Abstract
Histidine and cysteine residues, with their imidazole and thiol moieties that deprotonate at approximately physiological pH values, are primary binding sites for Zn(II), Ni(II) and Fe(II) ions and are thus ubiquitous both in peptidic metallophores and in antimicrobial peptides that may use nutritional immunity as a way to limit pathogenicity during infection. We focus on metal complex solution equilibria of model sequences encompassing Cys–His and His–Cys motifs, showing that the position of histidine and cysteine residues in the sequence has a crucial impact on its coordination properties. CH and HC motifs occur as many as 411 times in the antimicrobial peptide database, while similar CC and HH regions are found 348 and 94 times, respectively. Complex stabilities increase in the series Fe(II) < Ni(II) < Zn(II), with Zn(II) complexes dominating at physiological pH, and Ni(II) ones—above pH 9. The stabilities of Zn(II) complexes with Ac-ACHA-NH2 and Ac-AHCA-NH2 are comparable, and a similar tendency is observed for Fe(II), while in the case of Ni(II), the order of Cys and His does matter—complexes in which the metal is anchored on the third Cys (Ac-AHCA-NH2) are thermodynamically stronger than those where Cys is in position two (Ac-ACHA-NH2) at basic pH, at which point amides start to take part in the binding. Cysteine residues are much better Zn(II)-anchoring sites than histidines; Zn(II) clearly prefers the Cys–Cys type of ligands to Cys–His and His–Cys ones. In the case of His- and Cys-containing peptides, non-binding residues may have an impact on the stability of Ni(II) complexes, most likely protecting the central Ni(II) atom from interacting with solvent molecules.
1. Introduction
Cys–His and His–Cys motifs are quite abundant in antimicrobial peptides (AMPs), being present in 411 of such compounds (168 Cys–His and 243 His–Cys motifs, respectively). For the sake of comparison—348 Cys–Cys motifs and only 94 His–His motifs can be found in the AMP database [1].
Such peptide motifs are very tempting metal binding sites for metal ions such as Zn(II), Fe(II) or Ni(II), which play pivotal roles in bacterial and fungal cells. They are involved, for example in catalysis or structure functions in proteins, whose side chains or the terminal groups are able to bind metal ions [2,3]. The metal binding of the amino acids is selective and depends on the hard/soft character of the metal ion [2,4]. The most important metal anchoring sites are the side chains of histidine (imidazole) or cysteine (thiolate) residues and the carboxyl groups of glutamic and aspartic acids [5]. Thus, proteins that contain regions rich in histidine and cysteine residues are an interesting case of ligands due to a high affinity to bind Zn(II), Ni(II) and Fe(II) [6,7,8,9] and are chosen both by nature and by science as good metal chelators. Knowledge about the coordination mode, structure and function of specific binding sites in biomimetic artificial ligands with SH/NH groups and their metal complexes biomolecules provides us knowledge that can be transferred to a new generation of materials and applications, such as catalysis [10], bioassays [11] or general peptide-governed metal sorption [12,13]. It is important to note that often, seemingly very similar coordination motifs (as also in the case of this work) do not necessarily lead to the same coordination and functional outcomes, which are important factors to consider in the design of novel biomimetics, catalysts and materials [14].
Due to its structure and acid-base properties, histidine residue shows versatility in molecular interactions [15]. The functional group of the histidine residue is the aromatic imidazole ring (with a pKa slightly above 6 [16]), which contains two nitrogen atoms [17]. At physiological pH (about 7.4), the imidazole group of histidine is partially deprotonated—observed in both acidic and basic forms. In the case of the acidic imidazolium cation, the positive charge is evenly distributed between the two nitrogen atoms. In response to a change in pH values, each of them can release a proton and form equivalent tautomer forms: N1-H or N3-H, capable of binding metal ions [15].
Depending on the location and amount of histidine residues in the chain of proteins and peptides, we observe different coordination modes of metal ions [15,18,19,20,21]. Peptides that contain a histidine residue in the first position can form complexes with one of the nitrogen atoms of the histidine imidazole ring and the nitrogen atom of the N-terminal amino group [22]. This coordination mode is called the histamine-like model and its characteristic feature is the formation of a five-membered ring [23]. The histamine coordination mode for Zn(II) is maintained in the entire pH range, and the zinc coordination sphere can be filled by oxygen, nitrogen and sulfur atoms derived from side groups of other amino acids or water molecules.
In the case of Cu(II) and Ni(II) (and, theoretically, also Fe(II)), which are capable of deprotonating the amide bond at basic pH, the metal coordination sphere may include amide nitrogen atoms, resulting in stable, six-membered rings with a {NH2, 3N−} mode [19,22,24,25]. When the histidine residue is in the second position, it is possible to form stable five- or six-membered rings that stabilize the coordination of Ni(II) and Cu(II). The coordination sphere includes two nitrogen atoms derived from the histidine residue (one from the imidazole of the histidine residue and one amide nitrogen atom), as well as the nitrogen atom of the N-terminal amino group and the oxygen atom derived from the carboxyl group present in sequence or water molecule [19]. If the histidine residue is on the third position in the sequence, such a motif is called the ATCUN (amino terminal Cu(II) and Ni(II) binding site) motif or albumin-like coordination site [26,27]. This binding site is particularly attractive for Cu(II) and Ni(II). The coordination sphere includes an imidazole nitrogen atom, two amide nitrogen atoms and an amine N-terminal nitrogen atom, which results in the formation of three chelate rings: one six-membered ring and two five-membered ones.
The position and number of the histidine residues in peptides are very important and determine the coordination mode of metal ions. In addition to the previously mentioned coordination modes, it is also worth mentioning that at any place in the chain, there may be a specific sequence of amino acids—HEXXH—with a high affinity for Zn(II) [28]. Its presence has been found, among others, in metalloproteinases dependent on zinc ions [29]. In this case, the coordination sphere includes two imidazole nitrogen atoms, and the coordination sphere may be filled by oxygen atoms from the Glu carboxyl group or by water molecules [30]. Moreover, usually, the more histidine residues a peptide contains, the higher the thermodynamic stability of its imidazole-coordinated complexes with metal ions [24,31,32].
