960–972. [Google Scholar]
- Liu, S.; Liu, S.B.; Zuo, W.J.; Guo, Z.K.; Mei, W.L.; Dai, H.F. New sesquiterpenoids from Aglaia odorata var. microphyllina and their cytotoxic activity. Fitoterapia 2014, 92, 93–99. [Google Scholar] [CrossRef]
- Joycharat, N.; Plodpai, P.; Panthong, K.; Yingyongnarongkul, B.E.; Voravuthikunchai, S.P. Terpenoid constituents and antifungal activity of Aglaia forbessi seed against phytopathogens. Can. J. Chem. 2010, 88, 937–944. [Google Scholar] [CrossRef]
- Farabi, K.; Harneti, D.; Nurlelasari; Maharani, R.; Hidayat, A.C.; Awang, K.; Supratman, U.; Shiono, Y. New cytotoxic protolimonoids from the stem bark of Aglaia argentea (Meliaceae). Phytochem. Lett. 2017, 21, 211–215. [Google Scholar]
- Sun, Y.; Cui, L.; Sun, Y.; Li, Q.; Li, Y.; Wang, Z.; Xu, W.; Kong, L.; Luo, J. A/D-rings-seco limonoids from the fruits of Aglaia edulis and their bioactivities. Phytochemistry 2022, 195, 113049. [Google Scholar]
- Farabi, K.; Harneti, D.; Nurlelasari; Maharani, R.; Hidayat, A.C.; Awang, K.; Supratman, U.; Shiono, Y. New cytotoxic pregnane-type steroid from the stem bark of Aglaia elliptica (Meliaceae). Rec. Nat. Prod. 2018, 12, 121–127. [Google Scholar] [CrossRef]
- Wu, P.F.; Liu, J.; Li, Y.N.; Ding, R.; Tan, R.; Yang, X.M.; Yu, Y.; Hao, X.J.; Yuan, C.M.; Yi, P. Three new aglain derivatives from Aglaia odorata Lour. and their cytotoxic activities. Chem. Biodivers 2022, 19, e202101008. [Google Scholar]
- Zhang, L.; Wang, L.H.; Yang, Y.F.; Yang, S.M.; Zhang, J.H.; Tan, C.H. Aglaianine, a new bisamide from Aglaia abbreviata. Nat. Prod. Res. 2011, 25, 1676–1679. [Google Scholar] [CrossRef]
- Peng, L.; Fu, W.X.; Zeng, C.X.; Zhou, L.; Bao, M.F.; Cai, X.H. Two new lignans from twigs of Aglaia odorata. J. Asian Nat. Prod. Res. 2016, 18, 147–152. [Google Scholar]
- Wang, B.G.; Ebel, R.; Wang, C.Y.; Edrada, R.A.; Wray, V.; Proksch, P. Aglacins I–K, three highly methoxylated lignans from Aglaia cordata. J. Nat. Prod. 2004, 67, 682–684. [Google Scholar] [CrossRef]
- An, F.L.; Wang, X.B.; Wang, H.; Li, Z.R.; Yang, M.H.; Luo, J.; Kong, L.Y. Cytotoxic rocaglate derivatives from leaves of Aglaia perviridis. Sci. Rep. 2016, 28, 20045. [Google Scholar] [CrossRef]
- Pan, L.; Acuna, U.M.; Li, J.; Jena, N.; Ninh, T.N.; Pannel, C.M.; Chai, H.; Fuchs, J.R.; Carcache, E.J.; Soejarto, D.D.; et al. Bioactive flavaglines and other constituents isolated from Aglaia perviridis. J. Nat. Prod. 2013, 76, 394–404. [Google Scholar]
