Collision Cross Section Prediction Based on Machine Learning
Abstract
1. Introduction
2. Ion Mobility-Mass Spectrometry (IM-MS)
2.1. Ion Mobility Platforms with Different Separation Principles
2.2. Advantages of LC-IM-MS
- (1)
- LC-IM-MS provides four-dimensional information (tR, CCS, MS, and MS/MS). As a robust parameter for characterization and recognition, CCS provides orthogonal attributes for compound recognition, improving the confidence level of compound annotation [4,59]. IMS technology has proven that it can be used to separate various isomers, such as lipid isomers [60], steroid isomers [61], fatty acid isomers [62], amino acid isomers [22], and carbohydrate isomers [63]. Numerous strategies have been introduced to enhance the IMS characterization of isomers. A combination of chemical derivatization and IMS can improve the detection of steroid isomers [61], metabolites in nicotine [64], and carbohydrates [65]. The integration of dimers or polymers with IM-MS is another effective method for identifying isomers. More accurately predicting the relative differences in CCS between steroid epimers can be achieved through the energy characteristics of the sodium dimer configuration of epimers [66]. The enantiomers of aromatic amino acids can be differentiated by TWIM-MS through their cationization with copper (II) and multimer formation with D-proline (Pro) as a chiral reference compound [67]. The mobility of ions passing through IMS is affected by using different drift gases and/or by doping volatile chiral reagents in drift gases, which can also be used to separate isomers and enantiomers [68,69]. In addition, platforms such as cIMS [42,70,71,72], multiplexed ion mobility [26,28,73], and TIMS [74] have improved the separation of isomers by improving mobility resolution. IMS can distinguish between conformational isomers [75] and isotopic isomers [22]. By taking into account all relevant errors, N-glycan isomers with different conformations can be distinguished on the basis of the CCS gained from the IMS [75]. As we know, lipids have a wide range of structural diversity, with a large number of isomers. A recent study used IMS to analyze the relationship between lipid structure and its gas-phase conformation, providing accurate and comprehensive conformational lipid profiles [76]. IMS has been used in the separation of isomers with different isotopic atomic positions [77] and labeled/unlabeled isotope-substituted isomers [42]. Researchers have found that IMS can be incorporated into the standard LC-MS/MS isotope analysis process as an additional separation mechanism, which can provide broader separation space and higher identification confidence for metabolic characterization [22].
- (2)
- Thanks to the advantage of increasing peak capacity and improving the signal-to-noise ratio, IMS can improve the exposure rate of trace components in complex samples [58,78]. Configuring ion mobility technology in MS studies with different ionization principles (ESI, MSI, and MALDI) can increase the peak capacity by at least two times compared with using MS alone [79,80,81]. It has been reported that when the mass resolution is 35,000 (fwhm), 860 independent ions can be measured, accounting for 15% of the total 5639 counted ions, while the addition of IMS adds 3911 features for signal recognition [79]. Because IMS is used as a separation module between LC and MS, the number of MS features detected in the metabolite composition characterization experiment has significantly increased [82]. IM-MSI can reduce chemical noise and transfer target signals from congested spectral regions, thereby increasing the S/N of metabolites and lipid peaks by nearly 10 times and doubling the image contrast [83]. Some studies have shown that compared to the traditional lipidomics methods, LC-IM-MS analysis has an increased S/N and can detect a low abundance of phospholipids in highly complex brain lipoid samples [43]. In the experiment of adding IMS to MS imaging, it was concluded that lipids with different CCS values can be spatially separated, highlighting their spatial positioning and achieving more-accurate lipid recognition [79].
- (3)
- In addition to IMS’s direct use or combining IMS with LC, it can also combine with gas chromatography (GC), mass spectrometry imaging (MSI), or supercritical fluid chromatography (SFC) technologies. As a result, multidimensional analytical information is provided, and the selection of methods increases. IMS and LC can provide orthogonal separation, with IMS separation occurring within milliseconds, and it is compatible with modern MS that is running at microsecond scanning speeds, allowing maximum separation of metabolite ions prior to MS characterization. IMS is often used in series with reverse-phase liquid chromatography (RPLC) [84,85,86] and hydrophilic interaction liquid chromatography (HILIC) [87,88,89]. Some researchers have also proposed an offline two-dimensional liquid chromatography coupled with an ion mobility-quadrupole time-of-flight mass spectrometry (2D-LC/IM-QTOF-MS) analysis strategy, achieving a comprehensive characterization of multiple components in traditional Chinese medicine [8,58,90]. In addition, a study that coupled IMS with MSI technology achieved the spatial localization of bile acids in sample tissues [91]. One study integrated ultrahigh performance supercritical fluid chromatography/quadrupole time-of-flight mass spectrometry (UHPSFC/QTOF-MS) and ion mobility spectroscopy/time-of-flight mass spectrometry (IMS/QTOF-MS) to establish a lipid omics platform for CCS measurement, which has improved the analytical performance and recognition reliability of lipids [92].
- (4)
- IM can improve the overall resolution of the spectrum and obtain high-quality MS1 and MS2 spectra. Double-charged ion clusters make the types of precursors thoroughly complex and can easily generate false positives when annotating MS2 data. IM is capable of separating dimers or double-charged ions in a full scan spectrum and generating high-resolution spectra of MS1 and MS2 that are close to the standards [58,84]. Wang [58] used an LC-IM-MS system to comprehensively characterize the multicomponents of compound Danshen dripping pills (CDDPs) and elucidated the advantages of IM. IM can improve the overall resolution of the spectrum of CDDPs and effectively distinguish the doubly charged saponins or the dimers of salvianolic acids, to obtain high-quality MS1 and MS2 spectra and reduce the false positives of multicomponent characterization.
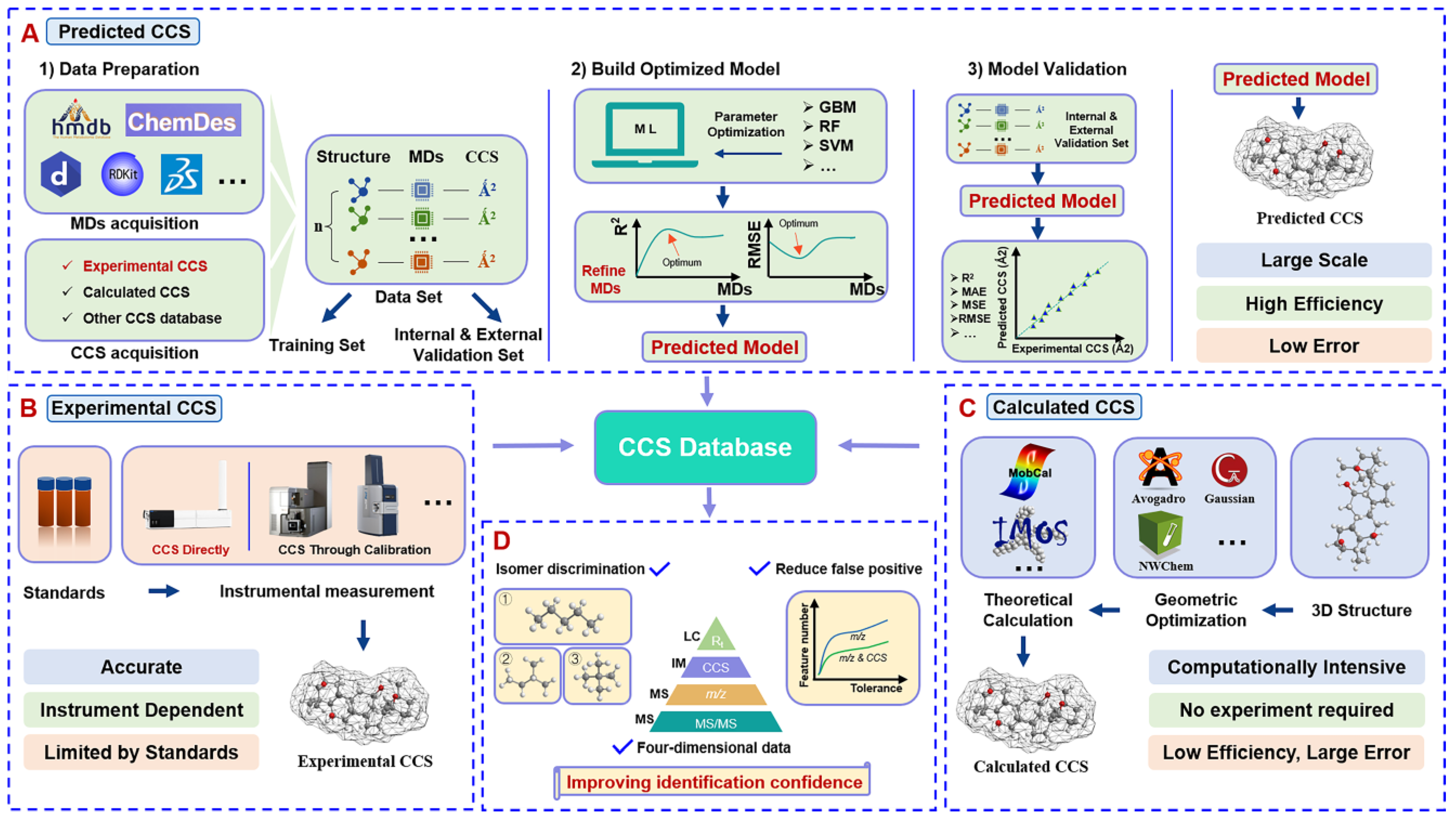
3. Collision Cross Section Value: Dependent Variable of the Model
3.1. Acquisition of CCS Values
3.2. Stability Evaluation of CCS Values
- (1)
- CCS values are consistent among instruments and laboratories. Numerous studies [79,95,99,117,118] have demonstrated that the measurement of CCS values for metabolites with different molecular weights on multiple TWIMS in independent laboratories (between different Vion IMSs and different SynaptG2 HDMSs, as well as between Vion IMS and SynaptG2 HDMS) is repeatable, with an RSD of CCS values within ±3%. Sarah [112] studied the reproducibility of CCS values obtained from DTIMS. Upon the completion of the analysis of 51 biologically related standards (amino acids and lipids), it was found that the interlaboratory RSD was 0.30 ± 0.16%. Some studies [23,133] have compared the CCS values measured by TWIMS and DTIMS and found that the absolute percentage error (APE) of the CCS values was within 2%.