Cysteine is an exceptional proteinogenic amino acid. It contains a reactive sulfhydryl group [33], and two cysteines may form a disulfide bond (cystine) [34]. The disulfide links between various residues present in different parts of the protein or between separated polypeptide chains induce the folding of proteins [33] and play a structural role in proteins. Due to the rigid planar nature of the peptide bond, two cysteine residues present next to each other or separated by only one residue are unable to interact. The separation of two cysteine residues by two amino acid residues causes their closest proximity in the α-helix and β-turn. This motif, C-(X)2-C, is commonly present in many proteins such as oxidoreductases (X may stand for any amino acid) [35]. In the case of peptides that contain both cysteine and histidine residues, such as Ac-HAAC-NH2 or Ac-CGAH-NH2 [5], the C-terminal donor group is the primary anchor group, and at higher pH values, the N-terminal group is replaced by a third amide nitrogen atom [36].
Zinc, after iron, is the second most important trace element in the human organism; [37] it is necessary for growth and cell division, not only in the human body, but also in all other forms of life. In contrast to other trace elements (such as copper or iron), Zn(II) does not change its oxidation state in solution (due to its electron configuration, [Ar]3d10, it does not show d-d transitions and, therefore, is not visible in absorption spectroscopy) [38]. Zn(II) also plays a significant structural role [28], for example, in proteins with zinc finger motifs [39]. The geometry of zinc complexes may be different, but the most common one is tetrahedral [40]. Zn(II)’s most preferred donor atoms are nitrogen, sulfur and oxygen present in the side chains of histidine, cysteine, glutamic or aspartic acid residues, respectively. The coordination sphere can be filled with water molecules.
Another biologically relevant metal ion is nickel(II). It is essential for many eubacteria, archaebacteria, fungi and plants because it has important functions in diverse metabolic processes (metabolism and virulence) [41]. Ni(II) plays a catalytic role in the cofactors of nine enzymes [42]. Humans do not have any Ni-dependent enzymes; however, this metal is present in negligible amounts in human bodies, most likely since it is necessary for commensal bacteria [43]. Large doses or prolonged contact with nickel could be genotoxic, haematotoxic, teratogenic, cancerogenic and immunotoxic [44]. From a chemical point of view, Ni(II) and Fe(II) are very interesting, because they could theoretically compete with Zn(II) for its binding sites.
Iron is an essential trace metal for most living organisms. It serves as a prosthetic group for proteins involved in central cellular processes, such as respiration, oxygen transport and DNA synthesis [45]. Non-heme iron is bound by a large number of enzymes found in the cytosol. There are two classes of these proteins, those containing iron-sulfur clusters and those without iron-sulfur clusters. In the case of non-sulfur cluster proteins, Fe(II) is coordinated by six donor atoms forming octahedral complexes or five donor atoms which form a trigonal bipyramidal geometry. For complexes with a coordination number equal to five, the coordination frequently involves open coordination sites for the simultaneous binding of substrates. Most often, nitrogen or oxygen donor atoms (e.g., histidyl residues, aspartic or glutamic acid residues) are involved in Fe(II) binding [46].
The results of our recent work [47], in which potentiometric and spectroscopic techniques were used to study the interactions of Ac-PNCHTHEGGQLHCT, the C-terminal part of the Aspf2 protein, with Zn(II) and Ni(II), encouraged us to pursue further studies on the topic of peptides with cysteine and histidine residues in different positions in the model peptides. In this work, we report results obtained in systematic studies of Zn(II), Ni(II) and Fe(II) complexes of two model peptides containing histidine, cysteine and alanine residues in their sequences. We checked the thermodynamic parameters of Ac-AHCA-NH2 and Ac-ACHA-NH2 peptides. The protonation constants and the stability constants of formed complexes were determined using potentiometry, and the obtained results allowed us to identify the species that are present in the solutions at different pH values. Independent spectroscopic methods (UV-Vis spectroscopy and CD spectroscopy) allowed us to determine precise Ni(II) binding sites. In the scope of this paper, we try to answer three most important questions. (i) His or Cys—does it matter for the stability of their complexes which one goes first? (ii) Do His–Cys and Cys–His ligands have a metal preference? The comparison with some previously studied peptides which contain histidine and/or cysteine residues in different positions will allow us to answer the last and equally important question: (iii) Do non-binding residues affect complex stability? These data may provide an overview of the metal binding ability of -HC- and -CH- motifs and may help to better understand how Zn(II) is able to compete with other metal ions for binding sites in proteins.
2. Results and Discussion
The structural and thermodynamic properties of Zn(II), Ni(II) and Fe(II) complexes with Ac-AHCA-NH2 and Ac-ACHA-NH2 were studied using potentiometry, UV-Vis and CD spectroscopy. Potentiometric titrations allowed us to determine the ligand protonation constants, the stability constants of the formed complexes and the pH-dependent distribution diagrams. The combined UV-Vis and CD spectroscopic results clarified the binding mode of Ni(II) and the geometry of the species present in solution. Consequently, the combination of all these methods allowed us to explain coordination geometries and perform a detailed thermodynamic analysis of Ac-AHCA-NH2 and Ac-ACHA-NH2 peptides.
2.1. Protonation Constants of the Ac-AHCA-NH2 and Ac-ACHA-NH2
Both peptides are protected in the N-terminus through acetylation and in the C-terminus through amidation and contain two possible sites of protonation—a His and a Cys residue; their pKa values are reported in Table 1. The first protonation constant for Ac-AHCA-NH2 corresponds to the imidazole nitrogen atom of histidine residue (6.42) and the second one, with the value 8.72, to the thiolate of the cysteine residue. The Ac-ACHA-NH2 peptide also behaves as an H2L acid with the deprotonating groups corresponding to the His and Cys residues with pKa values: 6.42 and 8.76, respectively (Table 1).