- Nugroho, B.W.; Gussregen, B.; Wray, V.; Witte, L.; Bringmann, G.; Proksch, P. Insecticidal rocaglamide derivatives from Aglaia elliptica and A. harmsiana. Phytochemistry 1997, 45, 1579–1585. [Google Scholar] [CrossRef]
- Engelmeier, D.; Hadacek, F.; Pacher, T.; Vajrodaya, S.; Greger, H. Cyclopenta[b] benzofurans from Aglaia species with pronounced antifungal activity against Rice Blast Fungus (Pyricularia grisea). J. Agric. Food. Chem. 2000, 48, 1400–1404. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, H.H.; Sonf, Z.J.; Chen, L.Y.; Wen, H.J. Molluscidal activity of Aglaia duperreana and the constituents of its twigs and leaves. Fitoterapia 2012, 83, 1081–1086. [Google Scholar]
- Shang, Y.; Huang, S. Multi-omics data-driven investigations of metabolic diversity of plant triterpenoids. Plant J. 2019, 97, 101–111. [Google Scholar]
- Chen, Y.; Zhou, B.; Li, J.; Tang, H.; Tang, J.; Yang, Z. Formation and change of chloroplast-located plant metabolites in response to light conditions. Int. J. Mol. Sci. 2018, 19, 654. [Google Scholar] [PubMed]
- Shady, N.H.; Youssif, K.A.; Sayed, A.M.; Belbahri, L.; Oszako, T.; Hassan, H.M.; Abdelmohsen, U.R. Sterols and triterpenes: Antiviral potential supported by in-silico analysis. Plants 2021, 10, 41. [Google Scholar]
- Innocente, A.; Casanova, B.B.; Klein, F.; Lana, A.D.; Pereira, D.; Muniz, M.N.; Sonnet, P.; Gosmann, G.; Fuentefria, A.M.; Gnoatto, S.C.B. Synthesis of isosteric triterpenoid derivatives and antifungal activity. Chem. Biol. Drug Des. 2014, 83, 344–349. [Google Scholar] [PubMed]
- Oprean, C.; Zambori, C.; Borcan, F.; Soica, C.; Zupko, I.; Minorics, R.; Bojin, F.; Ambrus, R.; Muntean, D.; Danciu, C.; et al. Anti-proliferative and antibacterial in vitro evaluation of the polyurethane nanostructures incorporating pentacyclic triterpenes. Pharm. Biol. 2016, 54, 2714–2722. [Google Scholar]
- Muhammad, D.; Hubert, J.; Lalun, N.; Renault, J.H.; Bobichon, H.; Nour, M.; Voutquenne-Nazabadioko, L. Isolation of flavonoids and triterpenoids from the fruits of Alphitonia neocaledonica and evaluation of their anti-oxidant, antityrosinase and cytotoxic activities. Phytochem. Anal. 2015, 26, 137–144. [Google Scholar]
- Zhang, J.; Zhang, Q.; Xu, Y.; Li, H.; Wang, C.; Liu, Z.; Liu, P.; Liu, Y.; Meng, Q.; Zhao, F.; et al. Synthesis and in vitro anti-inflammatory activity of C20 epimeric ocotillol-type triterpenes and protopanaxadiol. Planta Med. 2019, 85, 292–301. [Google Scholar] [CrossRef]