- (2)
- CCS has stability in different substrates. Giuseppe [94] found through experimental measurements that 97% of CCS values had a mean RSD of less than 2%, which demonstrates the repeatability of CCS values in various biological matrices. To test the accuracy and precision of CCS measurements in different matrices, one study [79] compared the CCS values in the database with CCS values measured from a series of lipid extracts such as porcine brain, E. coli, and yeast. The results showed that CCS measurements were highly stable in different matrices.
- (3)
- CCS values have long-term robustness. One study [117] evaluated the reproducibility of the CCS values of steroid compounds after 1.5 years, and the results showed that 95.7% of the CCS values had an RSD within ±1.0%.
- (4)
- CCS also has stability at different sample concentrations. In addition, some studies have proposed some insights into how to improve the repeatability of CCS measurements, especially the high reproducibility between different ion mobility platforms [1,19,134]. For example, consistent instruments, configurations, calibration procedures, etc. are used to achieve measurement standardization; the physical theory behind ion mobility is improved so that different platforms can provide the same, physically correctly calculated CCS values without requiring calibration.
4. Molecular Descriptors: Independent Variable of the Model
4.1. Molecular Representation
4.2. Access to Molecular Descriptors
4.3. Preprocessing and Optimization of Molecular Descriptors
5. Machine-Learning Algorithms
5.1. Different Prediction Algorithms and Prediction Platforms
| Algorithm | Method Type | Tools | Features | Refs. |
|---|---|---|---|---|
| Stepwise multiple linear analysis (SMLR) | linear | R package MLRMPA | Data need to be normalized to reduce the impact of overfitting | [156] |
| Principal component regression (PCR) | linear | R package MASS | Can reduce the dimensionality of the data set while maintaining the features with the maximum variance contribution in the data set | [156] |
| Partial least squares regression (PLS) | linear | Matlab with the PLS toolbox/R package pls | Not sensitive to multicollinearity issues caused by the use of simple linear regression models | [150,156] |
| Least absolute shrinkage and selection operator (LASSO) | linear | Open-source R programming | Have powerful ability to perform both variable selection and regularization | [60] |
| Support vector machine (SVM) | nonlinear | R package e1071 | Wide application; relatively small sample size; can effectively avoid overfitting | [10,14,16,145,147,158] |
| Artificial neural network (ANN) | nonlinear | Alyuda NeuroIntelligence 2.2 | Can perform supervised learning, unsupervised learning, and semisupervised learning | [15,110,133,159] |
| Random forest (RF) | nonlinear | The scikit-learn Python package | Low variance; low susceptibility to overfitting; poor model applicability | [137,165] |
| Gradient-boosting machine (GBM) | nonlinear | XGBoost library | Overfitting often occurs | [98,152,153,162] |
5.2. Evaluation and Verification of Prediction Algorithms
6. CCS Prediction Applications
6.1. In Multiomics
6.2. In Natural Products
6.3. In Foods
6.4. In Other Fields
7. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gabelica, V.; Shvartsburg; Alexandre, A.; Afonso, C.; Barran, P.; Benesch, J.J.P.J.; Bleiholder, C.; Bowers, M.T.; Bilbao, A.; Bush, M.F.; et al. Recommendations for reporting ion mobility mass spectrometry measurements. Mass Spectrom. Rev. 2019, 38, 291–320. [Google Scholar] [CrossRef]
- Lanucara, F.; Holman, S.W.; Gray, C.J.; Eyers, C.E. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 2014, 6, 281–294. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Sherrod, S.D.; McLean, J.A. Improving the discovery of secondary metabolite natural products using ion mobility-mass spectrometry. Curr. Opin. Chem. Biol. 2018, 42, 160–166. [Google Scholar] [CrossRef]
- Moran-Garrido, M.; Camunas-Alberca, S.M.; Gil-de-la Fuente, A.; Mariscal, A.; Gradillas, A.; Barbas, C.; Sáiz, J. Recent developments in data acquisition, treatment and analysis with ion mobility-mass spectrometry for lipidomics. Proteomics 2022, 22, 2100328. [Google Scholar] [CrossRef]
- Gabelica, V.; Marklund, E. Fundamentals of ion mobility spectrometry. Curr. Opin. Chem. Biol. 2018, 42, 51–59. [Google Scholar] [CrossRef]
- Thomson, J.J.; Rutherford, E.X.L. On the passage of electricity through gases exposed to Röntgen rays. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1896, 42, 392–407. [Google Scholar] [CrossRef]
- Ross, D.H.; Seguin, R.P.; Krinsky, A.M.; Xu, L.B. High-throughput measurement and machine learning-based prediction of collision cross sections for drugs and drug metabolites. J. Am. Soc. Mass Spectrom. 2022, 33, 1061–1072. [Google Scholar] [CrossRef]
- Zuo, T.T.; Zhang, C.X.; Li, W.W.; Wang, H.D.; Hu, Y.; Yang, W.Z.; Jia, L.; Gao, X.M.; Guo, D. Offline two-dimensional liquid chromatography coupled with ion mobility-quadrupole time-of-flight mass spectrometry enabling four-dimensional separation and characterization of the multicomponents from white ginseng and red ginseng. J. Pharm. Anal. 2022, 10, 597–609. [Google Scholar] [CrossRef]
- Liu, W.B.; Zhang, W.D.; Li, T.Z.; Zhou, Z.W.; Luo, M.D.; Chen, X.; Cai, Y.P.; Zhu, Z.J. Four-dimensional untargeted profiling of N-Acylethanolamine lipids in the mouse brain using ion mobility–mass spectrometry. Anal. Chem. 2022, 94, 12472–12480. [Google Scholar] [CrossRef]
- Zhou, Z.W.; Shen, X.T.; Tu, J.; Zhu, Z.J. Large-scale prediction of collision cross-section values for metabolites in ion mobility-mass spectrometry. Anal. Chem. 2016, 88, 11084–11091. [Google Scholar] [CrossRef]
- Ross, D.H.; Cho, J.H.; Xu, L.B. Breaking down structural diversity for comprehensive prediction of ion-neutral collision cross sections. Anal. Chem. 2020, 92, 4548–4557. [Google Scholar] [CrossRef]
- Colby, S.M.; Thomas, D.G.; Nuñez, J.R.; Baxter, D.J.; Glaesemann, K.R.; Brown, J.M.; Pirrung, M.A.; Govind, N.; Teeguarden, J.G.; Metz, T.O.; et al. ISiCLE: A quantum chemistry pipeline for establishing in silico collision cross section libraries. Anal. Chem. 2019, 91, 4346–4356. [Google Scholar] [CrossRef]
- Ross, D.H.; Xu, L.B. Determination of drugs and drug metabolites by ion mobility-mass spectrometry: A review. Anal. Chim. Acta 2021, 1154, 338270. [Google Scholar] [CrossRef]
- Zhou, Z.W.; Tu, J.; Xiong, X.; Shen, X.T.; Zhu, Z.J. LipidCCS: Prediction of collision cross-section values for lipids with high precision to support ion mobility-mass spectrometry-based lipidomics. Anal. Chem. 2017, 89, 9559–9566. [Google Scholar] [CrossRef]
- Plante, P.L.; Francovic-Fontaine, É.; May, J.C.; McLean, J.A.; Baker, E.S.; Laviolette, F.; Marchand, M.; Corbeil, J. Predicting ion mobility collision cross-sections using a deep neural network: DeepCCS. Anal. Chem. 2019, 91, 5191–5199. [Google Scholar] [CrossRef]
- Zhou, Z.W.; Luo, M.D.; Chen, X.; Yin, Y.D.; Xiong, X.; Wang, R.H.; Zhu, Z.J. Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nat. Commun. 2020, 111, 4334. [Google Scholar] [CrossRef]
- Koopman, J.; Grimme, S. From QCEIMS to QCxMS: A tool to routinely calculate CID mass spectra using molecular dynamics. J. Am. Soc. Mass Spectr. 2021, 32, 1735–1751. [Google Scholar] [CrossRef]
- May, J.C.; McLean, J.A. Ion mobility-mass spectrometry: Time-dispersive instrumentation. Anal. Chem. 2015, 87, 1422–1436. [Google Scholar] [CrossRef]
- Brinke, E.T.; Arrizabalaga-Larrañaga, A.; Blokland, M.H. Insights of ion mobility spectrometry and its application on food safety and authenticity: A review. Anal. Chim. Acta 2022, 1222, 340039. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Wojcik, R.; Zhang, X.; Ibrahim, Y.H.M.; Burnum-Johnson, K.E.; Orton, D.J.; Monroe, M.E.; Moore, R.J.; Smith, R.D.; Baker, E.S. Coupling front-end separations, ion mobility spectrometry, and mass spectrometry for enhanced multidimensional biological and environmental analyses. Annu. Rev. Anal. Chem. 2017, 10, 71–92. [Google Scholar] [CrossRef]
