Combating Antimicrobial Resistance in the Post-Genomic Era: Rapid Antibiotic Discovery
Abstract
1. Introduction
2. Genome Mining for Key Enzymes in Biosynthesis Gene Clusters
2.1. Polyketides
2.2. Nonribosomal Peptides
2.3. Ribosomally Synthesized and Post-Translationally Modified Peptides
2.4. Phenazines
2.5. Terpenes/Terpenoids
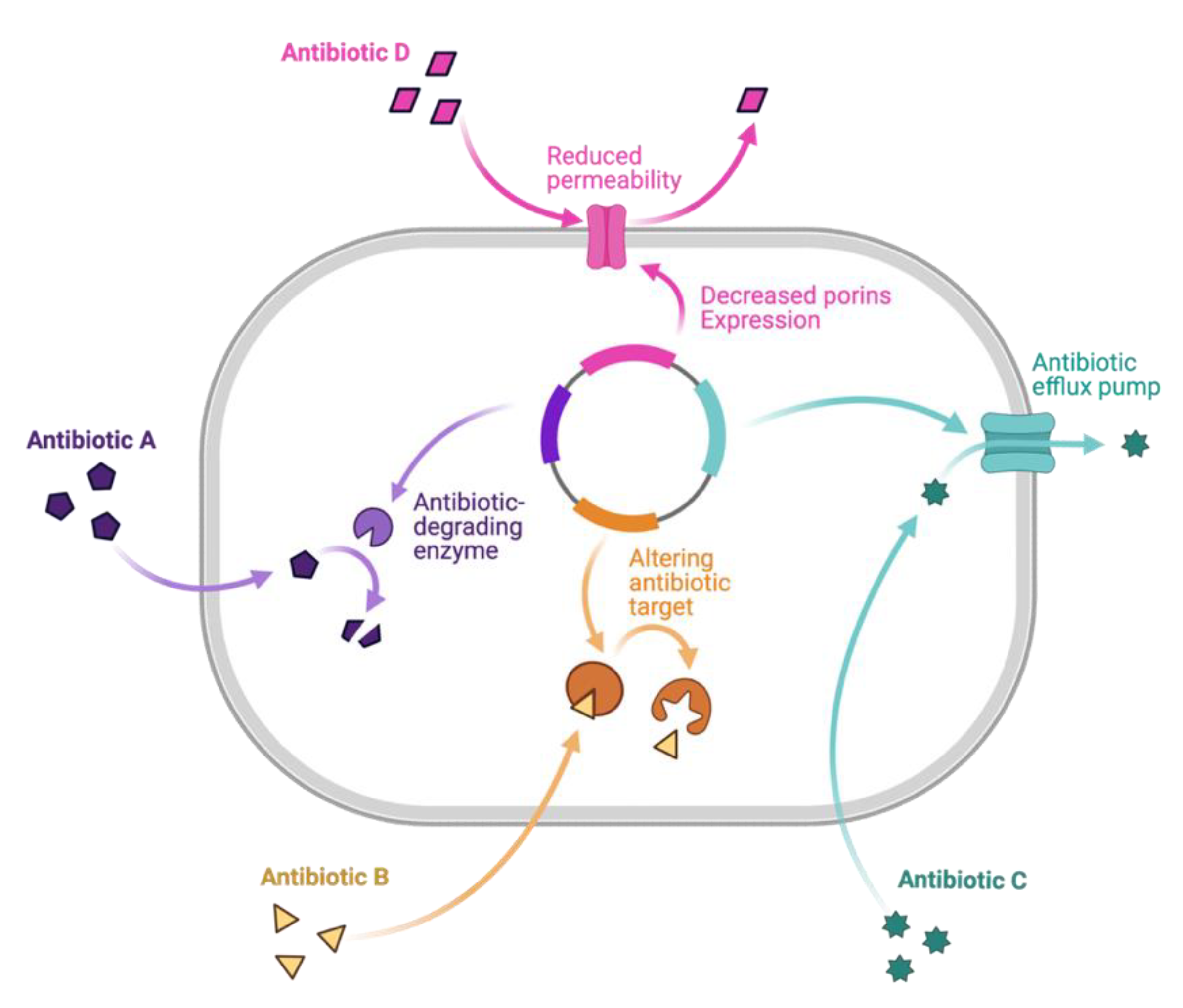
3. Mechanisms of Resistance and Promising Drug Leads
3.1. Combating Antimicrobial-Resistant Bacteria
3.2. Combating Antimicrobial-Resistant Fungi
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Micarelli, I.; Paine, R.; Giostra, C.; Tafuri, M.A.; Profico, A.; Boggioni, M.; Di Vincenzo, F.; Massani, D.; Papini, A.; Manzi, G. Survival to amputation in pre-antibiotic era: A case study from a Longobard necropolis (6th-8th centuries AD). J. Anthropol. Sci. 2018, 96, 185–200. [Google Scholar] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- CDC. Antibiotic Resistance Threats in the United States 2019; Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases, Division of Healthcare Quality Promotion, Antibiotic Resistance Coordination; Strategy Unit within the Division of Healthcare Quality Promotion, U.S. Department of Health and Human Services: Atlanta, GA, USA, 2019.
- CDC. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases, Division of Healthcare Quality Promotion, U.S. Department of Health and Human Services: Atlanta, GA, USA, 2022.
- Clardy, J.; Fischbach, M.A.; Walsh, C.T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Li, M.H.; Ung, P.M.; Zajkowski, J.; Garneau-Tsodikova, S.; Sherman, D.H. Automated genome mining for natural products. BMC Bioinform. 2009, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Albarano, L.; Esposito, R.; Ruocco, N.; Costantini, M. Genome mining as new challenge in natural products discovery. Mar. Drugs 2020, 18, 199. [Google Scholar] [CrossRef]
- Baltz, R.H. Genome mining for drug discovery: Progress at the front end. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab044. [Google Scholar] [CrossRef]
- Banskota, A.H.; McAlpine, J.B.; Sørensen, D.; Ibrahim, A.; Aouidate, M.; Piraee, M.; Alarco, A.-M.; Farnet, C.M.; Zazopoulos, E. Genomic analyses lead to novel secondary metabolites. J. Antiobiot. 2006, 59, 533–542. [Google Scholar] [CrossRef]
- Shen, B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 2003, 7, 285–295. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Reiter, S.; Piel, J. Recent advances in genome-based polyketide discovery. Curr. Opin. Biotechnol. 2014, 29, 107–115. [Google Scholar] [CrossRef]
- Bessa, L.J.; Buttachon, S.; Dethoup, T.; Martins, R.; Vasconcelos, V.; Kijjoa, A.; Martins da Costa, P. Neofiscalin A and fiscalin C are potential novel indole alkaloid alternatives for the treatment of multidrug-resistant Gram-positive bacterial infections. FEMS Microbiol. Lett. 2016, 363, fnw150. [Google Scholar] [CrossRef]
- Hensler, M.E.; Jang, K.H.; Thienphrapa, W.; Vuong, L.; Tran, D.N.; Soubih, E.; Lin, L.; Haste, N.M.; Cunningham, M.L.; Kwan, B.P.; et al. Anthracimycin activity against contemporary methicillin-resistant Staphylococcus aureus. J. Antibiot. 2014, 67, 549–553. [Google Scholar] [CrossRef]
- Panter, F.; Krug, D.; Baumann, S.; Müller, R. Self-resistance guided genome mining uncovers new topoisomerase inhibitors from myxobacteria. Chem. Sci. 2018, 9, 4898–4908. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, J.; Millan-Aguinaga, N.; Zhang, J.J.; O’Neill, E.C.; Ugalde, J.A.; Jensen, P.R.; Mantovani, S.M.; Moore, B.S. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 2015, 10, 2841–2849. [Google Scholar] [CrossRef]
- Thong, W.L.; Shin-ya, K.; Nishiyama, M.; Kuzuyama, T. Discovery of an antibacterial isoindolinone-containing tetracyclic polyketide by cryptic gene activation and characterization of its biosynthetic gene cluster. ACS Chem. Biol. 2018, 13, 2615–2622. [Google Scholar] [CrossRef]
- Fuoco, D. Classification framework and chemical biology of tetracycline-structure-based drugs. Antibiotics 2012, 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.X.; Chen, Y.Y.; Ge, L.; Fang, T.T.; Meng, J.; Liu, Z.; Fang, X.Y.; Ni, S.; Lin, C.; Wu, Y.Y.; et al. PCR screening reveals considerable unexploited biosynthetic potential of ansamycins and a mysterious family of AHBA-containing natural products in actinomycetes. J. Appl. Microbiol. 2013, 115, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Y.; Lu, C.; Zhang, J.; Zhu, J.; Wang, H.; Shen, Y. Activating a cryptic ansamycin biosynthetic gene cluster to produce three new naphthalenic octaketide ansamycins with n-pentyl and n-butyl side chains. Org. Lett. 2015, 17, 3706–3709. [Google Scholar] [CrossRef]
- Liu, M.; Lu, C.; Tang, R.; Li, S.; Wang, H.; Shen, Y. Neoansamycins from Streptomyces sp. LZ35. RSC Adv. 2017, 7, 35460–35465. [Google Scholar] [CrossRef]
- Adnani, N.; Chevrette, M.G.; Adibhatla, S.N.; Zhang, F.; Yu, Q.; Braun, D.R.; Nelson, J.; Simpkins, S.W.; McDonald, B.R.; Myers, C.L.; et al. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, Keyicin. ACS Chem. Biol. 2017, 12, 3093–3102. [Google Scholar] [CrossRef]
- Chanana, S.; Thomas, C.S.; Zhang, F.; Rajski, S.R.; Bugni, T.S. hcapca: Automated hierarchical clustering and principal component analysis of large metabolomic datasets in R. Metabolites 2020, 10, 297. [Google Scholar] [CrossRef]
- Brkljača, R.; Urban, S. Recent advancements in HPLC-NMR and applications for natural product profiling and identification. J. Liq. Chromatogr. Relat. Technol. 2011, 34, 1063–1076. [Google Scholar] [CrossRef]
- Danelius, E.; Halaby, S.; van der Donk, W.A.; Gonen, T. MicroED in natural product and small molecule research. Nat. Prod. Rep. 2021, 38, 423–431. [Google Scholar] [CrossRef]
- Hansen, P.E. NMR of natural products as potential drugs. Molecules 2021, 26, 3763. [Google Scholar] [CrossRef]
- Kim, L.J.; Ohashi, M.; Zhang, Z.; Tan, D.; Asay, M.; Cascio, D.; Rodriguez, J.A.; Tang, Y.; Nelson, H.M. Prospecting for natural products by genome mining and microcrystal electron diffraction. Nat. Chem. Biol. 2021, 17, 872–877. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, M.; Braun, D.R.; Ericksen, S.S.; Piotrowski, J.S.; Nelson, J.; Peng, J.; Ananiev, G.E.; Chanana, S.; Barns, K.; et al. A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science 2020, 370, 974–978. [Google Scholar] [CrossRef]