- Nazaruk, J.; Borzym-Kluczyk, M. The role of triterpenes in the management of diabetes mellitus and its complications. Phytochem. Rev. 2015, 14, 675–690. [Google Scholar]
- Farabi, K.; Harneti, D.; Darwati; Nurlelasari; Mayanti, T.; Maharani, R.; Supratman, U.; Fajriah, S.; Kuncoro, H.; Azmi, M.N.; et al. New dammarane-type triterpenoids from Aglaia elliptica (C.DC.) blume. Nat. Prod. Res. 2022, in press. [Google Scholar] [CrossRef]
- Farabi, K.; Harneti, D.; Darwati; Mayanti, T.; Nurlelasari; Maharani, R.; Sari, A.P.; Herlina, T.; Hidayat, A.T.; Supratman, U.; et al. Dammarane-Type Triterpenoid from the Stem Bark of Aglaia elliptica (Meliaceae) and Its Cytotoxic Activities. Molecules 2022, 27, 6757. [Google Scholar]
- Zhang, F.; Wang, J.S.; Gu, Y.C.; Kong, L.Y. Triterpenoids from Aglaia abbreviata and their cytotoxic activities. J. Nat. Prod. 2010, 73, 2042–2046. [Google Scholar] [CrossRef]
- Harneti, D.; Supriadin, A.; Ulfah, M.; Safari, A.; Supratman, U.; Awang, K.; Hayashi, H. Cytotoxic constituents from the bark of Aglaia eximia (Meliaceae). Phytochem. Lett. 2014, 8, 28–31. [Google Scholar]
- Oktaviani, D.; Sukmawati, W.; Farabi, K.; Harneti, D.; Nurlelasari; Darwati; Maharani, R.; Mayanti, T.; Safari, A.; Supratman, U. Terpenoids from the stem bark of Aglaia elaeagnoidea and their cytotoxic activity against HeLa and DU145 cancer cell lines. Molekul 2022, 17, 76–84. [Google Scholar] [CrossRef]
- Meepol, W.; Maxwell, G.S.; Havanond, S. Aglaia cucullata: A little-known mangrove with big potential for research. ISME/GLOMIS Electron. 2020, 18, 4–9. [Google Scholar]
- Aglaia cucullata. The IUCN Red List of Threatened Species. Available online: https://www.gbif.org/species/176776850 (accessed on 1 May 2023).
- Ahmed, F.; Toume, K.; Sadhu, S.K.; Ohtsuki, T.; Arai, M.A.; Ishibashi, M. Constituents of Amoora cucullata with TRAIL resistance-overcoming activity. Org. Biomol. Chem. 2010, 8, 3696–3703. [Google Scholar]
- Pancharoen, R.; Sommeechai, M.; Maelim, S.; Suanpaga, W.; Srichaichana, J.; Barber, P.; Dell, B. Phenology of urban trees in a tropical urban forest in Thailand. Songklanakarin J. Sci. Technol. 2021, 43, 87–95. [Google Scholar]
- Chumkaew, P.; Kato, S.; Chantrapromma, K. Potent cytotoxic rocaglamide derivatives from the fruits of Amoora cucullata. Chem. Pharm. Bull. 2006, 54, 1344–1346. [Google Scholar] [CrossRef]
- DeFilipps, R.A.; Krupnick, G.A. The medicinal plants of Myanmar. PhytoKeys 2018, 102, 1–341. [Google Scholar]
