Abstract
Human deaths caused by Gram-negative bacteria keep rising due to the multidrug resistance (MDR) phenomenon. Therefore, it is a priority to develop novel antibiotics with different mechanisms of action. Several bacterial zinc metalloenzymes are becoming attractive targets since they do not show any similarities with the human endogenous zinc-metalloproteinases. In the last decades, there has been an increasing interest from both industry and academia in developing new inhibitors against those enzymes involved in lipid A biosynthesis, and bacteria nutrition and sporulation, e.g., UDP-[3-O-(R)-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase (LpxC), thermolysin (TLN), and pseudolysin (PLN). Nevertheless, targeting these bacterial enzymes is harder than expected and the lack of good clinical candidates suggests that more effort is needed. This review gives an overview of bacterial zinc metalloenzyme inhibitors that have been synthesized so far, highlighting the structural features essential for inhibitory activity and the structure–activity relationships. Our discussion may stimulate and help further studies on bacterial zinc metalloenzyme inhibitors as possible novel antibacterial drugs.
1. Introduction
1.1. Bacteremia
The World Health Organization (WHO) has warned about the possibility of a new era where infections and minor wounds will be a primary cause of mortality. This emergency is correlated to the nosocomial infections (NIs) caused by the “ESKAPE” bacteria such as Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonie, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species [1]. The nosocomial infections, according to the definition of WHO, indicate diseases that appear after 48 h from admission to an acute-care hospital or 30 days after receiving medical assistance, or up to 90 days after surgery [2]. Generally, the hospitalized infections are divided into four main disease-correlated sections depending on etiology: catheter-associated urinary tract infections (CAUTIs), hospital-acquired pneumonias (HAPs), bloodstream infections (BSIs), and surgical site infections (SSIs). In all these cases, pathogens may infect different districts of the human body, becoming the main cause of death of several patients [3].
The microbes constitute a real threat for both healthy individuals and patients affected by different pathologies. In order to express their pathogenic activity, bacteria firstly need to invade and colonize the potential infection sites and then to survive during blood migration. The presence of bacteria in the bloodstream is denominated bacteremia, and this phenomenon can be classified by its origin in primary and secondary bacteremia [4]. The primary bacteremia is caused by unknown sources, whereas the secondary bacteremia is a consequence of prior pneumonia or urinary infections, or of contamination due to the use of infected medical devices [5,6].
Patients affected by bacteremia often show mild (e.g., fever) or no symptoms. Nevertheless, severe infections can lead to a sepsis condition, which requires an immediate treatment in order to avoid severe consequences including death. Bacteremia is mainly caused by bacterial infection, even though several fungi could also provoke this condition [4].
It is well known that bacteria are generally classified on the basis of differences in cell membrane structure into two main categories: Gram-negative and Gram-positive. Lately, diseases associated with Gram-negative bacteria are increasing due to the growing resistance towards classical antibiotics [7]. Although different strategies have been adopted to contrast the emergency of multiple antibiotic-resistant Gram-positive and Gram-negative bacteria in hospital and community settings, novel approaches for rapid diagnosis and treatment are required.
1.2. Multidrug Resistance
In the last decades, microbial infections have rapidly increased due to the multidrug-resistance (MDR) phenomenon, which is the microorganism insensitivity or resistance to antibiotics [8]. Nowadays, several antibiotics are not able to combat bacterial infections (e.g., Escherichia coli, Klebsiella pneumonia, Staphylococcus aureus and pneumonia, Pseudomonas aeruginosa) (Table 1), leading to an increase of mortality and morbidity [9]. Moreover, antimicrobial resistance (AMR) can lead to high medical costs, since patients cannot be treated with classical drugs but only with more expensive therapies.

Table 1.
Bacteria and their typical diseases. Adapted from [10].
1.3. MDR Classification
There are three types of MDR: primary resistance, secondary resistance and clinical resistance (Figure 1) [7].
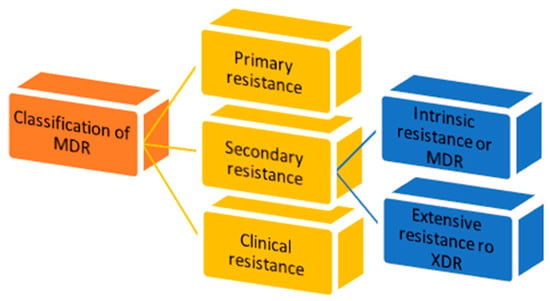
Figure 1.
Classification of multidrug resistance.
- 1.
- Primary resistance occurs when the host organism interacts for the first time with the drug of interest;
- 2.
- Secondary resistance refers to the aversion which onsets after an exposure to the drug. It is also known as “acquired resistance” and can be classified into:
- intrinsic resistance: a single species of microorganisms results in insensitivity towards a first-time drug administration [11];
- extensive resistance: microorganisms are able to resist one or more of the most potent drugs. It is also named “extended detection and response” (XDR) which arises after the exposure to first-line drugs [12];
- 3.
- Clinical resistance is described when, in order to have efficacy, the drug concentration must be significantly increased, due to dysregulation of the host immune system function. This imbalance can lead to a failure of the therapy or an outbreak of different infections [11].
1.4. MDR Mechanism
Microbes use different strategies in order to interfere with the activity of antimicrobial agents. These mechanisms can be intrinsic or could be acquired by gene exchange/transfer methods or genetic mutations [13].
1.4.1. Modifications in the Uptake of Drugs
In order to carry out their antimicrobial activity, drugs need to reach the target site, crossing the outer membrane of Gram-negative bacteria. Bacteria are able to modulate their outer membrane functions through specific proteins named porins. Indeed, most of Gram-negative bacteria modify porin selectivity, frequency and size, interfering with the uptake of drugs [14].
1.4.2. Inactivation of Drugs
Drug inactivation is one the most efficient mechanisms used by pathogens. Several bacteria use β-lactamases in order to hydrolase the β-lactam ring of classical antibiotics (penicillins and cephalosporins) into inactive compounds. Gram-negative bacteria use this mechanism to resist antibacterial agents, developing resistance. Moreover, some pathogens produce toxins as a defense mechanism, e.g., Streptomyces against neomycin and streptomycin [15].
1.4.3. Efflux Pumps
The use of efflux pumps as a mechanism to combat the antibacterial activity of drugs is common for both Gram-negative and Gram-positive bacteria such as Escherichia coli, Staphylococci and Streptococcus aureus and pneumoniae. By overexpressing efflux pumps, these pathogens expel drugs outside the membrane, thus decreasing the intracellular drug concentration to a level not sufficient for activity. Moreover, some efflux pumps showed substrate-specificity against antibiotics [16].
1.5. Zinc Metalloenzymes
Gram-negative bacterial resistance to antibiotics is recognized as one of the main health threats in the world. Gram-negative pathogens such as Pseudomonas aeruginosa, Escherichia coli, Acinetobacter baumannii, and Klebsiella pneumoniae are associated with urinary, pneumonia and bloodstream infections [10].
The increased drug resistance in the last decades is due to the abuse of antibiotics, which enhanced the evolution of different kinds of resistance mechanisms. To date, only a few drugs have been developed against antibiotic-resistant Gram-negative pathogens [17]. Thus, there is an urgent need to develop new antibacterial agents with alternative mechanisms of action. Over the last few years, academia and industry have focused their attention on enzymes such as UDP-[3-O-(R)-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase (LpxC), thermolysin (TLN), and pseudolysin (PLN). LpxC is a metalloenzyme involved in the biosynthesis of a key component of the Gram-negative bacterial outer membrane [1]. Otherwise, PLN and TLN are metalloproteinases secreted by Gram-negative bacteria which are involved in bacterial nutrition and sporulation [18]. Other bacterial zinc metalloenzymes taken in consideration as novel antibacterial targets are carbonic anhydrases [19,20] and histone deacetylases [21], which, however, are not considered in this review.
1.5.1. Gram-Negative Outer Membrane and Its Pathogenicity
The Gram-negative bacterial cell envelope is a complex multilayered structure constituted by two main membranes, the inner and the outer, which are fundamental elements in several bacterial mechanisms. Indeed, the outer membrane comprises integral membrane proteins, phospholipids, lipopolysaccharides (LPS) and lipoproteins [22]. Interestingly, the pathogenicity of Gram-negative bacteria is correlated with the components of their outer membrane [23]. In particular, LPS are essential for Gram-negative bacteria in order to protect them from chemicals and antibiotics [23,24].
LPS are constituted by three elements: lipid A, O-antigen, and core oligosaccharides. Nowadays, the research focus is on lipid A, which is the anchor for LPS in the outer membrane [25]. Lipid A is a potent immunogen that activates a severe hyperimmune response, leading to a septic shock in the host [26]. The inhibition of lipid A biosynthesis represents an attractive way to treat the Gram-negative infections and to sensitize bacteria to other drugs. The biosynthesis of lipid A involves nine unique enzymes and takes place in the cytosol of the inner membrane. Moreover, the first step of lipid A biosynthesis is thermodynamically unfavorable and it involves the LpxA enzyme [27]. Thus, the pivotal step is the second one, which requires the LpxC metalloenzyme. In particular, LpxC catalyzes the deacetylation of UDP-3-O-acyl-GlcNAc (1) through a general acid/base mechanism (Figure 2) [28].
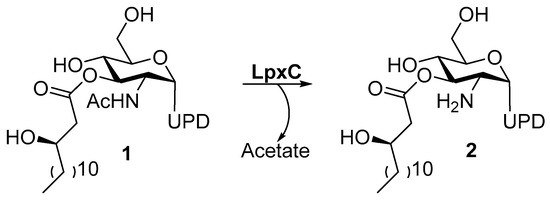
Figure 2.
Deacetylation of UDP-3-O-Acyl-GlcNAc by LpxC.
All the enzymes involved in this pathway could be potential targets. However, LpxC is the most promising due to its role in lipid A biosynthesis, and its modulation is fundamental for E. coli survival. Moreover, LpxC is conserved among the Gram-negative bacteria and does not show similarities or homologies with any mammalian protein. This uniqueness made it an attractive target in order to develop selective inhibitors devoid of off-target toxicity [27].
1.5.2. M4 Enzymes
Generally, proteases are able to hydrolyze proteins and peptides, and based on their activity, are classified into exo-proteases and endo-proteases. Considering their catalytic mechanism, the endo-protease family is constituted by aspartic proteases, cysteine proteases, metalloproteases, serine proteases and threonine proteases [29].
Metalloproteinases are a family of endopeptidases responsible for extracellular matrix degradation. The metalloproteinase catalytic site contains one or two metal ions (most commonly Zn2+). The divalent ions play a fundamental role in the catalytic mechanism by activating a nucleophilic water molecule, exploiting the protease mechanism. Furthermore, several calcium ions are located in the binding site and do not participate in the catalytic activity [30].
According to the MEROPS database (http://MEROPS.sanger.ac.uk/ accessed on 9 March 2023), metalloproteinases are classified into 76 families (M1-M76), which are divided into 16 clans (MA, MC, MD, ME, MF, MG, MH, MJ, MM, MN, MO, MP, MQ, MS, MT and unassigned) based on their similarities to the 3D structure and metal ion motifs. The clan is indicated by two letters; the first letter indicates the family which the clan is referred to ((M) Metalloproteinase). However, sometimes some families cannot yet be assigned to clans and when this happens the family is described as belonging to clan A-, C-, M-, S-, T- or U-, according to the catalytic type. Moreover, some clans are divided into sub-clans due to the age differences within the clan [31].
Most of the virulence factors involved in bacterial nutrition and sporulation (e.g., TLN and PLN) belong to the M4 family, in particular to the clan MA, sub-clan E. M4 metalloproteinases are extracellular proteases secreted by bacteria, and are fundamental in the protein degradation to provide nutrients for bacteria. Among MA metalloproteinases, Thermolysin (TNL) from Bacillus thermoproteolyticus has been the first M4 protease to be identified and it represents the prototype of this family [18].
The M4 enzymes are characterized by the presence of a Zn2+ ion in the catalytic site coordinated by two histidine residues, one glutamic acid and one water molecule constituting the HEXXH motif. These proteases are secreted as pre-propeptides constituted by a signal peptide, a propeptide sequence and a peptidase unit. Furthermore, the 3D structure is composed of two domains and the binding site is located between them [32]. The N-terminal domain includes α-helices and β-sheets, while the C-terminal is formed by α-helices connected by a turn. In this region, the glutamic acid residue coordinates the zinc ion [33].
Generally, the binding site of a metalloproteinase contains different subsites (S1, S2, S3 and S1′, S2′ and S3′). Among them, the S1′ subsite is crucial in substrate recognition because it presents a different shape in each metalloproteinase. In particular, the S1′ pocket interacts with hydrophobic or bulky side chains of amino acids constituting a hydrophobic site. The S2′ subsite is a hydrophobic pocket, whereas the S1 subsite is characterized by hydrophilic residues [34]. Generally, the hydrolysis of the substrates is based on an acid-base mechanism. In particular, a histidine residue present in the binding site reacts as a base that activates the water molecule. The formed hydroxyl group attacks the carbonyl group of the substrate, leading to the cleavage.