- Pidot, S.; Ishida, K.; Cyrulies, M.; Hertweck, C. Discovery of clostrubin, an exceptional polyphenolic polyketide antibiotic from a strictly anaerobic bacterium. Angew. Chem. Int. Ed. Engl. 2014, 53, 7856–7859. [Google Scholar] [CrossRef]
- Shabuer, G.; Ishida, K.; Pidot, S.J.; Roth, M.; Dahse, H.-M.; Hertweck, C. Plant pathogenic anaerobic bacteria use aromatic polyketides to access aerobic territory. Science 2015, 350, 670–674. [Google Scholar] [CrossRef]
- Süssmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Ed. Engl. 2017, 56, 3770–3821. [Google Scholar] [CrossRef]
- Engelhardt, K.; Degnes, K.F.; Kemmler, M.; Bredholt, H.; Fjaervik, E.; Klinkenberg, G.; Sletta, H.; Ellingsen, T.E.; Zotchev, S.B. Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl. Environ. Microbiol. 2010, 76, 4969–4976. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, X.; Shukla, R.; Kumar, R.; Weingarth, M.; Breukink, E.; Kuipers, O.P. Brevibacillin 2V, a novel antimicrobial lipopeptide with an exceptionally low hemolytic activity. Front. Microbiol. 2021, 12, 693725. [Google Scholar] [CrossRef]
- Zhou, L.; de Jong, A.; Yi, Y.; Kuipers, O.P. Identification, isolation, and characterization of medipeptins, antimicrobial peptides from Pseudomonas mediterranea EDOX. Front. Microbiol. 2021, 12, 732771. [Google Scholar] [CrossRef]
- Hover, B.M.; Kim, S.H.; Katz, M.; Charlop-Powers, Z.; Owen, J.G.; Ternei, M.A.; Maniko, J.; Estrela, A.B.; Molina, H.; Park, S.; et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018, 3, 415–422. [Google Scholar] [CrossRef]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef]
- Reynolds, K.A.; Luhavaya, H.; Li, J.; Dahesh, S.; Nizet, V.; Yamanaka, K.; Moore, B.S. Isolation and structure elucidation of lipopeptide antibiotic taromycin B from the activated taromycin biosynthetic gene cluster. J. Antibiot. (Tokyo) 2018, 71, 333–338. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
- Wang, Z.; Koirala, B.; Hernandez, Y.; Zimmerman, M.; Park, S.; Perlin, D.S.; Brady, S.F. A naturally inspired antibiotic to target multidrug-resistant pathogens. Nature 2022, 601, 606–611. [Google Scholar] [CrossRef]
- Chu, J.; Vila-Farres, X.; Inoyama, D.; Ternei, M.; Cohen, L.J.; Gordon, E.A.; Reddy, B.V.; Charlop-Powers, Z.; Zebroski, H.A.; Gallardo-Macias, R.; et al. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat. Chem. Biol. 2016, 12, 1004–1006. [Google Scholar] [CrossRef] [PubMed]
- Vila-Farres, X.; Chu, J.; Ternei, M.A.; Lemetre, C.; Park, S.; Perlin, D.S.; Brady, S.F. An optimized synthetic-bioinformatic natural product antibiotic sterilizes multidrug-resistant Acinetobacter baumannii-infected wounds. mSphere 2018, 3, e00528-17. [Google Scholar] [CrossRef]
- Vila-Farres, X.; Chu, J.; Inoyama, D.; Ternei, M.A.; Lemetre, C.; Cohen, L.J.; Cho, W.; Reddy, B.V.B.; Zebroski, H.A.; Freundlich, J.S.; et al. Antimicrobials inspired by nonribosomal peptide synthetase gene clusters. J. Am. Chem. Soc. 2017, 139, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, P.; Chen, G.; Li, C.; Cao, Z.; Zhang, Y.; Ren, J.; Xiang, H.; Lin, S.; Ju, J.; et al. Design and biosynthesis of dimeric alboflavusins with biaryl linkages via regiospecific C–C bond coupling. J. Am. Chem. Soc. 2018, 140, 18009–18015. [Google Scholar] [CrossRef]
- Poorinmohammad, N.; Bagheban-Shemirani, R.; Hamedi, J. Genome mining for ribosomally synthesised and post-translationally modified peptides (RiPPs) reveals undiscovered bioactive potentials of actinobacteria. Antonie Van Leeuwenhoek 2019, 112, 1477–1499. [Google Scholar] [CrossRef] [PubMed]
- Der Torossian Torres, M.; de la Fuente-Nunez, C. Reprogramming biological peptides to combat infectious diseases. Chem. Commun. 2019, 55, 15020–15032. [Google Scholar] [CrossRef]
- Donia, M.S.; Cimermancic, P.; Schulze, C.J.; Wieland Brown, L.C.; Martin, J.; Mitreva, M.; Clardy, J.; Linington, R.G. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 2014, 158, 1402–1414. [Google Scholar] [CrossRef]
- Dischinger, J.; Josten, M.; Szekat, C.; Sahl, H.G.; Bierbaum, G. Production of the novel two-peptide lantibiotic lichenicidin by Bacillus licheniformis DSM 13. PLoS ONE 2009, 4, e6788. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Finkina, E.I.; Nurmukhamedova, E.K.; Balandin, S.V.; Mineev, K.S.; Nadezhdin, K.D.; Yakimenko, Z.A.; Tagaev, A.A.; Temirov, Y.V.; Arseniev, A.S.; et al. Isolation, structure elucidation, and synergistic antibacterial activity of a novel two-component lantibiotic lichenicidin from Bacillus licheniformis VK21. Biochemistry 2010, 49, 6462–6472. [Google Scholar] [CrossRef] [PubMed]
- Hudson, G.A.; Zhang, Z.; Tietz, J.I.; Mitchell, D.A.; van der Donk, W.A. In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J. Am. Chem. Soc. 2015, 137, 16012–16015. [Google Scholar] [CrossRef]
- Kaweewan, I.; Hemmi, H.; Komaki, H.; Harada, S.; Kodani, S. Isolation and structure determination of a new lasso peptide specialicin based on genome mining. Bioorg. Med. Chem. 2018, 26, 6050–6055. [Google Scholar] [CrossRef] [PubMed]
- Gavrish, E.; Clarissa, S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; Bissell, A.; et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef]
- Cheung-Lee, W.L.; Parry, M.E.; Jaramillo Cartagena, A.; Darst, S.A.; Link, A.J. Discovery and structure of the antimicrobial lasso peptide citrocin. J. Biol. Chem. 2019, 294, 6822–6830. [Google Scholar] [CrossRef]
- Horbal, L.; Marques, F.; Nadmid, S.; Mendes, M.V.; Luzhetskyy, A. Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab. Eng. 2018, 49, 299–315. [Google Scholar] [CrossRef]
- Shimamura, H.; Gouda, H.; Nagai, K.; Hirose, T.; Ichioka, M.; Furuya, Y.; Kobayashi, Y.; Hirono, S.; Sunazuka, T.; Ōmura, S. Structure determination and total synthesis of bottromycin A2: A potent antibiotic against MRSA and VRE. Angew. Chem. 2009, 121, 932–935. [Google Scholar] [CrossRef]
- Goto, Y.; Li, B.; Claesen, J.; Shi, Y.; Bibb, M.J.; van der Donk, W.A. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 2010, 8, e1000339. [Google Scholar] [CrossRef] [PubMed]
- van Staden, A.D.P.; van Zyl, W.F.; Trindade, M.; Dicks, L.M.T.; Smith, C. Therapeutic application of lantibiotics and other lanthipeptides: Old and new findings. Appl. Environ. Microbiol. 2021, 87, e0018621. [Google Scholar] [CrossRef]
- Zhao, X.; Yin, Z.; Breukink, E.; Moll, G.N.; Kuipers, O.P. An engineered double lipid II binding motifs-containing lantibiotic displays potent and selective antimicrobial activity against Enterococcus faecium. Antimicrob. Agents Chemother. 2020, 64, e02050-19. [Google Scholar] [CrossRef]