- Abdelfattah, M.S.; Toume, K.; Ahmed, F.; Sadhu, S.K.; Ishibashi, M. Cucullamide, a new putrescine bisamide from Amoora cucullata. Chem. Pharm. Bull. 2010, 58, 1116–1118. [Google Scholar]
- Phongmaykin, J.; Kumamoto, T.; Ishikawa, T.; Suttisri, R.; Saifah, E. A new sesquiterpene and other terpenoid constituents of Chisocheton penduliflorus. Arch. Pharm. Res. 2008, 31, 21–27. [Google Scholar]
- Hidayat, A.T.; Farabi, K.; Harneti, D.; Maharani, R.; Darwati; Nurlelasari; Mayanti, T.; Setiawan, A.S.; Supratman, U.; Shiono, Y. Cytotoxicity and structure activity relationship of dammarane-type triterpenoids from the bark of Aglaia elliptica against P-388 murine leukemia cells. Nat. Prod. Sci. 2017, 23, 291–298. [Google Scholar]
- Roux, D.; Martin, M.T.; Adeline, M.T.; Sevenet, T.; Hadi, A.H.A.; Pais, M. Foveolins A dan B, dammarane triterpenes from Aglaia foveolata. Phytochemistry 1998, 49, 1745–1748. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, I.E.; Do Thi Thu, H.; Kazakova, O.B.; Tolstikov, G.A.; Kukovinets, O.S.; Lobov, A.N.; Suponitskii, K.Y. Ozonolysis of dipterocarpol and its derivatives. Russ. J. Org. Chem. 2012, 48, 1370–1376. [Google Scholar]
- Huong, D.T.T.; Thuy, T.T.T.; Hien, T.T.; Tra, N.T.; Tien, N.Q.; Smirnova, I.E.; Kazakova, O.B.; Minnibaeva, E.M.; Tolstikov, G.A. Synthesis and cytotoxicity of derivatives of dipterocarpol, a metabolite of Dipterocarpus alatus. Chem. Nat. Compd. 2013, 49, 58–65. [Google Scholar] [CrossRef]
- Präbst, K.; Engelhardt, H.; Ringgeler, S.; Hübner, H. Chapter 2 of Cell Viability Assays. Basic Colorimetric Proliferation Assays: MTT, WST, and Resazurin. Methods Mol. Biol. 2017, 1601, 1–17. [Google Scholar]
- Cao, J.; Zhang, X.; Qu, F.; Guo, Z.; Zhao, Y. Dammarane triterpenoids for pharmaceutical use: A patent review (2005–2014). Expert. Opin. Ther. Pat. 2015, 25, 805–817. [Google Scholar] [CrossRef]
- Ruan, J.; Zheng, C.; Qu, L.; Liu, Y.; Han, L.; Yu, H.; Zhang, Y.; Wang, T. Plant resources, 13C-NMR spectral characteristic and pharmacological activities of dammarane-type triterpenoids. Molecules 2016, 21, 1047. [Google Scholar]
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1 | 33.7 | 40 | 39.9 | 34.3 | 39.9 | 34.3 | 34.4 |
| 2 | 25.4 | 34.1 | 34.1 | 28.2 | 32.8 | 25.1 | 23 |
| 3 | 76.4 | 218.2 | 218 | 179.3 | 218.4 | 78.4 | 78.5 |
| 4 | 37.3 | 47.4 | 47.4 | 147.5 | 47.5 | 37.2 | 36.8 |