The M4 enzymes are structurally similar to the endogenous Metzincins, i.e., Matrix metalloproteinases (MMPs) and A disintegrin and metalloproteinases (ADAMs) [35,36]), which are classified into the M10 family. Proteases of the M10 family have a Zn2+ in the catalytic site coordinated by three histidine residues and one water molecule. Moreover, in the binding pocket the MMPs present the same subsites of the M4 enzymes [37]. However, the particular depth of the S1′ subsite might represent a selectivity factor between the M4 and M10 enzymes [38].
1.6. Targets of Interest
1.6.1. UDP-[3-O-(R)-3-hydroxymyristoyl]-N-acetylglucosamine Deacetylase
UDP-[3-O-(R)-3-hydroxymyristoyl]-N-acetylglucosamine deacetylase (LpxC) is a cytosolic zinc metalloenzyme which catalyzes the second step in the biosynthesis of lipid A (Figure 2), a key component of all Gram-negative bacterial outer membranes. Thus, the inhibition of its biosynthesis can lead to an increase of cell membrane permeability and leakage of cell contents up to cell death, as previously described [28].
Structure of LpxC
Structurally, LpxC is constituted by two topologically similar domains, presenting two α-helices packed against a five-stranded β-sheet, the active site being located at the interface between them [17]. The Zn2+ displayed at the bottom of the site is coordinated by two histidines (His79 and His238), one aspartate residue (Asp242) and one water molecule, preserving its classical tetrahedral coordination geometry. A crystal structure of E. coli LpxC in complex with the deacetylated compound 2 (Figure 2 and Figure 3) shows the different binding regions of the conical-shaped active site. The fatty acyl moiety of compound 2 interacts with the hydrophobic tunnel composed of Met195, Ile198, Phe212 and Val217. The hexose ring fragment of 2 is placed in a slightly hydrophobic pocket comprising Phe161, Phe162 and Phe194 residues. Lastly, the UDP-binding pocket is composed of a phosphate binding region surrounded by polar residues (Lys143, Lys239, Lys 262 and His265) and a nucleotide binding site, where the UDP moiety forms polar (Asp160) and π-stacking interactions (Lys262 and Phe161) [39].
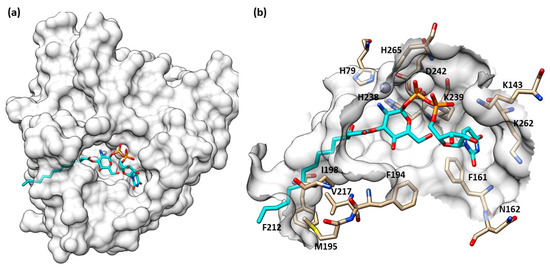
Figure 3.
(a) Crystal structure of compound 2 in LpxC (PDB ID: 4MDT) of E. coli. (b) Compound 2 into LpxC (PDB ID: 4MDT) binding site of E. coli. Image generated with Chimera, version 1.16.
1.6.2. Pseudolysin
Pseudolysin (LasB elastase or elastase B) is a virulence factor produced by P. aeruginosa and can be considered a novel target in the fight of nosocomial infections. This protease belongs to the thermolysin-like family of metallopeptidase (M4 family) and displays a crucial role in the pathogenesis of infections caused by P. aeruginosa [40]. Pseudolysin hydrolyses several host proteins useful for bacterial nutrition, leading to a disorder of the immune response and encouraging inflammatory processes.
Pseudolysin is involved in different diseases, such as the destruction of laminae of the arteriae in vasculitis combined with the action of staphylolysin [41]. Moreover, it causes hemorrhage, destroying the basement membrane of vessels [42]. Pseudolysin causes septic shock in septicemia and supports the growth and invasiveness of microbes in burns [43,44]. In lung diseases, this enzyme is involved in the damage of alveolar septal cells and, by destroying the alveolar epithelial cell junctions, it supports the epithelial permeability to macromolecules [45]. Moreover, pseudolysin is involved in the cystic fibrosis caused by P. aeruginosa [46]. Thus, it can be considered a good target to combat several diseases.
Structure of Pseudolysin
Pseudolysin is synthesized as a preproelastase with 498 residues and is composed of three domains. The N-terminal prepeptide or signal peptide (aa 1–23) is cleaved across the cytoplasmic membrane, giving the proenzyme. The second domain, the propeptide domain (aa 24–197), shows an intramolecular chaperone (ICM) activity for the C-terminal domain (aa 198–498) in order to encode the mature enzyme. The mature elastase has 67% homology with thermolysin from Bacillus thermoproteolyticus. Furthermore, the expression of pseudolysin is regulated by four sophisticated gene regulation systems (LasI-LasR, Rh1I-Rh1R, PqsABCDE-PqsR and AmbCDE-IqsR) that depend on the bacterial cell density. These systems belong to a complex Quorum sensing (QS) regulatory network [47]. Pseudomonas is also usually associated with biofilm infections because the biofilm formation is regulated by the QS [48].
The secondary structure of the protein is constituted by seven α-helices and two antiparallel β-sheets. The binding site is allocated among two helices and their connective loop (Figure 4a). Pseudolysin contains a zinc and a calcium ion [49]. The Ca2+ is useful for protein stability, whereas the Zn2+ close to the cleavage site plays a catalytic role [50]. Two disulfide bridges (Cys30-Cys58 and Cys270-Cys297) are useful for the protein stabilization. The tetrahedral coordination of Zn2+ is formed by two histidines (His140 and His144), one glutamate (Glu164) and a catalytic water molecule. Furthermore, the protease presents three significant pockets, as already mentioned in paragraph 1.5.2: S1′, S2′ and S1 (Figure 4b). The S1′ pocket recognizes aromatic and hydrophobic amino acids including Leu132 and Val137. The S2′ subsite contains Asn112, Phe129, Ile186 and Leu197 and interacts with hydrophobic residues too. In sharp contrast, the largest pocket, S1, composed of hydrophilic residues (Tyr114, Trp115, Tyr 155), interacts with hydrophilic moieties. The S1′ subsite is the most important for selectivity and activity; it is smaller than the S1′ subsites of human MMPs due to the Arg198 placed between the S1′ and S2′ pockets, which regulates the size and the entrance of the pocket [51].
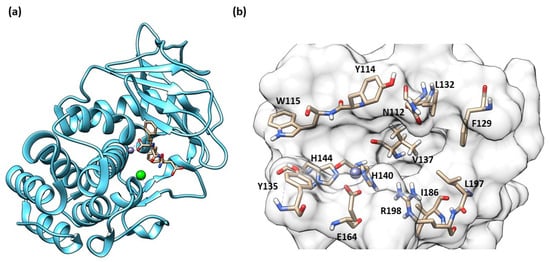
Figure 4.
(a) Secondary structure of Pseudolysin (PDB ID: 1U4G). (b) Amino-acids constituting the subsites of pseudolysin (PDB ID: 1U4G). Image generated with Chimera, version 1.16.
1.6.3. Thermolysin
Thermolysin (TLN) from Bacillus thermoproteolyticus is the enzyme prototype for the M4 family of metalloproteinases and represents another innovative target against bacterial infections. This enzyme is used in the biotechnology industry to cleave amino-peptide bonds and it is also utilized for the production of aspartame. It has peculiar characteristics, being a thermozyme: it is stable in extreme temperatures, pH values and is also stable in organic solvents [32]. For these reasons, it is used for different industrial applications. Thermolysin is involved in the development of peptic ulcer, gastritis, gastric carcinoma and cholera. The thermolysin-like proteinase from Legionella is suggested to be responsible for Legionnaires’ disease and pneumonia [18].
Structure of Thermolysin
Thermolysin’s three-dimensional structure presents one catalytic zinc ion in the binding site and four calcium ions (Figure 5a). The catalytic zinc, like all the M4 enzymes, has a tetrahedral coordination composed of two histidines, a glutamatic acid and one water molecule (Figure 5b). Pseudolysin’s structure shares 67% of homology with thermolysin. However, the binding site cleft of pseudolysin is slightly wider than the thermolysin binding site cleft. Moreover, the physics profile to interact with hydrophilic or hydrophobic residues of the subpockets (S1′, S2′ and S1) are the same between the two proteinases. The size of the S1′ subpocket of thermolysin, the main subsite for the inhibitor recognition, is slightly smaller than pseudolysin.
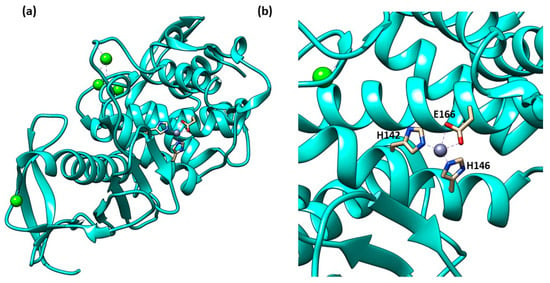
Figure 5.
(a) Secondary structure of TLN (PDB ID: 1PE5); (b) Tetrahedral coordination of catalytic zinc ion in TLN (PDB ID: 1PE5). Image generated with Chimera, version 1.16.
2. Inhibitors
The increase of nosocomial infections due to the “ESKAPE” pathogens is becoming an emergency and the antibiotic resistance phenomenon is its main cause. According to the World Health Organization (WHO), the discovery of novel enzyme inhibitors able to interfere with bacterial infections by avoiding the classical resistance mechanism could have an important role [1].
The high level of similarities among these metalloproteinases makes it challenging to selectively target M4 metalloproteinases. In fact, the crucial features for an effective inhibitor of metalloproteinases are the presence of a zinc-binding group (ZBG) and a hydrophobic group able to interact with the S1′ pocket. The ZBG is a moiety able to coordinate the catalytic zinc ion and the most reported ZBG is hydroxamate, even if it is responsible for important adverse effects and low selectivity [52]. More often, a good fitting with the S1′ pocket allows the use of alternative ZBGs such as carboxylate [53] or allows the development of non-zinc-binding inhibitors [54].
These elements are common to all the already reported inhibitors of metalloproteinases either of bacterial (such as M4) or human origin, including MMPs [55,56,57] and ADAMs [58,59]. Nowadays, the real challenge is to obtain selective M4 metalloproteinase inhibitors sparing the human endogenous metalloproteinase as the key factor to avoid several side effects.
This review aims to collect and classify the potential inhibitors developed from 2015 to the present against the most studied bacterial zinc metalloenzymes (LpxC, PLN and TLN), correlated to Gram-negative bacteria. In particular, the structural features of ligands essential for achieving inhibitory activity and their structure-activity relationships will be described. All reported compounds will be classified based on their chemical structure and ZBG.
2.1. LpxC inhibitors
2.1.1. Hydroxamate-Based LpxC inhibitors
CHIR-090 and Its Analogues
The development of LpxC inhibitors took place following the discovery of deacetylase inhibitor L-573,655 (3, Figure 6a) at Merck in 1996. Then, different pharmaceutical companies such as Chiron, Astra-Zeneca, and Pfizer accepted the challenge to develop inhibitors against the deacetylase enzyme [60].
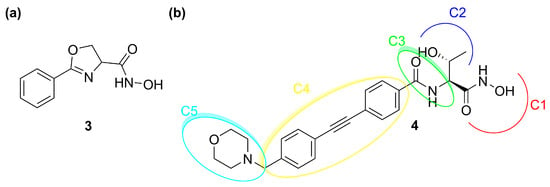
Figure 6.
(a) Structure of 3 (L-573,655). (b) Structure of 4 (CHIR-090).
So far, none of the developed compounds have reached the market and the most studied inhibitor of LpxC is CHIR-090 (4), which showed excellent in vitro potency against several Gram-negative pathogens. The analysis of its chemical structure allowed researchers to identify a pharmacophore with five necessary sites for the enzyme–inhibitor interaction (Figure 6b) [61].
C1 moiety is the ZBG which coordinates the Zn2+ in the catalytic site. C2 is the site for several substituents and C3 is a linker between the “head” of the inhibitor and the lipophilic “tail” (C4). The latter interacts with the hydrophobic tunnel of the enzyme. The last site is C5, which interacts with a solvent-exposed pocket and can influence the ability of the molecule to cross the bacterial membrane.
Moreover, the efficacy of 4 was tested alone and in co-administration with colistin against P. aeruginosa biofilm. Colistin (polymyxin E) is a natural antibiotic which interacts with the LPS, causing increasing penetration of substances from the outer membrane of the Gram-negative bacteria. The combination of colistin and 4 demonstrated a synergistic activity against both colistin-susceptible and colistin-non-susceptible P. aeruginosa biofilms. The mechanism of this synergism has not been explained yet [62].
Titecat et al. in 2016 evaluated the antibiotic activity of three deacetylase inhibitors (5, 6 and 7) against Gram-negative bacilli and investigated the effect of these inhibitors in combination with conventional antibiotics (Figure 7) [63].