- Mason, E.A.; Schamp, H.W., Jr. Mobility of gaseous ions in weak electric fields. Ann. Phys. 1958, 4, 233–270. [Google Scholar] [CrossRef]
- Dodds, J.N.; May, J.C.; McLean, J.A. Investigation of the complete suite of the leucine and isoleucine isomers: Toward prediction of ion mobility separation capabilities. Anal. Chem. 2017, 89, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Hinnenkamp, V.; Klein, J.; Meckelmann, S.W.; Balsaa, P.; Schmidt, T.C.; Schmitz, O.J. Comparison of CCS values determined by traveling wave ion mobility mass spectrometry and drift tube ion mobility mass spectrometry. Anal. Chem. 2018, 90, 12042–12050. [Google Scholar] [CrossRef]
- Kemper, P.R.; Dupuis, N.F.; Bowers, M.T. A new, higher resolution, ion mobility mass spectrometer. Int. J. Mass Spectrom. 2009, 287, 46–57. [Google Scholar] [CrossRef]
- Kirk, A.T.; Allers, M.; Cochems, P.; Langejuergen, J.; Zimmermann, S. A compact high resolution ion mobility spectrometer for fast trace gas analysis. Analyst 2013, 138, 5200–5207. [Google Scholar] [CrossRef] [PubMed]
- Demelenne, A.; Nys, G.; Nix, C.; Fjeldsted, J.C.; Crommen, J.; Fillet, M. Separation of phosphorothioated oligonucleotide diastereomers using multiplexed drift tube ion mobility mass spectrometry. Anal. Chim. Acta 2022, 1191, 339297. [Google Scholar] [CrossRef] [PubMed]
- Sipe, S.N.; Sanders, J.D.; Reinecke, T.; Clowers, B.H.; Brodbelt, J.S. Separation and collision cross section measurements of protein complexes afforded by a modular drift tube coupled to an orbitrap mass spectrometer. Anal. Chem. 2022, 94, 9434–9441. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.D.; Shields, S.W.; Escobar, E.E.; Lanzillotti, M.B.; Butalewicz, J.P.; James, V.K.; Blevins, M.S.; Sipe, S.N.; Brodbelt, J.S. Enhanced ion mobility separation and characterization of isomeric phosphatidylcholines using absorption mode frontier transform multiplexing and ultraviolet photodissociation mass spectrometry. Anal. Chem. 2022, 94, 4252–4259. [Google Scholar] [CrossRef]
- Lippmann, M.; Kirk, A.T.; Hitzemann, M.; Zimmermann, S. Compact and sensitive dual drift tube ion mobility spectrometer with a new dual field switching ion shutter for simultaneous detection of both ion polarities. Anal. Chem. 2020, 92, 11834–11841. [Google Scholar] [CrossRef]
- George, A.C.; Schmitz-Afonso, I.; Marie, V.; Colsch, B.; Fenaille, F.; Afonso, C.; Loutelier-Bourhis, C. A re-calibration procedure for interoperable lipid collision cross section values measured by traveling wave ion mobility spectrometry. Anal. Chem. 2022, 1226, 340236. [Google Scholar] [CrossRef]
- D’Atri, V.; Causon, T.; Hernandez-Alba, O.; Mutabazi, A.; Veuthey, J.L.; Cianferani, S.; Guillarme, D. Adding a new separation dimension to MS and LC-MS: What is the utility of ion mobility spectrometry? J. Sep. Sci. 2018, 41, 20–67. [Google Scholar] [CrossRef] [PubMed]
- Ruotolo, B.T.; Benesch, J.L.P.; Sandercock, A.M.; Hyung, S.J.; Robinson, C.V. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008, 3, 1139–1152. [Google Scholar] [CrossRef] [PubMed]
- Gelb, A.S.; Jarratt, R.E.; Huang, Y.; Dodds, E.D. A study of calibrant selection in measurement of carbohydrate and peptide ion-neutral collision cross sections by traveling wave ion mobility spectrometry. Anal. Chem. 2014, 86, 11396–11402. [Google Scholar] [CrossRef] [PubMed]
- Bush, M.F.; Hall, Z.; Giles, K.; Hoyes, J.; Robinson, C.V.; Ruotolo, B.T. Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem. 2010, 82, 9557–9565. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Conant, C.R.; Zheng, X.; Bloodsworth, K.J.; Orton, D.J.; Garimella, S.V.B.; Attah, I.K.; Nagy, G.; Smith, R.D.; Ibrahim, Y.M. Assessing collision cross section calibration strategies for traveling wave-based ion mobility separations in structures for lossless ion manipulations. Anal. Chem. 2020, 92, 14976–14982. [Google Scholar] [CrossRef] [PubMed]
- May, J.C.; Leaptrot, K.L.; Rose, B.S.; Moser, K.L.W.; Deng, L.L.; Maxon, L.; Debord, D.; McLean, J.A. Resolving power and collision cross section measurement accuracy of a prototype high-resolution ion mobility platform incorporating structures for lossless ion manipulation. J. Am. Soc. Mass Spectrom. 2021, 32, 1126–1137. [Google Scholar] [CrossRef] [PubMed]
- Giles, K.; Ujma, J.; Wildgoose, J.; Pringle, S.; Richardson, K.; Langridge, D.; Green, M. A cyclic ion mobility-mass spectrometry system. Anal. Chem. 2019, 91, 8564–8573. [Google Scholar] [CrossRef]
- Ropartz, D.; Fanuel, M.; Ujma, J.; Palmer, M.; Giles, K.; Rogniaux, H. Structure determination of large isomeric oligosaccharides of natural origin through multipass and multistage cyclic traveling-wave ion mobility mass spectrometry. Anal. Chem. 2019, 91, 12030–12037. [Google Scholar] [CrossRef]
- Colson, E.; Decroo, C.; Cooper-Shepherd, D.; Caulier, G.; Henoumont, C.; Laurent, S.; Winter, J.D.; Flammang, P.; Palmer, M.; Claereboudt, J.; et al. Discrimination of regioisomeric and stereoisomeric saponins from Aesculus hippocastanum seeds by ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30, 2228–2237. [Google Scholar] [CrossRef]
- Rüger, C.P.; Le Maître, J.; Maillard, J.; Riches, E.; Palmer, M.; Afonso, C.; Giusti, P. Exploring complex mixtures by cyclic ion mobility high-resolution mass spectrometry: Application toward petroleum. Anal. Chem. 2021, 93, 5872–5881. [Google Scholar] [CrossRef]
- Cavallero, G.J.; Zaia, J. Resolving heparan sulfate oligosaccharide positional isomers using hydrophilic interaction liquid chromatography-cyclic ion mobility mass spectrometry. Anal. Chem. 2022, 94, 2366–2374. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.L.; Bergman, A.E.; Heider, E.C.; Nagy, G. Experimental measurements of relative mobility shifts resulting from isotopic substitutions with high-resolution cyclic ion mobility separations. Anal. Chem. 2022, 94, 2988–2995. [Google Scholar] [CrossRef] [PubMed]
- Basit, A.; Pontis, S.; Piomelli, D.; Armirotti, A. Ion mobility mass spectrometry enhances low-abundance species detection in untargeted lipidomics. Metabolomics 2016, 12, 50. [Google Scholar] [CrossRef]
- Luo, M.D.; Zhou, Z.W.; Zhu, Z.J. The application of ion mobility-mass spectrometry in untargeted metabolomics: From separation to identification. J. Anal. Test. 2020, 4, 163–174. [Google Scholar] [CrossRef]
- Michelmann, K.; Silveira, J.A.; Ridgeway, M.E.; Park, M.A. Fundamentals of trapped ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 2014, 26, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Silveira, J.A.; Ridgeway, M.E.; Park, M.A. High resolution trapped ion mobility spectrometry of peptides. Anal. Chem. 2014, 86, 5624–5627. [Google Scholar] [CrossRef]
- Hernandez, D.R.; DeBord, J.D.; Ridgeway, M.E.; Kaplan, D.A.; Park, M.A.; Fernandez-Lima, F. Ion dynamics in a trapped ion mobility spectrometer. Analyst 2014, 139, 1913–1921. [Google Scholar] [CrossRef]
- Vasilopoulou, C.G.; Sulek, K.; Brunner, A.D.; Meitei, N.S.; Schweiger-Hufnagel, U.; Meyer, S.W.; Barsch, A.; Mann, M.; Meier, F. Trapped ion mobility spectrometry and PASEF enable in-depth lipidomics from minimal sample amounts. Nat. Commun. 2020, 11, 331. [Google Scholar] [CrossRef]
- Meier, F.; Brunner, A.D.; Koch, S.; Koch, H.; Lubeck, M.; Krause, M.; Goedecke, N.; Decker, J.; Kosinski, T.; Park, M.A.; et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteom. 2018, 17, 2534–2545. [Google Scholar] [CrossRef]
- Charkow, J.; Rost, H.L. Trapped ion mobility spectrometry reduces spectral complexity in mass spectrometry-based proteomics. Anal. Chem. 2021, 93, 16751–16758. [Google Scholar] [CrossRef]
