Protein Interactions in Rhodopseudomonas palustris TIE-1 Reveal the Molecular Basis for Resilient Photoferrotrophic Iron Oxidation
Abstract
1. Introduction
2. Results and Discussion
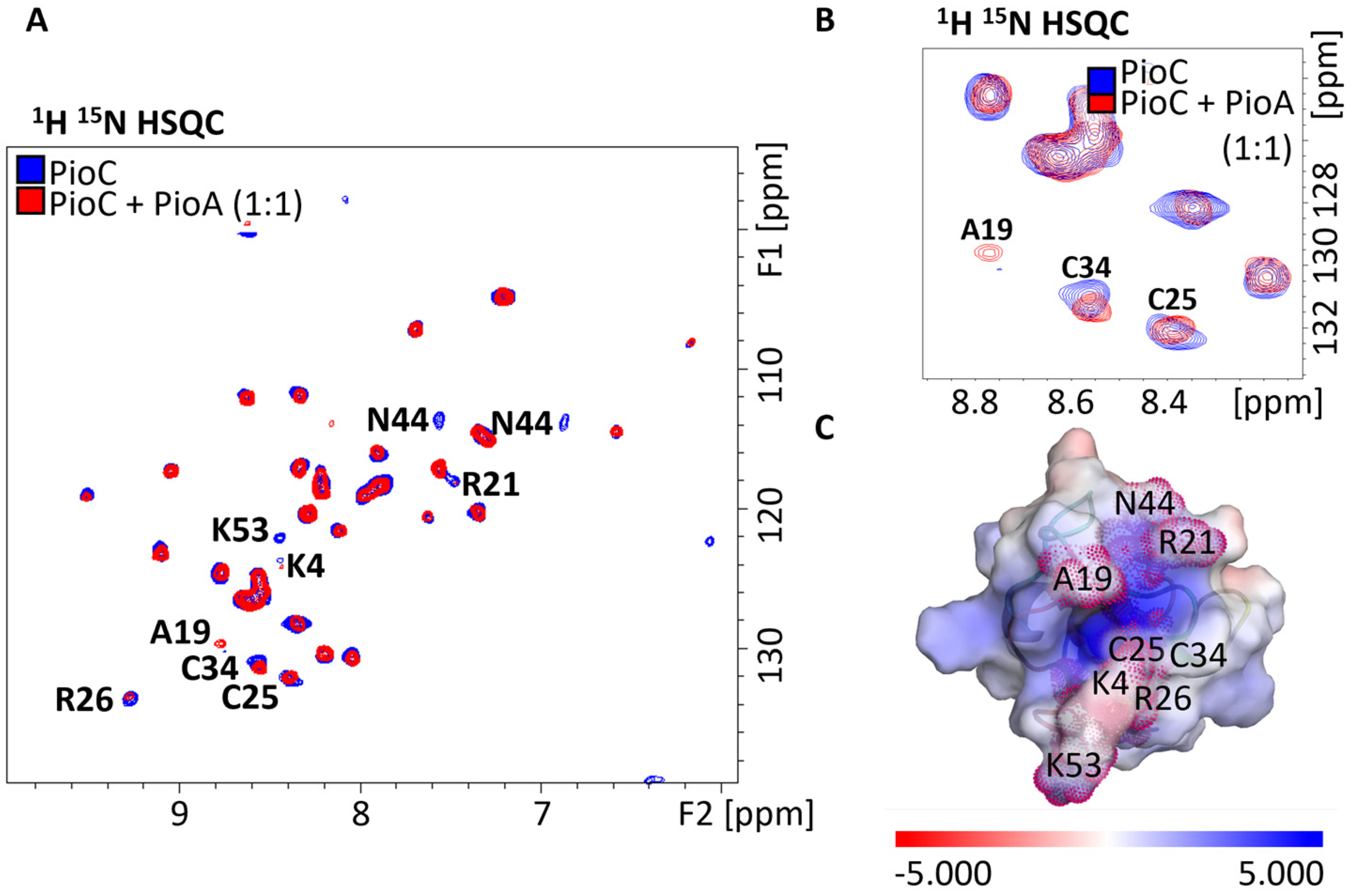
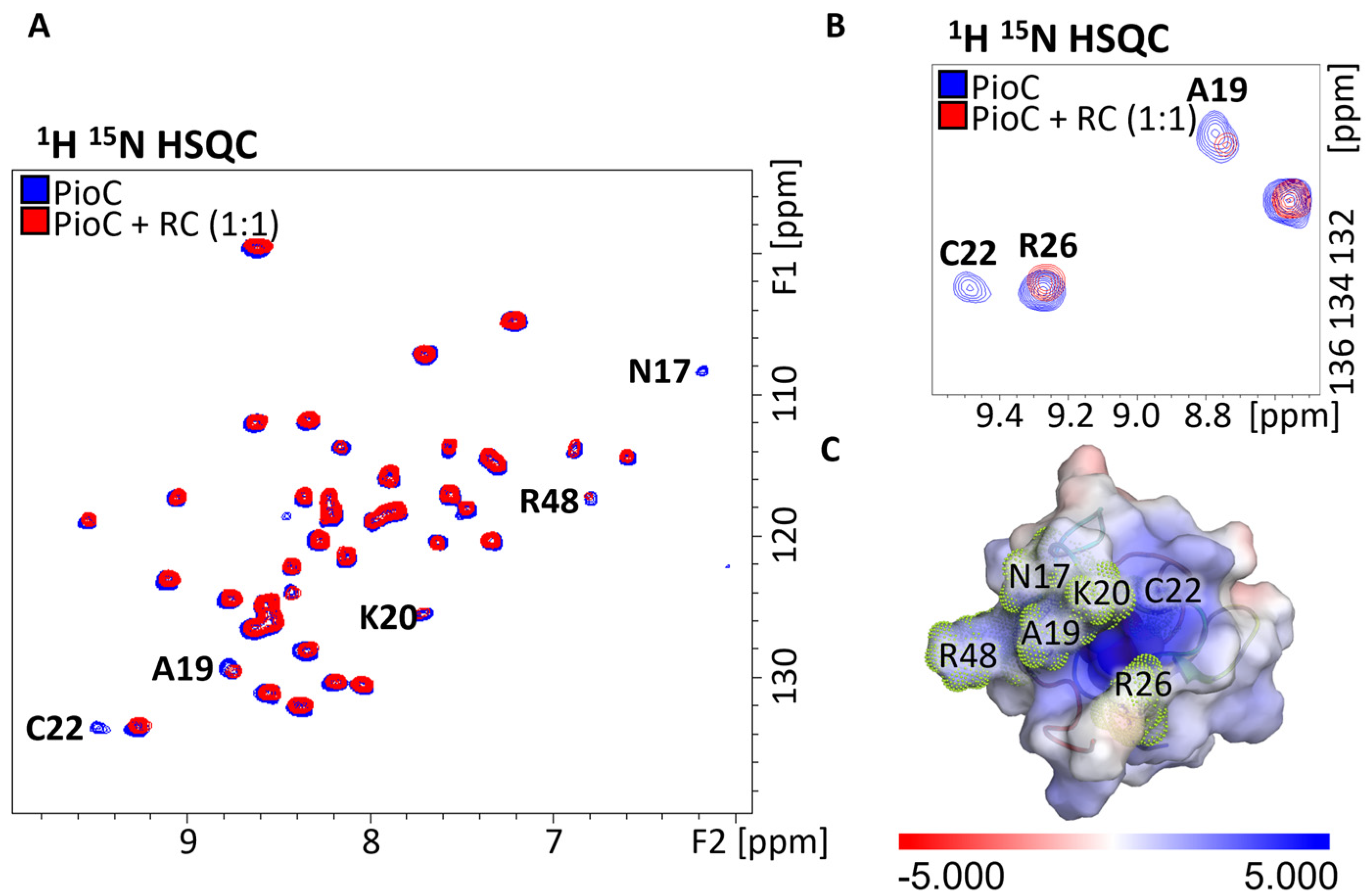
2.1. Probing Photoferrotrophic Iron Oxidation Interactions Using NMR Spectroscopy
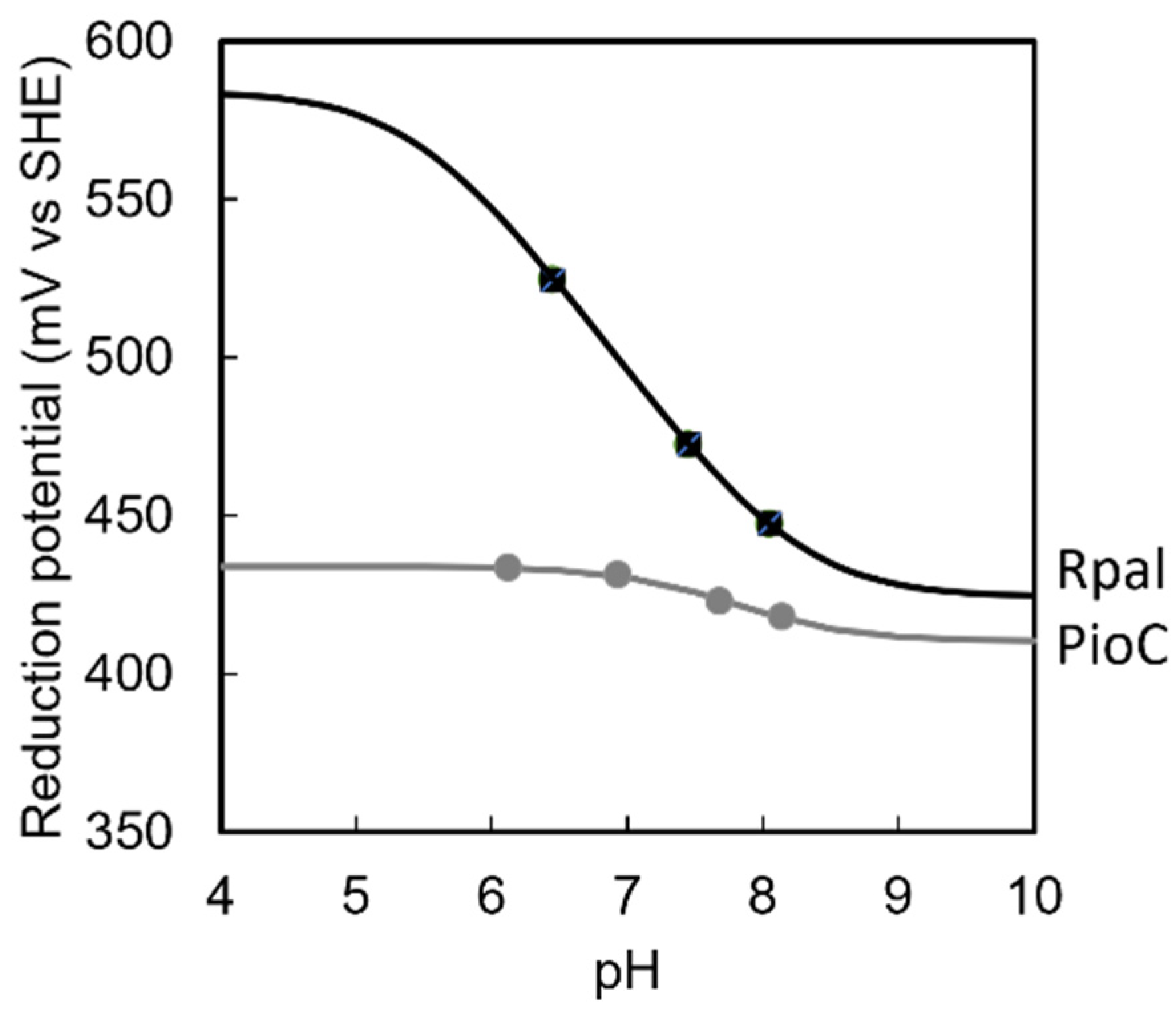
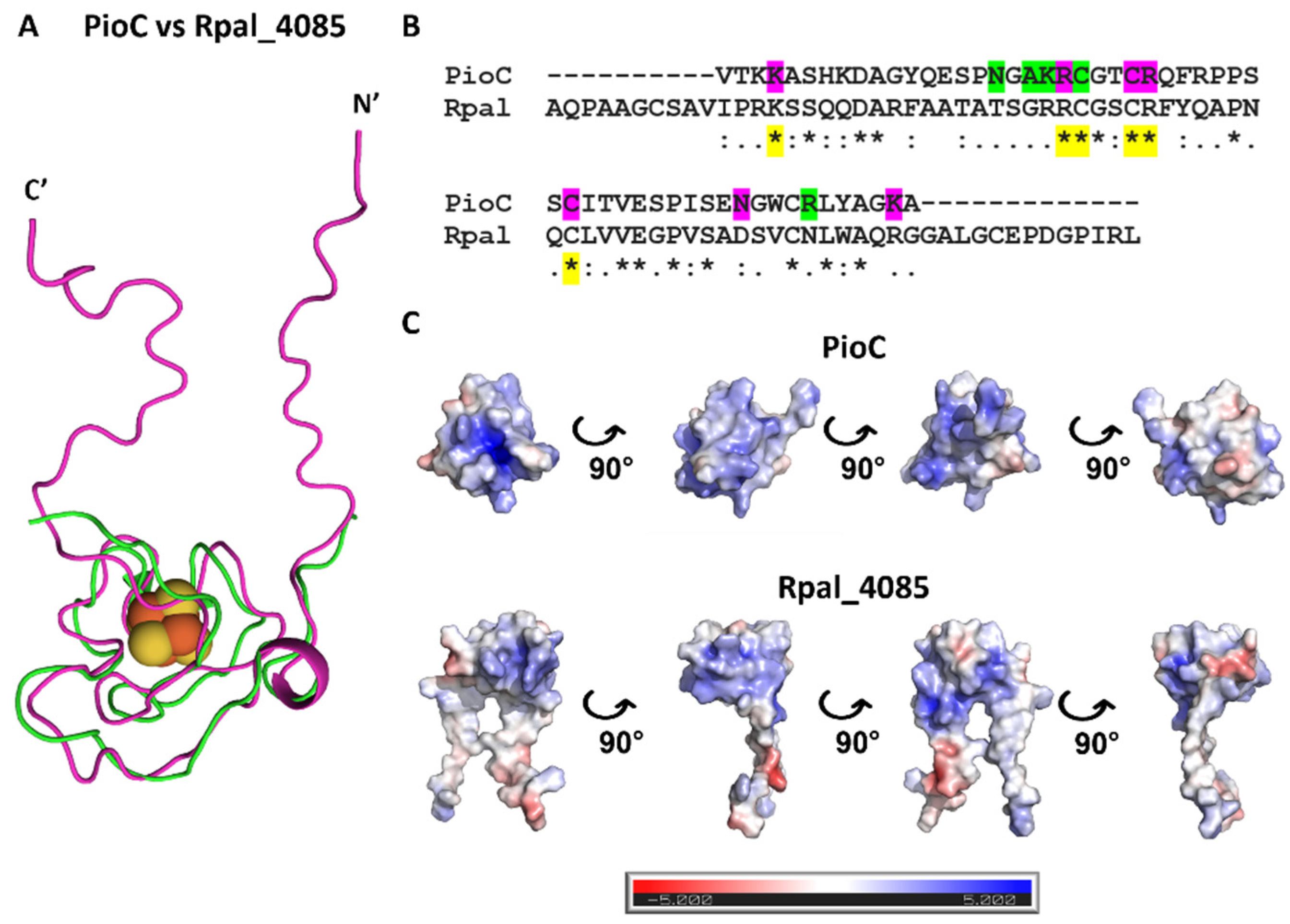
2.2. Rpal_4085—Why Is It Unable to Substitute PioC?
2.3. PioA: A Direct and Versatile Electron Transfer Pathway to the LH-RC
3. Materials and Methods
3.1. Expression and Purification of 15N-PioC
3.2. Expression and Purification of the LH-RC
3.3. Expression and Purification of PioA
3.4. Expression and Purification of Rpal_4085
3.5. Protein Film Voltammetry of PioC and Rpal_4085
3.6. 1H NMR Temperature Dependence Experiments
3.7. 15N PioC: NMR Binding Experiments
3.8. Reduction of LH-RC by PioA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Konhauser, K.O.; Newman, D.K.; Kappler, A. The Potential Significance of Microbial Fe(III) Reduction during Deposition of Precambrian Banded Iron Formations. Geobiology 2005, 3, 167–177. [Google Scholar] [CrossRef]
- Camacho, A.; Walter, X.A.; Picazo, A.; Zopfi, J. Photoferrotrophy: Remains of an Ancient Photosynthesis in Modern Environments. Front. Microbiol. 2017, 8, 323. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.S.; Rengasamy, K.; Binkley, M.M.; Jones, C.; Ranaivoarisoa, T.O.; Singh, R.; Fike, D.A.; Meacham, J.M.; Bose, A. Phototrophic Extracellular Electron Uptake Is Linked to Carbon Dioxide Fixation in the Bacterium Rhodopseudomonas palustris. Nat. Commun. 2019, 10, 1355. [Google Scholar] [CrossRef]