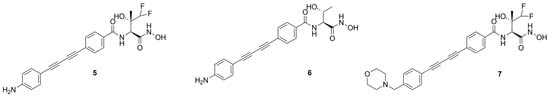
Figure 7.
Structures of 5, 6 and 7.
The best compound of this series and the first inhibitor to have activity against both A. baumannii and B. cepacian was identified as 5 [64]. Moreover, 5 was also active against MDR/XDR Enterobacteriaceae and P. aeruginosa strains and demonstrated a synergy with conventional antibiotics. The greater potency and wider spectrum of activity displayed by these compounds were due to the presence of the biphenyl–acetylene moiety which is supposed to interact within the hydrophobic tunnel of the enzyme. The difluoromethyl fragment might increase the ability to cross the lipid barrier and it might justify the activity of both 5 and 7 against A. baumannii. Furthermore, Gram-negative bacteria are known to be capable of excreting compounds containing a morpholine group through the efflux pump, which could explain the major activity of 5 with respect to morpholino-derivative 7.
In 2020, Surivet et al. [65] tried to identify novel methylsulfone hydroxamate LpxC inhibitors using three different cores (Figure 8a). The pyrrolo-imidazole series (core 2) displayed a good activity against E. coli in a murine infection model, especially compounds 8 and 9 (Figure 8b). Unfortunately, unattainable human doses of these compounds were necessary in order to exhibit their efficacy in vivo. Moreover, compounds 8 and 9 displayed solubility issues unfavorable to a future intravenous administration. Therefore, the terminus of the hydrophobic fragment of 8 was modified with polar substituents to improve its bioavailability by intravenous administration [66]. As expected, compounds 10 and 11 (Figure 8b), containing an amine and a diol group, respectively, showed high solubility and potency against Gram-negative bacteria in animal infection models (Table 2).
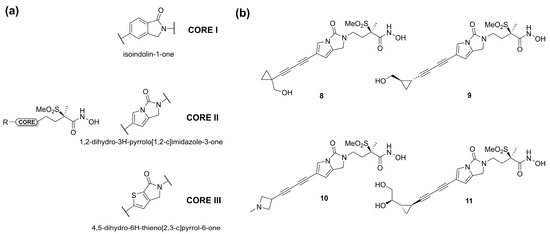
Figure 8.
(a) Core structures of methylsulfone hydroxamate LpxC inhibitors. (b) Chemical structure of Surivet inhibitors.

Table 2.
MIC values of Surivet LpxC inhibitors.
Ding et al. in 2018 synthesized several series of derivatives of compound 4 (Figure 9a) and tested their antibacterial activities against Escherichia coli (PA01, PA14) and P. aeruginosa (AB1157, DH5α) in vitro strains. Moreover, their metabolic stability was evaluated in liver microsomes. Due to its weak metabolism, the hydroxamic acid was replaced by acyl o-phenylenediamine and reversed hydroxamic acid. Unfortunately, these modifications turned out to be unsuccessful. Then, the introduction of a sulfonamide in the C3 site did not increase the antibacterial activity; instead, the metabolic stability in the human liver microsomes was improved with the replacement of the 2-hydroxy ethyl group by the 2-amino isopropyl group in the C2 site [61]. Moreover, in the C4 site the introduction of substituents on the benzene ring or its replacement with oxalidinone or pyridine ring decreased the inhibitory activity. Furthermore, the lengths of the hydrophobic scaffold and the hydrophilic terminus were investigated and the compounds bearing diphenylacetylene moiety in C4 and a methylsulfone group in C5 exhibited comparable activities to 4 against the classical Gram-negative pathogen strains (Figure 9a) [67].
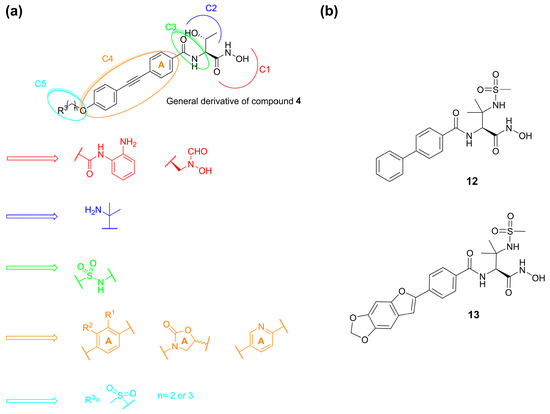
Figure 9.
(a) SAR developed by Ding et al. (b) Structures of compounds 12 and 13.
Another study by Kawai et al. identified a sulfonamide-based hydroxamic acid 12 (Figure 9b) that was more active on E. coli and P. aeruginosa than on K. pneumonia. The docking of 12 in E. coli LpxC showed the chelation of the Zn2+ by the hydroxamic acid. Furthermore, the oxygen atom of the amide made dual contacts with the backbone NH and the thiol on side chain of Cys63. Instead, the NH of the amide interacted with the hydroxyl group of Thr191 and the backbone carbonyl of Phe192. Finally, the biphenyl moiety was inserted in the hydrophobic tunnel (Figure 10). In order to improve the potency and the antibacterial spectrum of 12, the biphenyl group was replaced by different longer non-alkyne hydrophobic moieties and the analogue compound, 13, bearing a tricyclic fragment displayed the best activity against E. coli, K. pneumoniae and P. aeruginosa (Table 3) [68].
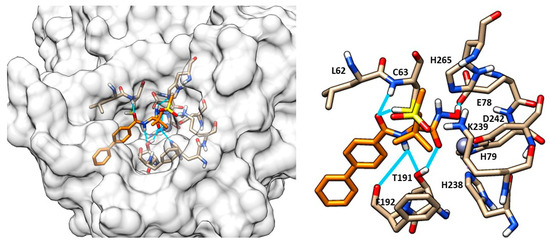
Figure 10.
Docking of compound 12 (orange) into LpxC (PDB ID: 3P3G). Image generated with Chimera, version 1.16.

Table 3.
Inhibitory activity of compounds 4, 12 and 13.
At Achaogen Inc., different LpxC inhibitors with a bis-alkyne scaffold against multidrug-resistant (MDR) Pseudomonas aeruginosa strains were studied. This bacterium is the most common cause of chronic infections in patients with cystic fibrosis (CF). The newly developed compounds (14–17, Figure 11) were tested in healthy volunteers to evaluate their safety, tolerability, and pharmacokinetics (PK).
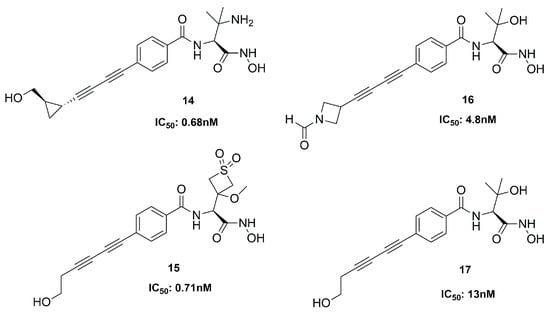
Figure 11.
Inhibitors developed at Achaogen.
Compound 14 was the first inhibitor to be evaluated in Phase I clinical trials, but its therapeutic window was insufficient. Then, three analogues were analyzed, and they showed a wider therapeutic window than 14. All of them were potent inhibitors of the deacetylase; in particular, 15 was the best, due to its favorable activity, safety and PK profile [69]. Unfortunately, since all these compounds displayed cardiovascular toxicity in preclinical models at any efficacious dose, the study could not be further developed [70].
The bicyclic derivative 19 (Figure 12) was developed by Zhang et al., starting from the promising threonyl compound 18, which showed an IC50 < 100 nM towards different Gram-negative pathogens. Compound 19 kept the broad-spectrum activity against Gram-negative bacteria, showing a good PK profile in three different animal species (rat, mice and monkey, IV and SC). However, despite this positive profile, 19 was not a good candidate for further clinical trials due to the observed acute tolerability problem associated with the inhibition of sodium channels [71].
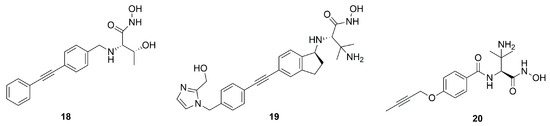
Figure 12.
Structures of compounds 18, 19 and 20.
LpxC inhibitors with a short hydrophobic group as a tail were synthesized by Novartis researchers. These new compounds should have a low PPB (plasma protein binding) and high solubility, with significant activity and low cytotoxicity. Moreover, the Novartis inhibitors should be selective against the P. aeruginosa LpxC enzyme in order to fight its infections and at the same time prevent the antibacterial cross-resistance and the unintentional disturbance of the human microbiome usually caused by broad-spectrum antibiotics. Indeed, the lack of the di-acetylene moiety together with its replacement with a short propargyl group led to derivative 20 (Figure 12) endowed with enhanced specific inhibition of P. aeruginosa LpxC. In particular, the specificity was achieved thanks to the “gatekeeper” helix and neighboring residues (aa 192−205) of the hydrophobic tunnel region, which adapted the channel to different inhibitor “tails”. Inhibitor 20, the best of the series (IC50 = 1.5 nM), showed a good efficacy in vitro and excellent results in in vivo studies too. These results would warrant further in vivo investigations to fully understand PK/PD parameters and subsequent human dose predictions [60].
Oxazolidinones Derivatives
Kurasaki et al. in 2016, starting from 4 (Figure 6b) and 21 (Figure 13), developed a series of hydroxamate-based compounds endowed with a good activity against E. coli and K. pneumoniae LpxC enzymes. Based on the crystal structure of 21 in the binding site of E. coli LpxC, the authors showed that the carbonyl group of the benzamide moiety lay in the same plane of the threonyl fragment. Thus, they combined the threonyl group and the benzamide moiety of 4 into an oxazolidinone cycle while the S-configuration of the ZBG was kept. In order to increase the affinity with the hydrophobic tunnel of the enzyme, the biphenyl acetylene moiety was replaced with a diene fragment. Moreover, the effect of substituents on the central phenyl ring led to a decrease in the activity against Pseudomonas aeruginosa and Acinetobacter baumannii [72]. Based on these considerations, the best compound of the series was the inhibitor 22 (IC50 = 6 nM, Figure 13), which showed a nanomolar activity against E. coli and K. pneumoniae and a low efflux ratio [73].
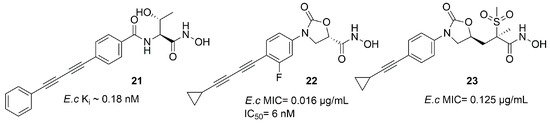
Figure 13.
Compound 21, 22 and 23 structures (E.c: Escherichia coli).
Moreover, compounds bearing an oxazolidinone or isoxazoline fragment as a linker between the ZBG and the hydrophobic moieties were selected by computational approach. Thus, two series of compounds were synthesized and tested against Lpxc enzymes from E. coli, P. aeruginosa and K. pneumoniae and both of them displayed similar inhibitory activity. However, the goal was reached by methylsulfone 23, which contained the oxazolidinone moiety (Figure 13) and proved to have a good activity against all the three bacteria. Furthermore, the activity against K. pneumoniae could be improved by a combination with Gram-positive antibiotics such as rifampicin and vancomycin, both in in vitro and in vivo experiments [74].
C-Furanose Derivatives as LpxC Inhibitors
To this class of LpxC inhibitors belong compounds with a furanose ring or an acyclic analogue moiety which bear different long hydrophobic fragments in 5 position (R3) in order to interact with the hydrophobic tunnel of the enzyme. The 1 position is occupied by the ZBG, which is usually a hydroxamic acid. The 3 and 4 positions present hydrophilic moieties (Figure 14). Moreover, the importance of each chiral carbon was investigated by different research groups.
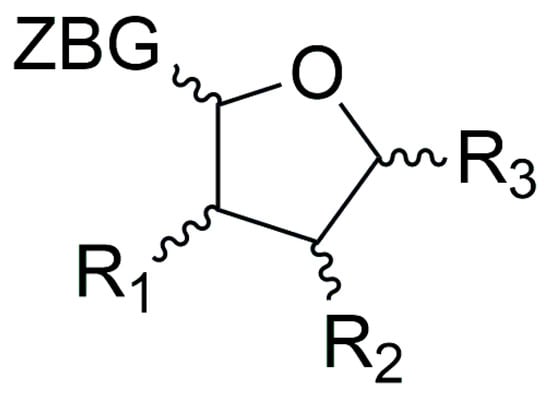
Figure 14.
General structure of furanoside inhibitors.
In 2015, Jana et al. synthesized a series of C-ethynyl furanoside LpxC inhibitors based on compound 4 structure. The 1-hydroxyethyl fragment of 4 was incorporated in the sugar moiety and the amide group was replaced by an ether group. Starting from compound 24 (Figure 15) (IC50 = 30.5 ± 4.2 µM, Ki = 4.2 ± 0.6 µM), the lipophilic side chain length and its correct orientation were analyzed focusing on the anomeric center of the sugar core. Compound 25 (Figure 15) was the only one active against E. coli (D22) LpxC with a Ki = 14.7 µM and IC50 = 107 ± 12 µM. Its β-anomer 26 (Figure 15) (IC50 > 200 µM) did not show any inhibitory activity (Figure 15). Thus, the α configuration proved to be necessary for efficacy [75].