| 5 | 49.6 | 55.4 | 55.4 | 50.8 | 55.4 | 50.4 | 50.6 |
| 6 | 18.3 | 19.7 | 19.7 | 24.6 | 19.7 | 18.1 | 18.2 |
| 7 | 34.8 | 34.8 | 34.6 | 33.9 | 34.6 | 35.1 | 34.7 |
| 8 | 40.7 | 40.4 | 40.3 | 40 | 40.3 | 50.3 | 40.6 |
| 9 | 50.7 | 50.3 | 50 | 41.2 | 50.1 | 50.7 | 50.8 |
| 10 | 37.7 | 36.9 | 36.8 | 39.1 | 36.9 | 36.8 | 37.2 |
| 11 | 21.7 | 21.9 | 22 | 22.3 | 26.7 | 26.8 | 21.7 |
| 12 | 27.1 | 25 | 27.5 | 26.9 | 22 | 21.3 | 27.1 |
| 13 | 42.8 | 45.4 | 42.4 | 42.9 | 43.6 | 43.2 | 42.9 |
| 14 | 50.2 | 49.4 | 50.3 | 50.4 | 50.1 | 40.6 | 50.2 |
| 15 | 31.5 | 31.4 | 31.2 | 31.5 | 31.3 | 31.1 | 31.6 |
| 16 | 25.9 | 28.9 | 24.8 | 25.8 | 25.5 | 22.9 | 25.9 |
| 17 | 49.8 | 47.8 | 49.8 | 49.7 | 51.2 | 49.4 | 49.9 |
| 18 | 16.2 | 15.8 | 15.2 | 16.3 | 15.3 | 15.6 | 15.6 |
| 19 | 16.6 | 16.1 | 16 | 20.2 | 16.3 | 16 | 16.1 |
| 20 | 86.7 | 152.6 | 75.4 | 86.6 | 88.1 | 90.3 | 86.7 |
| 21 | 27.3 | 107.6 | 25.5 | 27.2 | 23.5 | 25.5 | 27.3 |
| 22 | 35.3 | 34.2 | 40.5 | 34.7 | 35.1 | 31.3 | 35.2 |
| 23 | 26.4 | 27.1 | 22.6 | 26.4 | 34.2 | 29.3 | 26.4 |
| 24 | 86.3 | 124.4 | 124.7 | 86.4 | 104.7 | 176.9 | 86.4 |
| 25 | 70.3 | 131.5 | 131.6 | 70.3 | - | - | 70.3 |
| 26 | 27.9 | 25.7 | 25.7 | 27.9 | - | - | 27.9 |
| 27 | 24.1 | 17.7 | 17.7 | 23.2 | - | - | 24.1 |
| 28 | 28.4 | 26.8 | 26.7 | 113.5 | 26.8 | 27.9 | 27.8 |
| 29 | 22.2 | 21 | 21 | 24 | 21.1 | 21.8 | 21.8 |
| 30 | 15.6 | 15.4 | 16.4 | 15.3 | 16.1 | 16.5 | 16.7 |
| 1′ | 54.5 | 170.9 | 171 | ||||
| 2′ | 21.5 | 21.5 |
| Position | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 1 | 1.42 (2H, m) | 1.42, 1.94 (each 1H, m) | 1.86 (2H, m) | 1.97, 1.53 (each 1H, m) |
| 2 | 1.55 (2H, m) | 2.42, 2.48 (each 1H, m) | 2.37, 2.42 (each 1H, m) | 2.39, 2.18 (each 1H, m) |
| 3 | 3.38 (1H, t, 3) | - | - | - |
| 4 | - | - | - | - |
| 5 | 1.24 (1H, m) | 1.37 (1H, m) | 1.32 (1H, m) | - |
| 6 | 1.39 (2H, m) | 1.35, 1.50 (each 1H, m) | 1.49, 1.40 (each 1H, m) | 1.55 (2H, m) |
| 7 | 1.63 (2H, m) | 1.31, 1.63 (each 1H, m) | 1.50, 1.26 (each 1H, m) | 1.35 (2H, m) |
| 8 | - | - | - | - |
| 9 | 1.44 (1H, m) | 1.40 (1H, m) | 1.36 (1H, m) | 1.45 (1H, m) |
| 10 | - | - | - | - |
| 11 | 1.53 (2H, m) | 1.40 (2H, m) | 1.45, 1.25 (each 1H, m) | 1.41 (2H, m) |
| 12 | 1.75 (2H, m) | 1.55, 1.84 (each 1H, m) | 1.79, 1.23 (each 1H, m) | 1.89 (2H, m) |
| 13 | 1.62 (1H, m) | 1.67 (1H, m) | 1.60 (1H, m) | 1.62 (1H, m) |
| 14 | - | - | - | - |
| 15 | 1.04 (2H, m) | 1.10, 1.57 (each 1H, m) | 1.40, 1.03 (each 1H, m) | 1.43 (2H, m) |