- Ilbert, M.; Bonnefoy, V. Insight into the Evolution of the Iron Oxidation Pathways. Biochim. Biophys. Acta 2013, 1827, 161–175. [Google Scholar] [CrossRef]
- Saraiva, I.H.; Newman, D.K.; Louro, R.O. Functional Characterization of the FoxE Iron Oxidoreductase from the Photoferrotroph Rhodobacter Ferrooxidans SW2. J. Biol. Chem. 2012, 287, 25541–25548. [Google Scholar] [CrossRef] [PubMed]
- Schink, B. Extracellular Redox Chemistry. In Metals, Microbes, and Minerals—The Biogeochemical Side of Life; De Gruyter: Berlin, Germany, 2021; pp. 33–58. [Google Scholar]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Lu, A.; Liu, J.; Yu, H.-Q.; Fredrickson, J.K. Extracellular Electron Transfer Mechanisms between Microorganisms and Minerals. Nat. Rev. Microbiol. 2016, 14, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.J.; Butt, J.N.; Fredrickson, J.K.; Zachara, J.M.; Shi, L.; Edwards, M.J.; White, G.; Baiden, N.; Gates, A.J.; Marritt, S.J.; et al. The “porin-Cytochrome” Model for Microbe-to-Mineral Electron Transfer. Mol. Microbiol. 2012, 85, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Edwards, M.J.; Richardson, D.J.; Paquete, C.M.; Clarke, T.A. Role of Multiheme Cytochromes Involved in Extracellular Anaerobic Respiration in Bacteria. Protein Sci. 2020, 29, 830–842. [Google Scholar] [CrossRef]
- Costa, N.L.; Clarke, T.A.; Philipp, L.A.; Gescher, J.; Louro, R.O.; Paquete, C.M. Electron Transfer Process in Microbial Electrochemical Technologies: The Role of Cell-Surface Exposed Conductive Proteins. Bioresour. Technol. 2018, 255, 308–317. [Google Scholar] [CrossRef]
- Faustino, M.M.; Fonseca, B.M.; Costa, N.L.; Lousa, D.; Louro, R.O.; Paquete, C.M. Crossing the Wall: Characterization of the Multiheme Cytochromes Involved in the Extracellular Electron Transfer Pathway of Thermincola Ferriacetica. Microorganisms 2021, 9, 293. [Google Scholar] [CrossRef]