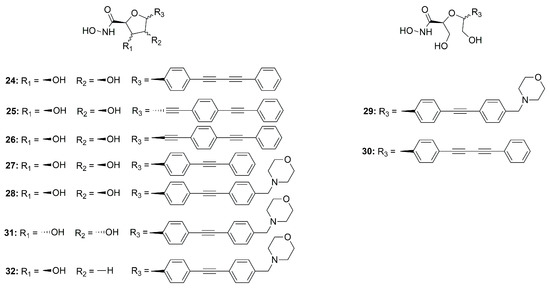
Figure 15.
Sugar-derivative inhibitors 24–32.
Müller et al. developed a SAR study on C-furanoside LpxC inhibitors, synthesizing and analyzing different series of compounds among which are derivatives 24, 27 and 28 (Figure 15 and Figure 16). It is known that compounds with a diphenylacetylene group as a lipophilic side chain have shown the worst inhibitory activity due to their shorter hydrophobic fragments that cannot occupy the entire hydrophobic tunnel of the enzyme. Morpholinomethyl-substituted diphenylacetylene derivatives displayed the highest antibacterial activities evaluated through the disc diffusion test, although their inhibitory activity was slightly weaker than that of the respective diphenyldiacetylene analogues. Furthermore, the data suggested that the 2,5-trans-configured C-glycosides (A series were more efficient than their respective cis-stereoisomers (C series). In particular, the best configuration was 2R, 5S. An improvement of the activity was pointed out by methylation of the hydroxyl group in 4 position and formation of the isopropylidene acetal (B series). However, the series of the acyclic diols displayed better biological activities than the respective C-glycosides, although they were not rigid compounds (B and C series). The open-chain (2S,5S)-configured compounds 29 and 30 (Figure 15) were the most potent LpxC inhibitors of these series of inhibitors (Table 4). Moreover, an improvement of the inhibitory activity was observed with the removal of the hydroxyl group in 3 position too (E series) [76].
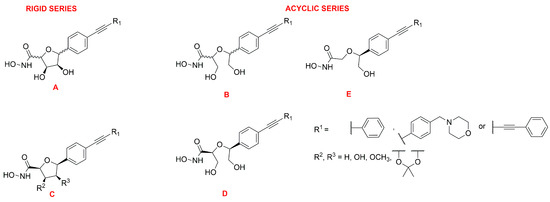
Figure 16.
General representation of cyclic (A and C series) and acyclic (B, D and E series) sugar-based inhibitors.

Table 4.
Inhibition activity of compounds 28–32.
The C-glycosides compounds were slightly weaker than the acyclic analogues because a hydroxyl group was located in a non-polar region of the binding site. Hence, the structure of compound 28 (Figure 15) was optimized by inverting the configuration in positions 3 and 4 of the rigid system. The new stereoisomer 31 (Figure 15) exhibited the best inhibitory activity against E. coli BL21 (DE3) and the D22 strain [39]. This improvement was due to the orientation of the hydroxyl group towards the entrance of the catalytic site of the enzyme, where it formed water-mediated hydrogen bonds with surrounding amino acids. The stereoisomer 31 showed the same activity of the respective (S, S)-acyclic compound 29 (Figure 15), suggesting that modification in positions 3 and 4 will be necessary in order to improve the activity (Table 4).
Dreger et al. have investigated the capacity of C-glycosides inhibitors to reach their cytoplasmatic target by crossing the bacterial cell wall. The study revealed that further optimization would be necessary to improve the permeability of the sugar-core inhibitors [39].
Later, monohydroxy-tetrahydrofuran analogues of the rigid series were analyzed by the same group. The removal of the hydroxyl group in 3 position did not increase the inhibitory activity, whereas the elimination of the hydroxyl group in position 4 led to an improvement of the efficacy. Compound 32 (Figure 15), with a (2S, 3R, 5R) configuration, exhibited the highest antibacterial activity against E. coli BL21 (DE3) and the D22 strain (Table 4) [77].
The acyclic series E was investigated by Tangherlini et al. in 2016, starting from compound 29 (Figure 15). The researchers demonstrated that the shift of the hydroxylmethyl group from the α-hydroxamic acid position (37–40, Figure 17) to the benzylic fragment (33–36, Figure 17) slightly increased the efficacy against the E. coli LpxC enzyme, whereas configuration in α-position to the hydroxamic acid did not have a relevant influence. Indeed, the Ki values of 37 and 38 were similar to those of their respective enantiomers 39 and 40. (Figure 17, Table 5) [78].
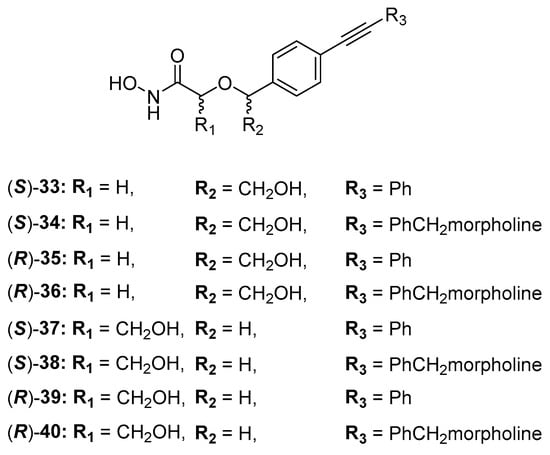
Figure 17.
Structures of acyclic derivatives of series E.

Table 5.
Inhibition activity of acyclic series E.
Further optimization of compound 33 was carried out by replacing its hydroxyl group with a triazole ring in order to keep the possibility of forming hydrogen bonds in the catalytic site of the enzyme. Moreover, the triazole ring was used as a linker to connect other substituents which could interact with the unoccupied region of the UDP-binding site. Compounds 41 (IC50 = 10.4 ± 9.7 µM) and 42 (IC50 = 8.5 ± 1.3 µM) (Figure 18) bearing a phenyl ring and a hydroxymethyl group, respectively, in position 4 of the triazole, and inhibitor 43 (IC50 = 23.4 ± 5.8 µM) with a hydroxymethyl group in position 5 of the triazole, were the best of the series, showing the biggest halos of inhibition in the disc diffusion assays [79].
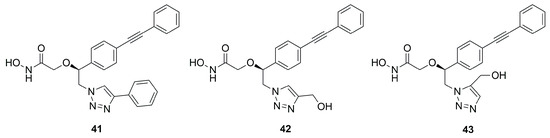
Figure 18.
Inhibitors bearing a triazole ring as a linker.
In particular, docking studies revealed that the increased activity should be attributed either to the aromatic interactions between the tetrazole ring and Phe192 or the interactions between the tetrazole ring substituent with Lys239. However, these compounds displayed a lower activity with respect to the most studied LpxC inhibitor 4.
In a following study, the hydroxamic acid of compound 33 (Figure 17) was replaced by different ZBGs due to its unfavorable pharmacokinetic properties. However, all new derivatives displayed a loss of LpxC inhibitory and antibacterial activity [80].
2.1.2. Non-Hydroxamate LpxC Inhibitors
The discovery of inhibitors without a hydroxamate group as ZBG represents an interesting approach in order to avoid side effects due to the lack of specificity previously seen in many matrix metalloproteinase (MMP) inhibitors [38]. Moreover, the hydroxamate group is typical of HDAC inhibitors [81] but also represents the main cause of their mutagenicity [82]. The strategies to avoid the use of hydroxamic acids are fundamentally three: use of alternative ZBGs, binding to exosites [52,83] and exploiting an excellent fitting with the S1′, S1 and S2 pockets of the catalytic site.
Furuya et al. in 2020 synthesized a series of LpxC inhibitors bearing a N-hydroxyformamide (NHF) as ZBG. Based on the structure of compound 4, they developed different derivatives with a benzotriazole as a core and several substituents in α-position to the NHF warhead. The best compound of the series was inhibitor 44 (Figure 19), which showed hypotensive side effects in a rat study despite its good data in in vitro studies (IC50 < 1 nM against P. aeruginosa and IC50 < 10 nM against E. coli) [84].
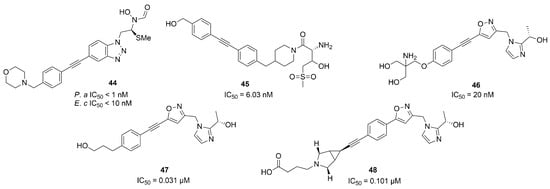
Figure 19.
Non-Hydroxamate LpxC inhibitors (P.a: Pseudomonas aeruginosa, E.c: Escherichia coli).
Two non-hydroxamate compounds selective for LpxC (45 and 46, Figure 19) were identified through fragment-based discovery methods. These compounds belong, respectively, to the glycine series and to the imidazole series, fragments that are able to interact with the zinc ion in the catalytic site. Compound 46 showed a promising activity (IC50 = 20 nM), and further optimization followed by in vivo studies is ongoing [85].
The imidazole core was also used as starting point by Ushiyama et al. to synthesize a series of inhibitors lacking the hydroxamic acid (HA) moiety. The authors optimized the pharmacokinetic parameters of compound 47 (Figure 19), keeping or improving its antibacterial activity. The study led to the synthesis of compound 48 (Figure 19) and a series of compounds bearing a primary amine in a position adjacent to the imidazole ring. Compound 48 presented inhibitory activity against K. pneumonie, including resistant strains, and the onset of cardiovascular diseases decreased with 48 with respect to those compounds bearing a primary amine in a position adjacent to the imidazole [86]. The latter enhanced the antibacterial activity of several antibiotics against Enterobacteriaceae, increasing the bacterial membrane permeability. This synergism could be useful in the treatment of carbapenem-resistant Enterobacteriaceae (CRE) infections [87]. Furthermore, 48 benefited patients with sepsis by acting on the release of lipopolysaccharides (LPS). In any case, further studies will be necessary in order to explain the effect due to the inhibition of LpxC on immune disorders and excessive inflammatory responses [88].
Biological results for the main LpxC inhibitors are summarized in Table 6.

Table 6.
Inhibitory activity (IC50 nM) of the main LpxC inhibitors.
2.2. Inhibitors of Pseudolysin and Thermolysin
Inhibitors of pseudolysin also always presented activity on thermolysin due to their high structural homology. For this reason, thermolysin and pseudolysin inhibitors are described in the same section.
2.2.1. Dipicolylamine (DPA) and Tripicolylamine (TPA)-Based Inhibitors
Rahman et al. tested different known compounds as putative inhibitors both on bacterial enzymes (thermolysin (TLN), pseudolysin (PLN) and aureolysin (ALN)) and on human metalloproteinases (matrix metalloproteinase-9 (MMP-9) and matrix metalloproteinase-14 (MMP-14) (Table 7). The tested compounds were DPA (49, Figure 20) or TPA (50, Figure 20) derivatives already shown to inhibit irreversibly metallo β-lactamases (MBLs) interacting with the Zn2+. An initial screening revealed that all of the inhibitors showed a better inhibition of MMP-14 and PLN than of the other enzymes. Furthermore, 50 and 53 (Figure 20) presented strong binding to all enzymes. Compound 51 (Figure 20) and its methyl-ester 52 (Figure 20) presented the same activity against MMP-14 and PLN. Compound 51 was able to coordinate the zinc ion with the carboxylic acid, while 52 coordinated the Zn2+ through the nitrogen of the pyridine ring and the methyl-ester group was exposed to solvent. Compounds 53, 54 and 55 (Figure 20) bearing a hydrophilic methyl-amino glycosyl side chain linked to the 50 core through different amide bonds showed to be weaker than 50, except for PLN and MMP-14 inhibition. Moreover, Rahman et al. noticed that the substitution of one of the 2-methylpyridine groups of 50 led to a decreasing activity against PLN and MMP-14. Compounds of this series turned out to be reversible inhibitors, in contrast with what is observed for MBLs [37].

Table 7.
Ki ± s.d. (µM) values of TPA, DPA and their derivatives for MMP-14, MMP-9, ALN, PLN and TLN.
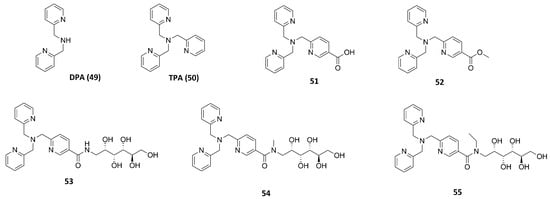
Figure 20.
TPA, DPA and their derivatives.