- Adams, K.J.; Montero, D.; Aga, D.; Fernandez-Lima, F. Isomer separation of polybrominated diphenyl ether metabolites using nanoESI-TIM-MS. Int. J. Ion Mobil. Spectrom. 2016, 19, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Brunner, A.D.; Frank, M.; Ha, A.; Bludau, I.; Voytik, E.; Kaspar-Schoenefeld, S.; Lubeck, M.; Raether, O.; Bache, N.; et al. diaPASEF: Parallel accumulation-serial fragmentation combined with data-independent acquisition. Nat. Methods 2020, 17, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Helmer, P.O.; Nordhorn, I.D.; Korf, A.; Behrens, A.; Buchholz, R.; Zubeil, F.; Karst, U.; Hayen, H. Complementing matrix-assisted laser desorption ionization-mass spectrometry imaging with chromatography data for improved assignment of isobaric and isomeric phospholipids utilizing trapped ion mobility-mass spectrometry. Anal. Chem. 2021, 93, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
- Drakopoulou, S.K.; Damalas, D.E.; Baessmann, C.; Thomaidis, N.S. Trapped ion mobility incorporated in LC-HRMS workflows as an integral analytical platform of high sensitivity: Targeted and untargeted 4D-metabolomics in extra virgin olive oil. J. Agri. Food Chem. 2021, 69, 15728–15737. [Google Scholar] [CrossRef]
- Zhang, J.D.; Kabir, K.M.M.; Lee, H.E.; Donald, W.A. Chiral recognition of amino acid enantiomers using high-definition differential ion mobility mass spectrometry. Int. J. Mass Spectrom. 2018, 428, 1–7. [Google Scholar] [CrossRef]
- Dodds, J.N.; Baker, E.S. Ion mobility spectrometry: Fundamental concepts, instrumentation, applications, and the road ahead. J. Am. Soc. Mass Spectrom. 2019, 30, 2185–2195. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Smith, A.J.; Astarita, G. Ion mobility mass spectrometry in the omics era: Challenges and opportunities for metabolomics and lipidomics. Mass Spectrom. Rev. 2022, 41, 722–765. [Google Scholar] [CrossRef]
- Wang, H.D.; Wang, H.M.; Wang, X.Y.; Xu, X.Y.; Hu, Y.; Li, X.; Shi, X.J.; Wang, S.M.; Liu, J.; Qian, Y.X.; et al. A novel hybrid scan approach enabling the ion-mobility separation and the alternate data-dependent and data-independent acquisitions (HDDIDDA): Its combination with off-line two-dimensional liquid chromatography for comprehensively characterizing the multicomponents from Compound Danshen Dripping Pill. Anal. Chim. Acta 2022, 1193, 339320. [Google Scholar]
- Picache, J.A.; Rose, B.S.; Balinski, A.; Leaptrot, K.L.; Sherrod, S.D.; May, J.C.; McLean, J.A. Collision cross section compendium to annotate and predict multi-omic compound identities. Chem. Sci. 2019, 10, 983–993. [Google Scholar] [CrossRef]
- Wang, J.Y.; Yin, Y.H.; Zheng, J.Y.; Liu, L.F.; Yao, Z.P.; Xin, G.Z. Least absolute shrinkage and selection operator-based prediction of collision cross section values for ion mobility mass spectrometric analysis of lipids. Analyst 2022, 147, 1236–1244. [Google Scholar] [CrossRef]
- Ahonen, L.; Fasciotti, M.; af Gennäs, G.B.; Kotiaho, T.; Daroda, R.J.; Eberlin, M.; Kostiainen, R. Separation of steroid isomers by ion mobility mass spectrometry. J. Chromatogr. A 2013, 1310, 133–137. [Google Scholar] [CrossRef]
- Wu, F.L.; Wu, X.S.; Chi, C.X.; Ding, C.F. Simultaneous differentiation of C = C position isomerism in fatty acids through ion mobility and theoretical calculations. Anal. Chem. 2022, 94, 12213–12220. [Google Scholar] [CrossRef]
- Hofmann, J.; Hahm, H.S.; Seeberger, P.H.; Pagel, K. Identification of carbohydrate anomers using ion mobility-mass spectrometry. Nature 2015, 526, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, M.L.; Harrington, P.B. Detection of methamphetamine in the presence of nicotine using in situ chemical derivatization and ion mobility spectrometry. Anal. Chem. 2004, 76, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Fenn, L.S.; McLean, J.A. Enhanced carbohydrate structural selectivity in ion mobility-mass spectrometry analyses by boronic acid derivatization. Chem. Commun. 2008, 43, 5505–5507. [Google Scholar] [CrossRef] [PubMed]
- Chouinard, C.D.; Cruzeiro, V.W.D.; Roitberg, A.E.; Yost, R.A. Experimental and theoretical investigation of sodiated multimers of steroid epimers with ion mobility-mass spectrometry. J. Am. Soc. Mass Spectrom. 2016, 28, 323–331. [Google Scholar] [CrossRef]
- Domalain, V.; Hubert-Roux, M.; Tognetti, V.; Joubert, L.; Lange, C.M.; Rouden, J.; Afonso, C. Enantiomeric differentiation of aromatic amino acids using traveling wave ion mobility-mass spectrometry. Chem. Sci. 2014, 5, 3234–3239. [Google Scholar] [CrossRef]
- Asbury, G.R.; Hill, H.H. Using different drift gases to change separation factors (α) in ion mobility spectrometry. Anal. Chem. 2000, 72, 580–584. [Google Scholar] [CrossRef]
- Dwivedi, P.; Wu, C.; Hill, H.H., Jr. Gas phase chiral separations by ion mobility spectrometry. Anal. Chem. 2006, 78, 8200–8206. [Google Scholar] [CrossRef]
- Higton, D.; Palmer, M.E.; Vissers, J.P.C.; Mullin, L.G.; Plumb, R.S.; Wilson, I.D. Use of cyclic ion mobility spectrometry (cIM)-mass spectrometry to study the intramolecular transacylation of diclofenac acylglucuronide. Anal. Chem. 2021, 93, 7413–7421. [Google Scholar] [CrossRef]
- Cooper-Shepherd, D.A.; Olivos, H.J.; Wu, Z.X.; Palmer, M.E. Exploiting self-association to evaluate enantiomeric composition by cyclic ion mobility-mass spectrometry. Anal. Chem. 2022, 94, 84418448. [Google Scholar] [CrossRef] [PubMed]
- Oganesyan, I.; Hajduk, J.; Harrison, J.A.; Marchand, A.; Czar, M.F.; Zenobi, R. Exploring gas-phase MS methodologies for structural elucidation of branched N-Glycan isomers. Anal. Chem. 2022, 94, 10531–10539. [Google Scholar] [CrossRef] [PubMed]
- May, J.C.; Knochenmuss, R.; Fjeldsted, J.C.; McLean, J.A. Resolution of isomeric mixtures in ion mobility using a combined demultiplexing and peak deconvolution technique. Anal. Chem. 2020, 92, 9482–9492. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yin, Y.D.; Luo, M.D.; Zhou, Z.W.; Cai, Y.P.; Zhu, Z.J. Trapped ion mobility spectrometry-mass spectrometry improves the coverage and accuracy of four-dimensional untargeted lipidomics. Anal. Chim. Acta 2022, 1210, 339886. [Google Scholar] [CrossRef]
- Kevin, P. Ion mobility-mass spectrometry of complex carbohydrates: Collision cross sections of sodiated N-linked glycans. Anal. Chem. 2013, 85, 5138–5145. [Google Scholar]
- Leaptrot, K.L.; May, J.C.; Dodds, J.N.; McLean, J.A. Ion mobility conformational lipid atlas for high confidence lipidomics. Nat. Commun. 2019, 10, 985. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, R.; Nagy, G.; Attah, I.K.; Webb, I.K.; Garimella, S.V.B.; Weitz, K.K.; Hollerbach, A.; Monroe, M.E.; Ligare, M.R.; Nielson, F.F.; et al. SLIM ultrahigh resolution ion mobility spectrometry separations of isotopologues and isotopomers reveal mobility shifts due to mass distribution changes. Anal. Chem. 2019, 91, 11952–11962. [Google Scholar] [CrossRef]
- Alves, T.O.; D’Almeida, C.T.S.; Victorio, V.C.M.; Souza, G.H.M.F.; Cameron, L.C.; Ferreira, M.S. L Immunogenic and allergenic profile of wheat flours from different technological qualities revealed by ion mobility mass spectrometry. J. Food Compos. Anal. 2018, 73, 67–75. [Google Scholar] [CrossRef]
- Paglia, G.; Angel, P.; Williams, J.P.; Richardson, K.; Olivos, H.J.; Thompson, J.W.; Menikarachchi, L.; Lai, S.; Walsh, C.; Moseley, A.; et al. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem. 2015, 87, 1137–1144. [Google Scholar] [CrossRef]
- Dwivedi, P.; Puzon, G.; Tam, M.; Langlais, D.; Jackson, S.; Kaplan, K.; Siems, W.F.; Schultz, A.J.; Xun, L.Y.; Woods, A.; et al. Metabolic profiling of Escherichia coli by ion mobility-mass spectrometry with MALDI ion source. J. Mass Spectrom. 2010, 45, 1383–1393. [Google Scholar] [CrossRef]
- Djambazova, K.V.; Klein, D.R.; Migas, L.G.; Neumann, E.K.; Rivera, E.S.; Van de Plas, R.; Caprioli, R.M.; Spraggins, J.M. Resolving the complexity of spatial lipidomics using MALDI TIMS imaging mass spectrometry. Anal. Chem. 2020, 92, 13290–13297. [Google Scholar] [CrossRef] [PubMed]