- Jiang, J.; Guiza Beltran, D.; Schacht, A.; Wright, S.; Zhang, L.; Du, L. Functional and structural analysis of Phenazine O-Methyltransferase LaPhzM from Lysobacter antibioticus OH13 and one-pot enzymatic synthesis of the antibiotic myxin. ACS Chem. Biol. 2018, 13, 1003–1012. [Google Scholar] [CrossRef]
- Lin, C.; Yang, D. DNA recognition by a novel bis-intercalator, potent anticancer drug XR5944. Curr. Top. Med. Chem. 2015, 15, 1385–1397. [Google Scholar] [CrossRef]
- Guttenberger, N.; Blankenfeldt, W.; Breinbauer, R. Recent developments in the isolation, biological function, biosynthesis, and synthesis of phenazine natural products. Bioorg. Med. Chem. 2017, 25, 6149–6166. [Google Scholar] [CrossRef]
- Heine, D.; Martin, K.; Hertweck, C. Genomics-guided discovery of endophenazines from Kitasatospora sp. HKI 714. J. Nat. Prod. 2014, 77, 1083–1087. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Shi, Y.-M.; Brachmann, A.O.; Westphalen, M.A.; Neubacher, N.; Tobias, N.J.; Bode, H.B. Dual phenazine gene clusters enable diversification during biosynthesis. Nat. Chem. Biol. 2019, 15, 331–339. [Google Scholar] [CrossRef]
- Giddens, S.R.; Bean, D.C. Investigations into the in vitro antimicrobial activity and mode of action of the phenazine antibiotic d-alanylgriseoluteic acid. Int. J. Antimicrob. Agents. 2007, 29, 93–97. [Google Scholar] [CrossRef]
- Wu, C.; van Wezel, G.P.; Hae Choi, Y. Identification of novel endophenaside antibiotics produced by Kitasatospora sp. MBT66. J. Antibiot. 2015, 68, 445–452. [Google Scholar] [CrossRef]
- Li, Y.; Han, L.; Rong, H.; Li, L.; Zhao, L.; Wu, L.; Xu, L.; Jiang, Y.; Huang, X. Diastaphenazine, a new dimeric phenazine from an endophytic Streptomyces diastaticus subsp. ardesiacus. J. Antibiot. 2015, 68, 210–212. [Google Scholar] [CrossRef]
- Rui, Z.; Ye, M.; Wang, S.; Fujikawa, K.; Akerele, B.; Aung, M.; Floss, H.G.; Zhang, W.; Yu, T.W. Insights into a divergent phenazine biosynthetic pathway governed by a plasmid-born esmeraldin gene cluster. Chem. Biol. 2012, 19, 1116–1125. [Google Scholar] [CrossRef]
- Zhao, Y.; Qian, G.; Ye, Y.; Wright, S.; Chen, H.; Shen, Y.; Liu, F.; Du, L. Heterocyclic aromatic N-Oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org. Lett. 2016, 18, 2495–2498. [Google Scholar] [CrossRef]
- Mitova, M.I.; Lang, G.; Wiese, J.; Imhoff, J.F. Subinhibitory concentrations of antibiotics induce phenazine production in a marine Streptomyces sp. J. Nat. Prod. 2008, 71, 824–827. [Google Scholar] [CrossRef]
- Gomez-Escribano, J.P.; Bibb, M.J. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: From genome mining to manipulation of biosynthetic pathways. J. Ind. Microbiol. Technol. 2014, 41, 425–431. [Google Scholar] [CrossRef]
- Saleh, O.; Flinspach, K.; Westrich, L.; Kulik, A.; Gust, B.; Fiedler, H.-P.; Heide, L. Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663. Beilstein J. Org. Chem. 2012, 8, 501–513. [Google Scholar] [CrossRef]
- Saleh, O.; Bonitz, T.; Flinspach, K.; Kulik, A.; Burkard, N.; Mühlenweg, A.; Vente, A.; Polnick, S.; Lämmerhofer, M.; Gust, B.; et al. Activation of a silent phenazine biosynthetic gene cluster reveals a novel natural product and a new resistance mechanism against phenazines. Med. Chem. Commun. 2012, 3, 1009–1019. [Google Scholar] [CrossRef]
- Lechartier, B.; Cole, S.T. Mode of action of clofazimine and combination therapy with benzothiazinones against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2015, 59, 4457–4463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Lu, Y.; Liu, K.; Liu, B.; Wang, J.; Zhang, G.; Zhang, H.; Liu, Y.; Wang, B.; Zheng, M.; et al. Identification of less lipophilic riminophenazine derivatives for the treatment of drug-resistant Tuberculosis. J. Med. Chem. 2012, 55, 8409–8417. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, Y.; Zhang, C.; Zhang, H.; Wang, B.; Xu, J.; Fu, L.; Yin, D.; Cooper, C.B.; Ma, Z.; et al. Synthesis and biological evaluation of novel 2-Methoxypyridylamino-substituted riminophenazine derivatives as antituberculosis agents. Molecules 2014, 19, 4380–4394. [Google Scholar] [CrossRef]
- Liu, B.; Liu, K.; Lu, Y.; Zhang, D.; Yang, T.; Li, X.; Ma, C.; Zheng, M.; Wang, B.; Zhang, G.; et al. Systematic evaluation of structure-activity relationships of the riminophenazine class and discovery of a C2 pyridylamino series for the treatment of multidrug-resistant Tuberculosis. Molecules 2012, 17, 4545–4559. [Google Scholar] [CrossRef]
- Coelho, T.S.; Silva, R.S.F.; Pinto, A.V.; Pinto, M.C.F.R.; Scaini, C.J.; Moura, K.C.G.; Almeida da Silva, P. Activity of β-lapachone derivatives against rifampicin-susceptible and -resistant strains of Mycobacterium tuberculosis. Tuberculosis 2010, 90, 293–297. [Google Scholar] [CrossRef]
- Borrero, N.V.; Bai, F.; Perez, C.; Duong, B.Q.; Rocca, J.R.; Jin, S.; Huigens Iii, R.W. Phenazine antibiotic inspired discovery of potent bromophenazine antibacterial agents against Staphylococcus aureus and Staphylococcus epidermidis. Org. Biomol. Chem. 2014, 12, 881–886. [Google Scholar] [CrossRef]
- Garrison, A.T.; Abouelhassan, Y.; Norwood, V.M.I.V.; Kallifidas, D.; Bai, F.; Nguyen, M.T.; Rolfe, M.; Burch, G.M.; Jin, S.; Luesch, H.; et al. Structure–activity relationships of a diverse class of halogenated phenazines that targets persistent, antibiotic-tolerant bacterial biofilms and Mycobacterium tuberculosis. J. Med. Chem. 2016, 59, 3808–3825. [Google Scholar] [CrossRef]
- Garrison, A.T.; Abouelhassan, Y.; Kallifidas, D.; Bai, F.; Ukhanova, M.; Mai, V.; Jin, S.; Luesch, H.; Huigens, R.W., 3rd. Halogenated phenazines that potently eradicate biofilms, MRSA persister cells in non-biofilm cultures, and Mycobacterium tuberculosis. Angew. Chem. Int. Ed. Engl. 2015, 54, 14819–14823. [Google Scholar] [CrossRef]
- Huigens, R.W., 3rd; Abouelhassan, Y.; Yang, H. Phenazine antibiotic-inspired discovery of bacterial biofilm-eradicating agents. Chembiochem 2019, 20, 2885–2902. [Google Scholar] [CrossRef] [PubMed]
- Reusch, W. Textbook of Organic Chemistry. Available online: https://chem.libretexts.org/Bookshelves/Organic_Chemistry/Supplemental_Modules_(Organic_Chemistry)/Lipids/Properties_and_Classification_of_Lipids/Terpenes (accessed on 6 January 2023).
- Soderberg, T.; Reusch, W.; Farmer, S.; Kennepohl, D. Terpenoids. Available online: https://chem.libretexts.org/Courses/Athabasca_University/Chemistry_360%3A_Organic_Chemistry_II/Chapter_27%3A_Biomolecules_-_Lipids/27.05_Terpenoids (accessed on 6 January 2023).