2.2.2. Inhibitors with Sulfur-Based ZBGs
Thiol Derivatives
Through a virtual screening study, Zhu et al. identified an aromatic core with a mercaptoacetamide side chain which was able to interact with PLN at non-toxic concentrations. Indeed, they synthesized several compounds and three of them displayed a good activity (56, 57 and 58) (Figure 21). The metalloenzyme selectivity was evaluated in addition to their in vitro activity. Thiol derivative 58 was the only compound inactive against MMP-2 and a Zn2+-dependent histone deacetylase (HDAC, encompassing class I and IIb) at a dose range up to 50 µM. The adsorption and metabolism in Caenorhabditis elegans were investigated and compounds 56, 57 and 58 with their respective acetamido prodrugs (59, 60 and 61) were examined. Interestingly, prodrug 61 was the only derivative of this series to be accumulated in the C. elegans, metabolized into its activated form (58) and then oxidized into the corresponding disulfide analogue. Therefore, prodrug 61 could be considered a good starting point for future studies [89].
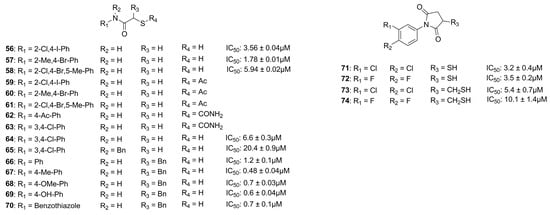
Figure 21.
Pseudolysin Inhibitors with sulfur-based ZBGs.
In 2018, Kany et al. discovered a thiocarbamate derivative (62, Figure 21) able to inhibit clostridial collagenase with high selectivity over MMPs. Compound 62 was able to release in vivo its thiol-activated form and to inhibit pseudolysin. Based on the structure of compound 62, a series of thiocarbamate derivatives has been developed. The best compound was the prodrug 63 (Figure 21), which liberated in vivo the active compound 64 (IC50 = 6.6 ± 0.3 µM) (Figure 21). The binding mode of compound 64 is reported in Figure 22. The thiol derivative 64 presented two different binding modes to the pseudolysin catalytic site due to the ambiguous electron density of the binding site. In the pink pose, the zinc ion was not coordinated by the thiol group. Instead, in the green pose the thiol group displaced the water to complete the tetrahedral coordination of the Zn2+. Furthermore, the green pose formed a bidentate hydrogen bond with Arg198 in the S1′ subsite and formed another hydrogen bond with the side chain of His223. Otherwise, the amide nitrogen of the pink pose was linked to the Asn112 by a hydrogen bond and its aromatic ring was placed into the S2′ pocket. Neither of the 64 poses led to the closure of the binding site, and for this reason a series of compounds bearing different groups on the amide nitrogen were synthesized, such as compound 65 (IC50 = 20.4 ± 0.9 µM) (Figure 21). Unfortunately, none of these derivatives showed improved activity with respect to 64.
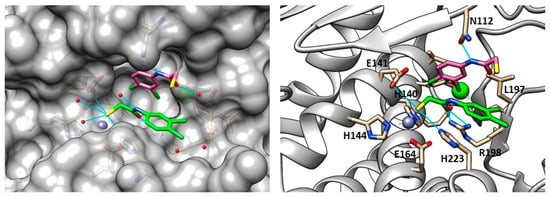
Figure 22.
Poses of compound 64 (PDB ID: 6F8B). The green pose chelated the zinc ion, the pink one is linked to a molecular water. Image generated with Chimera, version 1.16.
Furthermore, 64 with its prodrug 63 showed an efficacy against IMP-7, a metallo β-lactamase present in clinical isolates of P. aeruginosa, with an IC50 = 0.86 ± 0.06 µM for 64 and an IC50 = 1.16 ± 0.07 µM for 63, ensuring the future development of novel anti-infective compounds. Finally, the in vitro efficacy towards Galleria mellonella infection models was investigated, and the results suggested that the prodrug 63 did not sufficiently release the active thiol 64 [90].
Recently, Kaya et al. synthesized a series of α-benzyl N-heteroaryl mercaptoacetamide derivatives as pseudolysin inhibitors. The authors used a structure-based approach on compound 66 (Figure 21), which was previously developed in order to create the new series. Firstly, the binding mode of compound 66 (Figure 23) prompted the idea of increasing the inhibitory activity against pseudolysin by adding a hydrophobic group in the para position of the ring linked to the amide moiety. The introduction of the methyl group led to compound 67 (Figure 21 and Figure 23) with improved hydrophobic interactions with the S1′ pocket. Then, the methyl group of 67 was replaced by a –OMe (compound 68) and –OH (compound 69), leading to an improvement of the inhibitory activity.
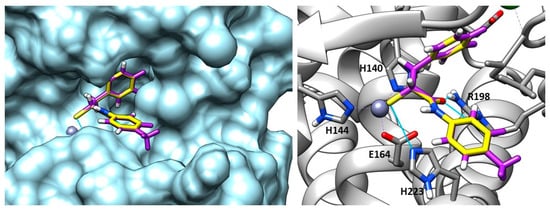
Figure 23.
Overlapping between compound 67 (purple) and compound 66 (yellow) into pseudolysin (PDB ID: 7OC7). Image generated with Chimera, version 1.16.
Furthermore, the same research group replaced the N-aryl group with several heterocycles in order to discover new interactions with the surrounding Arg and Asn residues or π–π interactions with the histidine residues. Unfortunately, only benzothiazole derivative 70 (Figure 21) showed a better activity compared to the parent compound 66, maintaining the selectivity profile over MMPs. The activity of 70 improved thanks to the additional π–π stacking interaction with the histidine residues. In conclusion, the replacement of the N-aryl ring with heterocycles did not lead to a significant improvement, so further optimization will be necessary.
Moreover, these inhibitors reduced the pathogenicity of P. aeruginosa without affecting the integrity of the lung and skin cells. In addition, 70 (Ki: 0.1 ± 0.01 µM) inhibited Clostridium histolyticum, which degrades the collagen fibers, causing several diseases. Further optimization will be necessary to improve the inhibitory efficacy and the physicochemical and pharmacokinetic profiles of thiol derivatives [91].
Mercaptosuccinimide Analogues
Konstantinovíc et al. developed a series of compounds bearing a mercaptosuccinimide group as ZBG in order to reduce the flexibility of thiols. These compounds were tested against both pseudolysin and Clostridium histolyticum collagenase (ColH). The mercaptosuccinimide analogues were obtained through a structure-based approach starting from compound 64 (Figure 21). The latter showed a nanomolar affinity to ColH in addition to its pseudolysin inhibitory activity. The best compounds, 71 and 72 (Figure 21), showed a better activity than 64, and compound 72 also displayed a lower toxicity profile than 64. According to these data, further modification and studies will be relevant in order to obtain good candidates for clinical trials [92]. Imai et al. also proposed two compounds (73 and 74, Figure 21) featuring a thiol group not directly linked to the succinimide ring endowed with a lower activity against pseudolysin than their analogues 71 and 72 [51].
2.2.3. Hydroxamic Acid-Based Inhibitors
Given the promising results obtained with thiol derivatives in terms of activity and selectivity, Kany et al. maintained the scaffold but replaced the thiol group of their compounds with an hydroxamic acid in order to avoid the oxidation of thiols and their inactivation as disulfide compounds. The introduction of a hydroxamic acid as ZBG led to a decrease of selectivity over MMPs, as expected. However, the group of researchers identified compound 75 (Figure 24), a derivative of 64 (Figure 21), which maintained both the activity against pseudolysin (IC50: 17.4 ± 0.8 µM and a Ki: 12.3 ± 0.6 µM) and the selectivity over MMPs. Compound 75 was oriented in the binding site in order to chelate the zinc ion with both the hydroxyl group and the carbonyl group of the hydroxamates moiety. Moreover, the oxygen atom of the hydroxamic acid formed a hydrogen bond with Glu141 and the amide nitrogen interacted with Ala113. The bidentate hydrogen bond between 75 and Arg198 was missing, and this explained the weaker activity of 75 with respect to 64. Furthermore, the selectivity was retained because 75 did not occupy the S1′ pocket like the classical inhibitors of MMPs. Hence, from these findings it can be deduced that sparing the S1′ pocket represents an element to achieve the selectivity over MMPs [93].
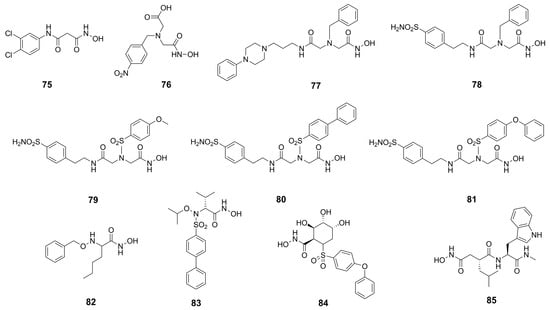
Figure 24.
Hydroxamic acid-based inhibitors.
Recently, several MMP inhibitors were tested against pseudolysin and thermolysin by Adekoya et al. In particular, three compounds (76, 77 and 78, Figure 24) displayed a higher affinity for the bacterial enzymes than for human matrix metalloproteinases, as shown in Table 8. These results were confirmed by docking studies where the aforementioned strong affinity was explained by the interaction of the ligands with the S1 and S2′ subpockets. In these regions, there are pivotal differences between the M4 enzymes and the MMPs, demonstrated by the three-dimensional alignment of the enzymes. Indeed, the conserved residue of tyrosine placed in an α-helix of M4 enzymes is located in a loop in the MMPs. Moreover, the Tyr114 (Phe114 for thermolysin) and Trp115 in the bacterial enzymes are replaced by a leucine in the MMPs. The side chain of this leucine as well as the side chain of the previously mentioned conserved tyrosine are pointed outward into the binding site of the MMPs, leading to a narrow access to the S1′ subpocket. This is specific of matrix metalloproteinases compared to M4 enzymes. Hence, these differences in the three-dimensional structure justified the low affinity of compounds 77 and 78 for MMPs. In sharp contrast, the interactions within the S1′ subpocket of compounds 79–81 (Figure 24, Table 8) into MMPs contributed to the high affinity to them. In general, these compounds bound better MMPs than bacterial enzymes, and between thermolysin and pseudolysin, the inhibitors presented higher affinity for pseudolysin [18]. These considerations were confirmed later by the same group when they compared hydroxamic acid compounds belonging to different chemical classes.

Table 8.
IC50 (µM) values of hydroxamic acid derivatives 76–83.
The best compound of the series was the N-benzyloxy-amino acid hydroxamate 82 (Figure 24, Table 8) which showed high activity against pseudolysin and thermolysin compared to MMPs. In particular, 82 displayed better activity against pseudolysin than against thermolysin. In pseudolysin, 82 chelated the Zn2+ with the hydroxamic acid group and its benzyl ring was placed within the S2′ pocket while its n-butane chain was directed towards the S1′ subsite. The hydrogen of the hydroxyl group participated in a hydrogen bond with Glu141, and the nitrogen atom of the hydroxamate moiety also formed a hydrogen bond with the backbone of Ala113 (Figure 25).
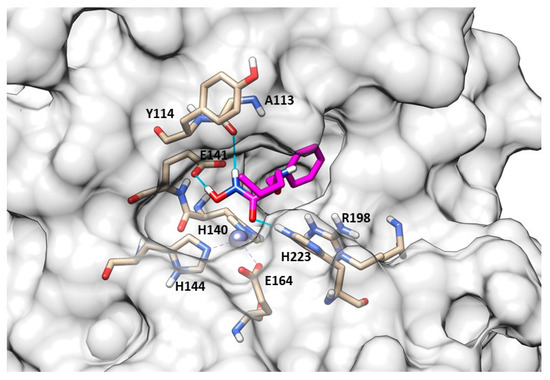
Figure 25.
82 (pink) binding-mode into pseudolysin (PDB ID: 1U4G). Image generated with Chimera, version 1.16.
Instead, the binding study did not predict a unique pose for 82 into thermolysin. Compound 82 had two possible poses to inhibit TLN. In one pose, the ring system was in the S2′ pocket (Figure 26b); in the other, the ring system was placed within the S1 subpocket (Figure 26a). However, in both of these poses the zinc ion was coordinated by the hydroxamates through a bidentate chelation. When the ring system was in the S1 subpocket, 82 showed interactions with Phe114 and Asp116 and through its carbonyl group had hydrogen bonds with the Arg203. Additionally, the NH-group interacted with Glu143. Moreover, 82 retained the selectivity towards M4 because it did not fit so deeply into the S1′ pocket or into the S2′ subsite of the MMPs, and the hydroxyl group of the hydroxamic acid also did not form a hydrogen bond with the enzyme (Figure 27). In sharp contrast, sulfonamide derivative 83 showed good IC50s towards the endogenous MMPs. These results were validated by docking studies where the hydroxamic acid of 83 chelated the zinc ion and the long ring system was placed deeply in the S1′ pocket (Figure 27).
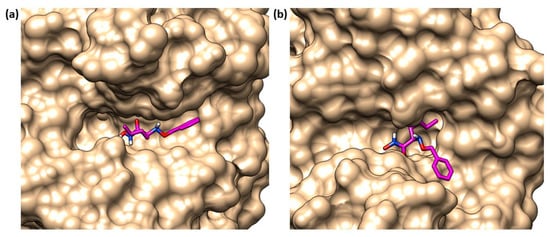
Figure 26.
