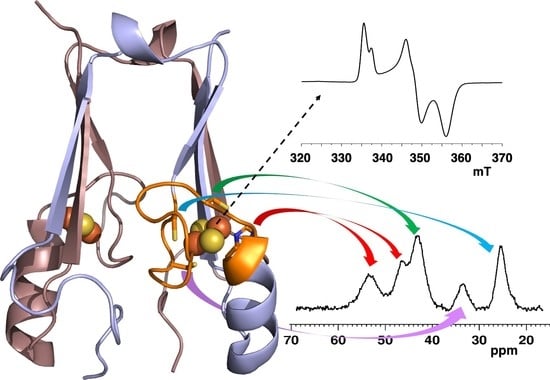
The Intriguing mitoNEET: Functional and Spectroscopic Properties of a Unique [2Fe-2S] Cluster Coordination Geometry
Abstract
1. Introduction
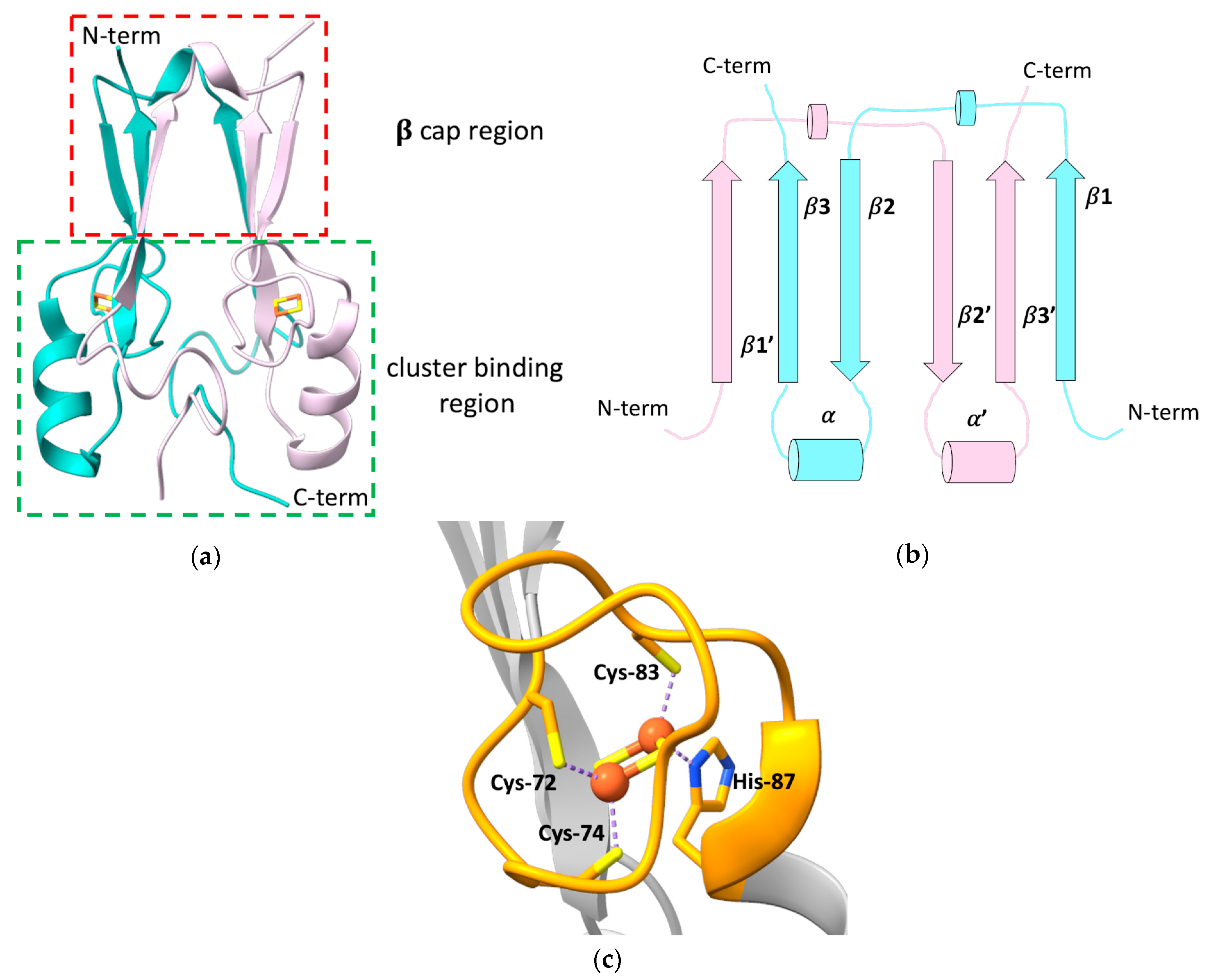
2. The Peculiar Properties of Human mitoNEET: A Unique Folding for a Multiplicity of Functions
3. Spectroscopic Characterization of the Reduced and Oxidized Forms of mitoNEET
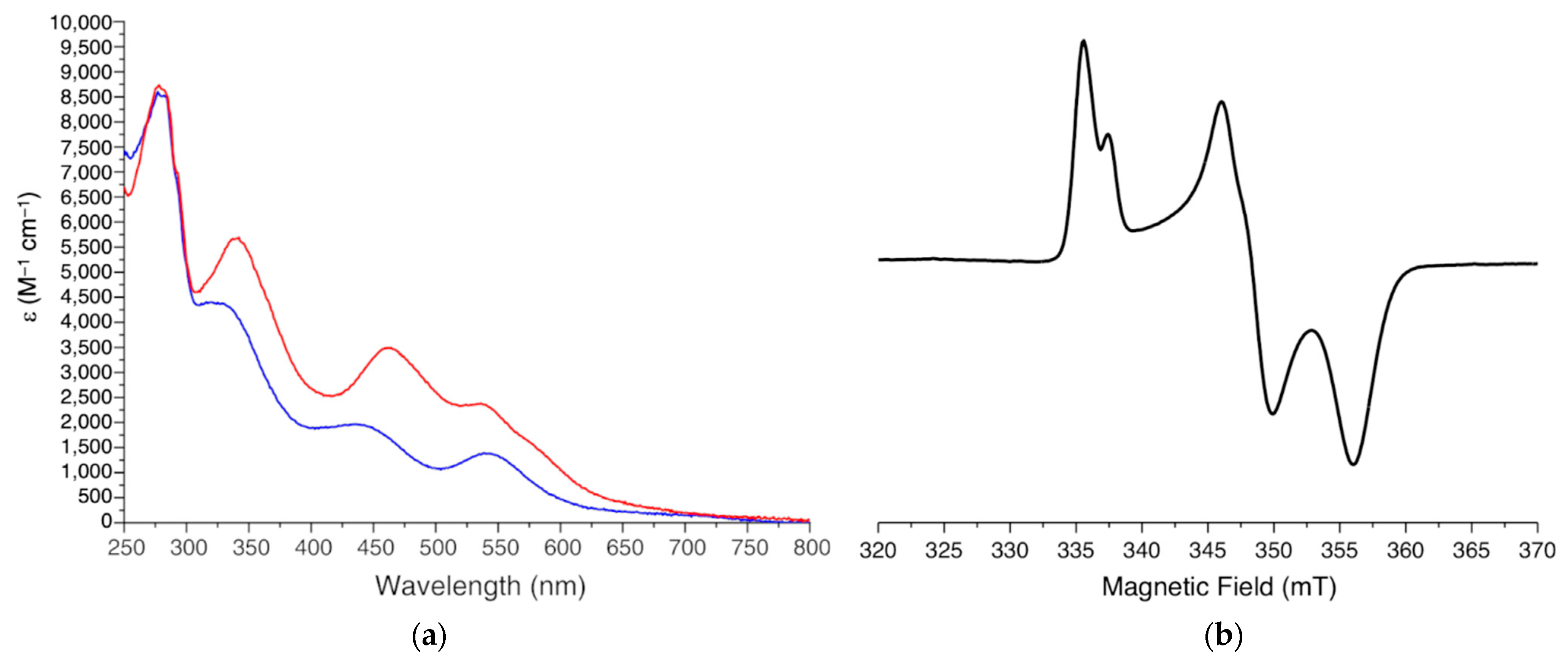
3.1. Electronic Spectroscopy
3.2. EPR Spectroscopy
3.3. Mössbauer Spectroscopy
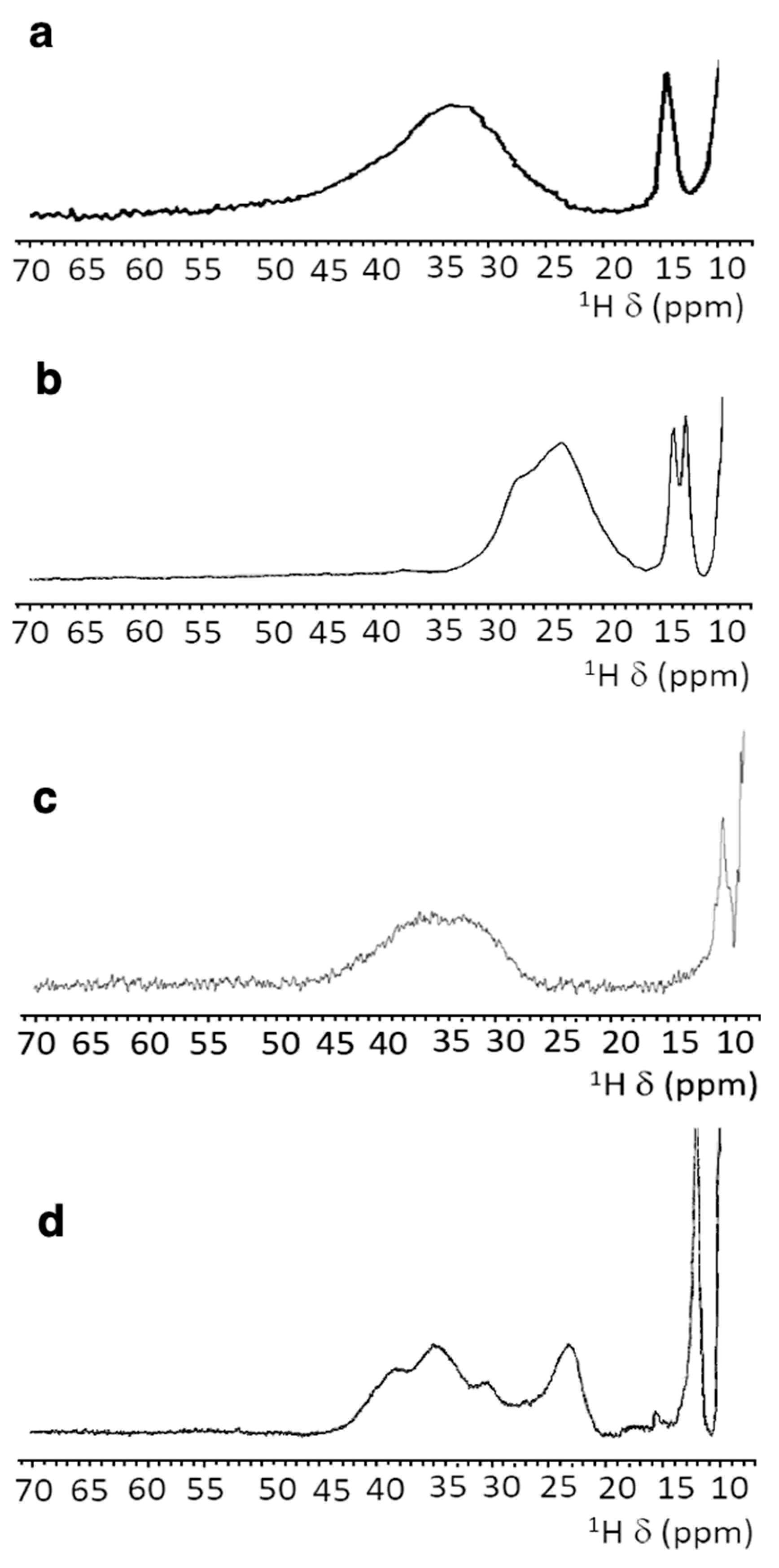
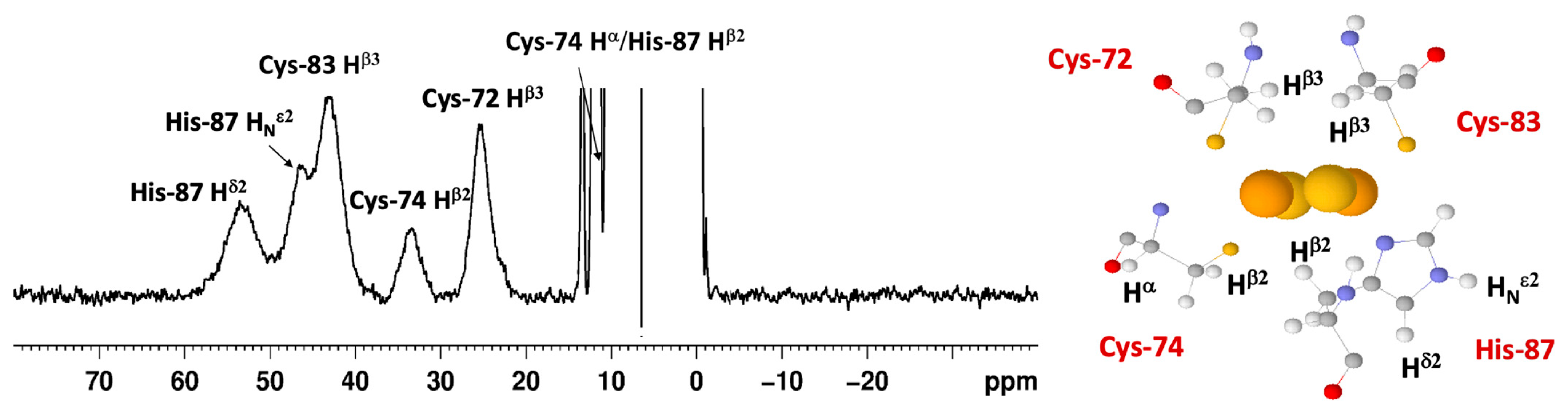
3.4. NMR Spectroscopy
3.5. Paramagnetic NMR and Antiferromagnetic Coupling Properties
4. Hints for Future Studies: Targeting mitoNEET to Fight Cancer
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beinert, H. Iron-Sulfur Proteins: Ancient Structures, Still Full of Surprises. J. Biol. Inorg. Chem. 2000, 5, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Lill, R. Function and Biogenesis of Iron-Sulphur Proteins. Nature 2009, 460, 831–838. [Google Scholar] [CrossRef]
- Rouault, T.A. The Indispensable Role of Mammalian Iron Sulfur Proteins in Function and Regulation of Multiple Diverse Metabolic Pathways. Biometals 2019, 32, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, Function, and Formation of Biological Iron-Sulfur Clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef] [PubMed]
- Golinelli-Cohen, M.-P.; Bouton, C. Fe-S Proteins Acting as Redox Switch: New Key Actors of Cellular Adaptive Responses. Curr. Chem. Biol. 2017, 11, 70–88. [Google Scholar] [CrossRef]
- Fuss, J.O.; Tsai, C.-L.; Ishida, J.P.; Tainer, J.A. Emerging Critical Roles of Fe-S Clusters in DNA Replication and Repair. Biochim. Biophys. Acta 2015, 1853, 1253–1271. [Google Scholar] [CrossRef]
- Colca, J.R.; McDonald, W.G.; Waldon, D.J.; Leone, J.W.; Lull, J.M.; Bannow, C.A.; Lund, E.T.; Mathews, W.R. Identification of a Novel Mitochondrial Protein (“mitoNEET”) Cross-Linked Specifically by a Thiazolidinedione Photoprobe. Am. J. Physiol. Endocrinol. Metab. 2004, 286, E252–E260. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, L.; Lai, S.; Ye, K. Structure and Molecular Evolution of CDGSH Iron-Sulfur Domains. PLoS ONE 2011, 6, e24790. [Google Scholar] [CrossRef]
- Mittler, R.; Darash-Yahana, M.; Sohn, Y.S.; Bai, F.; Song, L.; Cabantchik, I.Z.; Jennings, P.A.; Onuchic, J.N.; Nechushtai, R. NEET Proteins: A New Link Between Iron Metabolism, Reactive Oxygen Species, and Cancer. Antioxid. Redox Signal. 2019, 30, 1083–1095. [Google Scholar] [CrossRef]
- Nechushtai, R.; Karmi, O.; Zuo, K.; Marjault, H.-B.; Darash-Yahana, M.; Sohn, Y.-S.; King, S.D.; Zandalinas, S.I.; Carloni, P.; Mittler, R. The Balancing Act of NEET Proteins: Iron, ROS, Calcium and Metabolism. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118805. [Google Scholar] [CrossRef]
- Wiley, S.E.; Murphy, A.N.; Ross, S.A.; van der Geer, P.; Dixon, J.E. MitoNEET Is an Iron-Containing Outer Mitochondrial Membrane Protein That Regulates Oxidative Capacity. Proc. Natl. Acad. Sci. USA 2007, 104, 5318–5323. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Liu, R.; Ross, S.; Smart, E.J.; Zhu, H.; Gong, W. Crystallographic Studies of Human MitoNEET. J. Biol. Chem. 2007, 282, 33242–33246. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhou, T.; Ye, K.; Wang, J. Crystal Structure of Human MitoNEET Reveals Distinct Groups of Iron–Sulfur Proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 14640–14645. [Google Scholar] [CrossRef]