- Li, H.; Li, H.; Chen, S.; Wu, W.; Sun, P. Isolation and identification of pentalenolactone analogs from Streptomyces sp. NRRL S-4. Molecules 2021, 26, 7377. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Xu, B.; Adpressa, D.A.; Rudolf, J.D.; Loesgen, S. Discovery and biosynthesis of a structurally dynamic antibacterial diterpenoid. Angew Chem. Int. Ed. Engl. 2021, 60, 14163–14170. [Google Scholar] [CrossRef] [PubMed]
- Al-Footy, K.O.; Alarif, W.M.; Asiri, F.; Aly, M.M.; Ayyad, S.-E.N. Rare pyrane-based cembranoids from the Red Sea soft coral Sarcophyton trocheliophorum as potential antimicrobial–antitumor agents. Med. Chem. Res. 2015, 24, 505–512. [Google Scholar] [CrossRef]
- Kumar, R.; Subramani, R.; Aalbersberg, W. Three bioactive sesquiterpene quinones from the Fijian marine sponge of the genus Hippospongia. Nat. Prod. Res. 2013, 27, 1488–1491. [Google Scholar] [CrossRef] [PubMed]
- Kudo, F.; Matsuura, Y.; Hayashi, T.; Fukushima, M.; Eguchi, T. Genome mining of the sordarin biosynthetic gene cluster from Sordaria araneosa Cain ATCC 36386: Characterization of cycloaraneosene synthase and GDP-6-deoxyaltrose transferase. J. Antibiot. 2016, 69, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Hanadate, T.; Tomishima, M.; Shiraishi, N.; Tanabe, D.; Morikawa, H.; Barrett, D.; Matsumoto, S.; Ohtomo, K.; Maki, K. FR290581, a novel sordarin derivative: Synthesis and antifungal activity. Bioorg. Med. Chem. Lett. 2009, 19, 1465–1468. [Google Scholar] [CrossRef]
- Bueno, J.M.; Chicharro, J.; Fiandor, J.M.; Gómez de las Heras, F.; Huss, S. Antifungal sordarins. Part 4: Synthesis and structure–activity relationships of 3′,4′-fused alkyl-tetrahydrofuran derivatives. Bioorg. Med. Chem. Lett. 2002, 12, 1697–1700. [Google Scholar] [CrossRef]
- Bommineni, G.R.; Kapilashrami, K.; Cummings, J.E.; Lu, Y.; Knudson, S.E.; Gu, C.; Walker, S.G.; Slayden, R.A.; Tonge, P.J. Thiolactomycin-based inhibitors of bacterial β-ketoacyl-ACP synthases with in vivo activity. J. Med. Chem. 2016, 59, 5377–5390. [Google Scholar] [CrossRef]
- Kondratyuk, T.P.; Park, E.-J.; Yu, R.; Van Breemen, R.B.; Asolkar, R.N.; Murphy, B.T.; Fenical, W.; Pezzuto, J.M. Novel marine phenazines as potential cancer chemopreventive and anti-inflammatory agents. Mar. Drugs. 2012, 10, 451–464. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. β-Lactams and β-lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef]
- Worthington, R.J.; Melander, C. Overcoming resistance to β-lactam antibiotics. J. Org. Chem. 2013, 78, 4207–4213. [Google Scholar] [CrossRef]
- Dinos, G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017, 174, 2967–2983. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.P. Resistance to rifampicin: A review. J. Antibiot. 2014, 67, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Javid, B.; Sorrentino, F.; Toosky, M.; Zheng, W.; Pinkham, J.T.; Jain, N.; Pan, M.; Deighan, P.; Rubin, E.J. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 1132–1137. [Google Scholar] [CrossRef]