| 16 | 1.51 (2H, m) | 1.89 (2H, m) | 1.69, 1.44 (each 1H, m) | 1.78 (2H, m) |
| 17 | 1.44 (1H, m) | 2.18 (1H, m) | 1.68 (1H, m) | 1.81 (1H, m) |
| 18 | 0.84 (3H, s) | 0.86 (3H, s) | 0.93 (3H, s) | 0.89 (3H, s) |
| 19 | 0.87 (3H, s) | 0.92 (3H, s) | 0.88 (3H, s) | 0.86 (3H, s) |
| 20 | - | - | - | - |
| 21 | 1.13 (3H, s) | 4.69, 4.72 (each 1H, br.s) | 1.08 (3H, s) | 1.15 (3H, s) |
| 22 | 1.22 (2H, m) | 1.95 (2H, m) | 1.42 (2H, m) | 1.54 (2H, m) |
| 23 | 1.85 (2H, m) | 2.10 (2H, q, 7.5) | 1.99 (2H, m) | 1.86 (2H, m) |
| 24 | 3.62 (1H, dd, 4.8, 10.2) | 5.11 (1H, dd, 7.0, 1.5) | 5.05 (1H, t, 5.0) | 3.64 (1H, dd, 9.5, 5.5) |
| 25 | - | - | - | - |
| 26 | 1.17 (3H, s) | 1.60 (3H, s) | 1.62 (3H, s) | 1.20 (3H, s) |
| 27 | 1.09 (3H, s) | 1.67 (3H, s) | 1.56 (3H, s) | 1.12 (3H, s) |
| 28 | 0.92 (3H, s) | 1.02 (3H, s) | 1.01 (3H, s) | 4.66, 4.85 (each 1H, br.s) |
| 29 | 0.82 (3H, s) | 1.06 (3H, s) | 0.97 (3H, s) | 1.73 (3H, s) |
| 30 | 0.95 (3H, s) | 0.99 (3H, s) | 0.82 (3H, s) | 1.02 (3H, s) |
| Position | 5 | 6 | 7 |
|---|---|---|---|
| 1 | 1.92, 1.45 (each 1H, m) | 1.31, 1.36 (each 1H, m) | 1.40, 1.45 (each 1H, m) |
| 2 | 1.87, 2.05 (each 1H, m) | 1.57, 1.92 (each 1H, m) | 1.61 (2H, m) |
| 3 | - | 4.60 (1H, br.s) | 4.61 (1H, t, 3.5) |
| 4 | - | - | - |
| 5 | 1.36 (1H, m) | 1.26 (1H, m) | 1.43 (1H, m) |
| 6 | 1.44, 1.55 (each 1H, m) | 1.41 (1H, m) | 1.37 (2H, m) |
| 7 | 1.89, 1.54 (each 1H, m) | 1.21, 1.52 (each 1H, m) | 1.65, 1.85 (each 1H, m) |
| 8 | - | - | - |
| 9 | 1.39 (1H, m) | 1.42 (1H, m) | 1.21 (1H, m) |
| 10 | - | - | - |
| 11 | 1.79 (2H, m) | 1.75, 1.19 (each 1H, m) | 1.54 (2H, m) |
| 12 | 1.23, 1.47 (each 1H, m) | 1.13, 1.50 (each 1H, m) | 1.78 (2H, m) |
| 13 | 1.58 (1H, m) | 1.55 (1H, m) | 1.58 (1H, m) |
| 14 | - | - | - |
| 15 | 1.46 (2H, m) | 1.11, 1.48 (each 1H, m) | 1.06, 1.42 (each 1H, m) |
| 16 | 1.31 (each 1H, m) | 1.39, 1.82 (each 1H, m) | 1.85 (2H, m) |
| 17 | 1.85 (1H, m) | 1.98 (1H, m) | 1.88 (1H, m) |
| 18 | 0.99 (3H, s) | 0.95 (3H, s) | 0.96 (3H, s) |
| 19 | 0.92 (3H, s) | 0.84 (3H, s) | 0.85 (3H, s) |
| 20 | - | - | - |
| 21 | 1.12 (3H, s) | 1.36 (3H, s) | 1.14 (3H, s) |
| 22 | 1.88, 1.82 (each 1H, m) | 1.94 2.10 (each 1H, m) | 1.24 (2H, m) |
| 23 | 2.40, 2.50 (each 1H, m) | 2.53 2.63 (each 1H, m) | 1.75 (2H, m) |
| 24 | 4.91 (1H, br.s) | - | 3.63 (1H, dd, 4.7, 10.0) |
| 25 | - | - | - |
| 26 | - | - | 1.18 (3H, s) |
| 27 | - | - | 1.10 (3H, s) |
| 28 | 1.07 (3H, s) | 0.91 (3H, s) | 0.82 (3H, s) |