- Conlan, A.R.; Paddock, M.L.; Axelrod, H.L.; Cohen, A.E.; Abresch, E.C.; Wiley, S.; Roy, M.; Nechushtai, R.; Jennings, P.A. The Novel 2Fe–2S Outer Mitochondrial Protein MitoNEET Displays Conformational Flexibility in Its N-Terminal Cytoplasmic Tethering Domain. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Paddock, M.L.; Wiley, S.E.; Axelrod, H.L.; Cohen, A.E.; Roy, M.; Abresch, E.C.; Capraro, D.; Murphy, A.N.; Nechushtai, R.; Dixon, J.E.; et al. MitoNEET Is a Uniquely Folded 2Fe 2S Outer Mitochondrial Membrane Protein Stabilized by Pioglitazone. Proc. Natl. Acad. Sci. USA 2007, 104, 14342–14347. [Google Scholar] [CrossRef]
- Baxter, E.L.; Jennings, P.A.; Onuchic, J.N. Interdomain Communication Revealed in the Diabetes Drug Target MitoNEET. Proc. Natl. Acad. Sci. USA 2011, 108, 5266–5271. [Google Scholar] [CrossRef]
- Tamir, S.; Paddock, M.L.; Darash-Yahana-Baram, M.; Holt, S.H.; Sohn, Y.S.; Agranat, L.; Michaeli, D.; Stofleth, J.T.; Lipper, C.H.; Morcos, F.; et al. Structure–Function Analysis of NEET Proteins Uncovers Their Role as Key Regulators of Iron and ROS Homeostasis in Health and Disease. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 1294–1315. [Google Scholar] [CrossRef]
- Ferecatu, I.; Gonçalves, S.; Golinelli-Cohen, M.-P.; Clémancey, M.; Martelli, A.; Riquier, S.; Guittet, E.; Latour, J.-M.; Puccio, H.; Drapier, J.-C.; et al. The Diabetes Drug Target MitoNEET Governs a Novel Trafficking Pathway to Rebuild an Fe-S Cluster into Cytosolic Aconitase/Iron Regulatory Protein 1. J. Biol. Chem. 2014, 289, 28070–28086. [Google Scholar] [CrossRef]
- Lipper, C.H.; Karmi, O.; Sohn, Y.S.; Darash-Yahana, M.; Lammert, H.; Song, L.; Liu, A.; Mittler, R.; Nechushtai, R.; Onuchic, J.N.; et al. Structure of the Human Monomeric NEET Protein MiNT and Its Role in Regulating Iron and Reactive Oxygen Species in Cancer Cells. Proc. Natl. Acad. Sci. USA 2018, 115, 272–277. [Google Scholar] [CrossRef]
- Iwasaki, T.; Samoilova, R.I.; Kounosu, A.; Ohmori, D.; Dikanov, S.A. Continuous-Wave and Pulsed EPR Characterization of the [2Fe-2S](Cys)3(His)1 Cluster in Rat MitoNEET. J. Am. Chem. Soc. 2009, 131, 13659–13667. [Google Scholar] [CrossRef]
- Kusminski, C.M.; Holland, W.L.; Sun, K.; Park, J.; Spurgin, S.B.; Lin, Y.; Askew, G.R.; Simcox, J.A.; McClain, D.A.; Li, C.; et al. MitoNEET-Driven Alterations in Adipocyte Mitochondrial Activity Reveal a Crucial Adaptive Process That Preserves Insulin Sensitivity in Obesity. Nat. Med. 2012, 18, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Seok, B.G.; Lee, S.-J.; Chung, S.W. Inhibition of MitoNEET Attenuates LPS-Induced Inflammation and Oxidative Stress. Cell Death Dis. 2022, 13, 127. [Google Scholar] [CrossRef] [PubMed]
- Yonutas, H.M.; Hubbard, W.B.; Pandya, J.D.; Vekaria, H.J.; Geldenhuys, W.J.; Sullivan, P.G. Bioenergetic Restoration and Neuroprotection after Therapeutic Targeting of MitoNEET: New Mechanism of Pioglitazone Following Traumatic Brain Injury. Exp. Neurol. 2020, 327, 113243. [Google Scholar] [CrossRef]
- Vernay, A.; Marchetti, A.; Sabra, A.; Jauslin, T.N.; Rosselin, M.; Scherer, P.E.; Demaurex, N.; Orci, L.; Cosson, P. MitoNEET-Dependent Formation of Intermitochondrial Junctions. Proc. Natl. Acad. Sci. USA 2017, 114, 8277–8282. [Google Scholar] [CrossRef]
- Sohn, Y.-S.; Tamir, S.; Song, L.; Michaeli, D.; Matouk, I.; Conlan, A.R.; Harir, Y.; Holt, S.H.; Shulaev, V.; Paddock, M.L.; et al. NAF-1 and MitoNEET Are Central to Human Breast Cancer Proliferation by Maintaining Mitochondrial Homeostasis and Promoting Tumor Growth. Proc. Natl. Acad. Sci. USA 2013, 110, 14676–14681. [Google Scholar] [CrossRef] [PubMed]
- Salem, A.F.; Whitaker-Menezes, D.; Howell, A.; Sotgia, F.; Lisanti, M.P. Mitochondrial Biogenesis in Epithelial Cancer Cells Promotes Breast Cancer Tumor Growth and Confers Autophagy Resistance. Cell Cycle 2012, 11, 4174–4180. [Google Scholar] [CrossRef]