The two possible poses of 82 (pink) in TLN binding site. (a) 82 (pink) with its ring in S1 pocket of thermolysin (PDB ID: 1PE5). (b) 82 (pink) with its ring in S2′ pocket of thermolysin (PDB ID: 1PE5). Image generated with Chimera, version 1.16.
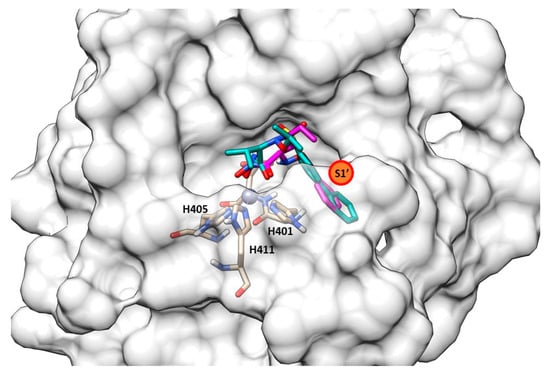
Figure 27.
Superposition between 82 (pink) and 83 (light sea-green) into MMP-9 (PDB ID: 1GKC). Image generated with Chimera, version 1.16.
Thus, it can be deduced that the inhibitory activity against bacterial proteases increases when the ligand has groups that interact strongly within the S2′ subpocket of M4 enzymes [34]. Compounds 84 and 85 (Figure 24, Table 8) were also tested, showing that the size and shape of the inhibitor fragment binding within the S1′ subpocket was a key factor for selectivity. Azasugar sulfonamide 84 was not able to bind the bacterial enzymes due to its big aromatic system, while the known broad-spectrum MMP inhibitor galardin, 85, inhibited the bacterial enzymes (PLN and TLN) in the nanomolar range (Ki: 20 nM) [94].
2.2.4. Bisphosphonate-, Catechol- and Carboxylate-Based Inhibitors
With the aim of inhibiting bacterial metalloproteases, compounds with different ZBGs were tested. Rahman et al. screened bisphosphonate- and catechol-based inhibitors, previously synthesized to inhibit MMPs, against MMPs (MMP-9 and MMP-14), thermolysin (TLN), pseudolysin (PLN) and aureolysin (ALN). The catechol structures, 86–89 (Figure 28, Table 9), did not show strong activity against the bacterial proteases or a good selectivity between bacterial and human metalloproteinases. Instead, the bisphosphonate derivatives (90–93, Figure 28, Table 9) displayed a high affinity for pseudolysin and thermolysin compared to the catechol analogues. This inhibitory activity was achieved by the interaction between one of the bisphosphonate groups and the arginine residue (Arg198 for PLN, Arg203 for TLN) located at the entrance of the S1 subpocket [95].
Figure 28.
Bisphosphonate-, catechol-based inhibitors.

Table 9.
Ki values of catechol and bisphosphonate compounds.
Leiris et al., through a virtual screening and computer-assisted drug design, identified two carboxylic acids (94 and 95, Figure 29a) that were able to inhibit the P. aeruginosa elastase. These indane carboxylic acids, 94 and 95, inhibited pseudolysin with a Ki of 0.16 µM and 0.12 µM, respectively. Compound 94 formed a strong interaction both with Arg198 and Asn112 through a bidentate hydrogen bond. Additionally, the benzothiazole was placed into a cleft closed to the S2′ pocket by π–π interactions with Phe129. Furthermore, carboxylic acid 94 extended its interaction to the base of the S2′ pocket mediated by a binding with a water molecule (Figure 29b) [96]. Further investigations will be necessary for delivering an innovative antibacterial drug.
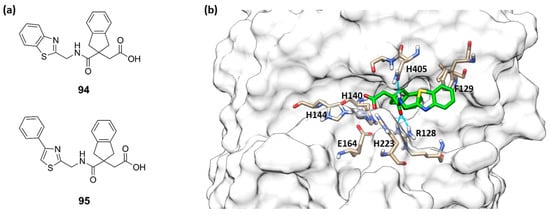
Figure 29.
(a) Structures of 94 and 95. (b) Compound 94 (green) within the catalytic site of pseudolysin (PDB ID: 7AJR). Image generated with Chimera, version 1.16.
Biological results for the main PLN and TLN inhibitors are summarized in Table 10.

Table 10.
Inhibitory activity (IC50 µM) of the main zinc metalloproteinase inhibitors against PLN, TLN, MMP-2 and MMP-9.
3. Conclusions
Nowadays, infections due to Gram-negative bacteria are emphasized by the increasing multidrug-resistance phenomenon. Inhibition of bacterial metalloenzymes LpxC, pseudolysin and thermolysin may represent an alternative strategy to combat the development of infections caused by Gram-negative bacteria, avoiding the resistance mechanism. The inhibition of LpxC interferes with the bacterial outer membrane permeability, enhancing drug absorption, whereas the inhibition of pseudolysin and thermolysin affects the bacterial nutrition and sporulation.
In this review, we presented all the synthetic metalloenzymes inhibitors against LpxC, PLN and TLN reported in literature from 2015 to the present. These inhibitors have been classified based on their target protein and according to their chemical structure.
LpxC inhibitors have been divided into hydroxamic acid-based (HA) inhibitors and non-hydroxamate inhibitors. Among HA, compound 4 was the hit compound which inspired another series of inhibitors.
The hydroxamic acid 14 with a di-acetylene tail (IC50 = 0.68 nM) was the first inhibitor to reach Phase I of clinical trials. Moreover, among the series developed by Novartis, compound 20 (IC50 = 1.5 nM), bearing a propargyl group in its tail, displayed good results both in vitro and in vivo, and further studies are ongoing.
Several compounds belonging to the C-furanose class have also shown good activity. The best compound of this series was 42 (IC50 = 8.5 ± 1.3 µM), and its higher activity was due to the interaction between the triazole ring and Lys239 and Phe192 residues. However, more studies will be required to analyze the safety of these promising compounds. Furthermore, among compounds without hydroxamic acid as ZBG, the best compound 48 (IC50 = 0.101 µM) showed several benefits in patients with cardiovascular diseases or in sepsis condition. Biological results for the main LpxC inhibitors have finally been summarized in Table 6.
Inhibitors against PLN and TLN have been classified based on their ZBG. Among the thiol derivatives, the pro-drug 61 was shown to be a promising candidate due to its capacity to release compound 58 (IC50 = 5.94 ± 0.02 µM) active against C. elegans. Moreover, the α-benzyl N-heteroaryl mercaptoacetamide compound 67 (IC50 = 0.48 ± 0.04 µM) showed a good activity against PLN due to the interaction of the ring linked to the amide moiety within the S1′ subpocket.
The best results were reached by those inhibitors bearing the HA as ZBG. Several MMP inhibitors, previously synthesized, were tested against M4 enzymes. Among these, compound 82 showed higher affinity against PLN (IC50 = 1.9 µM) and TLN (IC50 = 9.5 µM) than against MMPs. This selectivity was achieved because 82 did not fit so deeply into the S1′ pocket or into S2′ subsite of the MMPs. Biological results for the main PLN and TLN inhibitors have been summarized in Table 10.
Based on the results shown, LpxC inhibitors hold promising potential as new antibacterial agents. Future research should aim to optimize the properties of LpxC inhibitors to ensure good bioavailability and pharmacokinetics. Otherwise, research involving PLN and TLN inhibitors is less developed, and further studies are required to improve both enzymatic efficacy and pharmacokinetic properties of these agents.
Author Contributions
Conceptualization, E.N. and D.C.; writing—original draft preparation, R.D.L.; writing—review and editing, E.N. and D.C.; supervision, A.R.; funding acquisition, A.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Northern Norway Health Authorities (Helse Nord) (grant number HNF 1514-20).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rodríguez-Alvarado, M.; Russo, R.; Connell, N.D.; Brenner-Moyer, S.E. Design, Organocatalytic Synthesis, and Bioactivity Evaluation of Enantiopure Fluorinated LpxC Inhibitors. Org. Biomol. Chem. 2020, 18, 5867–5878. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Sartelli, M.; McKimm, J.; Abu Bakar, M.B. Health Care-Associated Infections – an Overview. Infect. Drug Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef]
- Khan, H.A.; Baig, F.K.; Mehboob, R. Nosocomial Infections: Epidemiology, Prevention, Control and Surveillance. Asian Pac. J. Trop. Biomed. 2017, 7, 478–482. [Google Scholar] [CrossRef]
- Holmes, C.L.; Anderson, M.T.; Mobley, H.L.T.; Bachman, M.A. Pathogenesis of Gram-Negative Bacteremia. Clin. Microbiol. Rev. 2021, 34, e00234-20. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Lancet Infect. Dis. 2010, 10, 303–304. [Google Scholar] [CrossRef]
- Seifert, H. The Clinical Importance of Microbiological Findings in the Diagnosis and Management of Bloodstream Infections. Clin. Infect. Dis. 2009, 48, S238–S245. [Google Scholar] [CrossRef]
- Cerceo, E.; Deitelzweig, S.B.; Sherman, B.M.; Amin, A.N. Multidrug-Resistant Gram-Negative Bacterial Infections in the Hospital Setting: Overview, Implications for Clinical Practice, and Emerging Treatment Options. Microb. Drug Resist. 2016, 22, 412–431. [Google Scholar] [CrossRef]
- Iacopetta, D.; Rosano, C.; Sirignano, M.; Mariconda, A.; Ceramella, J.; Ponassi, M.; Saturnino, C.; Sinicropi, M.S.; Longo, P. Is the Way to Fight Cancer Paved with Gold? Metal-Based Carbene Complexes with Multiple and Fascinating Biological Features. Pharmaceuticals 2020, 13, 91. [Google Scholar] [CrossRef]
- Nikaido, H. Multidrug Resistance in Bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef]
- Tanwar, J.; Das, S.; Fatima, Z.; Hameed, S. Multidrug Resistance: An Emerging Crisis. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 541340. [Google Scholar] [CrossRef]
- Loeffler, J.; Stevens, D.A. Antifungal Drug Resistance. Clin. Infect. Dis. 2003, 36, S31–S41. [Google Scholar] [CrossRef]
- Lee, C.-R.; Cho, I.H.; Jeong, B.C.; Lee, S.H. Strategies to Minimize Antibiotic Resistance. Int. J. Environ. Res. Public Health 2013, 10, 4274–4305. [Google Scholar] [CrossRef]
- Yang, X.; Ye, W.; Qi, Y.; Ying, Y.; Xia, Z. Overcoming Multidrug Resistance in Bacteria Through Antibiotics Delivery in Surface-Engineered Nano-Cargos: Recent Developments for Future Nano-Antibiotics. Front. Bioeng. Biotechnol. 2021, 9, 696514. [Google Scholar] [CrossRef]
- Malouin, F.; Bryan, L.E. Modification of Penicillin-Binding Proteins as Mechanisms of Beta-Lactam Resistance. Antimicrob. Agents Chemother. 1986, 30, 1–5. [Google Scholar] [CrossRef]
- Bockstael, K.; Aerschot, A. Antimicrobial Resistance in Bacteria. Open Med. 2009, 4, 141–155. [Google Scholar] [CrossRef]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Scumaci, D.; Giuzio, F.; Saturnino, C.; Aquaro, S.; Rosano, C.; Sinicropi, M.S. Multidrug Resistance (MDR): A Widespread Phenomenon in Pharmacological Therapies. Molecules 2022, 27, 616. [Google Scholar] [CrossRef]
- Kalinin, D.V.; Holl, R. LpxC Inhibitors: A Patent Review (2010–2016). Expert Opin. Ther. Pat. 2017, 27, 1227–1250. [Google Scholar] [CrossRef]