Table 1.
Potentiometric data for the proton, Zn(II), Ni(II), Fe(II) complexes of the Ac-AHCA-NH2 and Ac-ACHA-NH2 in a water solution of 4 mM HClO4 with I = 0.1 M NaClO4, T = 298 K.
2.2. Metal Complexes of the Ac-AHCA-NH2 Peptide
In the case of the Zn(II)-Ac-AHCA-NH2 complex, the first detected species, with a maximum at pH 7, is ZnL (Table 1 and Figure S1A). Most probably, in this form, the Zn(II) is coordinated to one imidazole nitrogen and one Cys thiol. The remaining coordination positions are likely occupied by water molecules. The next observed species, ZnH−1L (maximum at pH 8.90), has a {Nim, S−, OH−} binding mode, and it forms with pKa = 7.84, corresponding to the deprotonation of a coordinated water molecule. Above pH 10.0, ZnH−2L dominates in solution. This case is most likely related to the deprotonation of the second water molecule (pKa = 9.98), and in this form, the metal ion coordination sphere includes the imidazole, the sulfur and two oxygen atoms from the water molecules (Figure 1A).
The first Ni(II) complex observed at low pH, NiL reaches its maximum at pH 7.5 (Table 1 and Figure S1B). Most probably, in this complex, Ni(II) is bound to the His imidazole and Cys sulfur. CD spectra confirm this finding with pronounced bands around 270 nm and 350 nm, typical for Nim → Ni(II) [48] and S− → Ni(II) [49] charge transfer transitions, respectively (Figure S3A). The next deprotonation leads to the formation of NiH−1L species, in which an amide nitrogen binds to Ni(II). The loss of the next proton leads to the NiH−2L form and is most likely related to the deprotonation and binding of a second amide. The last observed form, NiH−3L, most likely corresponds to the deprotonation of a water molecule bound to the central Ni(II) ion. In this form, one cysteine thiolate, one histidine imidazole and two amide nitrogens already participate in binding, forming a square planar nickel complex (Figure 1B). The hypothesis is supported also by the UV-Vis spectra (Figure S4A), where no changes in the wavelength of maximum absorption are observed above pH 8.5.
The coordination of Fe(II) ions begins at a pH of about 6, (Table 1, Figure S1C). The first species observed at this pH, FeL, is related to the simultaneous deprotonation of two functional groups (His imidazole and Cys thiol). As Fe(II) ions have a preference for both nitrogen and sulfur donor atoms [50], both of these amino acid residues are most likely involved in coordination. As pH values increase, another complex, FeH−2L, is formed due to the simultaneous loss of two protons. Most likely, it is related to the deprotonation and participation in the binding of the metal ion of the amide nitrogen of the peptide backbone [50,51] (Figure 1C), although it is also possible that at this point, two water molecules bound to the central iron atom deprotonate; advanced studies on the possibility of involving amides in binding are forthcoming. In the last deprotonation step, an FeH−3L species is formed and, as in the case of the Ni(II) complex, the deprotonation of a water molecule bound to the metal ion possibly takes place.
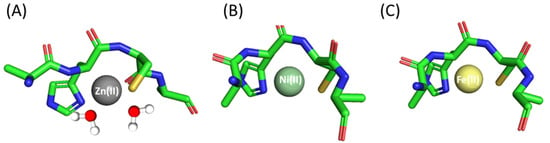
Figure 1.
Proposed binding mode for the (A) Zn(II), (B) Ni(II) and (C) Fe(II) complexes with Ac-AHCA-NH2—a schematic model. The figure was generated using PyMOL [52].
2.3. Metal Complexes of the Ac-ACHA-NH2 Peptide
Ac-ACHA-NH2 coordination to Zn(II) results in three different species in the studied pH range: ZnL, ZnH−1L and ZnH−2L. ZnL is the first detected species, where most probably histidine and cysteine residues are already bound to Zn(II). This form has a maximum of around pH 7.0 (Table 1 and Figure S2A). The second one, ZnH−1L, which dominates in solution in the pH range 8.0–10.0, is most likely related to the deprotonation of the first water molecule bound to the central Zn(II). Most probably, in the ZnH−2L complex, a subsequent coordinated water molecule deprotonates (Figure 2A).
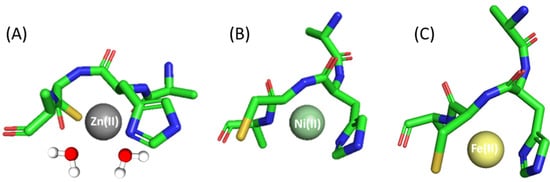
Figure 2.
Proposed binding mode for the (A) Zn(II), (B) Ni(II) and (C) Fe(II) complexes with Ac-ACHA-NH2—a schematic model. The figure was generated using PyMOL [52].
In the studied pH range (2.5–10.5) and at a metal:ligand molar ratio of 1:1, we observed four different species of Ni(II) complex (Table 1 and Figure S2B). The first species is a bis-complex with two ligand molecules bound to the metal (NiL2). This species occurs at pH 6.0 and achieves maximum concentration at around pH 8.0. Spectroscopic data obtained in this pH range, in particular the shoulders in the spectra at around 500 and 410 nm, suggest that in NiL2, Ac-ACHA-NH2 binds Ni(II) via four donor atoms in a {2S−, 2Nim} binding mode. This is consistent with previously studied Ni(II) complexes with Cys- and His-containing peptides [53]. CD spectra also confirm this finding with pronounced d-d bands at 305 nm, which can be assigned to S−→Ni(II) charge transfer transitions [49]. In the NiH−1L form, apart from Cys and His side chains, an amide also takes part in metal binding, as confirmed by the presence of negative and positive bands at 550 and 480 nm in the CD spectra (Figure S3B) [53,54,55]. Above pH 8.5, the square planar nickel complex reorganizes; the next two forms, NiH−2L and NiH−3L, most likely involve additional amide nitrogen atoms and a water molecule in binding, which can be confirmed via d-d transitions in the UV-Vis spectra at around 430 nm (Figure S4B).