- Rainville, P.D.; Wilson, I.D.; Nicholson, J.K.; Isaac, G.; Mullin, L.; Langridge, J.I.; Plumb, R.S. Ion mobility spectrometry combined with ultra performance liquid chromatography/mass spectrometry for metabolic phenotyping of urine: Effects of column length, gradient duration and ion mobility spectrometry on metabolite detection. Anal. Chim. Acta 2017, 982, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.V.; Gamage, C.M.; Galhena, A.S.; Fernández, F.M. Contrast-enhanced differential mobility-desorption electrospray ionization-mass spectrometry imaging of biological tissues. Anal. Chem. 2014, 86, 3756–3763. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.X.; Zuo, T.T.; Wang, X.Y.; Wang, H.D.; Hu, Y.; Li, Z.; Li, W.W.; Jia, L.; Qian, Y.X.; Yang, W.Z.; et al. Integration of data-dependent acquisition (DDA) and data-independent high-definition MSE (HDMSE) for the comprehensive profiling and characterization of multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS. Molecules 2019, 24, 2708. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Yang, X.N.; Chen, B.X.; Zhao, D.X.; Wang, H.D.; Sun, M.X.; Li, X.Y.; Liu, J.; Wang, S.M.; Mi, Y.G.; et al. Ultra-high performance liquid chromatography/ion mobility time-of-flight mass spectrometry-based untargeted metabolomics combined with quantitative assay unveiled the metabolic difference among the root, leaf, and flower bud of Panax notoginseng. Arab. J. Chem. 2021, 1411, 103409. [Google Scholar] [CrossRef]
- Jeanne Dit Fouque, K.; Ramirez, C.E.; Lewis, R.L.; Koelmel, J.P.; Garrett, T.J.; Yost, R.A.; Fernandez-Lima, F. Effective liquid chromatography-trapped ion mobility spectrometry-mass spectrometry separation of isomeric lipid species. Anal. Chem. 2019, 918, 5021–5027. [Google Scholar] [CrossRef]
- King, A.M.; Mullin, L.G.; Wilson, I.D.; Coen, M.; Rainville, P.D.; Plumb, R.S.; Gethings, L.A.; Maker, G.; Trengove, R. Development of a rapid profiling method for the analysis of polar analytes in urine using HILIC-MS and ion mobility enabled HILIC-MS. Metabolomics 2019, 15, 17. [Google Scholar] [CrossRef]
- Szykuła, K.M.; Meurs, J.; Turner, M.A.; Creaser, C.S.; Reynolds, J.C. Combined hydrophilic interaction liquid chromatography-scanning field asymmetric waveform ion mobility spectrometry-time-of-flight mass spectrometry for untargeted metabolomics. Anal. Bioanal. Chem. 2019, 411, 6309–6317. [Google Scholar] [CrossRef]
- Li, A.; Hines, K.M.; Xu, L.B. Lipidomics by HILIC-ion mobility-mass spectrometry. Methods Mol. Biol. 2020, 2084, 119–132. [Google Scholar]
- Qian, Y.X.; Li, W.W.; Wang, H.M.; Hu, W.D.; Wang, H.D.; Zhao, D.X.; Hu, Y.; Li, X.; Gao, X.M.; Yang, W.Z. A four-dimensional separation approach by offline 2D-LC/IM-TOF-MS in combination with database-driven computational peak annotation facilitating the in-depth characterization of the multicomponents from Atractylodis Macrocephalae Rhizoma (Atractylodes macrocephala). Arab. J. Chem. 2021, 142, 102957. [Google Scholar]
- Genangeli, M.; Heijens, A.M.M.; Rustichelli, A.; Schuit, N.D.; Micioni Di Bonaventura, M.V.M.D.; Cifani, C.; Vittori, S.; Siegel, T.P.; Heeren, R.M.A. MALDI-mass spectrometry imaging to investigate lipid and bile acid modifications caused by lentil extract used as a potential hypocholesterolemic treatment. J. Am. Soc. Mass Spectrom. 2019, 3010, 2041–2050. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.J.; Yang, W.Z.; Qiu, S.; Hou, J.J.; Wu, W.Y.; Guo, D. Systematic profiling and comparison of the lipidomes from Panax ginseng, P. quinquefolius, and P. notoginseng by ultrahigh performance supercritical fluid chromatography/high-resolution mass spectrometry and ion mobility-derived collision cross section measurement. J. Chromatogr. A 2018, 1548, 64–75. [Google Scholar] [PubMed]
- Kurulugama, R.T.; Darland, E.; Kuhlmann, F.; Stafford, G.; Fjeldsted, J. Evaluation of drift gas selection in complex sample analyses using a high performance drift tube ion mobility-QTOF mass spectrometer. Analyst 2015, 140, 6834–6844. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Williams, J.P.; Menikarachchi, L.; Thompson, J.W.; Tyldesley-Worster, R.; Halldόrsson, S.; Rolfsson, O.; Moseley, A.; Grant, D.; Langridge, J.; et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal. Chem. 2014, 868, 3985–3993. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Aly, N.A.; Zhou, Y.X.; Dupuis, K.T.; Bilbao, A.; Paurus, V.L.; Orton, D.J.; Wilson, R.; Payne, S.H.; Smith, R.D.; et al. A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chem. Sci. 2017, 811, 7724–7736. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.M.; Dodds, J.N.; Rose, B.S.; Picache, J.A.; Morris, C.B.; Codreanu, S.G.; May, J.C.; Sherrod, S.D.; McLean, J.A. Untargeted molecular discovery in primary metabolism: Collision cross section as a molecular descriptor in ion mobility-mass spectrometry. Anal. Chem. 2018, 9024, 14484–14492. [Google Scholar] [CrossRef]
- Hernández-Mesa, M.; Bizec, B.L.; Monteau, F.; García-Campaña, A.M.; Dervilly-Pinel, G. Collision cross section CCS database: An additional measure to characterize steroids. Anal. Chem. 2018, 907, 4616–4625. [Google Scholar] [CrossRef]
- Nye, L.C.; Williams, J.P.; Munjoma, N.C.; Letertre, M.P.M.; Coen, M.; Bouwmeester, R.; Martens, L.; Swann, J.R.; Nicholson, J.K.; Plumb, R.S.; et al. A comparison of collision cross section values obtained via travelling wave ion mobility-mass spectrometry and ultra high performance liquid chromatography-ion mobility-mass spectrometry: Application to the characterisation of metabolites in rat urine. J. Chromatogr. A 2019, 1602, 386–396. [Google Scholar] [CrossRef]
- Poland, J.C.; Leaptrot, K.L.; Sherrod, S.D.; Flynn, C.R.; McLean, J.A. Collision cross section conformational analyses of bile acids via ion mobility-mass spectrometry. J. Am. Soc. Mass Spectrom. 2020, 318, 1625–1631. [Google Scholar] [CrossRef]
- May, J.C.; Goodwin, C.R.; Lareau, N.M.; Leaptrot, K.L.; Morris, C.B.; Kurulugama, R.T.; Mordehai, A.; Klein, C.; Barry, W.; Darland, E.; et al. Conformational ordering of biomolecules in the gas phase: Nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal. Chem. 2014, 864, 2107–2116. [Google Scholar] [CrossRef]
- Stephan, S.; Hippler, J.; Köhler, T.; Deeb, A.A.; Schmidt, T.C.; Schmitz, O.J. Contaminant screening of wastewater with HPLC-IM-qTOF-MS and LC+ LC-IM-qTOF-MS using a CCS database. Anal. Bioanal. Chem. 2016, 408, 6545–6555. [Google Scholar] [CrossRef] [PubMed]
- Hines, K.M.; Ross, D.H.; Davidson, K.L.; Bush, M.F.; Xu, L.B. Large-scale structural characterization of drug and drug-like compounds by high-throughput ion mobility-mass spectrometry. Anal. Chem. 2017, 89, 9023–9030. [Google Scholar] [CrossRef] [PubMed]
- Mesleh, M.F.; Hunter, J.M.; Shvartsburg, A.A.; Schatz, G.C.; Jarrold, M.F. Structural information from ion mobility measurements: Effects of the long-range potential. J. Phys. Chem. C 1996, 100, 16082–16086. [Google Scholar] [CrossRef]
- Bleiholder, C.; Wyttenbach, T.; Bowers, M.T. A novel projection approximation algorithm for the fast and accurate computation of molecular collision cross sections (I). Method. Int. J. Mass Spectrom. 2011, 308, 1–10. [Google Scholar] [CrossRef]
- Campuzano, I.; Bush, M.F.; Robinson, C.V.; Beaumont, C.; Richardson, K.; Kim, H.; Kim, H.I. Structural characterization of drug-like compounds by ion mobility mass spectrometry: Comparison of theoretical and experimentally derived nitrogen collision cross sections. Anal. Chem. 2012, 84, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Hadavi, D.; Borzova, M.; Siegel, T.P.; Honing, M. Uncovering the behavior of ions in the gas-phase to predict the ion mobility separation of isomeric steroid compounds. Anal. Chim. Acta 2022, 1200, 339617. [Google Scholar] [CrossRef]
- Przybylski, C.; Bonnet, V. Probing topology of supramolecular complexes between cyclodextrins and alkali metals by ion mobility-mass spectrometry. Carbohydr. Polym. 2022, 297, 120019. [Google Scholar] [CrossRef]