- Adekoya, O.A.; Sjøli, S.; Wuxiuer, Y.; Bilto, I.; Marques, S.M.; Santos, M.A.; Nuti, E.; Cercignani, G.; Rossello, A.; Winberg, J.-O.; et al. Inhibition of Pseudolysin and Thermolysin by Hydroxamate-Based MMP Inhibitors. Eur. J. Med. Chem. 2015, 89, 340–348. [Google Scholar] [CrossRef]
- Supuran, C.T.; Capasso, C. An Overview of the Bacterial Carbonic Anhydrases. Metabolites 2017, 7, 56. [Google Scholar] [CrossRef]
- Nocentini, A.; Cuffaro, D.; Ciccone, L.; Orlandini, E.; Nencetti, S.; Nuti, E.; Rossello, A.; Supuran, C.T. Activation of Carbonic Anhydrases from Human Brain by Amino Alcohol Oxime Ethers: Towards Human Carbonic Anhydrase VII Selective Activators. J. Enzym. Inhib. Med. Chem. 2021, 36, 48–57. [Google Scholar] [CrossRef]
- Yedery, R.D.; Jerse, A.E. Augmentation of Cationic Antimicrobial Peptide Production with Histone Deacetylase Inhibitors as a Novel Epigenetic Therapy for Bacterial Infections. Antibiotics 2015, 4, 44–61. [Google Scholar] [CrossRef] [PubMed]
- Konovalova, A.; Kahne, D.E.; Silhavy, T.J. Outer Membrane Biogenesis. Annu. Rev. Microbiol. 2017, 71, 539–556. [Google Scholar] [CrossRef] [PubMed]
- Clayton, G.M.; Klein, D.J.; Rickert, K.W.; Patel, S.B.; Kornienko, M.; Zugay-Murphy, J.; Reid, J.C.; Tummala, S.; Sharma, S.; Singh, S.B.; et al. Structure of the Bacterial Deacetylase LpxC Bound to the Nucleotide Reaction Product Reveals Mechanisms of Oxyanion Stabilization and Proton Transfer. J. Biol. Chem. 2013, 288, 34073–34080. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Garrett, T.A.; Reynolds, C.M.; Shaw, W.A.; Moore, J.D.; Smith, D.C.; Ribeiro, A.A.; Murphy, R.C.; Ulevitch, R.J.; Fearns, C.; et al. Kdo2-Lipid A of Escherichia Coli, a Defined Endotoxin That Activates Macrophages via TLR-4. J. Lipid Res. 2006, 47, 1097–1111. [Google Scholar] [CrossRef] [PubMed]
- Bürger, M.; Chory, J. Structural and Chemical Biology of Deacetylases for Carbohydrates, Proteins, Small Molecules and Histones. Commun. Biol. 2018, 1, 217. [Google Scholar] [CrossRef]
- Miller, S.I.; Ernst, R.K.; Bader, M.W. LPS, TLR4 and Infectious Disease Diversity. Nat. Rev. Microbiol. 2005, 3, 36–46. [Google Scholar] [CrossRef]
- Zhou, P.; Barb, A.W. Mechanism and Inhibition of LpxC: An Essential Zinc-Dependent Deacetylase of Bacterial Lipid A Synthesis. Curr. Pharm. Biotechnol. 2008, 9, 9–15. [Google Scholar] [CrossRef]
- Abdel-Magid, A.F. LpxC Inhibitors as Effective Therapy Against Multidrug Resistant Bacterial Infections. ACS Med. Chem. Lett. 2015, 6, 1095–1096. [Google Scholar] [CrossRef]
- Barrett, A.J. Proteases. Curr. Protoc. Protein Sci. 2000, 21, 21.1.1–21.1.12. [Google Scholar] [CrossRef]
- Cerdà-Costa, N.; Xavier Gomis-Rüth, F. Architecture and Function of Metallopeptidase Catalytic Domains: Metallopeptidase Catalytic Domains. Protein Sci. 2014, 23, 123–144. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS Database of Proteolytic Enzymes, Their Substrates and Inhibitors in 2017 and a Comparison with Peptidases in the PANTHER Database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Adekoya, O.A.; Sylte, I. The Thermolysin Family (M4) of Enzymes: Therapeutic and Biotechnological Potential. Chem. Biol. Drug Des. 2009, 73, 7–16. [Google Scholar] [CrossRef]
- Hasan, R.; Rony, M.N.H.; Ahmed, R. In Silico Characterization and Structural Modeling of Bacterial Metalloprotease of Family M4. J. Genet. Eng. Biotechnol. 2021, 19, 25. [Google Scholar] [CrossRef]
- Sjøli, S.; Nuti, E.; Camodeca, C.; Bilto, I.; Rossello, A.; Winberg, J.-O.; Sylte, I.; Adekoya, O.A. Synthesis, Experimental Evaluation and Molecular Modelling of Hydroxamate Derivatives as Zinc Metalloproteinase Inhibitors. Eur. J. Med. Chem. 2016, 108, 141–153. [Google Scholar] [CrossRef]
- Camodeca, C.; Cuffaro, D.; Nuti, E.; Rossello, A. ADAM Metalloproteinases as Potential Drug Targets. Curr. Med. Chem. 2019, 26, 2661–2689. [Google Scholar] [CrossRef]
- Calligaris, M.; Cuffaro, D.; Bonelli, S.; Spanò, D.P.; Rossello, A.; Nuti, E.; Scilabra, S.D. Strategies to Target ADAM17 in Disease: From Its Discovery to the IRhom Revolution. Molecules 2021, 26, 944. [Google Scholar] [CrossRef]
- Rahman, F.; Wushur, I.; Malla, N.; Åstrand, O.A.H.; Rongved, P.; Winberg, J.-O.; Sylte, I. Zinc-Chelating Compounds as Inhibitors of Human and Bacterial Zinc Metalloproteases. Molecules 2021, 27, 56. [Google Scholar] [CrossRef]
- Cuffaro, D.; Nuti, E.; D’Andrea, F.; Rossello, A. Developments in Carbohydrate-Based Metzincin Inhibitors. Pharmaceuticals 2020, 13, 376. [Google Scholar] [CrossRef]
- Dreger, A.; Hoff, K.; Agoglitta, O.; Hotop, S.-K.; Brönstrup, M.; Heisig, P.; Kirchmair, J.; Holl, R. Antibacterial Activity of Xylose-Derived LpxC Inhibitors–Synthesis, Biological Evaluation and Molecular Docking Studies. Bioorg. Chem. 2021, 107, 104603. [Google Scholar] [CrossRef]
- Odunuga, O.O.; Adekoya, O.A.; Sylte, I. High-Level Expression of Pseudolysin, the Extracellular Elastase of Pseudomonas aeruginosa, in Escherichia Coli and Its Purification. Protein Expr. Purif. 2015, 113, 79–84. [Google Scholar] [CrossRef]
- Galloway, D.R. Pseudomonas aeruginosa Elastase and Elastolysis Revisited: Recent Developments. Mol. Microbiol. 1991, 5, 2315–2321. [Google Scholar] [CrossRef] [PubMed]
- Komori, Y.; Nonogaki, T.; Nikai, T. Hemorrhagic Activity and Muscle Damaging Effect of Pseudomonas aeruginosa Metalloproteinase (Elastase). Toxicon 2001, 39, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Yamamoto, T. Pathogenic Mechanisms Induced by Microbial Proteases in Microbial Infections. Biol. Chem. Hoppe-Seyler 1996, 377, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Holder, I.A.; Neely, A.N. Pseudomonas Elastase Acts as a Virulence Factor in Burned Hosts by Hageman Factor-Dependent Activation of the Host Kinin Cascade. Infect. Immun. 1989, 57, 3345–3348. [Google Scholar] [CrossRef] [PubMed]
- Azghani, A.O.; Miller, E.J.; Peterson, B.T. Virulence Factors from Pseudomonas aeruginosa Increase Lung Epithelial Permeability. Lung 2000, 178, 261–269. [Google Scholar] [CrossRef]
- Wilson, R.; Dowling, R.B. Pseudomonas aeruginosa and Other Related Species. Thorax 1998, 53, 213–219. [Google Scholar] [CrossRef]
- Everett, M.J.; Davies, D.T. Pseudomonas aeruginosa Elastase (LasB) as a Therapeutic Target. Drug Discov. Today 2021, 26, 2108–2123. [Google Scholar] [CrossRef]
- Casilag, F.; Lorenz, A.; Krueger, J.; Klawonn, F.; Weiss, S.; Häussler, S. The LasB Elastase of Pseudomonas aeruginosa Acts in Concert with Alkaline Protease AprA To Prevent Flagellin-Mediated Immune Recognition. Infect. Immun. 2016, 84, 162–171. [Google Scholar] [CrossRef]
- Ezawa, T.; Sugiyama, S.; Ara, A.; Sylte, I.; Kurita, N. Design of Galardine Analogs as Putative Psudolysin Inhibitors Based on Ab Initio Fragment Molecular Orbital Calculations. J. Biomol. Struct. Dyn. 2020, 38, 3307–3317. [Google Scholar] [CrossRef]
- Ezawa, T.; Saito, R.; Suzuki, S.; Sugiyama, S.; Sylte, I.; Kurita, N. Protonation States of Central Amino Acids in a Zinc Metalloprotease Complexed with Inhibitor: Molecular Mechanics Optimizations and Ab Initio Molecular Orbital Calculations. Biophys. Chem. 2020, 261, 106368. [Google Scholar] [CrossRef]
- Imai, K.; Saito, R.; Ezawa, T.; Sugiyama, S.; Sylte, I.; Kurita, N. Proposal of Selective Inhibitor for Bacterial Zinc Metalloprotease: Molecular Mechanics and Ab Initio Molecular Orbital Calculations. J. Mol. Graph. Model. 2022, 110, 108047. [Google Scholar] [CrossRef]
- Cuffaro, D.; Ciccone, L.; Rossello, A.; Nuti, E.; Santamaria, S. Targeting Aggrecanases for Osteoarthritis Therapy: From Zinc Chelation to Exosite Inhibition. J. Med. Chem. 2022, 65, 13505–13532. [Google Scholar] [CrossRef]
- Nuti, E.; Cuffaro, D.; Bernardini, E.; Camodeca, C.; Panelli, L.; Chaves, S.; Ciccone, L.; Tepshi, L.; Vera, L.; Orlandini, E.; et al. Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies. J. Med. Chem. 2018, 61, 4421–4435. [Google Scholar] [CrossRef]
- Jacobsen, J.A.; Major Jourden, J.L.; Miller, M.T.; Cohen, S.M. To Bind Zinc or Not to Bind Zinc: An Examination of Innovative Approaches to Improved Metalloproteinase Inhibition. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 72–94. [Google Scholar] [CrossRef]
- Cuffaro, D.; Camodeca, C.; D’Andrea, F.; Piragine, E.; Testai, L.; Calderone, V.; Orlandini, E.; Nuti, E.; Rossello, A. Matrix Metalloproteinase-12 Inhibitors: Synthesis, Structure-Activity Relationships and Intestinal Absorption of Novel Sugar-Based Biphenylsulfonamide Carboxylates. Bioorg. Med. Chem. 2018, 26, 5804–5815. [Google Scholar] [CrossRef]
- Nuti, E.; Rossello, A.; Cuffaro, D.; Camodeca, C.; Van Bael, J.; van der Maat, D.; Martens, E.; Fiten, P.; Pereira, R.V.S.; Ugarte-Berzal, E.; et al. Bivalent Inhibitor with Selectivity for Trimeric MMP-9 Amplifies Neutrophil Chemotaxis and Enables Functional Studies on MMP-9 Proteoforms. Cells 2020, 9, 1634. [Google Scholar] [CrossRef]
- Nuti, E.; Rosalia, L.; Cuffaro, D.; Camodeca, C.; Giacomelli, C.; Da Pozzo, E.; Tuccinardi, T.; Costa, B.; Antoni, C.; Vera, L.; et al. Bifunctional Inhibitors as a New Tool To Reduce Cancer Cell Invasion by Impairing MMP-9 Homodimerization. ACS Med. Chem. Lett. 2017, 8, 293–298. [Google Scholar] [CrossRef]
- Cuffaro, D.; Camodeca, C.; Tuccinardi, T.; Ciccone, L.; Bartsch, J.W.; Kellermann, T.; Cook, L.; Nuti, E.; Rossello, A. Discovery of Dimeric Arylsulfonamides as Potent ADAM8 Inhibitors. ACS Med. Chem. Lett. 2021, 12, 1787–1793. [Google Scholar] [CrossRef]
- Pece, R.; Tavella, S.; Costa, D.; Varesano, S.; Camodeca, C.; Cuffaro, D.; Nuti, E.; Rossello, A.; Alfano, M.; D’Arrigo, C.; et al. Inhibitors of ADAM10 Reduce Hodgkin Lymphoma Cell Growth in 3D Microenvironments and Enhance Brentuximab-Vedotin Effect. Haematologica 2021, 107, 909–920. [Google Scholar] [CrossRef]
- Piizzi, G.; Parker, D.T.; Peng, Y.; Dobler, M.; Patnaik, A.; Wattanasin, S.; Liu, E.; Lenoir, F.; Nunez, J.; Kerrigan, J.; et al. Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC. J. Med. Chem. 2017, 60, 5002–5014. [Google Scholar] [CrossRef]