- Louro, R.O.; Costa, N.L.; Fernandes, A.P.; Silva, A.V.; Trindade, I.B.; Fonseca, B.M.; Paquete, C.M. Exploring the Molecular Mechanisms of Extracellular Electron Transfer for Harnessing Reducing Power in METs: Methodologies and Approaches. In Biomass, Biofuels, Biochemicals: Microbial Electrochemical Technology: Sustainable Platform for Fuels, Chemicals and Remediation; Elsevier: Amsterdam, The Netherlands, 2018; pp. 261–293. ISBN 9780444640529. [Google Scholar]
- Jiao, Y.; Newman, D.K. The Pio Operon Is Essential for Phototrophic Fe(II) Oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 2007, 189, 1765–1773. [Google Scholar] [CrossRef]
- Bird, L.J.; Saraiva, I.H.; Park, S.; Calcada, E.O.; Salgueiro, C.A.; Nitschke, W.; Louro, R.O.; Newman, D.K. Nonredundant Roles for Cytochrome C2 and Two High-Potential Iron-Sulfur Proteins in the Photoferrotroph Rhodopseudomonas palustris TIE-1. J. Bacteriol. 2014, 196, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Sutherland, M.C.; Rengasamy, K.; Meacham, J.M.; Kranz, R.G.; Bose, A. Photoferrotrophs Produce a PioAB Electron Conduit for Extracellular Electron Uptake. mBio 2019, 10, e02668-19. [Google Scholar] [CrossRef] [PubMed]
- Doud, D.F.R.; Holmes, E.C.; Richter, H.; Molitor, B.; Jander, G.; Angenent, L.T. Metabolic Engineering of Rhodopseudomonas palustris for the Obligate Reduction of N-Butyrate to n-Butanol. Biotechnol. Biofuels 2017, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.; Wilkins, M.; Saha, R. Rhodopseudomonas palustris: A Biotechnology Chassis. Biotechnol. Adv. 2022, 60, 108001. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ning, P.; Sun, Y.; Luo, J.; Yang, J. Characteristics and Application of Rhodopseudomonas palustris as a Microbial Cell Factory. Front. Bioeng. Biotechnol. 2022, 10, 897003. [Google Scholar] [CrossRef]