Fe(II)- Ac-ACHA-NH2 behaves similarly to the complex with a Ac-AHCA-NH2 ligand; only slight differences in logβ values were observed. Thus, the coordination process begins at pH 6 with the formation of an FeL species that reaches its maximum of formation at a pH of around 8.5, being also the only species present at physiological pH (pH 7.4). The Fe(II) ion is probably bound to sulfur and nitrogen donor atoms of cysteine and histidine residues, respectively, and the rest of the coordination sites are filled with either nitrogen amide or water molecules (Figure 2C).
To compare the strength of Zn(II), Ni(II) and Fe(II) binding to Ac-AHCA-NH2, Ac-ACHA-NH2 and other peptides described in the literature, we used their calculated stability constants to prepare so-called competition plots, which illustrate the comparison of the complexes’ stability. Such plots show a theoretical situation in which equimolar amounts of all reagents, metals and peptides are mixed. In the whole pH range, the Ac-ACHA-NH2 and Ac-AHCA-NH2 bind Zn(II) with comparable affinity (Figure 3A). In both cases, the coordination of Zn(II) involves one histidine imidazole and one cysteine thiolate, and the coordination sphere is filled with two water molecules. The results suggest that the location of the histidine and cysteine residues, which are next to each other, does not have a significant effect on the stability of the Zn(II) complexes—Zn(II) is equally ready to bind to the HC motif as it is to bind to the CH one.
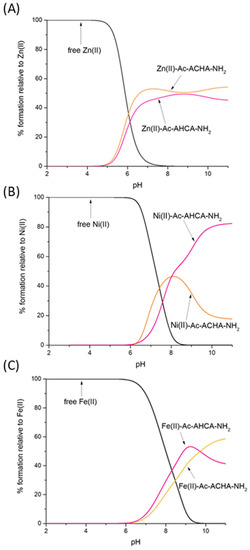
Figure 3.
Competition plot between (A) the Ac-AHCA-NH2, Ac-ACHA-NH2 peptides and Zn(II); (B) the Ac-AHCA-NH2, Ac-ACHA-NH2 peptides and Ni(II), and (C) the Ac-AHCA-NH2, Ac-ACHA-NH2 peptides and Fe(II). The plot describes complex formation at different pH values in a hypothetical situation in which equimolar amounts of the three reagents are mixed. Calculations are based on constants from Table 1.
Nickel’s preferences in binding Ac-ACHA-NH2 and Ac-AHCA-NH2 are substantially different (Figure 3B). Ni(II) binds to both ligands with comparable affinity up to pH 8, while at a higher pH, at which amides take part in binding, Ni(II) coordination to Ac-AHCA-NH2 is strongly preferred, suggesting that the {S−, 2N−} binding mode is more stable than the {Nim, 2N−} one.
In the case of Fe(II) complexes, the trend is very similar to that of Zn(II) complexes; Fe(II) seems to bind to both Ac-ACHA-NH2 and Ac-AHCA-NH2 with comparable affinity (Figure 3C).
Which is the preferred Zn(II) binding site—CH/HC or CC? Figure 4A gives us a straightforward answer—the ‘CC’ motif clearly binds Zn(II) with higher affinity, at least in the compared sequences: Ac-CC-NH2, Ac-GSCCHTGNHD-NH2, Ac-EEGCCHGHHE-NH2 and Ac-CCSTSDSHHQ-NH2 [56].
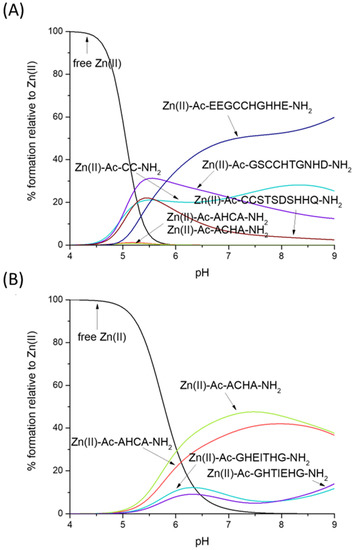
Figure 4.
Competition plot between the Zn(II), Ac-AHCA-NH2, Ac-ACHA-NH2 peptides and (A) Ac-CC-NH2, Ac-GSCCHTGNHD-NH2, Ac-EEGCCHGHHE-NH2, Ac-CCSTSDSHHQ-NH2 [56] vs. (B) Ac-GHEITHG-NH2, Ac-GHTIEHG-NH2 [57]. The plot describes complex formation at different pH values in a hypothetical situation in which equimolar amounts of all reagents are mixed.
Thiolates are clearly the most likely binding sites for Zn(II)—our HC/CH ligands (Ac-AHCA-NH2 and Ac-ACHA-NH2) bind this metal much more effectively than peptides which contain two His imidazoles (Ac-GHEITHG-NH2, Ac-GHTIEHG-NH2, Figure 4B), showing that the Zn(II)-Nim, S− complex turns out to be thermodynamically stronger than Zn(II)-2Nim—in our hypothetical situation, at physiological pH (pH 7.4), nearly 50% of Zn(II) is bound to Ac-ACHA-NH2, nearly 40% of Zn(II) is bound to Ac-AHCA-NH2 and only around 15% is bound to Ac-GHEITHG-NH2 and Ac-GHTIEHG-NH2 [57].
Nickel’s preferences for CH- and HC-type ligands differ somewhat from those of Zn(II). Ni(II) forms very stable square-planar complexes with the CC motif, binding to two thiolates and the amide in between. Ni(II)-CC complexes are, as Zn(II) complexes, more stable than CH and HC ones, but for different reasons—in the case of Zn(II), the affinity for sulfur seems to be the reason for the high affinity, while in the case of Ni(II), the planar geometry and additional stabilization by the amide seem to play the major role (the competition plot between Ni(II), Ac-AHCA-NH2, Ac-ACHA-NH2, Ac-CC-NH2, Ac-GSCCHTGNHD-NH2, Ac-EEGCCHGHHE-NH2 and Ac-CCSTSDSHHQ-NH2 [56] is shown on Figure 5A).