- Bialvaei, A.Z.; Samadi Kafil, H. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Hoenigl, M.; Seidel, D.; Sprute, R.; Cunha, C.; Oliverio, M.; Goldman, G.H.; Ibrahim, A.S.; Carvalho, A. COVID-19-associated fungal infections. Nat. Microbiol. 2022, 7, 1127–1140. [Google Scholar] [CrossRef]
- Armstrong-James, D.; Meintjes, G.; Brown, G.D. A neglected epidemic: Fungal infections in HIV/AIDS. Trends Microbiol. 2014, 22, 120–127. [Google Scholar] [CrossRef]
- Jenks, J.D.; Cornely, O.A.; Chen, S.C.; Thompson, G.R., 3rd; Hoenigl, M. Breakthrough invasive fungal infections: Who is at risk? Mycoses 2020, 63, 1021–1032. [Google Scholar] [CrossRef]
- Francois, I.E.; Aerts, A.M.; Cammue, B.P.; Thevissen, K. Currently used antimycotics: Spectrum, mode of action and resistance occurrence. Curr. Drug Targets 2005, 6, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Puumala, E.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Molecular mechanisms in Candida albicans and beyond. Chem. Rev. 2021, 121, 3390–3411. [Google Scholar] [CrossRef]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2015, 62, e1–e50. [Google Scholar] [CrossRef] [PubMed]
- Patterson, T.F.; Thompson, G.R., III; Denning, D.W.; Fishman, J.A.; Hadley, S.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Nguyen, M.H.; et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 63, e1–e60. [Google Scholar] [CrossRef]
- Ghannoum, M.A.; Rice, L.B. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999, 12, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, P.; Lynch, S.; Flood, E.; Finnan, S.; Oliynyk, M. Amphotericin biosynthesis in Streptomyces nodosus: Deductions from analysis of polyketide synthase and late genes. Chem. Biol. 2001, 8, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Carolus, H.; Pierson, S.; Lagrou, K.; Van Dijck, P. Amphotericin B and other polyenes-discovery, clinical use, mode of action and drug resistance. J. Fungi 2020, 6, 321. [Google Scholar] [CrossRef]
- Denning, D.W. Echinocandin antifungal drugs. Lancet 2003, 362, 1142–1151. [Google Scholar] [CrossRef]
- Hüttel, W. Echinocandins: Structural diversity, biosynthesis, and development of antimycotics. Appl. Microbiol. Biotechnol. 2021, 105, 55–66. [Google Scholar] [CrossRef]
- Ostrosky-Zeichner, L.; Casadevall, A.; Galgiani, J.N.; Odds, F.C.; Rex, J.H. An insight into the antifungal pipeline: Selected new molecules and beyond. Nat. Rev. Drug Discov. 2010, 9, 719–727. [Google Scholar] [CrossRef]
- Shao, Y.; Molestak, E.; Su, W.; Stankevič, M.; Tchórzewski, M. Sordarin- an anti-fungal antibiotic with a unique modus operandi. Br. J. Pharmacol. 2022, 179, 1125–1145. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zaleta-Rivera, K.; Zhu, X.; Huffman, J.; Millet, J.C.; Harris, S.D.; Yuen, G.; Li, X.-C.; Du, L. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 2007, 51, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, Y.; Yu, Y.; Chen, H.; Shen, Y.; Du, L. Indole-induced reversion of intrinsic multiantibiotic resistance in Lysobacter enzymogenes. Appl. Environ. Microbiol. 2017, 83, e00995-17. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, Z.; Li, Y.; Lu, C.; Wang, H.; Shen, Y.; Du, L. HSAF-induced antifungal effects in Candida albicans through ROS-mediated apoptosis. RSC Adv. 2016, 6, 30895–30904. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef]