- Harwood, C.S. Rhodopseudomonas palustris. Trends Microbiol. 2022, 30, 307–308. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gao, D.; Geng, H.; Xing, C. Enhancing Hydrogen Production by Photobiocatalysis through Rhodopseudomonas palustris coupled with Conjugated Polymers. J. Mater. Chem. A 2021, 9, 19788–19795. [Google Scholar] [CrossRef]
- Jiao, Y.; Kappler, A.; Croal, L.R.; Newman, K.; Newman, D.K. Isolation and Characterization of a Genetically Tractable Photoautotrophic Isolation and Characterization of a Genetically Tractable Photoautotrophic Fe(II)-Oxidizing Bacterium, Rhodopseudomonas palustris Strain TIE-1. Appl. Environ. Microbiol. 2005, 71, 4487–4496. [Google Scholar] [CrossRef]
- Bose, A.; Gardel, E.J.; Vidoudez, C.; Parra, E.A.; Girguis, P.R. Electron Uptake by Iron-Oxidizing Phototrophic Bacteria. Nat. Commun. 2014, 5, 3391. [Google Scholar] [CrossRef]
- Bax, A.; Clore, G.M. Protein NMR: Boundless Opportunities. J. Magn. Reson. 2019, 306, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Purslow, J.A.; Khatiwada, B.; Bayro, M.J.; Venditti, V. NMR Methods for Structural Characterization of Protein-Protein Complexes. Front. Mol. Biosci. 2020, 7, 9. [Google Scholar] [CrossRef]
- Trindade, I.B.; Invernici, M.; Cantini, F.; Louro, R.O.; Piccioli, M. 1H, 13C and 15N Assignment of the Paramagnetic High Potential Iron–Sulfur Protein (HiPIP) PioC from Rhodopseudomonas palustris TIE-1. Biomol. NMR Assign 2020, 14, 211–215. [Google Scholar] [CrossRef]