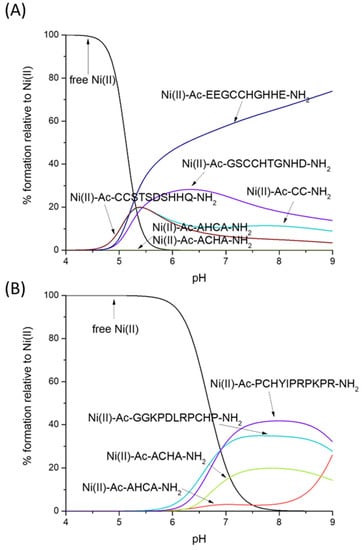
Figure 5.
Competition plot between the Ni(II), Ac-AHCA-NH2, Ac-ACHA-NH2 peptides and (A) Ac-CC-NH2, Ac-GSCCHTGNHD-NH2, Ac-EEGCCHGHHE-NH2, Ac-CCSTSDSHHQ-NH2 [56] vs. (B) GGKPDLRPCHP-NH2, PCHYIPRPKPR-NH2, PCHPPCHYIPR-NH2 [58]. The plot describes complex formation at different pH values in a hypothetical situation in which equimolar amounts of all reagents are mixed.
What is the impact of the non-binding residues on the Ni–CH complex stabilities with a {Nim, N−, S−} binding mode? As hypothesized, the non-binding residues are able to protect the complex from hydrolysis, most likely forming a network of hydrogen bonds that protects the central Ni(II) ion. In our hypothetical situation in Figure 5B, Ni(II) complexes with Ac-AHCA-NH2 and Ac-ACHA-NH2 are substantially weaker than those with Ac-GGKPDLRPCHP-NH2 and Ac-PCHYIPRPKPR-NH2 [58].
3. Experimental Section
3.1. Materials
The peptides (Ac-AHCA-NH2 and Ac-ACHA-NH2,) were purchased from Karebay Biochem (certified purity: 98%) and were used as received. Zn(II), Ni(II) perchlorate and ammonium iron(II) sulfate (Mohr’s salt) were extra pure products (Sigma-Aldrich, Poznań, Poland). The carbonate-free stock solution of 0.1 M KOH was purchased from Sigma-Aldrich and then potentiometrically standardized with potassium hydrogen phthalate.
3.2. Potentiometric Measurements
The stability constants for proton Zn(II) and Ni(II) complexes with ligands Ac-AHCA-NH2 and Ac-ACHA-NH2 were calculated from titration curves carried out over the pH range of 2–11 at 298 K and ionic strength 0.1 M NaClO4. The total volume of the solution used was 3.0 cm3. The potentiometric titrations were performed using a Dosimat 800 Metrohm Titrator connected to a Methrom 905 pH-meter and a Mettler Toledo pH inLab Science electrode. The thermostabilized glass cell was equipped with a magnetic stirring system, a microburet delivery tube and an inlet-outlet tube for argon. Solutions were titrated with 0.1 M carbonate-free NaOH. The electrodes were calibrated daily for hydrogen ion concentration through titrating HClO4 with NaOH using a total volume of 3.0 cm3. The purities and the exact concentrations of the ligand solutions were determined using the Gran method. The ligand concentration was 0.5 mM. The Zn(II), Ni(II) and Fe(II) to ligand ratio was 1:1. The HYPERQUAD 2008 program was used for the stability constant calculations. The standard deviations were computed using HYPERQUAD 2008 and referenced to random errors only. The constants for hydrolytic Zn(II) and Ni(II) species were used in these calculations. The speciation and competition diagrams were computed using the HYSS program [59].
In the case of measurements of Fe(II) complexes, all the experiments were carried out in degassed solvents. A freshly prepared Fe(II) ion stock solution was employed for each measurement. The argon with which the measuring cell is purged prevents both carbonate formation and Fe(II) oxidation.
3.3. Spectroscopic Studies
The absorption spectra were recorded on a Jasco-730 spectrophotometer, in the range 200–800 nm, using a quartz cuvette with an optical path of 1 cm. Circular dichroism spectra were recorded on a Jasco J-1500 CD spectrometer in the 200–800 nm range, using a quartz cuvette with an optical path of 1 cm in the visible and near-UV range. The concentration of sample solutions used for spectroscopic studies was similar to those employed in the potentiometric experiment. The metal:ligand ratio was 1:1. All spectroscopic measurements were recorded in the pH range 2.5–10.5. The pH of the samples was adjusted with appropriate amounts of concentrated HClO4 and KOH solutions. The UV-Vis and CD spectroscopy parameters were calculated from the spectra obtained at the pH values corresponding to the maximum concentration of each particular species, based on distribution diagrams. OrginPro 2016 was used to process and visualize the obtained spectra.
4. Conclusions
Zn(II), Ni(II) and Fe(II) are crucial for proper cell development. Therefore, it is important to understand which type of binding sites and which type of precise sequences bind them with the highest (or with ‘the most appropriate’) affinity. For this reason, we wanted to answer three questions: (i) His or Cys—does it matter (for metal complex stability) which one goes first? (ii) Do His–Cys and Cys–His ligands have a metal preference? (iii) Do non-binding residues have a large effect on complex stability?
The results of this work allowed us to come to the following conclusions: (i) The position of His and Cys residues affects the stability of the complexes with Ni(II), but not those with Zn(II) and Fe(II). Ni(II) will more likely bind to Ac-AHCA-NH2 than to Ac-ACHA-NH2 above pH 8.0, when amide nitrogen atoms begin to take part in coordination (Figure 6A,B). (ii) His–Cys and Cys–His ligands do have a strong metal preference; up to pH 8.5, Zn(II) complexes are clearly the strongest ones. At alkaline pH (above 9.0), in which amides start to bind Ni(II), the square planar Ni(II) species become strongest. Fe(II) complexes are the weakest ones, thus obeying the Irving–Williams series (Figure 6A,B). (iii) Non-binding residues have a clear enhancing effect on the stability of Ni(II) complexes, most likely through shielding the central Ni(II) ion from surrounding solvent molecules.