- Turzo, S.B.A.; Seffernick, J.T.; Rolland, A.D.; Donor, M.T.; Heinze, S.; Prell, J.S.; Wysocki, V.H.; Lindert, S. Protein shape sampled by ion mobility mass spectrometry consistently improves protein structure prediction. Nat. Commun. 2022, 13, 4377. [Google Scholar] [CrossRef]
- Song, X.C.; Dreolin, N.; Damiani, T.; Canellas, E.; Nerin, C. Prediction of collision cross section values: Application to non-intentionally added substance identification in food contact materials. J. Agric. Food Chem. 2022, 70, 1272–1281. [Google Scholar] [CrossRef]
- Bijlsma, L.; Bade, R.; Celma, A.; Mullin, L.; Cleland, G.; Stead, S.; Hernandez, F.; Sancho, J.V. Prediction of collision cross-section values for small molecules: Application to pesticide residue analysis. Anal. Chem. 2017, 89, 6583–6589. [Google Scholar] [CrossRef]
- Li, T.Z.; Yin, Y.D.; Zhou, Z.W.; Qiu, J.Q.; Liu, W.B.; Zhang, X.T.; He, K.W.; Zhu, Z.J. Ion mobility-based sterolomics reveals spatially and temporally distinctive sterol lipids in the mouse brain. Nat. Commun. 2021, 12, 4343. [Google Scholar] [CrossRef] [PubMed]
- Stow, S.M.; Causon, T.J.; Zheng, X.; Kurulugama, R.T.; Mairinger, T.; May, J.C.; Rennie, E.E.; Baker, E.S.; Smith, R.D.; McLean, J.A.; et al. An interlaboratory evaluation of drift tube ion mobility-mass spectrometry collision cross section measurements. Anal. Chem. 2017, 89, 9048–9055. [Google Scholar] [CrossRef] [PubMed]
- Hines, K.M.; May, J.C.; McLean, J.A.; Xu, L.B. Evaluation of collision cross section calibrants for structural analysis of lipids by traveling wave ion mobility-mass spectrometry. Anal. Chem. 2016, 88, 7329–7336. [Google Scholar] [CrossRef]
- Paglia, G.; Astarita, G. Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat. Protoc. 2017, 12, 797–813. [Google Scholar] [CrossRef] [PubMed]
- Plachká, K.; Pezzatti, J.; Musenga, A.; Nicoli, R.; Kuuranne, T.; Rudaz, S.; Nováková, L.; Guillarme, D. Ion mobility-high resolution mass spectrometry in anti-doping analysis. Part I: Implementation of a screening method with the assessment of a library of substances prohibited in sports. Anal. Chim. Acta 2021, 1152, 338257. [Google Scholar] [CrossRef]
- Jariyasopit, N.; Limjiasahapong, S.; Kurilung, A.; Sartyoungkul, S.; Wisanpitayakorn, P.; Nuntasaen, N.; Kuhakarn, C.; Reutrakul, V.; Kittakoop, P.; Sirivatanauksorn, Y.; et al. Traveling wave ion mobility-derived collision cross section database for plant specialized metabolites: An application to ventilago harmandiana pierre. J. Proteome Res. 2022, 21, 2481–2492. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Mesa, M.; D’atri, V.; Barknowitz, G.; Fanuel, M.; Pezzatti, J.; Dreolin, N.; Ropartz, D.; Monteau, F.; Vigneau, E.; Rudaz, S.; et al. Interlaboratory and interplatform study of steroids collision cross section by traveling wave ion mobility spectrometry. Anal. Chem. 2020, 92, 5013–5022. [Google Scholar] [CrossRef]
- Righetti, L.; Dreolin, N.; Celma, A.; McCullagh, M.; Barknowitz, G.; Sancho, J.V.; Dall’Asta, C. Travelling wave ion mobility-derived collision cross section for mycotoxins: Investigating interlaboratory and interplatform reproducibility. J. Agric. Food Chem. 2020, 68, 10937–10943. [Google Scholar] [CrossRef]
- Hofmann, J.; Struwe, W.B.; Scarff, C.A.; Scrivens, J.H.; Harvey, D.J.; Pagel, K. Estimating collision cross sections of negatively charged N-glycans using traveling wave ion mobility-mass spectrometry. Anal. Chem. 2014, 86, 10789–10795. [Google Scholar] [CrossRef]
- Das, S.; Tanemura, K.A.; Dinpazhoh, L.; Keng, M.; Schumm, C.; Leahy, L.; Asef, C.K.; Rainey, M.; Edison, A.S.; Fernández, F.M.; et al. In silico collision cross section calculations to aid metabolite annotation. J. Am. Soc. Mass Spectrom. 2022, 33, 750–759. [Google Scholar] [CrossRef]
- Boschmans, J.; Jacobs, S.; Williams, J.P.; Palmer, M.; Richardson, K.; Giles, K.; Lapthorn, C.; Herrebout, W.A.; Lemière, F.; Sobott, F. Combining density functional theory (DFT) and collision cross-section (CCS) calculations to analyze the gas-phase behavior of small molecules and their protonation site isomers. Analyst 2016, 141, 4044–4054. [Google Scholar] [CrossRef] [PubMed]
- Ewing, S.A.; Donor, M.T.; Wilson, J.W.; Prell, J.S. Collidoscope: An improved tool for computing collisional cross-sections with the trajectory method. J. Am. Soc. Mass Spectrom. 2017, 28, 587–596. [Google Scholar] [CrossRef]
- Bleiholder, C. A local collision probability approximation for predicting momentum transfer cross sections. Analyst 2015, 140, 6804–6813. [Google Scholar] [CrossRef] [PubMed]
- Shvartsburg, A.A.; Jarrold, M.F. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem. Phys. Lett. 1996, 261, 86–91. [Google Scholar] [CrossRef]
- Larriba, C.; Hogan, J.C.J. Ion mobilities in diatomic gases: Measurement versus prediction with non-specular scattering models. J. Phys. Chem. Lett. 2013, 117, 3887–3901. [Google Scholar] [CrossRef]
- Zanotto, L.; Heerdt, G.; Souza, P.C.T.; Araujo, G.; Araujo, M.S. High performance collision cross section calculation—HPCCS. J. Comput. Chem. 2018, 39, 1675–1681. [Google Scholar] [CrossRef]
- Myers, C.A.; D’Esposito, R.J.; Fabris, D.; Ranganathan, S.V.; Chen, A.A. CoSIMS: An optimized trajectory-based collision simulator for ion mobility spectrometry. J. Phys. Chem. B 2019, 123, 4347–4357. [Google Scholar] [CrossRef]
- Zhou, Z.W.; Xiong, X.; Zhu, Z.J. MetCCS predictor: A web server for predicting collision cross-section values of metabolites in ion mobility-mass spectrometry based metabolomics. Bioinformatics 2017, 33, 2235–2237. [Google Scholar] [CrossRef]
- Lalli, P.M.; Corilo, Y.E.; Fasciotti, M.; Riccio, M.F.; Sa, G.F.; Daroda, R.J.; Souza, G.H.; McCullagh, M.; Bartberger, M.D.; Eberlin, M.N.; et al. Baseline resolution of isomers by traveling wave ion mobility mass spectrometry: Investigating the effects of polarizable drift gases and ionic charge distribution. J. Mass Spectrom. 2013, 48, 989–997. [Google Scholar] [CrossRef]
- Lee, J.W.; Lee, H.H.L.; Davidson, K.L.; Bush, M.F.; Kim, H.I. Structural characterization of small molecular ions by ion mobility mass spectrometry in nitrogen drift gas: Improving the accuracy of trajectory method calculations. Analyst 2018, 143, 1786–1796. [Google Scholar] [CrossRef]
- Shrivastav, V.; Nahin, M.; Hogan C., J.; Larriba-Andaluz, C. Benchmark comparison for a multi-processing ion mobility calculator in the free molecular regime. J. Am. Soc. Mass Spectrom. 2017, 28, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Marklund, E.G.; Degiacomi, M.T.; Robinson, C.V.; Baldwin, A.J.; Benesch, J.L.P. Collision cross sections for structural proteomics. Structure 2015, 23, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Belova, L.; Celma, A.; Van Haesendonck, G.; Lemière, F.; Sancho, J.V.; Covaci, A.; van Nuijs, A.L.N.; Bijlsma, L. Revealing the differences in collision cross section values of small organic molecules acquired by different instrumental designs and prediction models. Anal. Chim. Acta 2022, 1229, 340361. [Google Scholar] [CrossRef] [PubMed]
- May, J.C.; McLean, J.A. Integrating ion mobility into comprehensive multidimensional metabolomics workflows: Critical considerations. Metabolomics 2022, 18, 104. [Google Scholar] [CrossRef] [PubMed]
- Carracedo-Reboredo, P.; Liñares-Blanco, J.; Rodríguez-Fernández, N.; Cedrón, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Comput. Struct. Biotechnol. J. 2021, 19, 4538–4558. [Google Scholar] [CrossRef]
- Mauri, A. alvaDesc: A tool to calculate and analyze molecular descriptors and fingerprints. In Ecotoxicological QSARs; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 801–820. [Google Scholar]
- Yang, F.; van Herwerden, D.; Preud’homme, H.; Samanipour, S. Collision cross section prediction with molecular fingerprint using machine learning. Molecules 2022, 27, 6424. [Google Scholar] [CrossRef] [PubMed]
- Cereto-Massagué, A.; Ojeda, M.J.; Valls, C.; Mulero, M.; Garcia-Vallvé, S.; Pujadas, G. Molecular fingerprint similarity search in virtual screening. Methods 2015, 71, 58–63. [Google Scholar] [CrossRef]