- Trindade, I.B.; Invernici, M.; Cantini, F.; Louro, R.O.; Piccioli, M. Sequence-Specific Assignments in NMR Spectra of Paramagnetic Systems: A Non-Systematic Approach. Inorg. Chim. Acta 2021, 514, 119984. [Google Scholar] [CrossRef]
- Konecny, R.; Baker, N.A.; McCammon, J.A. IAPBS: A Programming Interface to the Adaptive Poisson-Boltzmann Solver. Comput. Sci. Discov. 2012, 5, 015005. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; Version 2.3; Schrödinger LLC: New York, NY, USA, 2020. [Google Scholar]
- Bertini, I.; Briganti, F.; Luchinat, C.; Scozzafava, A.; Sola, M. Proton NMR Spectroscopy and the Electronic Structure of the High Potential Iron-Sulfur Protein from Chromatium Vinosum. J. Am. Chem. Soc. 1991, 113, 1237–1245. [Google Scholar] [CrossRef]
- Poe, M.; Phillips, W.D.; Mcdonald, C.C.; Lovenberg, W. Proton Magnetic Resonance Study of Ferredoxin from Clostridium Pasteurianum. Proc. Natl. Acad. Sci. USA 1970, 65, 797–804. [Google Scholar] [CrossRef]
- Trindade, I.B.; Coelho, A.; Cantini, F.; Piccioli, M.; Louro, R.O. NMR of Paramagnetic Metalloproteins in Solution: Ubi Venire, Quo Vadis? J. Inorg. Biochem. 2022, 234, 111871. [Google Scholar] [CrossRef]
- Bertini, I.; Dikiy, A.; Luchinat, C.; Macinai, R.; Viezzoli, M.S.; Vincenzini, M. An NMR Study of the 7Fe-8S Ferredoxin from Rhodopseudomonas palustris and Reinterpretation of Data on Similar Systems. Biochemistry 1997, 36, 3570–3579. [Google Scholar] [CrossRef]
- Louro, R.O.; Catarino, T.; Salgueiro, C.A.; LeGall, J.; Xavier, A.V. Redox-Bohr Effect in the Tetrahaem Cytochrome C3 from Desulfovibrio Vulgaris: A Model for Energy Transduction Mechanisms. J. Biol. Inorg. Chem. 1996, 1, 34–38. [Google Scholar] [CrossRef]