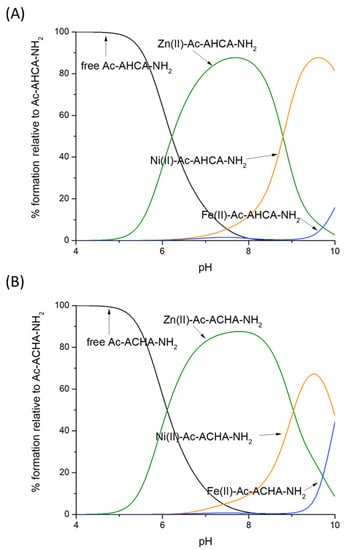
Figure 6.
Competition plot between (A) the Ac-AHCA-NH2 peptide, Zn(II), Ni(II) and Fe(II) vs. (B) the Ac-ACHA-NH2 peptide, Zn(II), Ni(II) and Fe(II). The plot describes complex formation at different pH values in a hypothetical situation in which equimolar amounts of the three reagents are mixed. Calculations are based on constants from Table 1.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28103985/s1.
Author Contributions
Conceptualization, M.R.-Ż.; Methodology, K.G. and V.D.; Validation, K.G. and J.W.; Formal analysis, J.W., J.Ś.-K., H.K. and D.B.; Investigation, K.G. and V.D.; Writing—original draft, K.G., V.D. and D.B.; Writing—review & editing, K.G., J.W., K.S.-S., J.Ś.-K., H.K., M.B.-O., D.B. and M.R.-Ż.; Visualization, K.G.; Supervision, M.R.-Ż.; Funding acquisition, J.W., H.K. and M.R.-Ż. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the National Science Centre (UMO2017/26/A/ST5/00363—H.K, UMO2017/26/E/ST5/00364—M.R-Ż and UMO-2021/41/B/ST4/02654—J.W.). Funding which allowed staff exchange COST Action CA18202, NECTAR—Network for Equilibria and Chemical Thermodynamics Advanced Research, supported by COST (European Cooperation in Science and Technology) is gratefully acknowledged.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Csire, G.; Kolozsi, A.; Gajda, T.; Pappalardo, G.; Várnagy, K.; Sóvágó, I.; Fábián, I.; Lihi, N. The ability of the NiSOD binding loop to chelate zinc(II): The role of the terminal amino group in the enzymatic functions. Dalton Trans. 2019, 48, 6217–6227. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, C.C.; Skaar, E.P. Nutritional immunity: The battle for nutrient metals at the host–pathogen interface. Nat. Rev. Microbiol. 2022, 20, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Pearson, G.R. Hard and Soft Acids and Bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Raics, M.; Lihi, N.; Laskai, A.; Kállay, C.; Várna, K.; Sóvágó, I. Nickel(II), zinc(II) and cadmium(II) complexes of hexapeptides containing separate histidyl and cysteinyl binding sites. New J. Chem. 2016, 40, 5420–5427. [Google Scholar] [CrossRef]
- Zhou, L.; Li, S.; Su, Y.; Yi, X.; Zheng, Z.; Deng, F. Interaction between Histidine and Zn(II) Metal Ions over a Wide pH as Revealed by Solid-State NMR Spectroscopy and DFT Calculations. J. Phys. Chem. B 2013, 117, 8954–8965. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Lin, Y.-F.; Pratihar, S.; Angel, L.A.; Hase, W.L. Direct Dynamics Simulations of Fragmentation of a Zn(II)-2Cys-2His Oligopeptide.Comparison with Mass Spectrometry Collision-Induced Dissociation. J. Phys. Chem. A 2019, 123, 6868–6885. [Google Scholar] [CrossRef] [PubMed]
- Giles, N.M.; Giles, G.I.; Jacob, C. Multiple roles of cysteine in biocatalysis. Biochem. Biophys. Res. Commun. 2003, 300, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Potocki, S.; Rowińska-Żyrek, M.; Witkowska, D.; Pyrkosz, M.; Szebesczyk, A.; Krzywoszynska, K.; Kozlowski, H. Specific Metal Transport and Homeostasis within the Human Body: Toxicity Associated with Transport Abnormalities. Curr. Med. Chem. 2012, 19, 2738–2759. [Google Scholar] [CrossRef] [PubMed]
- Banach, Ł.; Brykczyńska, D.; Gorczyński, A.; Wyrzykiewicz, B.; Skrodzki, M.; Pawluć, P. Markovnikov-selective double hydrosilylation of challenging terminal aryl alkynes under cobalt and iron catalysis. Chem. Commun. 2022, 58, 13763–13766. [Google Scholar] [CrossRef] [PubMed]
- Bocian, A.; Szymańska, M.; Brykczyńska, D.; Kubicki, M.; Wałęsa-Chorab, M.; Roviello, G.N.; Fik-Jaskółka, M.A.; Gorczyński, A.; Patroniak, V. New Artificial Biomimetic Enzyme Analogues based on Iron(II/III) Schiff Base Complexes: An Effect of (Benz)imidazole Organic Moieties on Phenoxazinone Synthase and DNA Reconition. Molecules 2019, 24, 3173. [Google Scholar] [CrossRef] [PubMed]
- Irankunda, R.; Camaño Echavarría, J.A.; Paris, C.; Stefan, L.; Desobry, S.; Katalin Selmeczi, K.; Laurence Muhr, L.; Canabady-Rochelle, L. Metal-Chelating Peptides Separation Using Immobilized Metal Ion Affinity Chromatography: Experimental Methodology and Simulation. Separations 2022, 9, 370. [Google Scholar] [CrossRef]
- Birdsall, R.E.; Kellett, J.; Ippoliti, S.; Ranbaduge, N.; Lauber, M.A.; Yu, Y.-Q.; Chen, W. Reducing metal-ion mediated adsorption of acidic peptides in RPLC-based assays using hybrid silica chromatographic surfaces. J. Chromatogr. B 2021, 1179, 122700. [Google Scholar] [CrossRef]
- Bocian, A.; Skrodzki, M.; Kubicki, M.; Gorczyński, A.; Pawluć, P.; Patroniak, V. The effect of Schiff base ligands on the structure and catalytic activity of cobalt complexes in hydrosilylation of olefins. Appl. Catal. A Gen. 2020, 602, 117665. [Google Scholar] [CrossRef]