- Geldenhuys, W.J.; Piktel, D.; Moore, J.C.; Rellick, S.L.; Meadows, E.; Pinti, M.V.; Hollander, J.M.; Ammer, A.G.; Martin, K.H.; Gibson, L.F. Loss of the Redox Mitochondrial Protein MitoNEET Leads to Mitochondrial Dysfunction in B-Cell Acute Lymphoblastic Leukemia. Free Radic. Biol. Med. 2021, 175, 226–235. [Google Scholar] [CrossRef]
- Molino, D.; Pila-Castellanos, I.; Marjault, H.-B.; Dias Amoedo, N.; Kopp, K.; Rochin, L.; Karmi, O.; Sohn, Y.-S.; Lines, L.; Hamaï, A.; et al. Chemical Targeting of NEET Proteins Reveals Their Function in Mitochondrial Morphodynamics. EMBO Rep. 2020, 21, e49019. [Google Scholar] [CrossRef]
- Lipper, C.H.; Stofleth, J.T.; Bai, F.; Sohn, Y.-S.; Roy, S.; Mittler, R.; Nechushtai, R.; Onuchic, J.N.; Jennings, P.A. Redox-Dependent Gating of VDAC by MitoNEET. Proc. Natl. Acad. Sci. USA 2019, 116, 19924–19929. [Google Scholar] [CrossRef]
- Karmi, O.; Marjault, H.-B.; Bai, F.; Roy, S.; Sohn, Y.-S.; Darash Yahana, M.; Morcos, F.; Ioannidis, K.; Nahmias, Y.; Jennings, P.A.; et al. A VDAC1-Mediated NEET Protein Chain Transfers [2Fe-2S] Clusters between the Mitochondria and the Cytosol and Impacts Mitochondrial Dynamics. Proc. Natl. Acad. Sci. USA 2022, 119, e2121491119. [Google Scholar] [CrossRef]
- Kusminski, C.M.; Park, J.; Scherer, P.E. MitoNEET-Mediated Effects on Browning of White Adipose Tissue. Nat. Commun. 2014, 5, 3962. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Navarrete, J.M.; Moreno, M.; Ortega, F.; Sabater, M.; Xifra, G.; Ricart, W.; Fernández-Real, J.M. CISD1 in Association with Obesity-Associated Dysfunctional Adipogenesis in Human Visceral Adipose Tissue. Obesity 2016, 24, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Geldenhuys, W.J.; Benkovic, S.A.; Lin, L.; Yonutas, H.M.; Crish, S.D.; Sullivan, P.G.; Darvesh, A.S.; Brown, C.M.; Richardson, J.R. MitoNEET (CISD1) Knockout Mice Show Signs of Striatal Mitochondrial Dysfunction and a Parkinson’s Disease Phenotype. ACS Chem. Neurosci. 2017, 8, 2759–2765. [Google Scholar] [CrossRef] [PubMed]
- Lipper, C.H.; Paddock, M.L.; Onuchic, J.N.; Mittler, R.; Nechushtai, R.; Jennings, P.A. Cancer-Related NEET Proteins Transfer 2Fe-2S Clusters to Anamorsin, a Protein Required for Cytosolic Iron-Sulfur Cluster Biogenesis. PLoS ONE 2015, 10, e0139699. [Google Scholar] [CrossRef]
- Zuris, J.A.; Harir, Y.; Conlan, A.R.; Shvartsman, M.; Michaeli, D.; Tamir, S.; Paddock, M.L.; Onuchic, J.N.; Mittler, R.; Cabantchik, Z.I.; et al. Facile Transfer of [2Fe-2S] Clusters from the Diabetes Drug Target MitoNEET to an Apo-Acceptor Protein. Proc. Natl. Acad. Sci. USA 2011, 108, 13047–13052. [Google Scholar] [CrossRef]
- Landry, A.P.; Wang, Y.; Cheng, Z.; Crochet, R.B.; Lee, Y.-H.; Ding, H. Flavin Nucleotides Act as Electron Shuttles Mediating Reduction of the [2Fe-2S] Clusters in Mitochondrial Outer Membrane Protein MitoNEET. Free Radic. Biol. Med. 2017, 102, 240–247. [Google Scholar] [CrossRef]
- Wang, Y.; Landry, A.P.; Ding, H. The Mitochondrial Outer Membrane Protein MitoNEET Is a Redox Enzyme Catalyzing Electron Transfer from FMNH2 to Oxygen or Ubiquinone. J. Biol. Chem. 2017, 292, 10061–10067. [Google Scholar] [CrossRef]
- Tasnim, H.; Landry, A.P.; Fontenot, C.R.; Ding, H. Exploring the FMN Binding Site in the Mitochondrial Outer Membrane Protein MitoNEET. Free Radic. Biol. Med. 2020, 156, 11–19. [Google Scholar] [CrossRef]
- Landry, A.P.; Cheng, Z.; Ding, H. Reduction of Mitochondrial Protein MitoNEET [2Fe-2S] Clusters by Human Glutathione Reductase. Free Radic. Biol. Med. 2015, 81, 119–127. [Google Scholar] [CrossRef]