- Lloyd, D.R.; Phillips, D.H. Oxidative DNA Damage Mediated by Copper II, Iron II and ž / Nickel II Fenton Reactions: Evidence for Site-Specific Mechanisms in the Formation of Double-Strand Breaks, 8-Hydroxydeoxyguanosine and Putative Intrastrand Cross-Links. Mutat. Res. 1999, 424, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Trindade, I.B.; Invernici, M.; Cantini, F.; Louro, R.O.; Piccioli, M. PRE-Driven Protein NMR Structures: An Alternative Approach in Highly Paramagnetic Systems. FEBS J. 2021, 288, 3010–3023. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pitts, K.E.; Dobbin, P.S.; Reyes-Ramirez, F.; Thomson, A.J.; Richardson, D.J.; Seward, H.E. Characterization of the Shewanella Oneidensis MR-1 Decaheme Cytochrome MtrA: Expression in Escherichia Coli Confers the Ability to Reduce Soluble FE(III) Chelates. J. Biol. Chem. 2003, 278, 27758–27765. [Google Scholar] [CrossRef] [PubMed]
- Li, D.B.; Edwards, M.J.; Blake, A.W.; Newton-Payne, S.E.; Piper, S.E.H.; Jenner, L.P.; Sokol, K.P.; Reisner, E.; Van Wonderen, J.H.; Clarke, T.A.; et al. His/Met Heme Ligation in the PioA Outer Membrane Cytochrome Enabling Light-Driven Extracellular Electron Transfer by Rhodopseudomonas palustris TIE-1. Nanotechnology 2020, 31, 354002. [Google Scholar] [CrossRef] [PubMed]
- Antonkine, M.L.; Koay, M.S.; Epel, B.; Breitenstein, C.; Gopta, O.; Gärtner, W.; Bill, E.; Lubitz, W. Synthesis and Characterization of de Novo Designed Peptides Modelling the Binding Sites of [4Fe-4S] Clusters in Photosystem I. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 995–1008. [Google Scholar] [CrossRef]