- Rowińska-Żyrek, M.; Witkowska, D.; Potocki, S.; Remelli, M.; Kozłowski, H. His-rich sequences—Is plagiarism from nature a good idea? New J. Chem. 2013, 37, 58–70. [Google Scholar] [CrossRef]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2015, 44, D1214–D1219. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, J.R.; Martin, R.B. Interactions of Histidine and Other Imidazole Derivatives with Transition Metal Ions in Chemical and Biological Systems. Chem. Rev. 1974, 74, 471–517. [Google Scholar] [CrossRef]
- Matera, A.; Brasuń, J.; Cebrat, M.; Świątek-Kozłowska, J. The role of the histidine residue in the coordination abilities of peptides with a multi-histidine sequence towards copper(II) ions. Polyhedron 2008, 27, 1539–1555. [Google Scholar] [CrossRef]
- Kozłowski, H.; Bal, W.; Dyba, M.; Kowalik-Jankowska, T. Specific structure—Stability relations in metallopeptides. Coord. Chem. Rev. 1999, 184, 319–346. [Google Scholar] [CrossRef]
- Murariu, M.; Dragan, E.S.; Drochioiu, G. Model peptide-based system used for the investigation of metal ions binding to histidine-containing polypeptides. Biopolymers 2010, 93, 497–508. [Google Scholar] [CrossRef]
- Kołkowska, P.; Hecel, A.; Kędzierska, D.; Ostrowska, M.; Walencik, P.K.; Wątły, J.; Zdyb, K.; Spodzieja, M.; Rodziewicz-Motowidło, S.; Potocki, S.; et al. HENRYK—An endless source of metal coordination surprises. J. Inorg. Biochem. 2016, 163, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Migliorini, C.; Witkowska, D.; Valensin, D.; Kamysz, W.; Kozlowski, H. Competition between histamine-like and poly-imidazole coordination sites for Cu2+ and Zn2+ ions in zebra-fish peptide of prion-like protein. Dalton Trans. 2010, 37, 8663–8670. [Google Scholar] [CrossRef] [PubMed]
- Török, I.; Gajda, T.; Gyurcsik, B.; Tóth, G.K.; Péter, A. Metal complexes of imidazole ligands containing histamine-like donor sets: Equilibrium, solution structure and hydrolytic activity. J. Chem. Soc. Dalton Trans. 1998, 7, 1205–1212. [Google Scholar] [CrossRef]
- Sóvágó, I.; Várnagy, K.; Lihi, N.; Grenács, Á. Coordinating properties of peptides containing histidyl residues. Coord. Chem. Rev. 2016, 327–328, 43–54. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Nurchi, V.M.; Zoroddu, M.A. The Involvement of Amino Acid Side Chains in Shielding the Nickel Coordination Site: An NMR Study. Molecules 2013, 18, 12396–12414. [Google Scholar] [CrossRef]
- Harford, C.; Sarkar, B. Amino Terminal Cu(II)- and Ni(II)-Binding (ATCUN) Motif of Proteins and Peptides: Metal Binding, DNA Cleavage, and Other Properties. Acc. Chem. Res. 1997, 30, 123–130. [Google Scholar] [CrossRef]
- Witkowska, D.; Szebesczyk, A.; Wątły, J.; Braczkowski, M.; Rowińska-Żyrek, M. A Comparative Study on Nickel Binding to Hpn-like Polypeptides from Two Helicobacter pylori Strains. Int. J. Mol. Sci. 2021, 22, 13210. [Google Scholar] [CrossRef]
- Wątły, J.; Potocki, S.; Rowińska-Żyrek, M. Zinc Homeostasis at the Bacteria/Host Interface—From Coordination Chemistry to Nutritional Immunity. Chemistry 2016, 22, 15992–16010. [Google Scholar] [CrossRef]
- Jongeneel, C.V.; Bouvier, J.; Bairoch, A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989, 242, 211–214. [Google Scholar] [CrossRef]
- Hooper, N.M. Families of zinc metalloproteases. FEBS Lett. 1994, 354, 1873–3468. [Google Scholar] [CrossRef]
- Remelli, M.; Nurchi, V.M.; Lachowicz, J.I.; Medici, S.; Zoroddu, M.A.; Peana, M. Competition between Cd(II) and other divalent transition metal ions during complex formation with amino acids, peptides, and chelating agents. Coord. Chem. Rev. 2016, 327–328, 55–69. [Google Scholar] [CrossRef]
- Sóvágó, I.; Várnagy, K. Cadmium(II) complexes of amino acids and peptides. Met. Ions Life Sci. 2013, 11, 275–302. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, H.; Potocki, S.; Remelli, M.; Rowińska-Żyrek, M.; Valensin, D. Specific metal ion binding sites in unstructured regions of proteins. Coord. Chem. Rev. 2013, 257, 2625–2638. [Google Scholar] [CrossRef]
- Muskal, S.M.; Holbrook, S.R.; Kim, S.H. Prediction of the disulfide-bonding state of cysteine in proteins. Protein Eng. Des. Sel. 1990, 3, 667–672. [Google Scholar] [CrossRef]
- Shuber, A.P.; Orr, E.C.; Recny, M.A.; Schendel, P.F.; May, H.D.; Schauer, N.L.; Ferry, J.G. Cloning, expression, and nucleotide sequence of the formate dehydrogenase genes from Methanobacterium formicicum. J. Biol. Chem. 1986, 261, 12942–12947. [Google Scholar] [CrossRef] [PubMed]