- Camponeschi, F.; Ciofi-Baffoni, S.; Banci, L. Anamorsin/Ndor1 Complex Reduces [2Fe-2S]-MitoNEET via a Transient Protein-Protein Interaction. J. Am. Chem. Soc. 2017, 139, 9479–9482. [Google Scholar] [CrossRef]
- Golinelli-Cohen, M.-P.; Lescop, E.; Mons, C.; Gonçalves, S.; Clémancey, M.; Santolini, J.; Guittet, E.; Blondin, G.; Latour, J.-M.; Bouton, C. Redox Control of the Human Iron-Sulfur Repair Protein MitoNEET Activity via Its Iron-Sulfur Cluster. J. Biol. Chem. 2016, 291, 7583–7593. [Google Scholar] [CrossRef] [PubMed]
- Bak, D.W.; Zuris, J.A.; Paddock, M.L.; Jennings, P.A.; Elliott, S.J. Redox Characterization of the FeS Protein MitoNEET and Impact of Thiazolidinedione Drug Binding. Biochemistry 2009, 48, 10193–10195. [Google Scholar] [CrossRef] [PubMed]
- Tirrell, T.F.; Paddock, M.L.; Conlan, A.R.; Smoll, E.J.; Nechushtai, R.; Jennings, P.A.; Kim, J.E. Resonance Raman Studies of the (His)(Cys)3 2Fe-2S Cluster of MitoNEET: Comparison to the (Cys)4 Mutant and Implications of the Effects of PH on the Labile Metal Center. Biochemistry 2009, 48, 4747–4752. [Google Scholar] [CrossRef]
- Landry, A.P.; Ding, H. Redox Control of Human Mitochondrial Outer Membrane Protein MitoNEET [2Fe-2S] Clusters by Biological Thiols and Hydrogen Peroxide. J. Biol. Chem. 2014, 289, 4307–4315. [Google Scholar] [CrossRef] [PubMed]
- Schröter, T.; Hatzfeld, O.M.; Gemeinhardt, S.; Korn, M.; Friedrich, T.; Ludwig, B.; Link, T.A. Mutational Analysis of Residues Forming Hydrogen Bonds in the Rieske [2Fe-2S] Cluster of the Cytochrome Bc1 Complex in Paracoccus Denitrificans. Eur. J. Biochem. 1998, 255, 100–106. [Google Scholar] [CrossRef]
- Bak, D.W.; Elliott, S.J. Conserved Hydrogen Bonding Networks of MitoNEET Tune FeS Cluster Binding and Structural Stability. Biochemistry 2013, 52, 4687–4696. [Google Scholar] [CrossRef]
- Pesce, L.; Calandrini, V.; Marjault, H.-B.; Lipper, C.H.; Rossetti, G.; Mittler, R.; Jennings, P.A.; Bauer, A.; Nechushtai, R.; Carloni, P. Molecular Dynamics Simulations of the [2Fe-2S] Cluster-Binding Domain of NEET Proteins Reveal Key Molecular Determinants That Induce Their Cluster Transfer/Release. J. Phys. Chem. B 2017, 121, 10648–10656. [Google Scholar] [CrossRef]
- Song, G.; Tian, F.; Liu, H.; Li, G.; Zheng, P. Pioglitazone Inhibits Metal Cluster Transfer of MitoNEET by Stabilizing the Labile Fe-N Bond Revealed at Single-Bond Level. J. Phys. Chem. Lett. 2021, 12, 3860–3867. [Google Scholar] [CrossRef]
- Zhou, T.; Lin, J.; Feng, Y.; Wang, J. Binding of Reduced Nicotinamide Adenine Dinucleotide Phosphate Destabilizes the Iron−Sulfur Clusters of Human MitoNEET. Biochemistry 2010, 49, 9604–9612. [Google Scholar] [CrossRef]
- Wiley, S.E.; Paddock, M.L.; Abresch, E.C.; Gross, L.; van der Geer, P.; Nechushtai, R.; Murphy, A.N.; Jennings, P.A.; Dixon, J.E. The Outer Mitochondrial Membrane Protein MitoNEET Contains a Novel Redox-Active 2Fe-2S Cluster. J. Biol. Chem. 2007, 282, 23745–23749. [Google Scholar] [CrossRef]
- Noodleman, L.; Baerends, E.J. Electronic Structure, Magnetic Properties, ESR, and Optical Spectra for 2-Iron Ferredoxin Models by LCAO-X.Alpha. Valence Bond Theory. J. Am. Chem. Soc. 1984, 106, 2316–2327. [Google Scholar] [CrossRef]
- Karlsson, A.; Parales, J.V.; Parales, R.E.; Gibson, D.T.; Eklund, H.; Ramaswamy, S. The Reduction of the Rieske Iron–Sulfur Cluster in Naphthalene Dioxygenase by X-Rays. J. Inorg. Biochem. 2000, 78, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Fujinaga, J.; Gaillard, J.; Meyer, J. Mutated Forms of a [2Fe-2S] Ferredoxin with Serine Ligands to the Iron-Sulfur Cluster. Biochem. Biophys. Res. Commun. 1993, 194, 104–111. [Google Scholar] [CrossRef]