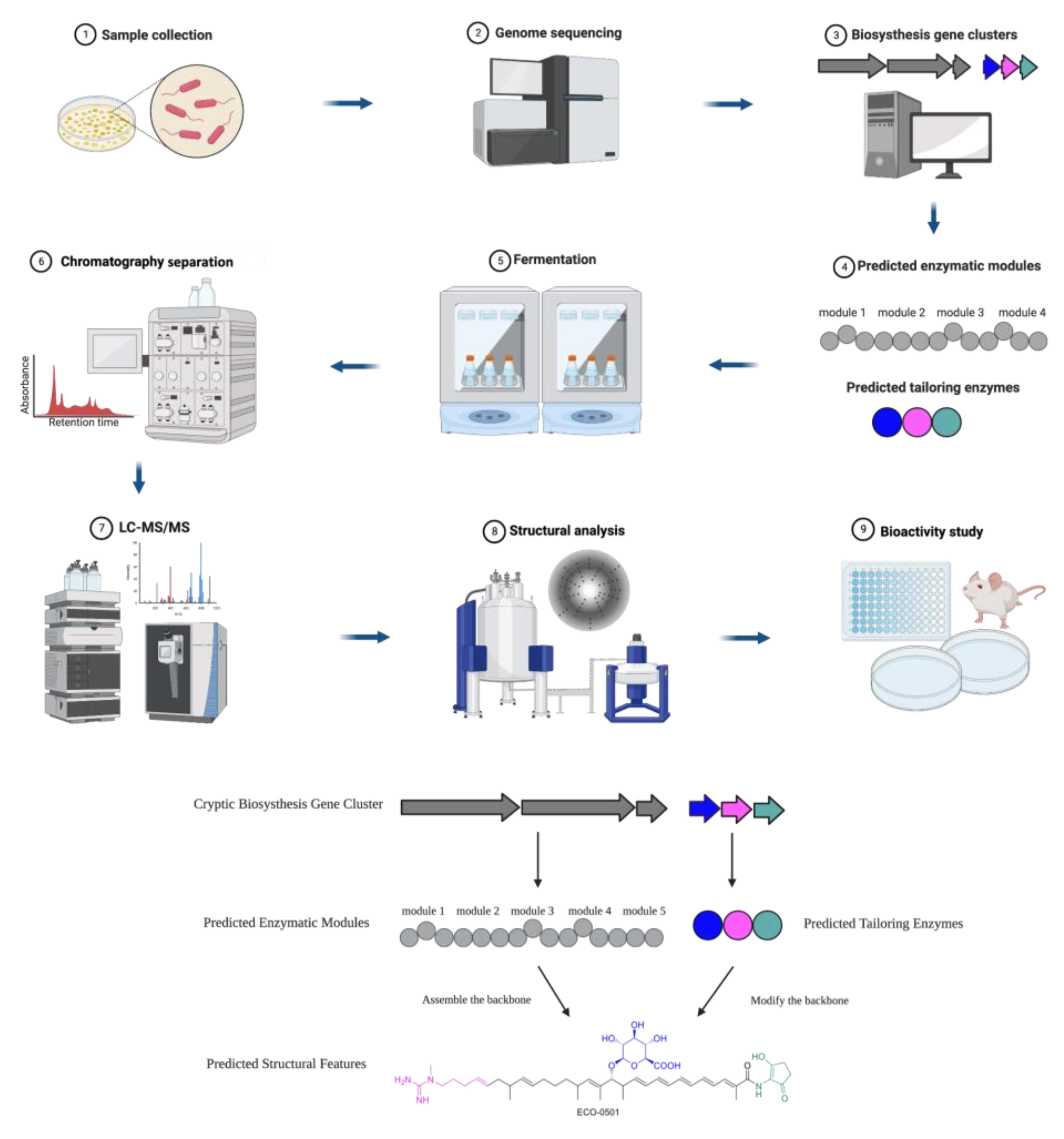
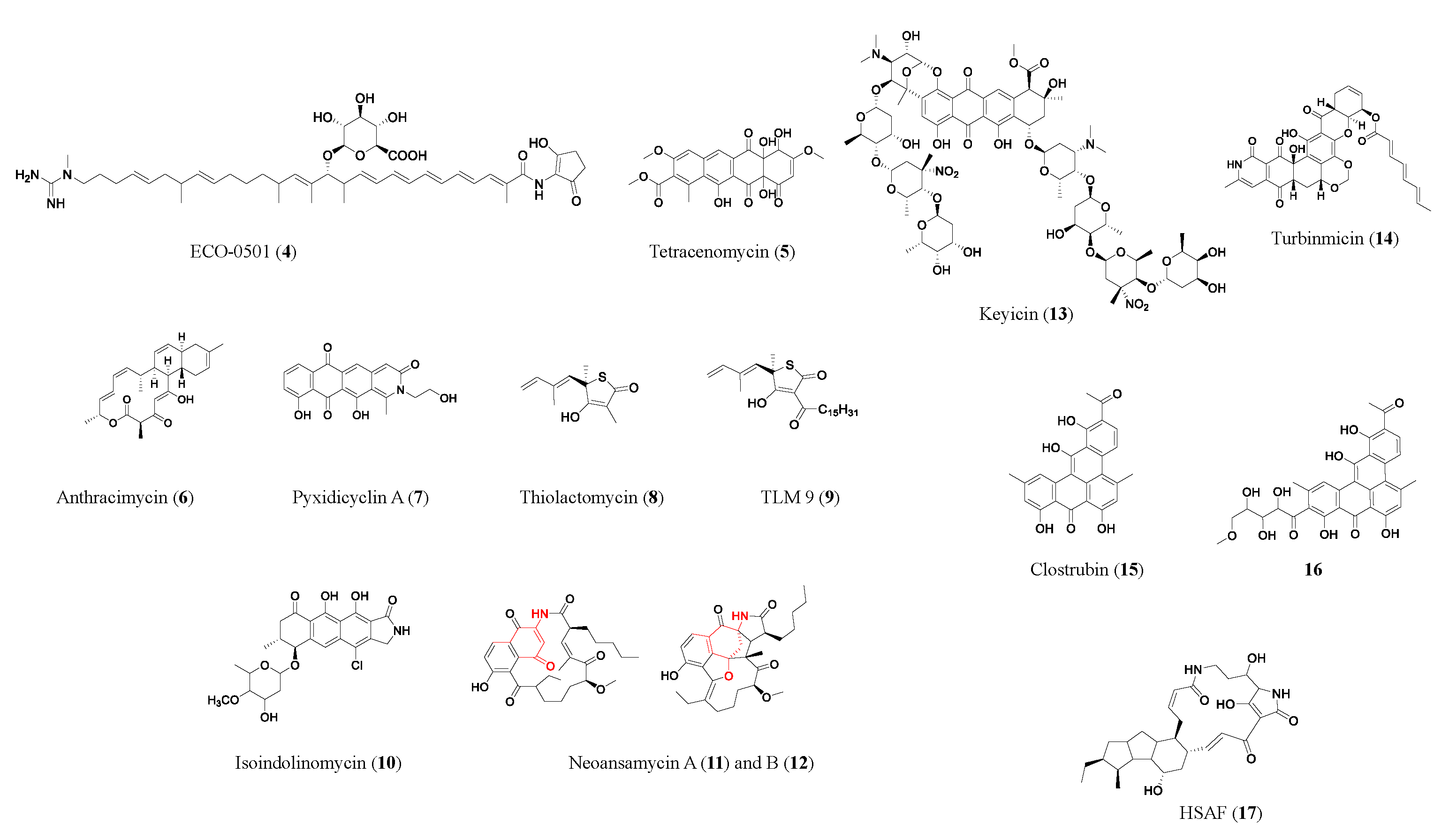
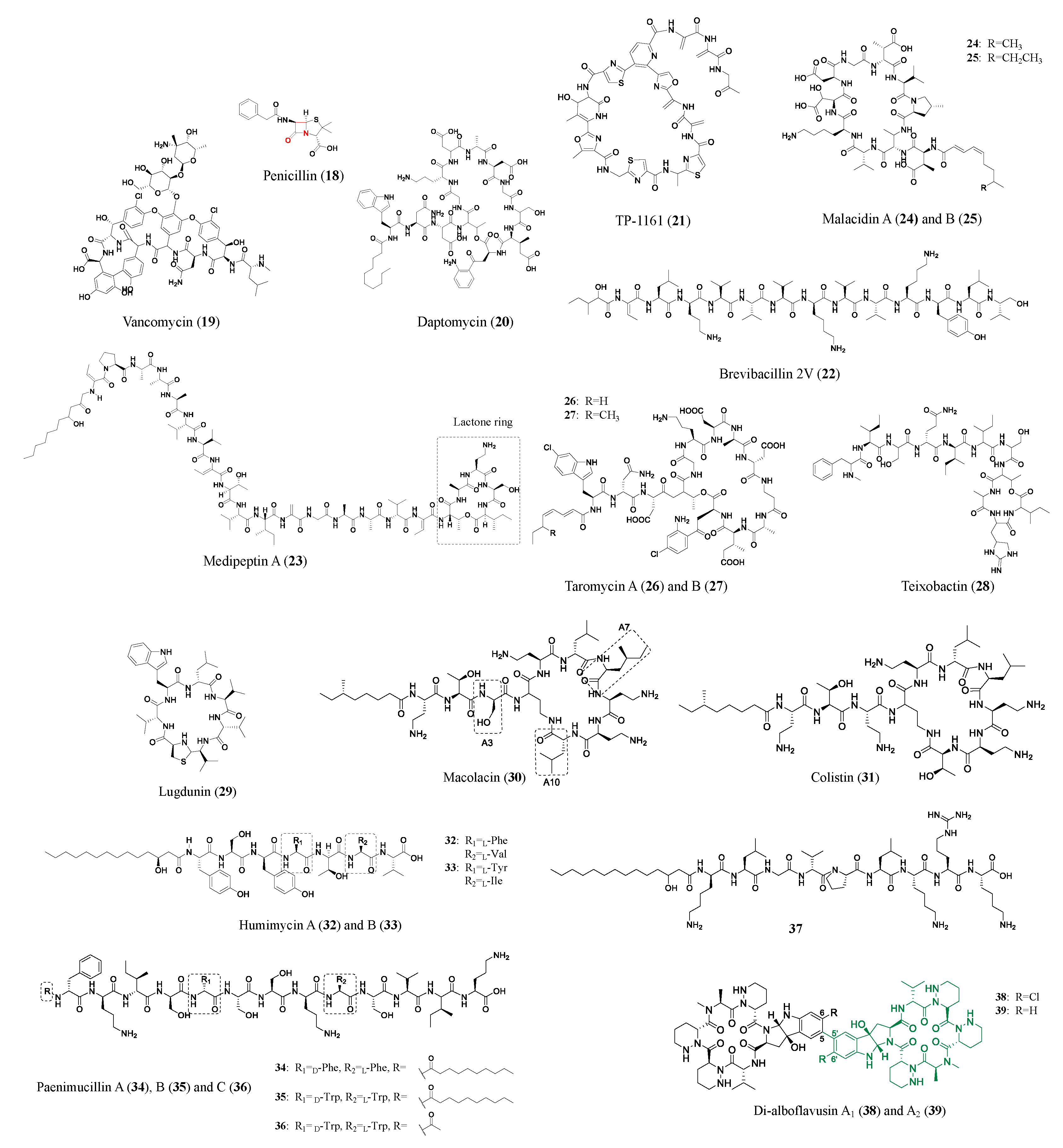
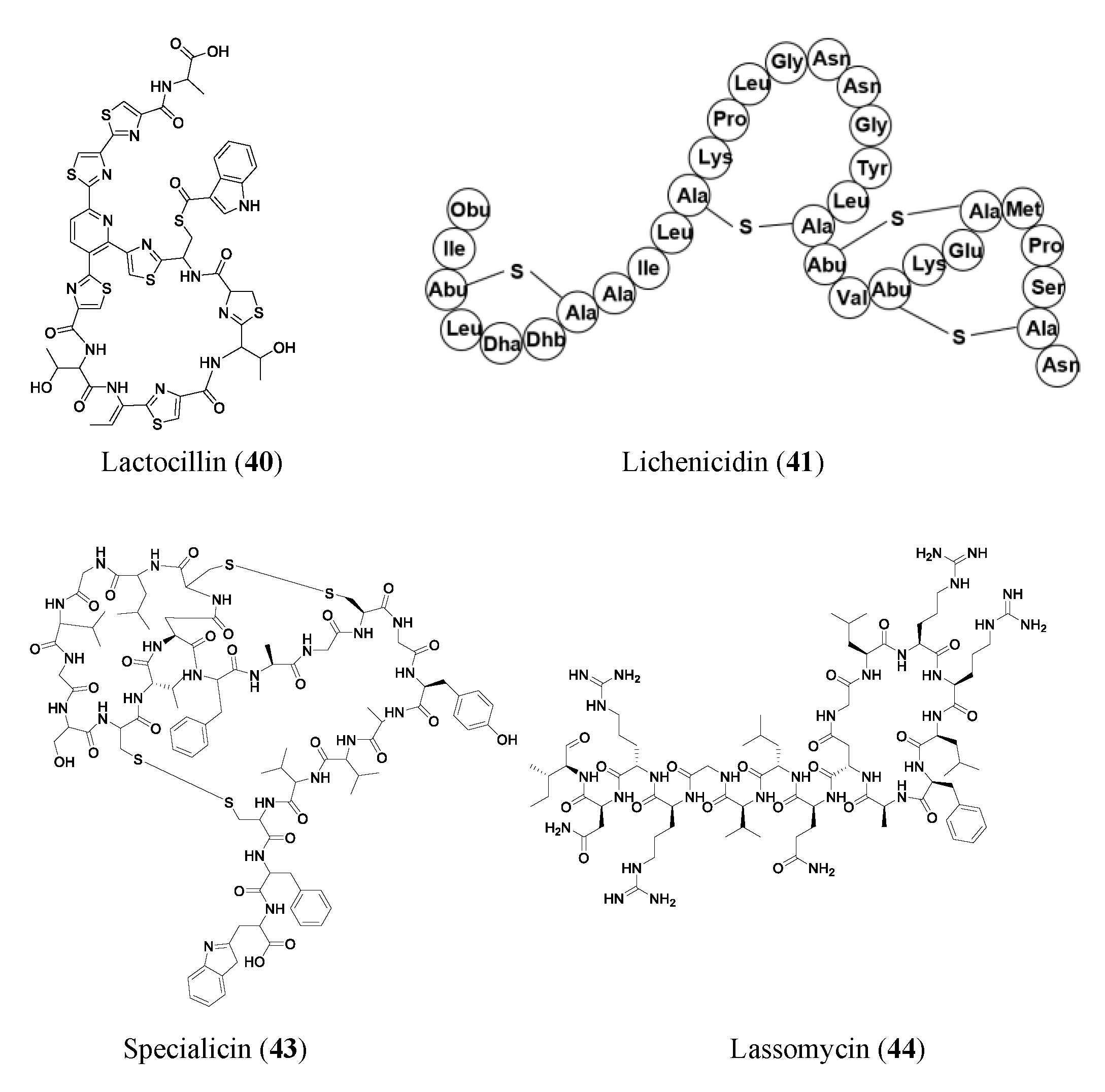


| Compound | Class | Source | Activity (MIC) | Mode of Action | Ref. |
|---|---|---|---|---|---|
| ECO-0501 (4) | PK | Amycolatopsis orientalis ATCC 43491 | MRSA (4.8 μM); VRE (10 μM) | Inhibits the bacteria cell membrane and/or cell wall | [9] |
| Anthracimycin (6) | PK | Streptomyces sp. CNH365 | MRSA (≥0.63 μM); vancomycin-resistant Staphylococcus aureus (≥0.63 μM) | Inhibits RNA and/or DNA synthesis | [13] |
| Pyxidicyclin A (7) | PK | Pyxidicoccus fallax An d48 | S. aureus newman (36 μM); E. coli ΔTolC (8 μM); C. albicans (>100 μM) | Topoisomerase IV inhibition | [14] |
| Thiolactomycin (8) | PK/NRP | Nocardia spp. | MRSA (357 μM); Mycobacterium tuberculosis (143 μM) | Inhibits type II fatty acid synthase | [15] |
| TLM 9 (9) | PK/NRP | Semi-synthetic derivative of thiolactomycin | MRSA (2.3 μM) | Inhibits type II fatty acid synthase | [92] |
| Isoindolinomycin (10) | PK | Streptomyces sp. SoC090715LN-16 | S. aureus (11.6 μM) | May bind to bacterial ribosomal subunits | [16] |
| Neoansamycin A (11) | PK | Streptomyces sp. LZ35 | S. aureus ATCC 25923 (6 μM) | -- | [18] |
| Keyicin (13) | PK | Micromonospora sp. | Bacillus subtilis (9.9 μM); Methicillin-sensitive Staphylococcus aureus (MSSA) (2.5 μM) | May modulate fatty acid metabolism | [21] |
| Turbinmicin (14) | PK | Micromonospora sp. WMMC-415 | Panresistant Candidas auris B11211 (0.4 μM); C. albicans (0.4 μM); C. glabrata (0.8 μM); Aspergillus fumigatus (0.4 μM) | Targeting fungal-specific Sec14 of the vesicular trafficking pathway | [27] |
| Clostrubin (15) | PK | Clostridium beijerincki | MRSA (0.12 μM); VRE111 (0.97 μM); Bacillus subtilis (0.075 μM); Mycobacterium smegmatis (0.48 μM) | -- | [28] |
| TP-1161 (21) | NRP | Norcardiopsis sp. TSF65-07 | VRE 560 and VRE 569 (0.84 μM) | -- | [31] |
| Brevibacillin 2V (22) | NRP | Brevibacillus laterosporus DSM 25 | MRSA (1.3 μM); VRE (1.3 μM); Bacillus cereus ATCC14579 (1.3 μM) | -- | [32] |
| Medipeptin A (23) | NRP | Pseudomonas sp. | Pseudomonas syringae pv. Tomato DC3000 (4 μM); Staphylococcus aureus subsp. Aureus 5334R4 (2 μM), Bacillus cereus ATCC14579 (2 μM); Xanthromonas translucens pv. Graminis LMG587 (2 μM) | Binding with lipoteichoic acids and forming pores in the bacterial membrane | [33] |
| Malacidin A (24) | NRP | Desert soil metagenome BGC heterologous expressed by Streptomyces albus | MRSA (0.16–0.64 μM); S. aureus NRS146 (0.32–0.64 μM); VRE (0.64–1.6 μM) | Calcium-dependent bacteria cell wall inhibition | [34] |