- Lukács, M.; Pálinkás, D.C.; Szunyog, G.; Várnagy, K. Metal Binding Ability of Small Peptides Containing. Chem. Open 2021, 10, 451–463. [Google Scholar] [CrossRef]
- Roohani, N.; Hurrell, R.; Kelishadi, R.; Schulin, R. Zinc and its importance for human health: An integrative review. J. Res. Med. Sci. 2013, 18, 144–157. [Google Scholar]
- Penner-Hahn, J.E. Characterization of “spectroscopically quiet” metals in biology. Coord. Chem. Rev. 2005, 249, 161–177. [Google Scholar] [CrossRef]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Dudev, T.; Lim, C. Tetrahedral vs Octahedral Zinc Complexes with Ligands of Biological Interest: A DFT/CDM Study. J. Am. Chem. Soc. 2000, 122, 11146–11153. [Google Scholar] [CrossRef]
- Żamojć, K.; Wyrzykowski, D.; Sabatino, G.; Papini, A.M.; Wieczorek, R.; Chmurzyński, L.; Makowska, J. Key role of histidine residues orientation in affinity binding of model pentapeptides with Ni2+ ions: A theoretical supported experimental study. J. Mol. Liq. 2021, 341, 117414. [Google Scholar] [CrossRef]
- Li, Y.; Zamble, D.B. Nickel Homeostasis and Nickel Regulation: An Overview. Chem. Rev. 2009, 109, 4617–4643. [Google Scholar] [CrossRef] [PubMed]
- Alfano, M.; Cavazza, C. Structure, function, and biosynthesis of nickel-dependent enzymes. Protein Sci. 2020, 29, 1071–1089. [Google Scholar] [CrossRef] [PubMed]
- Zdrojewicz, Z.; Popowicz, E.; Winiarski, J. Nickel—Role in human organism and toxic effects. Pol. Merkur. Lek. 2016, 41, 115–118. [Google Scholar]
- Anderson, C.; Shen, M.; Eisenstein, R.; Leibold, E. Mammalian iron metabolism and its control by iron regulatory proteins. Biochim. Biophys. Acta 2012, 1823, 1468–1483. [Google Scholar] [CrossRef] [PubMed]
- Hider, R.C.; Kong, X. Iron speciation in the cytosol: An overview. Dalton Trans. 2013, 42, 3220–3229. [Google Scholar] [CrossRef] [PubMed]
- Garstka, G.; Hecel, A.; Kozłowski, H.; Rowińska-Żyrek, M. Specific Zn(II)-binding site in the C-terminus of Aspf2, a zincophore from Aspergillus fumigatus. Metallomics 2022, 20, mfac042. [Google Scholar] [CrossRef]
- Bal, W.; Lukszo, J.; Jeżowska-Bojczuk, M.; Kasprzak, K.S. Interactions of Nickel(II) with Histones—Stability and Solution Structure of Complexes with CH3CO-Cys-Ala-Ile-His-NH2, a Putative Metal-Binding Sequence of Histone H3. Chem. Res. Toxicol. 1995, 8, 683–692. [Google Scholar] [CrossRef]
- Cherifi, K.; Reverend, B.D.L.; Loucheux, C.; Várna, K.; Kiss, T.; Sóvágó, I.; Kozłowski, H. Transition metal complexes of L-cysteine containing Di- and tripeptides. J. Inorg. Biochem. 1990, 38, 69–80. [Google Scholar] [CrossRef]
- Nemirovskiy, O.V.; Gross, M.L. Complexes of Iron(II) with Cysteine-Containing. Am. Soc. Mass Spectrom. 1996, 7, 977–980. [Google Scholar] [CrossRef]
- Guajardo, R.J.; Chavez, F.; Farinas, E.T.; Mascharak, P.M. Structural Features That Control Oxygen Activation at the Non-Heme Iron Site in Fe(II)-Bleomycin: An Analog Study. J. Am. Chem. Soc. 1995, 117, 3883–3884. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LLC: New York, NY, USA, 2015.
- Kozłowski, H.; Lebkiri, A.; Onindo, C.O.; Pettit, L.D.; Galey, J.F. The Influence of Aspartic or Glutamic-Acid Residues in Tetrapeptides on the Formation of Complexes with Nickel(II) And Zinc(II). Polyhedron 1995, 14, 211–218. [Google Scholar] [CrossRef]
- Martin, R.B. Nickel ion binding to amino acids and peptides. In Metal Ions in Biological Systems; Sigel, H., Sigel, A., Eds.; Marcel Dekker Inc.: New York, NY, USA, 1987; pp. 124–164. [Google Scholar]
- Kowalik-Jankowska, T.; Kozłowski, H.; Farkas, E.; Sóvágó, I. Nickel Ion Complexes of Amino Acids and Peptides. Nickel Surpris. Impact Nat. 2007, 2, 63–107. [Google Scholar] [CrossRef]
- Rowińska-Żyrek, M.; Witkowska, D.; Bielińska, S.; Kamysz, W.; Kozłowski, H. The–Cys–Cys–motif in Helicobacter pylori’s Hpn and HspA proteins is an essential anchoring site for metal ions. Dalton Trans. 2011, 40, 5604–5610. [Google Scholar] [CrossRef] [PubMed]
- Grasso, G.; Magrì, A.; Bellia, F.; Pietropaolo, A.; La Mendola, D.; Rizzarelli, E. The copper(II) and zinc(II) coordination mode of HExxH and HxxEH motif in small peptides: The role of carboxylate location and hydrogen bonding network. J. Inorg. Biochem. 2014, 130, 92–102. [Google Scholar] [CrossRef]
- Kulon, K.; Valensin, D.; Kamysz, W.; Nadolny, R.; Gaggelli, E.; Valensin, G.; Kozłowski, H. Binding of Ni2+ and Cu2+ ions to peptides with a Cys–His motif. Dalton Trans. 2008, 39, 5323–5330. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad simulation and speciation (HySS): A utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).