- Meyer, J.; Fujinaga, J.; Gaillard, J.; Lutz, M. Mutated Forms of the [2Fe-2S] Ferredoxin from Clostridium Pasteurianum with Noncysteinyl Ligands to the Iron-Sulfur Cluster. Biochemistry 1994, 33, 13642–13650. [Google Scholar] [CrossRef]
- Banci, L.; Ciofi-Baffoni, S.; Mikolajczyk, M.; Winkelmann, J.; Bill, E.; Pandelia, M.-E. Human Anamorsin Binds [2Fe-2S] Clusters with Unique Electronic Properties. J. Biol. Inorg. Chem. 2013, 18, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Brancaccio, D.; Ciofi-Baffoni, S.; Del Conte, R.; Gadepalli, R.; Mikolajczyk, M.; Neri, S.; Piccioli, M.; Winkelmann, J. [2Fe-2S] Cluster Transfer in Iron-Sulfur Protein Biogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 6203–6208. [Google Scholar] [CrossRef]
- Cai, K.; Tonelli, M.; Frederick, R.O.; Markley, J.L. Human Mitochondrial Ferredoxin 1 (FDX1) and Ferredoxin 2 (FDX2) Both Bind Cysteine Desulfurase and Donate Electrons for Iron-Sulfur Cluster Biosynthesis. Biochemistry 2017, 56, 487–499. [Google Scholar] [CrossRef]
- Heidrich, H.-G.; Albracht, S.P.J.; Bäckström, D. Two Iron—Sulfur Centers in Mitochondrial Outer Membranes from Beef Heart as Prepared by Free-Flow Electrophoresis. FEBS Lett. 1978, 95, 314–318. [Google Scholar] [CrossRef]
- Dicus, M.M.; Conlan, A.; Nechushtai, R.; Jennings, P.A.; Paddock, M.L.; Britt, R.D.; Stoll, S. Binding of Histidine in the (Cys)3(His)1-Coordinated [2Fe−2S] Cluster of Human MitoNEET. J. Am. Chem. Soc. 2010, 132, 2037–2049. [Google Scholar] [CrossRef]
- Guigliarelli, B.; Bertrand, P. Application of EPR Spectroscopy to the Structural and Functional Study of Iron-Sulfur Proteins. In Advances in Inorganic Chemistry; Sykes, A.G., Ed.; Academic Press: Cambridge, MA, USA, 1999; Volume 47, pp. 421–497. [Google Scholar]
- Bertrand, P.; More, C.; Guigliarelli, B.; Fournel, A.; Bennett, B.; Howes, B. Biological Polynuclear Clusters Coupled by Magnetic Interactions: From the Point Dipole Approximation to a Local Spin Model. J. Am. Chem. Soc. 1994, 116, 3078–3086. [Google Scholar] [CrossRef]
- Cline, J.F.; Hoffman, B.M.; Mims, W.B.; LaHaie, E.; Ballou, D.P.; Fee, J.A. Evidence for N Coordination to Fe in the [2Fe-2S] Clusters of Thermus Rieske Protein and Phthalate Dioxygenase from Pseudomonas. J. Biol. Chem. 1985, 260, 3251–3254. [Google Scholar] [CrossRef] [PubMed]
- Gurbiel, R.J.; Batie, C.J.; Sivaraja, M.; True, A.E.; Fee, J.A.; Hoffman, B.M.; Ballou, D.P. Electron-Nuclear Double Resonance Spectroscopy of 15N-Enriched Phthalate Dioxygenase from Pseudomonas Cepacia Proves That Two Histidines Are Coordinated to the [2Fe-2S] Rieske-Type Clusters. Biochemistry 1989, 28, 4861–4871. [Google Scholar] [CrossRef] [PubMed]
- Iwata, S.; Saynovits, M.; Link, T.A.; Michel, H. Structure of a Water Soluble Fragment of the “Rieske” Iron-Sulfur Protein of the Bovine Heart Mitochondrial Cytochrome Bc1 Complex Determined by MAD Phasing at 1.5 A Resolution. Structure 1996, 4, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Pandelia, M.-E.; Lanz, N.D.; Booker, S.J.; Krebs, C. Mössbauer Spectroscopy of Fe/S Proteins. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Fee, J.A.; Findling, K.L.; Yoshida, T.; Hille, R.; Tarr, G.E.; Hearshen, D.O.; Dunham, W.R.; Day, E.P.; Kent, T.A.; Münck, E. Purification and Characterization of the Rieske Iron-Sulfur Protein from Thermus Thermophilus. Evidence for a [2Fe-2S] Cluster Having Non-Cysteine Ligands. J. Biol. Chem. 1984, 259, 124–133. [Google Scholar] [CrossRef]
- Fleischhacker, A.S.; Stubna, A.; Hsueh, K.-L.; Guo, Y.; Teter, S.J.; Rose, J.C.; Brunold, T.C.; Markley, J.L.; Münck, E.; Kiley, P.J. Characterization of the [2Fe-2S] Cluster of Escherichia coli Transcription Factor IscR. Biochemistry 2012, 51, 4453–4462. [Google Scholar] [CrossRef]