| Taromycin A (26) | NRP | Saccharomonospora sp. CNQ-490 | With 50 μg ml−1 CaCl2: DapR MRSA A8817 (30.5 μM); DapS MRSA A8819 (1.89 μM); DapR VRE 447 (30.5 μM) | Calcium-dependent bacteria membrane disruption | [36] |
| Teixobactin (28) | NRP | Eleftheria terrae | MRSA (0.2 μM); VRE (0.4 μM); penicillin-resistant Streptococcus pneumoniae (≤24 nM); M. tuberculosis H37Rv (0.1 μM); Clostridium difficile (4 nM) | Binds to lipid II and III, inhibits bacteria cell wall synthesis | [37] |
| Lugdunin (29) | NRP | Staphylococcus lugdunensis | MRSA (1.92 μM); VRE (3.8 μM); S. aureus Mu50 (3.8 μM) | -- | [38] |
| Macolacin (30) | NRP | Paenibacillus xylanexedens mac BGC | Antimicrobial-resistant Acinetobacter baumannii (1.7–6.85 μM); colistin-resistant Neisseria gonorrhoeae (3.4 μM) | Binds to lipid A of LPS and disrupts the bacteria cell membrane | [39] |
| Humimycin A (32) | NRP | Rhodococcus equi BGC | MRSA (7.25 μM); Streptococcus pneumoniae (3.6 μM) | Inhibit bacteria cell wall synthesis by inhibiting peptidoglycan translocation | [40] |
| Paenimucillin A (34) | NRP | Paenibacillus mucilaginosus K02 BGC | Enterococcus faecium (2.5 μM); Staphylococcus aureus (9.8 μM); Acinetobacter baumannii (9.8 μM) | May bind lipid II | [41,42] |
| Paenimucillins C (36) | NRP | Paenibacillus mucilaginosus K02 BGC | Multidrug-resistant Acinetobacter baumannii (0.3–2.5 μM) | Bacteria cell membrane disruption | [41,42] |
| 37 | NRP | Xenorhabdus nematophila BGC | C. albicans (3.2 μM) | -- | [42] |
| Di-alboflavusin A1 (38) | NRP | Streptomyces alboflavus sp. 313 | MRSA 113 (0.78 μM); MRSA 09L098 (0.63 μM) | -- | [43] |
| Lactocillin (40) | RiPP | Lactobacillus paragasseri JV-V03 | Staphylococcus aureus (0.42 μM); Corynebacterium aurimucosum (0.42 μM); Streptococcus sobrinus (0.85 μM) | -- | [46] |
| Lichenicidin (41) | RiPP | Bacillus licheniformis D13 | Bacillus megaterium (0.48 μM); Bacillus subtilis (0.96 μM); Micrococcus luteus (0.17 μM); Rhodococcus spp. (>2.56 μM); S. aureus (0.96 μM) | -- | [47] |
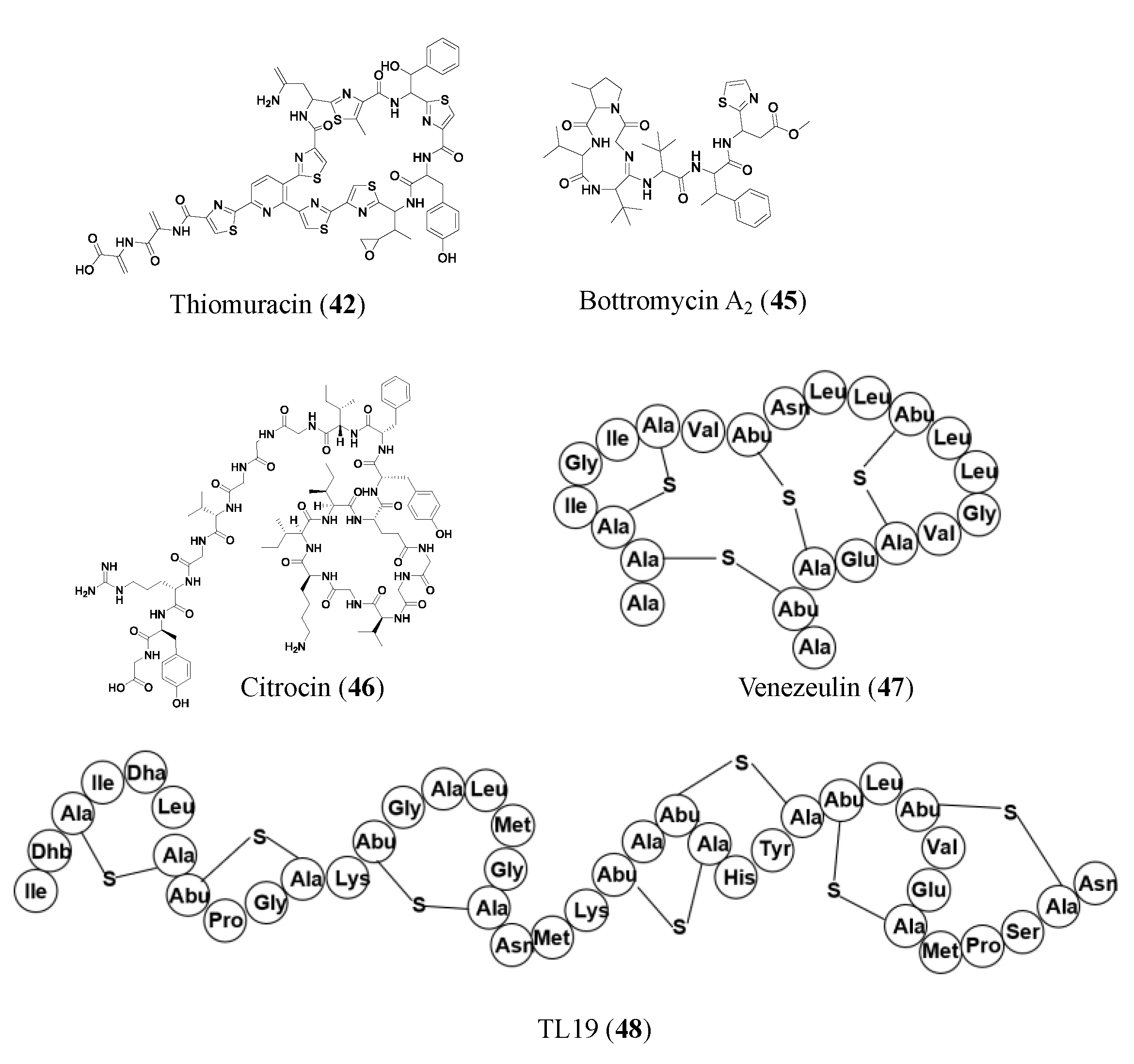
| Thiomuracin (42) | RiPP | Thermobispora bispora | MRSA (0.18 μM); VRE E. faecium U503 (0.046 μM); B. anthracis str. Sterne (1.5 μM) | Inhibits bacterial protein synthesis | [49] |
| Specialicin (43) | RiPP | Streptomyces specialis | M. luteus (3.75 μM); IC50 of 7.2 μM against HIV-1 | -- | [50] |
| Lassomycin (44) | RiPP | Lentzia kentuckyensis | Mycobacterium tuberculosis (0.41–1.65 μM); drug-resistant M. tuberculosis (0.21–1.65 μM) | Inhibits ATP-dependent protease of M. tuberculosis | [51] |
| Bottromycin A2 (45) | RiPP | Streptomyces sp. BC16019 | MRSA (1.2 μM); VRE (<1.2 μM) | Inhibits protein synthesis by blocking the binding site on the 50S ribosome | [53,54] |
| Citrocin (46) | RiPP | Citrobacter pasteurii and Citrobacter braakii | E. coli O157:H7 strain TUV93-0 (16 μM); Salmonella enterica serovar Newport (1000 μM); Citrobacter clinical isolate (125 μM) | -- | [52] |
| TL19 (48) | RiPP | Synthetically produced | MDR E. faecium (0.9 to 15 μM) | May bind to lipid II | [56] |