- Reymond, J.L.; Awale, M. Exploring chemical space for drug discovery using the chemical universe database. ACS Chem. Neurosci. 2012, 3, 649–657. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef]
- Aćimović, M.; Pezo, L.; Tešević, V.; Čabarkapa, I.; Todosijević, M. QSRR model for predicting retention indices of Satureja kitaibelii Wierzb. ex Heuff. essential oil composition. Ind. Crops Prod. 2020, 154, 112752. [Google Scholar] [CrossRef]
- Soper-Hopper, M.T.; Petrov, A.S.; Howard, J.N.; Yu, S.S.; Forsythe, J.G.; Grover, M.A.; Fernández, F.M. Collision cross section predictions using 2-dimensional molecular descriptors. Chem. Commun. 2017, 53, 7624–7627. [Google Scholar] [CrossRef] [PubMed]
- Soper-Hopper, M.T.; Vandegrift, J.; Baker, E.S.; Fernández, F.M. Metabolite collision cross section prediction without energy-minimized structures. Analyst 2020, 145, 5414–5418. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.L.; Venkatesh, R.; McBride, M.; Reichmanis, E.; Meredith, J.C.; Grover, M.A. Small data machine learning: Classification and prediction of poly (ethylene terephthalate) stabilizers using molecular descriptors. ACS Appl. Polym. Mater. 2020, 2, 5592–5601. [Google Scholar] [CrossRef]
- Song, X.C.; Dreolin, N.; Canellas, E.; Goshawk, J.; Nerin, C. Prediction of collision cross-section values for extractables and leachables from plastic products. Environ. Sci. Technol. 2022, 56, 9463–9473. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, H.; Tian, Y.S.; Kawashita, N.; Takagi, T. Mordred: A molecular descriptor calculator. J. Cheminf. 2018, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Rainey, M.A.; Watson, C.A.; Asef, C.K.; Foster, M.R.; Baker, E.S.; Fernández, F.M. CCS Predictor 2.0: An open-source jupyter notebook tool for filtering out false positives in metabolomics. Anal. Chem. 2022, 94, 17456–17466. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.S.; Xu, Q.S.; Hu, Q.N.; Liang, Y.Z. ChemoPy: Freely available python package for computational biology and chemoinformatics. Bioinformatics 2013, 29, 1092–1094. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Cao, D.S.; Miao, H.Y.; Liu, S.; Deng, B.C.; Yun, Y.H.; Wang, N.N.; Lu, A.P.; Zeng, W.B.; Chen, A.F. ChemDes: An integrated web-based platform for molecular descriptor and fingerprint computation. J. Cheminf. 2015, 7, 60. [Google Scholar] [CrossRef]
- Du, J.X.; Chang, Y.Z.; Zhang, X.; Hu, C.Q. Development of a method of analysis for profiling of the impurities in phenoxymethylpenicillin potassium based on the analytical quality by design concept combined with the degradation mechanism of penicillins. J. Pharm. Biomed. Anal. 2020, 186, 113309. [Google Scholar] [CrossRef]
- Willighagen, E.L.; Mayfield, J.W.; Alvarsson, J.; Berg, A.; Carlsson, L.; Jeliazkova, N.; Kuhn, S.; Pluskal, T.; Rojas-Chertó, M.; Spjuth, O.; et al. The Chemistry Development Kit (CDK) v2.0: Atom typing, depiction, molecular formulas, and substructure searching. J. Cheminf. 2017, 9, 33. [Google Scholar] [CrossRef]
- Steinbeck, C.; Han, Y.; Kuhn, S.; Horlacher, O.; Luttmann, E.; Willighagen, E. The Chemistry Development Kit (CDK): An open-source Java library for chemo-and bioinformatics. J. Chem. Inf. Model. 2003, 43, 493–500. [Google Scholar]
- Broeckling, C.D.; Yao, L.; Isaac, G.; Gioioso, M.; Ianchis, V.; Vissers, J.P. Application of predicted collisional cross section to metabolome databases to probabilistically describe the current and future ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2021, 32, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Sushko, I.; Novotarskyi, S.; Körner, R.; Pandey, A.K.; Rupp, M.; Teetz, W.; Brandmaier, S.; Abdelaziz, A.; Prokopenko, V.V.; Tanchuk, V.Y.; et al. Online chemical modeling environment (OCHEM): Web platform for data storage, model development and publishing of chemical information. J. Comput. Aided Mol. Des. 2011, 25, 533–554. [Google Scholar] [CrossRef]
- Nielson, F.F.; Colby, S.M.; Thomas, D.G.; Renslow, R.S.; Metz, T.O. Exploring the impacts of conformer selection methods on ion mobility collision cross section predictions. Anal. Chem. 2021, 93, 3830–3838. [Google Scholar] [CrossRef]
- Gonzales, G.B.; Smagghe, G.; Coelus, S.; Adriaenssens, D.; De Winter, K.; Desmet, T.; Raes, K.; Van Camp, J. Collision cross section prediction of deprotonated phenolics in a travelling-wave ion mobility spectrometer using molecular descriptors and chemometrics. Anal. Chim. Acta 2016, 924, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.H.; Cho, J.H.; Zhang, R.T.; Hines, K.M.; Xu, L.B. LiPydomics: A python package for comprehensive prediction of lipid collision cross sections and retention times and analysis of ion mobility-mass spectrometry-based lipidomics data. Anal. Chem. 2020, 92, 14967–14975. [Google Scholar] [CrossRef] [PubMed]
- Asef, C.K.; Rainey, M.A.; Garcia, B.M.; Gouveia, G.J.; Shaver, A.O.; Leach, F.E., III; Morse, A.M.; Edison, A.S.; McIntyre, L.M.; Fernández, F.M. Unknown metabolite identification using machine learning collision cross-section prediction and tandem mass spectrometry. Anal. Chem. 2023, 95, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Colby, S.M.; Nuñez, J.R.; Hodas, N.O.; Corley, C.D.; Renslow, R.R. Deep learning to generate in silico chemical property libraries and candidate molecules for small molecule identification in complex samples. Anal. Chem. 2019, 92, 1720–1729. [Google Scholar] [CrossRef]
- Malik, A.; Saggi, M.K.; Rehman, S.; Sajjad, H.; Inyurt, S.; Bhatia, A.S.; Farooque, A.A.; Oudah, A.Y.; Yaseen, Z.M. Deep learning versus gradient boosting machine for pan evaporation prediction. Eng. Appl. Comp. Fluid Mech. 2022, 16, 570–587. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Connolly, J.R.; Munoz-Muriedas, J.; Lapthorn, C.; Higton, D.; Vissers, J.P.; Webb, A.; Beaumont, C.; Dear, G.J. Investigation into small molecule isomeric glucuronide metabolite differentiation using in silico and experimental collision cross-section values. J. Am. Soc. Mass Spectrom. 2021, 32, 1976–1986. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Picache, J.A.; May, J.C.; McLean, J.A. Chemical class prediction of unknown biomolecules using ion mobility-mass spectrometry and machine learning: Supervised inference of feature taxonomy from ensemble randomization. Anal. Chem. 2020, 92, 10759–10767. [Google Scholar] [CrossRef] [PubMed]
- Ieritano, C.; Lee, A.; Crouse, J.; Bowman, Z.; Mashmoushi, N.; Crossley, P.M.; Friebe, B.F.; Campbell, J.L.; Hopkins, W.S. Determining collision cross sections from differential ion mobility spectrometry. Anal. Chem. 2021, 93, 8937–8944. [Google Scholar] [CrossRef] [PubMed]
- Song, X.C.; Canellas, E.; Dreolin, N.; Nerin, C.; Goshawk, J. Discovery and characterization of phenolic compounds in Bearberry (Arctostaphylos uva-ursi) leaves using liquid chromatography-ion mobility-high-resolution mass spectrometry. J. Agric. Food Chem. 2021, 69, 10856–10868. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xu, X.Y.; Wang, H.D.; Wang, H.M.; Liu, M.Y.; Hu, W.D.; Chen, B.X.; Jiang, M.T.; Jing, Q.; Li, X.H.; et al. A multi-dimensional liquid chromatography/high-resolution mass spectrometry approach combined with computational data processing for the comprehensive characterization of the multicomponents from Cuscuta chinensis. J. Chromatogr. A 2022, 1675, 463162. [Google Scholar] [CrossRef]
- Zhu, H.D.; Wu, X.D.; Huo, J.Y.; Hou, J.J.; Long, H.L.; Zhang, Z.J.; Wang, B.; Tian, M.H.; Chen, K.X.; Guo, D.A.; et al. A five-dimensional data collection strategy for multicomponent discovery and characterization in Traditional Chinese Medicine: Gastrodia Rhizoma as a case study. J. Chromatogr. A 2021, 1653, 462405. [Google Scholar] [CrossRef]