- Chandramouli, K.; Unciuleac, M.-C.; Naik, S.; Dean, D.R.; Huynh, B.H.; Johnson, M.K. Formation and Properties of [4Fe-4S] Clusters on the IscU Scaffold Protein. Biochemistry 2007, 46, 6804–6811. [Google Scholar] [CrossRef]
- Münck, E.; Debrunner, P.G.; Tsibris, J.C.; Gunsalus, I.C. Mössbauer Parameters of Putidaredoxin and Its Selenium Analog. Biochemistry 1972, 11, 855–863. [Google Scholar] [CrossRef]
- Meyer, J.; Clay, M.D.; Johnson, M.K.; Stubna, A.; Münck, E.; Higgins, C.; Wittung-Stafshede, P. A Hyperthermophilic Plant-Type [2Fe-2S] Ferredoxin from Aquifex Aeolicus Is Stabilized by a Disulfide Bond. Biochemistry 2002, 41, 3096–3108. [Google Scholar] [CrossRef]
- Wolfe, M.D.; Altier, D.J.; Stubna, A.; Popescu, C.V.; Münck, E.; Lipscomb, J.D. Benzoate 1,2-Dioxygenase from Pseudomonas Putida: Single Turnover Kinetics and Regulation of a Two-Component Rieske Dioxygenase. Biochemistry 2002, 41, 9611–9626. [Google Scholar] [CrossRef]
- Garcia-Serres, R.; Clémancey, M.; Latour, J.-M.; Blondin, G. Contribution of Mössbauer Spectroscopy to the Investigation of Fe/S Biogenesis. J. Biol. Inorg. Chem. 2018, 23, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mapolelo, D.T.; Dingra, N.N.; Naik, S.G.; Lees, N.S.; Hoffman, B.M.; Riggs-Gelasco, P.J.; Huynh, B.H.; Johnson, M.K.; Outten, C.E. The Yeast Iron Regulatory Proteins Grx3/4 and Fra2 Form Heterodimeric Complexes Containing a [2Fe-2S] Cluster with Cysteinyl and Histidyl Ligation. Biochemistry 2009, 48, 9569–9581. [Google Scholar] [CrossRef] [PubMed]
- Kristina Beilschmidt, L.; Choudens, S.; Fournier, M.; Sanakis, Y.; Hograindleur, M.-A.; Clémancey, M.; Blondin, G.; Schmucker, S.; Eisenmann, A.; Weiss, A.; et al. ISCA1 Is Essential for Mitochondrial Fe4S4 Biogenesis in Vivo. Nat. Comm. 2017, 8, 15124. [Google Scholar] [CrossRef] [PubMed]
- Camponeschi, F.; Gallo, A.; Piccioli, M.; Banci, L. The Long-Standing Relationship between Paramagnetic NMR and Iron–Sulfur Proteins: The MitoNEET Example. An Old Method for New Stories or the Other Way Around? Magn. Reson. 2021, 2, 203–221. [Google Scholar] [CrossRef]
- Banci, L.; Camponeschi, F.; Ciofi-Baffoni, S.; Piccioli, M. The NMR Contribution to Protein-Protein Networking in Fe-S Protein Maturation. J. Biol. Inorg. Chem. 2018, 23, 665–685. [Google Scholar] [CrossRef]
- Piccioli, M. Paramagnetic NMR Spectroscopy Is a Tool to Address Reactivity, Structure, and Protein–Protein Interactions of Metalloproteins: The Case of Iron–Sulfur Proteins. Magnetochemistry 2020, 6, 46. [Google Scholar] [CrossRef]
- Trindade, I.B.; Coelho, A.; Cantini, F.; Piccioli, M.; Louro, R.O. NMR of Paramagnetic Metalloproteins in Solution: Ubi Venire, Quo Vadis? J. Inorg. Biochem. 2022, 234, 111871. [Google Scholar] [CrossRef]
- Trindade, I.B.; Invernici, M.; Cantini, F.; Louro, R.O.; Piccioli, M. PRE-Driven Protein NMR Structures: An Alternative Approach in Highly Paramagnetic Systems. FEBS J. 2021, 288, 3010–3023. [Google Scholar] [CrossRef]
- Invernici, M.; Selvolini, G.; Silva, J.M.; Marrazza, G.; Ciofi-Baffoni, S.; Piccioli, M. Interconversion between [2Fe–2S] and [4Fe–4S] Cluster Glutathione Complexes. Chem. Commun. 2022, 58, 3533–3536. [Google Scholar] [CrossRef]
- Brancaccio, D.; Gallo, A.; Piccioli, M.; Novellino, E.; Ciofi-Baffoni, S.; Banci, L. [4Fe-4S] Cluster Assembly in Mitochondria and Its Impairment by Copper. J. Am. Chem. Soc. 2017, 139, 719–730. [Google Scholar] [CrossRef]