| 29 | 1.03 (3H, s) | 0.82 (3H, s) | 0.87 (3H, s) |
| 30 | 0.87 (3H, s) | 0.86 (3H, s) | 0.91 (3H, s) |
| 1′ | 3.30 (3H, s) | - | - |
| 2′ | 2.07 (3H, s) | 2.08 (3H, s) |
| Compounds | IC50 for MCF-7 (μg/mL) | IC50 for MCF-7(μM) | IC50 for B16-F10 (μg/mL) | IC50 for B16-F10 (μM) | IC50 for CV-1 (μg/mL) | IC50 for CV-1 (μM) |
|---|---|---|---|---|---|---|
| 20S,24S-epoxy-3α,25-dihydroxy-dammarane (1) | 65.54 | 142.25 | 44.84 | 97.33 | 80.08 | 173.82 |
| dammaradienone (2) | 56.15 | 132.21 | 54.77 | 128.96 | 82.85 | 195,07 |
| 20S-hydroxy-dammar-24-en-3-on (3) | 142.00 | >300 | 49.08 | 110.86 | >300 | >300 |
| eichlerianic acid (4) | >300 | >300 | 275.40 | >300 | 136.60 | 287.76 |
| (20S,24R,S)-23,24-epoxy-24-methoxy-25,26,27-tris-nordammar-3-one (5) | 57.34 | 133.13 | 45.36 | 105.32 | >300 | >300 |
| 3α-acetyl-cabraleahydroxy lactone (6) | 125.89 | 274.45 | 125.89 | 274.45 | 130.70 | 285.00 |
| 3α-acetyl-20S,24S-epoxy-3α,25-dihydroxydammarane (7) | 97.91 | 194.73 | 79.43 | 157.97 | >300 | >300 |
| Cisplatin (positive control) | 53.00 | 176.02 | 43.00 | 142.80 | 43.00 | 142.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Purnama; Farabi, K.; Runadi, D.; Kuncoro, H.; Harneti, D.; Nurlelasari; Mayanti, T.; Azmi, M.N.; Fajriah, S.; Supratman, U. The Cytotoxic Activity of Dammarane-Type Triterpenoids Isolated from the Stem Bark of Aglaia cucullata (Meliaceae). Molecules 2023, 28, 4946. https://doi.org/10.3390/molecules28134946
Purnama, Farabi K, Runadi D, Kuncoro H, Harneti D, Nurlelasari, Mayanti T, Azmi MN, Fajriah S, Supratman U. The Cytotoxic Activity of Dammarane-Type Triterpenoids Isolated from the Stem Bark of Aglaia cucullata (Meliaceae). Molecules. 2023; 28(13):4946. https://doi.org/10.3390/molecules28134946
Chicago/Turabian StylePurnama, Kindi Farabi, Dudi Runadi, Hadi Kuncoro, Desi Harneti, Nurlelasari, Tri Mayanti, Mohamad Nurul Azmi, Sofa Fajriah, and Unang Supratman. 2023. "The Cytotoxic Activity of Dammarane-Type Triterpenoids Isolated from the Stem Bark of Aglaia cucullata (Meliaceae)" Molecules 28, no. 13: 4946. https://doi.org/10.3390/molecules28134946
APA StylePurnama, Farabi, K., Runadi, D., Kuncoro, H., Harneti, D., Nurlelasari, Mayanti, T., Azmi, M. N., Fajriah, S., & Supratman, U. (2023). The Cytotoxic Activity of Dammarane-Type Triterpenoids Isolated from the Stem Bark of Aglaia cucullata (Meliaceae). Molecules, 28(13), 4946. https://doi.org/10.3390/molecules28134946