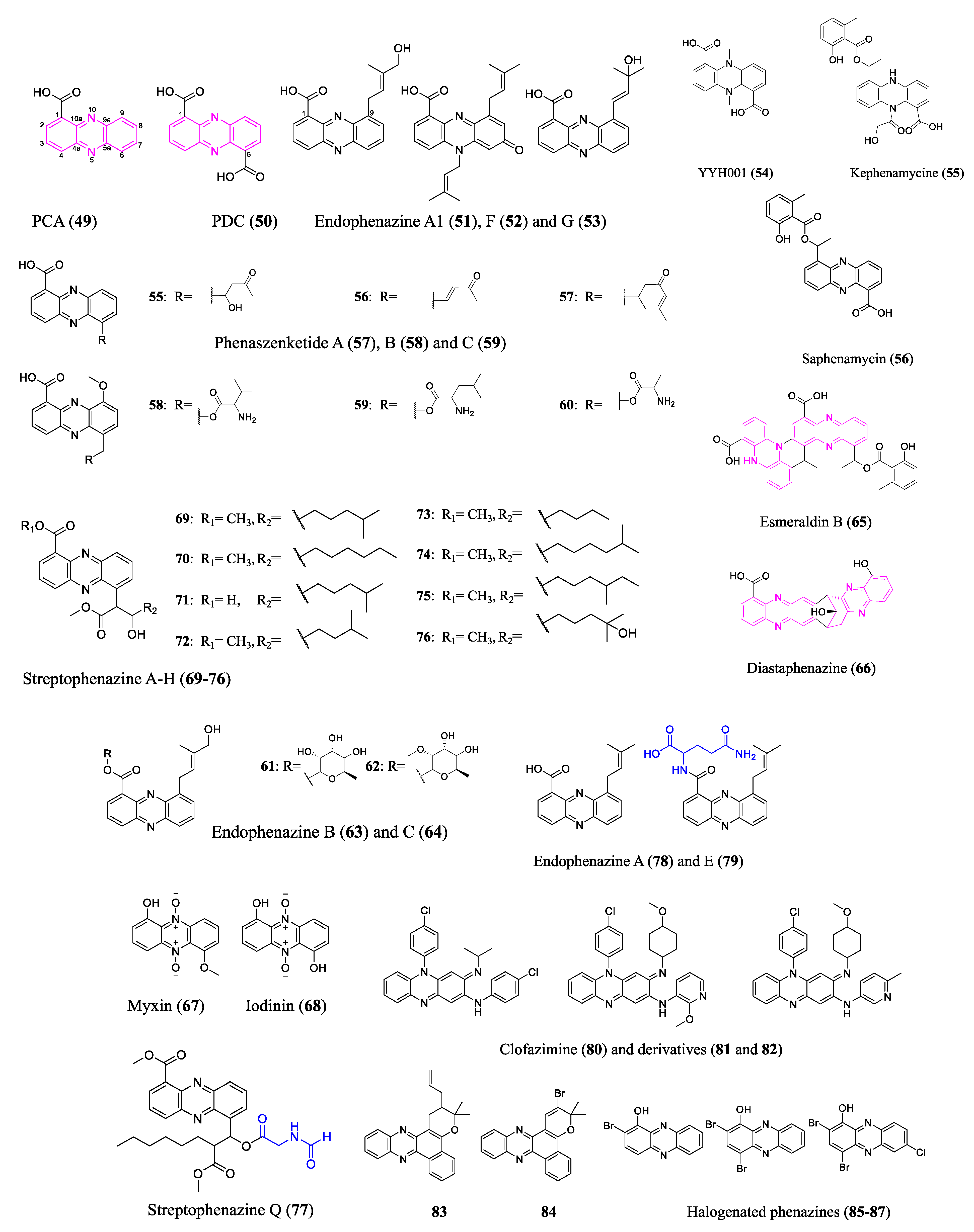
| Phenaszentines C (61) | Phz/NRP | Xenorhabdus szentirmaii | Board spectrum of Gram-positive and -negative bacteria (MIC not available) | -- | [64] |
| D-alanylgriseoluteic acid (62) | Phz/NRP | Pantoea agglomerans Eh1087 | Penicillin-resistant Streptococcus pneumoniae (1.4 µM); E. coli (2.8–5.6 µM) | Induces SOS response | [65] |
| Diastaphenazine (64) | Phz | Streptomyces diastaticus subsp. ardesiacus. | S. aureus (147 µM) | -- | [67] |
| Streptophenazine C (71) | Phz | Streptomyces sp. HB202 | Bacillus subtilis (38 µM); Staphylococcus lentus (114 µM) | -- | [70] |
| Streptophenazine H (76) | Phz | Streptomyces sp. HB202 | Bacillus subtilis (35 µM) | -- | [70] |
| Streptophenazine Q (77) | Phz/NRP/PK | Streptomyces sp. CNB-091 | MRSA TCH1516 (78 µM); Group A Streptococcus (4.9 µM); Acinetobacter baumannii 5075 (78 µM) | -- | [71] |
| Clofazimine derivative (81) | Phz | Organic synthesized | multidrug-resistant M. tuberculosis (0.3 µM) | May release reactive oxygen species | [75,76,77] |
| 83 | Phz | Organic synthesized | Rifampicin-resistant M. tuberculosis (2.2 μM); M. tuberculosis H37Rv (2.2 μM) | -- | [78] |
| 2-Br-1-OHPhz (85) | Phz | Streptomyces sp. CNS284 | MRSA (6.25 μM) | -- | [93] |
| 2,4-diBr-1-OHPhz (86) | Phz | Organic synthesized | MRSA (1.17 μM); MRSE (1.17 μM); VRE (6.25 μM) | Causes iron starvation in MRSA biofilms | [82] |
| 2,4-diBr-7-Cl-1-OHPhz (87) | Phz | Organic synthesized | MRSA (0.04 μM); MRSE (0.10 μM); VRE (0.39 μM) | Causes iron starvation in MRSA biofilms | [82] |
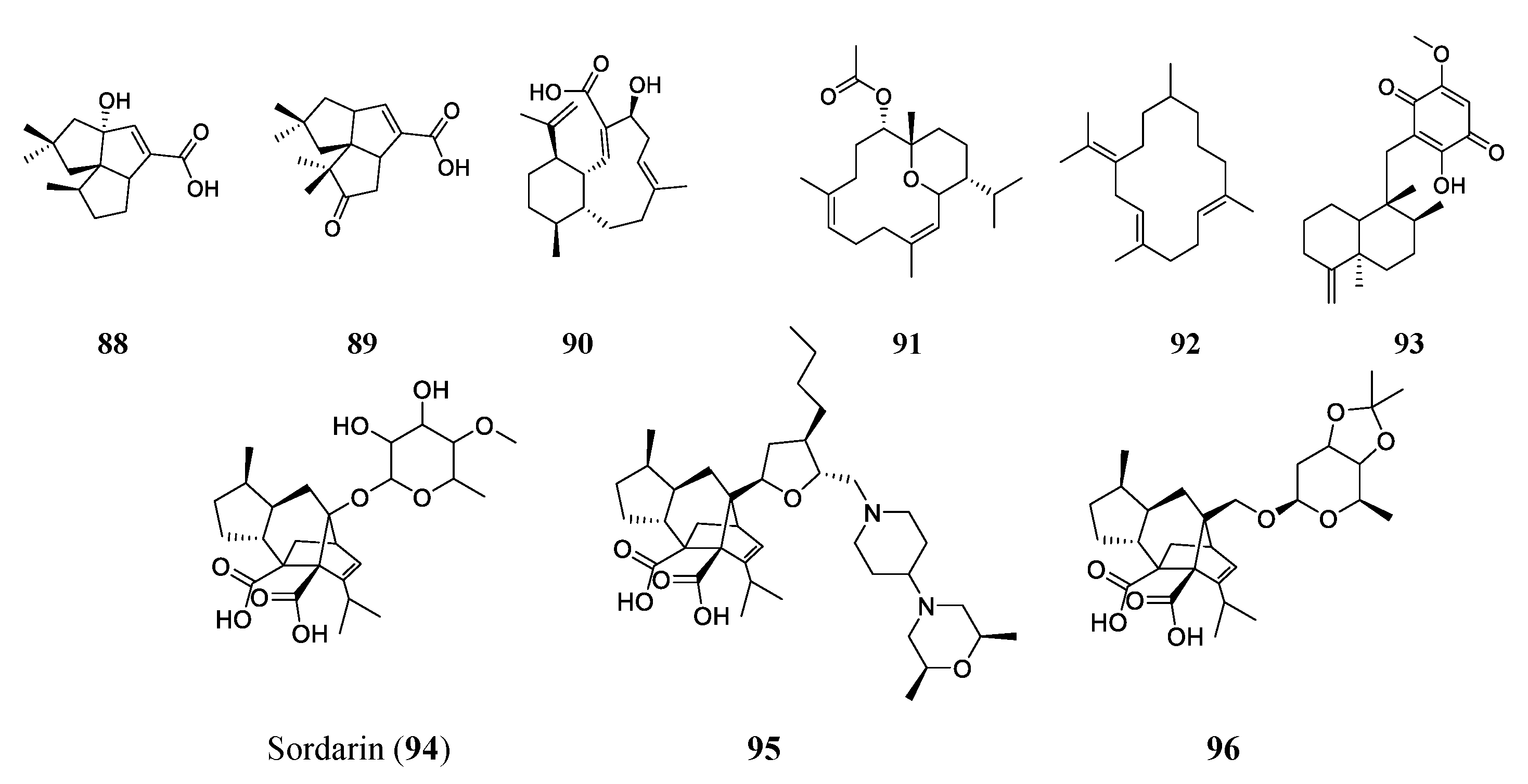
| Benditerpenoic acid (90) | TP | Streptomyces sp. CL12-4 | E. faecium (401μM); S. aureus (200 μM); MRSA (200 μM); MDR S. aureus (401 μM); B. subtilis (100 μM) | -- | [86] |
| Sarcotrocheliol (91) | TP | Sarcophyton. trocheliophorum | S. aureus (1.53 μM); MRSA (3.06 μM); Acinetobacter spp. (3.06 μM) | -- | [87] |
| Cembrene-C (92) | TP | S. trocheliophorum | Candida albicans (0.68 μM); Aspergillus flavus (0.68 μM) | -- | [87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Kessler, M.G.C.; Marchán-Rivadeneira, M.R.; Han, Y. Combating Antimicrobial Resistance in the Post-Genomic Era: Rapid Antibiotic Discovery. Molecules 2023, 28, 4183. https://doi.org/10.3390/molecules28104183
Yang Y, Kessler MGC, Marchán-Rivadeneira MR, Han Y. Combating Antimicrobial Resistance in the Post-Genomic Era: Rapid Antibiotic Discovery. Molecules. 2023; 28(10):4183. https://doi.org/10.3390/molecules28104183
Chicago/Turabian StyleYang, Yuehan, Mara Grace C. Kessler, Maria Raquel Marchán-Rivadeneira, and Yong Han. 2023. "Combating Antimicrobial Resistance in the Post-Genomic Era: Rapid Antibiotic Discovery" Molecules 28, no. 10: 4183. https://doi.org/10.3390/molecules28104183
APA StyleYang, Y., Kessler, M. G. C., Marchán-Rivadeneira, M. R., & Han, Y. (2023). Combating Antimicrobial Resistance in the Post-Genomic Era: Rapid Antibiotic Discovery. Molecules, 28(10), 4183. https://doi.org/10.3390/molecules28104183