- Camponeschi, F.; Prusty, N.R.; Heider, S.A.E.; Ciofi-Baffoni, S.; Banci, L. GLRX3 Acts as a [2Fe–2S] Cluster Chaperone in the Cytosolic Iron–Sulfur Assembly Machinery Transferring [2Fe–2S] Clusters to NUBP1. J. Am. Chem. Soc. 2020, 142, 10794–10805. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, D.; Gallo, A.; Mikolajczyk, M.; Zovo, K.; Palumaa, P.; Novellino, E.; Piccioli, M.; Ciofi-Baffoni, S.; Banci, L. Formation of [4Fe-4S] Clusters in the Mitochondrial Iron-Sulfur Cluster Assembly Machinery. J. Am. Chem. Soc. 2014, 136, 16240–16250. [Google Scholar] [CrossRef] [PubMed]
- Camponeschi, F.; Muzzioli, R.; Ciofi-Baffoni, S.; Piccioli, M.; Banci, L. Paramagnetic 1H NMR Spectroscopy to Investigate the Catalytic Mechanism of Radical S-Adenosylmethionine Enzymes. J. Mol. Biol. 2019, 431, 4514–4522. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Luchinat, C. The 1H NMR Parameters of Magnetically Coupled Dimers—The Fe2S2 Proteins as an Example. Struct. Bond. 1990, 72, 113–136. [Google Scholar] [CrossRef]
- Holz, R.C.; Small, F.J.; Ensign, S.A. Proton Nuclear Magnetic Resonance Investigation of the [2Fe-2S](1-)-Containing “Rieske-Type” Protein from Xanthobacter Strain Py2. Biochemistry 1997, 36, 14690–14696. [Google Scholar] [CrossRef]
- Skjeldal, L.; Markley, J.L.; Coghlan, V.M.; Vickery, L.E. 1H NMR Spectra of Vertebrate [2Fe-2S] Ferredoxins. Hyperfine Resonances Suggest Different Electron Delocalization Patterns from Plant Ferredoxins. Biochemistry 1991, 30, 9078–9083. [Google Scholar] [CrossRef]
- Maio, N.; Rouault, T.A. Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis. Trends Biochem. Sci. 2020, 45, 411–426. [Google Scholar] [CrossRef]
- Machonkin, T.E.; Westler, W.M.; Markley, J.L. Strategy for the Study of Paramagnetic Proteins with Slow Electronic Relaxation Rates by NMR Spectroscopy: Application to Oxidized Human [2Fe-2S] Ferredoxin. J. Am. Chem. Soc. 2004, 126, 5413–5426. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Jenk, D.; LeMaster, D.M.; Westler, W.M.; Markley, J.L. Electron-Nuclear Interactions in Two Prototypical [2Fe-2S] Proteins: Selective (Chiral) Deuteration and Analysis of (1)H and (2)H NMR Signals from the Alpha and Beta Hydrogens of Cysteinyl Residues That Ligate the Iron in the Active Sites of Human Ferredoxin and Anabaena 7120 Vegetative Ferredoxin. Arch. Biochem. Biophys. 2000, 373, 328–334. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Boscaro, F.; Chatzi, A.; Mikolajczyk, M.; Tokatlidis, K.; Winkelmann, J. Anamorsin Is a [2Fe-2S] Cluster-Containing Substrate of the Mia40-Dependent Mitochondrial Protein Trapping Machinery. Chem. Biol. 2011, 18, 794–804. [Google Scholar] [CrossRef]
- Spronk, C.A.E.M.; Żerko, S.; Górka, M.; Koźmiński, W.; Bardiaux, B.; Zambelli, B.; Musiani, F.; Piccioli, M.; Basak, P.; Blum, F.C.; et al. Structure and Dynamics of Helicobacter Pylori Nickel-Chaperone HypA: An Integrated Approach Using NMR Spectroscopy, Functional Assays and Computational Tools. J. Biol. Inorg. Chem. 2018, 23, 1309–1330. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Xia, B.; Reed, G.H.; Markley, J.L. Optical, EPR, and 1H NMR Spectroscopy of Serine-Ligated [2Fe-2S] Ferredoxins Produced by Site-Directed Mutagenesis of Cysteine Residues in Recombinant Anabaena 7120 Vegetative Ferredoxin. Biochemistry 1994, 33, 3155–3164. [Google Scholar] [CrossRef] [PubMed]
- Trindade, I.B.; Hernandez, G.; Lebègue, E.; Barrière, F.; Cordeiro, T.; Piccioli, M.; Louro, R.O. Conjuring up a Ghost: FhuF—A Ferric-Siderophore Reductase of Unknown Structure. J. Biol. Inorg. Chem. 2021, 26, 313–326. [Google Scholar] [CrossRef]
- Dugad, L.B.; La Mar, G.N.; Banci, L.; Bertini, I. Identification of Localized Redox States in Plant-Type Two-Iron Ferredoxins Using the Nuclear Overhauser Effect. Biochemistry 1990, 29, 2263–2271. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Capozzi, F.; Luchinat, C.; Piccioli, M.; Vila, A.J. The Fe4S4 Centers in Ferredoxins Studied through Proton and Carbon Hyperfine Coupling. Sequence-Specific Assignments of Cysteines in Ferredoxins from Clostridium Acidi Urici and Clostridium Pasteurianum. J. Am. Chem. Soc. 1994, 116, 651–660. [Google Scholar] [CrossRef]