- Saga, Y.; Hirota, K. Determination of the Molar Extinction Coefficients of the B800 and B850 Absorption Bands in Light-Harvesting Complexes Derived from Three Purple Photosynthetic Bacteria Rhodoblastus Acidophilus, Rhodobacter Sphaeroides, and Phaeospirillum Molischianum by Extraction of Bacteriochlorophyll a. Anal. Sci. 2016, 32, 801–805. [Google Scholar] [CrossRef]
- Francis, R.T.; And, J.R.; Becker, R.R. Specific Indication of Hemoproteins in Polyacrylamide Gels Using a Double-Staining Process. Anal. Biochem. 1984, 136, 509–514. [Google Scholar] [CrossRef]
- Massey, V. The Microestimation of Succinate and the Extinction Coefficient of Cytochrome c. Biochim. Biophys. Acta Bioenerg. 1959, 34, 255–256. [Google Scholar] [CrossRef]
- Friis, E.P.; Andersen, J.E.T.; Madsen, L.L.; Bonander, N.; Moller, P.; Ulstrup, J. Dynamics of Pseudomonas Aeruginosa Azurin and Its Cys3Ser Mutant at Single-Crystal Gold Surfaces Investigated by Cyclic Voltammetry and Atomic Force Microscopy. Electrochim. Acta 1998, 43, 1114–1122. [Google Scholar] [CrossRef]
- Ciofi-Baffoni, S.; Gallo, A.; Muzzioli, R.; Piccioli, M. The IR-15N-HSQC-AP Experiment: A New Tool for NMR Spectroscopy of Paramagnetic Molecules. J. Biomol. NMR 2014, 58, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Yousafzai, F.K.; Eady, R.R. Dithionite Reduction Kinetics of the Dissimilatory Copper-Containing Nitrite Reductase of Alcalegenes Xylosoxidans. The SO2.- Radical Binds to the Substrate Binding Type 2 Copper Site before the Type 2 Copper Is Reduced. J. Biol. Chem. 2002, 277, 34067–34073. [Google Scholar] [CrossRef] [PubMed]
- Verissimo, A.F.; Daldal, F. Cytochrome c Biogenesis System I: An Intricate Process Catalyzed by a Maturase Supercomplex? Biochim. Biophys. Acta Bioenerg. 