- Ding, S.; Dai, R.-Y.; Wang, W.-K.; Cao, Q.; Lan, L.-F.; Zhou, X.-L.; Yang, Y.-S. Design, Synthesis and Structure-Activity Relationship Evaluation of Novel LpxC Inhibitors as Gram-Negative Antibacterial Agents. Bioorg. Med. Chem. Lett. 2018, 28, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.H.; Vidaillac, C.; Yam, J.K.H.; Chua, S.L.; Givskov, M.; Yang, L. In Vitro and In Vivo Efficacy of an LpxC Inhibitor, CHIR-090, Alone or Combined with Colistin against Pseudomonas aeruginosa Biofilm. Antimicrob. Agents Chemother. 2017, 61, e02223-16. [Google Scholar] [CrossRef] [PubMed]
- Titecat, M.; Liang, X.; Lee, C.-J.; Charlet, A.; Hocquet, D.; Lambert, T.; Pagès, J.-M.; Courcol, R.; Sebbane, F.; Toone, E.J.; et al. High Susceptibility of MDR and XDR Gram-Negative Pathogens to Biphenyl-Diacetylene-Based Difluoromethyl-allo-Threonyl-Hydroxamate LpxC Inhibitors. J. Antimicrob. Chemother. 2016, 71, 2874–2882. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Gopalaswamy, R.; Navas, F.; Toone, E.J.; Zhou, P. A Scalable Synthesis of the Difluoromethyl-allo-Threonyl Hydroxamate-Based LpxC Inhibitor LPC-058. J. Org. Chem. 2016, 81, 4393–4398. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, P.; Surivet, J.-P.; Diethelm, S.; Blumstein, A.-C.; Gauvin, J.-C.; Jacob, L.; Masse, F.; Mathieu, G.; Mirre, A.; Schmitt, C.; et al. Optimization of LpxC Inhibitor Lead Compounds Focusing on Efficacy and Formulation for High Dose Intravenous Administration. J. Med. Chem. 2020, 63, 88–102. [Google Scholar] [CrossRef]
- Surivet, J.-P.; Panchaud, P.; Specklin, J.-L.; Diethelm, S.; Blumstein, A.-C.; Gauvin, J.-C.; Jacob, L.; Masse, F.; Mathieu, G.; Mirre, A.; et al. Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria. J. Med. Chem. 2020, 63, 66–87. [Google Scholar] [CrossRef]
- Ding, S.; Wang, W.-K.; Cao, Q.; Chu, W.-J.; Lan, L.-F.; Hu, W.-H.; Yang, Y.-S. Design, Synthesis and Biological Evaluation of LpxC Inhibitors with Novel Hydrophilic Terminus. Chin. Chem. Lett. 2015, 26, 763–767. [Google Scholar] [CrossRef]
- Kawai, T.; Kazuhiko, I.; Takaya, N.; Yamaguchi, Y.; Kishii, R.; Kohno, Y.; Kurasaki, H. Sulfonamide-Based Non-Alkyne LpxC Inhibitors as Gram-Negative Antibacterial Agents. Bioorg. Med. Chem. Lett. 2017, 27, 1045–1049. [Google Scholar] [CrossRef]
- Krause, K.M.; Haglund, C.M.; Hebner, C.; Serio, A.W.; Lee, G.; Nieto, V.; Cohen, F.; Kane, T.R.; Machajewski, T.D.; Hildebrandt, D.; et al. Potent LpxC Inhibitors with In Vitro Activity against Multidrug-Resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00977-19. [Google Scholar] [CrossRef]
- Cohen, F.; Aggen, J.B.; Andrews, L.D.; Assar, Z.; Boggs, J.; Choi, T.; Dozzo, P.; Easterday, A.N.; Haglund, C.M.; Hildebrandt, D.J.; et al. Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety. ChemMedChem 2019, 14, 1560–1572. [Google Scholar] [CrossRef]
- Zhang, J.; Chan, A.; Lippa, B.; Cross, J.B.; Liu, C.; Yin, N.; Romero, J.A.C.; Lawrence, J.; Heney, R.; Herradura, P.; et al. Structure-Based Discovery of LpxC Inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 1670–1680. [Google Scholar] [CrossRef]
- Ding, S.; Ji, J.; Zhang, M.; Yang, Y.; Wang, R.; Zhu, X.; Wang, L.; Zhong, Y.; Gao, L.; Lu, M.; et al. Exploration of the Structure–Activity Relationship and Druggability of Novel Oxazolidinone-based Compounds as Gram-negative Antibacterial Agents. Arch. Pharm. Chem. Life Sci. 2019, 352, 1900129. [Google Scholar] [CrossRef]
- Kurasaki, H.; Tsuda, K.; Shinoyama, M.; Takaya, N.; Yamaguchi, Y.; Kishii, R.; Iwase, K.; Ando, N.; Nomura, M.; Kohno, Y. LpxC Inhibitors: Design, Synthesis, and Biological Evaluation of Oxazolidinones as Gram-Negative Antibacterial Agents. ACS Med. Chem. Lett. 2016, 7, 623–628. [Google Scholar] [CrossRef]
- Lee, P.S.; Lapointe, G.; Madera, A.M.; Simmons, R.L.; Xu, W.; Yifru, A.; Tjandra, M.; Karur, S.; Rico, A.; Thompson, K.; et al. Application of Virtual Screening to the Identification of New LpxC Inhibitor Chemotypes, Oxazolidinone and Isoxazoline. J. Med. Chem. 2018, 61, 9360–9370. [Google Scholar] [CrossRef]
- Jana, S.K.; Löppenberg, M.; Daniliuc, C.G.; Holl, R. Synthesis and Biological Evaluation of C-Ethynyl Furanosides as LpxC Inhibitors. Tetrahedron 2015, 71, 956–966. [Google Scholar] [CrossRef]
- Müller, H.; Gabrielli, V.; Agoglitta, O.; Holl, R. Chiral Pool Synthesis and Biological Evaluation of C-Furanosidic and Acyclic LpxC Inhibitors. Eur. J. Med. Chem. 2016, 110, 340–375. [Google Scholar] [CrossRef]
- Dreger, A.; Hoff, K.; Agoglitta, O.; Bülbül, E.F.; Melesina, J.; Sippl, W.; Holl, R. Synthesis, Biological Evaluation, and Molecular Docking Studies of Deoxygenated C-Glycosides as LpxC Inhibitors. Bioorg. Chem. 2021, 117, 105403. [Google Scholar] [CrossRef]
- Tangherlini, G.; Torregrossa, T.; Agoglitta, O.; Köhler, J.; Melesina, J.; Sippl, W.; Holl, R. Synthesis and Biological Evaluation of Enantiomerically Pure Glyceric Acid Derivatives as LpxC Inhibitors. Bioorg. Med. Chem. 2016, 24, 1032–1044. [Google Scholar] [CrossRef]
- Hoff, K.; Mielniczuk, S.; Agoglitta, O.; Iorio, M.T.; Caldara, M.; Bülbül, E.F.; Melesina, J.; Sippl, W.; Holl, R. Synthesis and Biological Evaluation of Triazolyl-Substituted Benzyloxyacetohydroxamic Acids as LpxC Inhibitors. Bioorg. Med. Chem. 2020, 28, 115529. [Google Scholar] [CrossRef]
- Galster, M.; Löppenberg, M.; Galla, F.; Börgel, F.; Agoglitta, O.; Kirchmair, J.; Holl, R. Phenylethylene Glycol-Derived LpxC Inhibitors with Diverse Zn2+-Binding Groups. Tetrahedron 2019, 75, 486–509. [Google Scholar] [CrossRef]
- Nencetti, S.; Cuffaro, D.; Nuti, E.; Ciccone, L.; Rossello, A.; Fabbi, M.; Ballante, F.; Ortore, G.; Carbotti, G.; Campelli, F.; et al. Identification of Histone Deacetylase Inhibitors with (Arylidene)Aminoxy Scaffold Active in Uveal Melanoma Cell Lines. J. Enzym. Inhib. Med. Chem. 2021, 36, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Kozikowski, A.P. Why Hydroxamates May Not Be the Best Histone Deacetylase Inhibitors-What Some May Have Forgotten or Would Rather Forget? ChemMedChem 2016, 11, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Fields, G.B. New Strategies for Targeting Matrix Metalloproteinases. Matrix Biol. 2015, 44–46, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.; Shapiro, A.B.; Comita-Prevoir, J.; Kuenstner, E.J.; Zhang, J.; Ribe, S.D.; Chen, A.; Hines, D.; Moussa, S.H.; Carter, N.M.; et al. N-Hydroxyformamide LpxC Inhibitors, Their in Vivo Efficacy in a Mouse Escherichia Coli Infection Model, and Their Safety in a Rat Hemodynamic Assay. Bioorg. Med. Chem. 2020, 28, 115826. [Google Scholar] [CrossRef]
- Yamada, Y.; Takashima, H.; Walmsley, D.L.; Ushiyama, F.; Matsuda, Y.; Kanazawa, H.; Yamaguchi-Sasaki, T.; Tanaka-Yamamoto, N.; Yamagishi, J.; Kurimoto-Tsuruta, R.; et al. Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity. J. Med. Chem. 2020, 63, 14805–14820. [Google Scholar] [CrossRef]
- Ushiyama, F.; Takashima, H.; Matsuda, Y.; Ogata, Y.; Sasamoto, N.; Kurimoto-Tsuruta, R.; Ueki, K.; Tanaka-Yamamoto, N.; Endo, M.; Mima, M.; et al. Lead Optimization of 2-Hydroxymethyl Imidazoles as Non-Hydroxamate LpxC Inhibitors: Discovery of TP0586532. Bioorg. Med. Chem. 2021, 30, 115964. [Google Scholar] [CrossRef]
- Yoshida, I.; Takata, I.; Fujita, K.; Takashima, H.; Sugiyama, H. TP0586532, a Novel Non-Hydroxamate LpxC Inhibitor: Potentiating Effect on In Vitro Activity of Meropenem against Carbapenem-Resistant Enterobacteriaceae. Microbiol. Spectr. 2022, 10, e00828-22. [Google Scholar] [CrossRef]
- Fujita, K.; Takata, I.; Yoshida, I.; Takashima, H.; Sugiyama, H. TP0586532, a Non-Hydroxamate LpxC Inhibitor, Reduces LPS Release and IL-6 Production Both in Vitro and in Vivo. J. Antibiot. 2022, 75, 136–145. [Google Scholar] [CrossRef]
- Zhu, J.; Cai, X.; Harris, T.L.; Gooyit, M.; Wood, M.; Lardy, M.; Janda, K.D. Disarming Pseudomonas aeruginosa Virulence Factor LasB by Leveraging a Caenorhabditis Elegans Infection Model. Chem. Biol. 2015, 22, 483–491. [Google Scholar] [CrossRef]
- Kany, A.M.; Sikandar, A.; Haupenthal, J.; Yahiaoui, S.; Maurer, C.K.; Proschak, E.; Köhnke, J.; Hartmann, R.W. Binding Mode Characterization and Early in Vivo Evaluation of Fragment-Like Thiols as Inhibitors of the Virulence Factor LasB from Pseudomonas aeruginosa. ACS Infect. Dis. 2018, 4, 988–997. [Google Scholar] [CrossRef]
- Kaya, C.; Walter, I.; Alhayek, A.; Shafiei, R.; Jézéquel, G.; Andreas, A.; Konstantinović, J.; Schönauer, E.; Sikandar, A.; Haupenthal, J.; et al. Structure-Based Design of α-Substituted Mercaptoacetamides as Inhibitors of the Virulence Factor LasB from Pseudomonas aeruginosa. ACS Infect. Dis. 2022, 8, 1010–1021. [Google Scholar] [CrossRef]
- Konstantinović, J.; Yahiaoui, S.; Alhayek, A.; Haupenthal, J.; Schönauer, E.; Andreas, A.; Kany, A.M.; Müller, R.; Koehnke, J.; Berger, F.K.; et al. N-Aryl-3-Mercaptosuccinimides as Antivirulence Agents Targeting Pseudomonas aeruginosa Elastase and Clostridium Collagenases. J. Med. Chem. 2020, 63, 8359–8368. [Google Scholar] [CrossRef]
- Kany, A.M.; Sikandar, A.; Yahiaoui, S.; Haupenthal, J.; Walter, I.; Empting, M.; Köhnke, J.; Hartmann, R.W. Tackling Pseudomonas aeruginosa Virulence by a Hydroxamic Acid-Based LasB Inhibitor. ACS Chem. Biol. 2018, 13, 2449–2455. [Google Scholar] [CrossRef]
- Sylte, I.; Dawadi, R.; Malla, N.; von Hofsten, S.; Nguyen, T.-M.; Solli, A.I.; Berg, E.; Adekoya, O.A.; Svineng, G.; Winberg, J.-O. The Selectivity of Galardin and an Azasugar-Based Hydroxamate Compound for Human Matrix Metalloproteases and Bacterial Metalloproteases. PLoS ONE 2018, 13, e0200237. [Google Scholar] [CrossRef]
- Rahman, F.; Nguyen, T.-M.; Adekoya, O.A.; Campestre, C.; Tortorella, P.; Sylte, I.; Winberg, J.-O. Inhibition of Bacterial and Human Zinc-Metalloproteases by Bisphosphonate- and Catechol-Containing Compounds. J. Enzym. Inhib. Med. Chem. 2021, 36, 819–830. [Google Scholar] [CrossRef]
- Leiris, S.; Davies, D.T.; Sprynski, N.; Castandet, J.; Beyria, L.; Bodnarchuk, M.S.; Sutton, J.M.; Mullins, T.M.G.; Jones, M.W.; Forrest, A.K.; et al. Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors. ACS Med. Chem. Lett. 2021, 12, 217–227. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).