- Holt, S.H.; Darash-Yahana, M.; Sohn, Y.S.; Song, L.; Karmi, O.; Tamir, S.; Michaeli, D.; Luo, Y.; Paddock, M.L.; Jennings, P.A.; et al. Activation of Apoptosis in NAF-1-Deficient Human Epithelial Breast Cancer Cells. J. Cell Sci. 2016, 129, 155–165. [Google Scholar] [CrossRef]
- Darash-Yahana, M.; Pozniak, Y.; Lu, M.; Sohn, Y.-S.; Karmi, O.; Tamir, S.; Bai, F.; Song, L.; Jennings, P.A.; Pikarsky, E.; et al. Breast Cancer Tumorigenicity Is Dependent on High Expression Levels of NAF-1 and the Lability of Its Fe-S Clusters. Proc. Natl. Acad. Sci. USA 2016, 113, 10890–10895. [Google Scholar] [CrossRef] [PubMed]
| System | Cluster Type | Formal Valences | Fe Ligands | Stot | δ (mm/s) | |ΔEQ| (mm/s) | Refs. |
|---|---|---|---|---|---|---|---|
| FdI A.aeolicus | [2Fe-2S]2+ | 2 Fe3+ | 4 Cys | 0 | 0.27 | 0.60 | [70] |
| putidaredoxin P. putida | [2Fe-2S]2+ | 2 Fe3+ | 4 Cys | 0 | 0.27 | 0.60 | [69] |
| yeast Grx3 | [2Fe-2S]2+ | 2 Fe3+ | 4 Cys | 0 | 0.29 | 0.55–0.76 | [73] |
| human ISCA1 | [2Fe-2S]2+ | 2 Fe3+ | 4 Cys | 0 | 0.28 | 0.50 | [74] |
| human ISCA2 | [2Fe-2S]2+ | 2 Fe3+ | 4 Cys | 0 | 0.27 | 0.53 | [74] |
| anamorsin (site 1) | [2Fe-2S]2+ | 2 Fe3+ | 4 Cys | 0 | 0.26 | 0.57 | [55] |
| anamorsin (site 2) | [2Fe-2S]2+ | 2 Fe3+ | 4 Cys | 0 | 0.28 | 0.39 | [55] |
| Rieske thermus thermophilus | [2Fe-2S]2+ | Fe3+ Fe3+ | 2 Cys 2 His | 0 | 0.24 0.32 | 0.32 0.91 | [66] |
| mitoNEET | [2Fe-2S]2+ | Fe3+ Fe3+ | 2 Cys 1 Cys 1 His | 0 | 0.26 0.30 | 0.47 0.96 | [18,41] |
| E. coliIscU | [2Fe-2S]2+ | Fe3+ Fe3+ | 2 Cys 1 Cys 1 His | 0 | 0.27 0.32 | 0.66 0.94 | [68] |
| E. coliIscR | [2Fe-2S]2+ | Fe3+ Fe3+ | 2 Cys 1 Cys 1 His | 0 | 0.27 0.30 | 0.48 0.72 | [67] |
| yeast Fra2-Grx3 | [2Fe-2S]2+ | Fe3+ Fe3+ | 2 Cys 1 Cys 1 His | 0 | 0.30 0.32 | 0.50 0.82 | [73] |
| FdI A. aeolicus | [2Fe-2S]1+ | Fe3+ Fe2+ | 2 Cys 2 Cys | 1/2 | 0.30 0.62 | 1.0 3.0 | [70] |
| putidaredoxin P. putida | [2Fe-2S]1+ | Fe3+ Fe2+ | 2 Cys 2 Cys | 1/2 | 0.35 0.60 | 0.65 2.70 | [69] |
| anamorsin (site 1) | [2Fe-2S]1+ | 2 Fe2.5+ | 4 Cys | 1/2 | 0.26 | 0.57 | [55] |
| anamorsin (site 2) | [2Fe-2S]1+ | 2 Fe2.5+ | 4 Cys | 1/2 | 0.28 | 0.39 | [55] |
| Rieske thermus thermophilus | [2Fe-2S]1+ | Fe3+ Fe2+ | 2 Cys 2 His | 1/2 | 0.31 0.74 | 0.63 3.05 | [66] |
| E. coliIscR | [2Fe-2S]1+ | Fe3+ Fe2+ | 2 Cys 1 Cys 1 His | 1/2 | 0.33 0.70 | 1.09 3.4 | [67] |
| mitoNEET | [2Fe-2S]1+ | Fe3+ Fe2+ | 2 Cys 1 Cys, 1 His | 1/2 | 0.32 0.68 | 1.07 3.15 | [41] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camponeschi, F.; Piccioli, M.; Banci, L. The Intriguing mitoNEET: Functional and Spectroscopic Properties of a Unique [2Fe-2S] Cluster Coordination Geometry. Molecules 2022, 27, 8218. https://doi.org/10.3390/molecules27238218
Camponeschi F, Piccioli M, Banci L. The Intriguing mitoNEET: Functional and Spectroscopic Properties of a Unique [2Fe-2S] Cluster Coordination Geometry. Molecules. 2022; 27(23):8218. https://doi.org/10.3390/molecules27238218
Chicago/Turabian StyleCamponeschi, Francesca, Mario Piccioli, and Lucia Banci. 2022. "The Intriguing mitoNEET: Functional and Spectroscopic Properties of a Unique [2Fe-2S] Cluster Coordination Geometry" Molecules 27, no. 23: 8218. https://doi.org/10.3390/molecules27238218
APA StyleCamponeschi, F., Piccioli, M., & Banci, L. (2022). The Intriguing mitoNEET: Functional and Spectroscopic Properties of a Unique [2Fe-2S] Cluster Coordination Geometry. Molecules, 27(23), 8218. https://doi.org/10.3390/molecules27238218