2014, 1837, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Jackson, P.J.; Hitchcock, A.; Swainsbury, D.J.K.; Qian, P.; Martin, E.C.; Farmer, D.A.; Dickman, M.J.; Canniffe, D.P.; Hunter, C.N. Identification of Protein W, the Elusive Sixth Subunit of the Rhodopseudomonas palustris Reaction Center-Light Harvesting 1 Core Complex. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 119–128. [Google Scholar] [CrossRef]
- Nelson, K.E.; Fraser, C.M. Champions of Versatility. Trends Microbiol. 2004, 12, 106–111. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trindade, I.B.; Firmino, M.O.; Noordam, S.J.; Alves, A.S.; Fonseca, B.M.; Piccioli, M.; Louro, R.O. Protein Interactions in Rhodopseudomonas palustris TIE-1 Reveal the Molecular Basis for Resilient Photoferrotrophic Iron Oxidation. Molecules 2023, 28, 4733. https://doi.org/10.3390/molecules28124733
Trindade IB, Firmino MO, Noordam SJ, Alves AS, Fonseca BM, Piccioli M, Louro RO. Protein Interactions in Rhodopseudomonas palustris TIE-1 Reveal the Molecular Basis for Resilient Photoferrotrophic Iron Oxidation. Molecules. 2023; 28(12):4733. https://doi.org/10.3390/molecules28124733
Chicago/Turabian StyleTrindade, Inês B., Maria O. Firmino, Sander J. Noordam, Alexandra S. Alves, Bruno M. Fonseca, Mario Piccioli, and Ricardo O. Louro. 2023. "Protein Interactions in Rhodopseudomonas palustris TIE-1 Reveal the Molecular Basis for Resilient Photoferrotrophic Iron Oxidation" Molecules 28, no. 12: 4733. https://doi.org/10.3390/molecules28124733
APA StyleTrindade, I. B., Firmino, M. O., Noordam, S. J., Alves, A. S., Fonseca, B. M., Piccioli, M., & Louro, R. O. (2023). Protein Interactions in Rhodopseudomonas palustris TIE-1 Reveal the Molecular Basis for Resilient Photoferrotrophic Iron Oxidation. Molecules, 28(12), 4733. https://doi.org/10.3390/molecules28124733