- Yang, X.N.; Xiong, Y.; Wang, H.D.; Jiang, M.T.; Xu, X.Y.; Mi, Y.G.; Lou, J.; Li, X.H.; Sun, H.; Zhao, Y.Y.; et al. Multicomponent characterization of the flower bud of Panax notoginseng and its metabolites in rat plasma by ultra-high performance liquid chromatography/ion mobility quadrupole time-of-flight mass spectrometry. Molecules 2022, 27, 9049. [Google Scholar] [CrossRef]
- Mullin, L.; Jobst, K.; DiLorenzo, R.A.; Plumb, R.; Reiner, E.J.; Yeung, L.W.; Jogsten, I.E. Liquid chromatography-ion mobility-high resolution mass spectrometry for analysis of pollutants in indoor dust: Identification and predictive capabilities. Anal. Chem. Acta 2020, 1125, 29–40. [Google Scholar] [CrossRef]
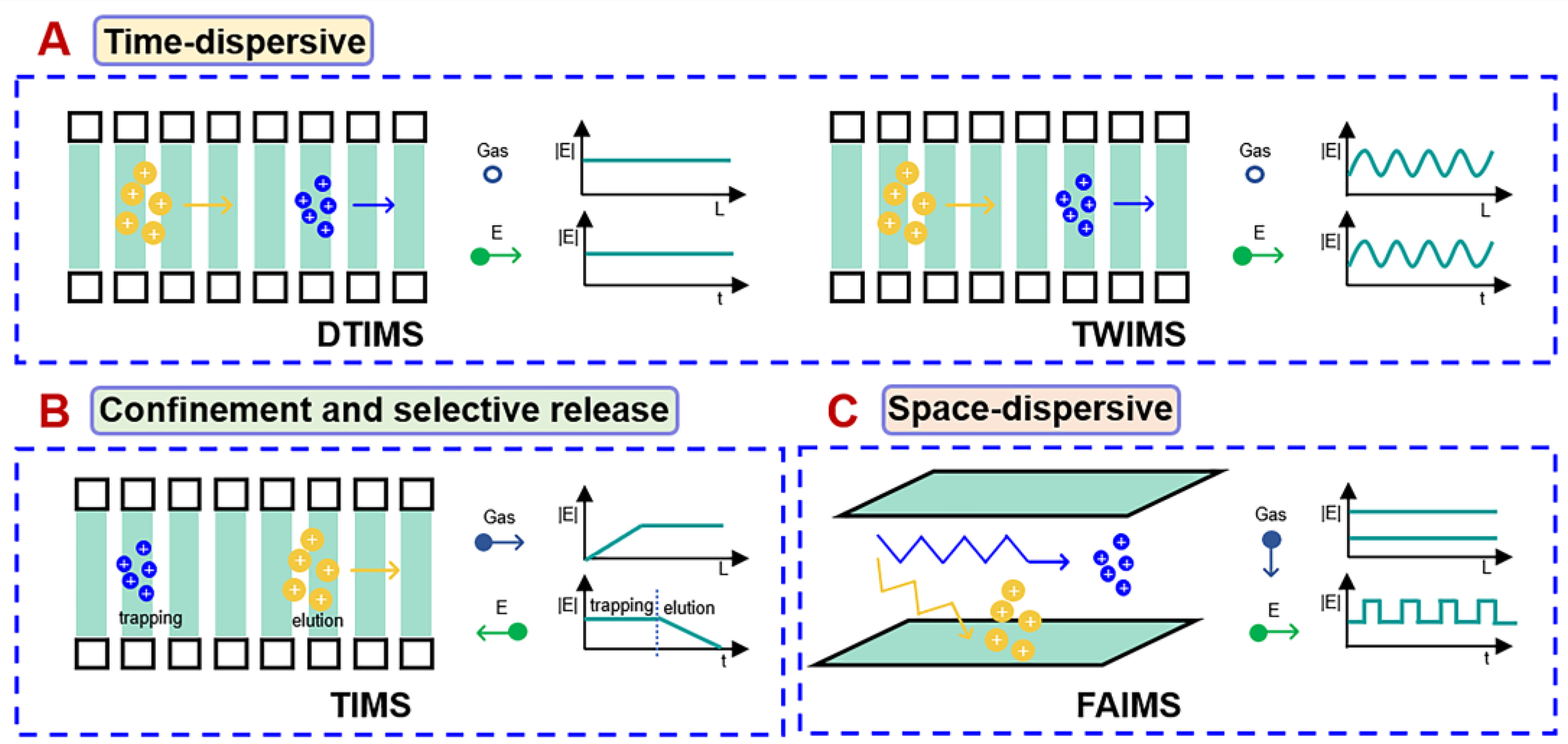

| IMS Technique | Gas State | Resolving Power | Year of Release | CCS Calibration | Available Device | Sort |
|---|---|---|---|---|---|---|
| DTIMS | Stationary | ~60–80 | 2014 | Not required | Agilent IM-QTOF | Time dispersive |
| TWIMS | Stationary | ~40–50 | 2006 | Required | Waters Synapt HDMS Waters Vion IMS-QTOF | Time dispersive |
| SLIMS | Parallel gas flow | ~200–300 | 2021 | Required | MOBILion | Time dispersive |
| TIMS | Parallel gas flow | ~200–400 | 2015 | Required | Bruker tims TOF Bruker tims TOF pro Bruker Impact Q-TOF | Confinement and selective release |
| cIMS | Parallel gas flow | ~750 | 2019 | Required | Waters SELECT SERIES cyclic IMS | Confinement and selective release |
| FAIMS/DMS | Parallel gas flow | Not comparable | 2012 | - | AB Sciex SelexION | Space dispersive |
| Source | Research Object | Number of Compounds | Number of CCS Values | Instrument Platform | Web | Ref. |
|---|---|---|---|---|---|---|
| Experimental CCS | Metabolites | 125 | 209 | TWIMS | / | [94] |
| Lipids | 244 | 244 | TWIMS | / | [79] | |
| Metabolites and xenobiotics | 459 | 826 | DTIMS | http://panomics.pnnl.gov/metabolites/ (accessed on 10 February 2023) | [95] | |
| Primary metabolites | 417 | 1246 | DTIMS | / | [96] | |
| Steroids | 300 | 1080 | TWIMS | / | [97] | |
| Metabolites | 1142 | 3271 | DTIMS | / | [59] | |
| Metabolites | 2193 | 5119 | DTIMS, TWIMS | http://allccs.zhulab.cn/ (accessed on 10 February 2023) | [16] | |
| Metabolites | 510 | 942 | TWIMS | / | [98] | |
| Bile acids | 47 | 400 | DTIMS | / | [99] | |
| Lipids | / | 594 | DTIMS | / | [100] | |
| Lipids | 1856 | 1856 | TIMS | / | [48] | |
| Drug-like compounds and pesticides | ~500 | ~500 | DTIMS | / | [101] | |
| Small molecules | 124 | 124 | DTIMS, TWIMS | / | [23] | |
| Drug or drug-like molecules | 1425 | 1440 | TWIMS | / | [102] | |
| Doping agents | 192 | 192 | TWIMS | / | [115] | |
| Metabolites | 112 | 207 | TWIMS | https://massive.ucsd.edu (accessed on 10 February 2023) | [116] | |
| Metabolites | 87 | 142 | TWIMS | / | [117] | |
| Mycotoxins | 53 | 219 | TWIMS | / | [118] | |
| Lipids | 217 | 456 | DTIMS | https://mcleanresearchgroup.shinyapps.io/CCS-Compendium/ (accessed on 10 February 2023) | [76] | |
| N-glycans | 500 | 500 | TWIMS | / | [119] | |
| Calculated CCS | ISiCLE: metabolites | / | ~1,000,000 | / | metabolomics.pnnll.gov | [12] |
| Metabolites | 125 | 205 | / | / | [94] | |
| POMICS | / | / | / | https://www.pomics.org/ (accessed on 10 February 2023) | [120] | |
| Predicted CCS | MetCCS: metabolites | 35,203 | 176,015 | DTIMS | http://www.metabolomics-shanghai.org/software.php (accessed on 10 February 2023) | [10] |
| LipidCCS: lipids | 15,646 | 63,434 | DTIMS | http://www.metabolomics-shanghai.org/LipidCCS/ (accessed on 10 February 2023) | [14] | |
| AllCCS: metabolites | 1,670,596 | 11,697,711 | / | http://allccs.zhulab.cn/ (accessed on 10 February 2023) | [16] | |
| Pesticide residues | 336 | 336 | / | / | [110] | |
| DeepCCS: metabolites | 2400 | 2400 | / | / | [15] | |
| Sterol lipids | 2068 | 2068 | / | / | [111] | |
| Food contact materials | 488 | 635 | TWIMS | / | [109] | |
| dmCCS: drugs and their metabolites | 3286 | 2068 | / | https://CCSbase.net/dmccs_predictions (accessed on 10 February 2023) | [7] | |
| CCSbase: lipids, metabolites, drugs | 4742 | 7669 | DTIMS, TWIMS | https://CCSbase.net (accessed on 10 February 2023) | [11] |
| Software | Year | Methods | Collision Gas | Open Source | Ref. |
|---|---|---|---|---|---|
| MobCal | 1996 | PA, EHSS, TM | He/N2 | Yes | [131] |
| IMoS | 2013 | DTM, DHSS | He/N2 | Yes | [125] |
| IMPACT | 2015 | PA | He | Yes | [132] |
| Collidoscope | 2017 | TM | He/N2 | Yes | [122] |
| HPCCS | 2018 | TM | He/N2 | Yes | [126] |
| CoSIMS | 2019 | TM | He | Yes | [127] |
| Software | Operating System | Number of Descriptors | Features | Ref. |
|---|---|---|---|---|
| PaDEL-Descriptor | Windows, Linux, MacOS | >1700 | Supports more than 90 molecular file formats | [140] |
| alvaDesc | Windows, Linux, MacOS | 5666 | Can handle full and non-full connection structures | [144] |
| OCHEM | Web | 5666 | Is a web version of alvaDesc | [154] |
| chemDes | Web | 3679 | Integrates with multiple advanced software packages | [149] |
| Dragon | Windows, Linux, web (e-Dragon) | 5270 | Has a fast calculation speed, allowing disconnected structures | [a] |
| Mordred | Windows, Linux, MacOS | >1800 | Can calculate macromolecule descriptor | [146] |
| BlueDesc | Windows, Linux, MacOS | 174 | Is only applicable to 3D structures | [b] |
| Chemopy | Windows, Linux | 1135 | Is applicable to 2D and 3D structures | [148] |
| Discovery Studio | Windows, Linux | Hundreds | Enables structural optimization | [150] |
| CDK | Development kit | 268 | Contains the chemical and bioinformatics Java library | [151] |
| RDkit | Development kit | 200 | Is based on the Python language, supporting multiple file formats | [153] |
| rcdk | Development kit | 221 | Has the CDK toolkit integrated under the R language | [106] |
| Object | Year | Effect | Ref. |
|---|---|---|---|
| Metabolites | 2016 | MRE < 3%; the identification accuracy can be improved | [10] |
| 2017 | MRE < 1%; the false-positive identifications of lipids can be effectively reduced | [14] | |
| 2019 | MRE < 3%; only SMILES notation and ion type are needed | [15] | |
| 2020 | MRE < 2%; the accuracy and coverage of both known metabolite and unknown metabolite annotation from biological samples can be improved | [16] | |
| 2022 | MRE < 1.1%; cis–trans and sn-positional isomers can be distinguished | [60] | |
| 2022 | MRE < 2%; the false positives can be filtered out | [147] | |
| Natural products | 2021 | a higher identification confidence level can be obtained | [166] |
| 2022 | more possibilities to distinguish isomers can be provided | [167] | |
| Foods | 2020 | a certain degree of credibility can be obtained | [118] |
| 2022 | MRE < 2%; the identification confidence of 11 oligomers can be improved | [109] | |
| Drugs | 2017 | MRE < 2%; the confidence in the tentative identification of suspect and nontarget pesticides can be notably improved | [110] |
| 2022 | MRE < 2.2%; sufficient precision to differentiate isomers and conformers can be obtained | [7] | |
| Environment | 2020 | identification confidence can be increased | [170] |
| 2022 | MRE < 2%; the false positives were reduced, and the recognition confidence levels can be improved | [145] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, H.; Jiang, M.; Ding, M.; Xu, X.; Xu, B.; Zou, Y.; Yu, Y.; Yang, W. Collision Cross Section Prediction Based on Machine Learning. Molecules 2023, 28, 4050. https://doi.org/10.3390/molecules28104050
Li X, Wang H, Jiang M, Ding M, Xu X, Xu B, Zou Y, Yu Y, Yang W. Collision Cross Section Prediction Based on Machine Learning. Molecules. 2023; 28(10):4050. https://doi.org/10.3390/molecules28104050
Chicago/Turabian StyleLi, Xiaohang, Hongda Wang, Meiting Jiang, Mengxiang Ding, Xiaoyan Xu, Bei Xu, Yadan Zou, Yuetong Yu, and Wenzhi Yang. 2023. "Collision Cross Section Prediction Based on Machine Learning" Molecules 28, no. 10: 4050. https://doi.org/10.3390/molecules28104050
APA StyleLi, X., Wang, H., Jiang, M., Ding, M., Xu, X., Xu, B., Zou, Y., Yu, Y., & Yang, W. (2023). Collision Cross Section Prediction Based on Machine Learning. Molecules, 28(10), 4050. https://doi.org/10.3390/molecules28104050






