Green Synthesized Zinc Oxide Nanoparticles Based on Cestrum diurnum L. of Potential Antiviral Activity against Human Corona 229-E Virus
Abstract
1. Introduction
2. Results and Discussion
2.1. HPLC Analysis of Phenolic Constituents
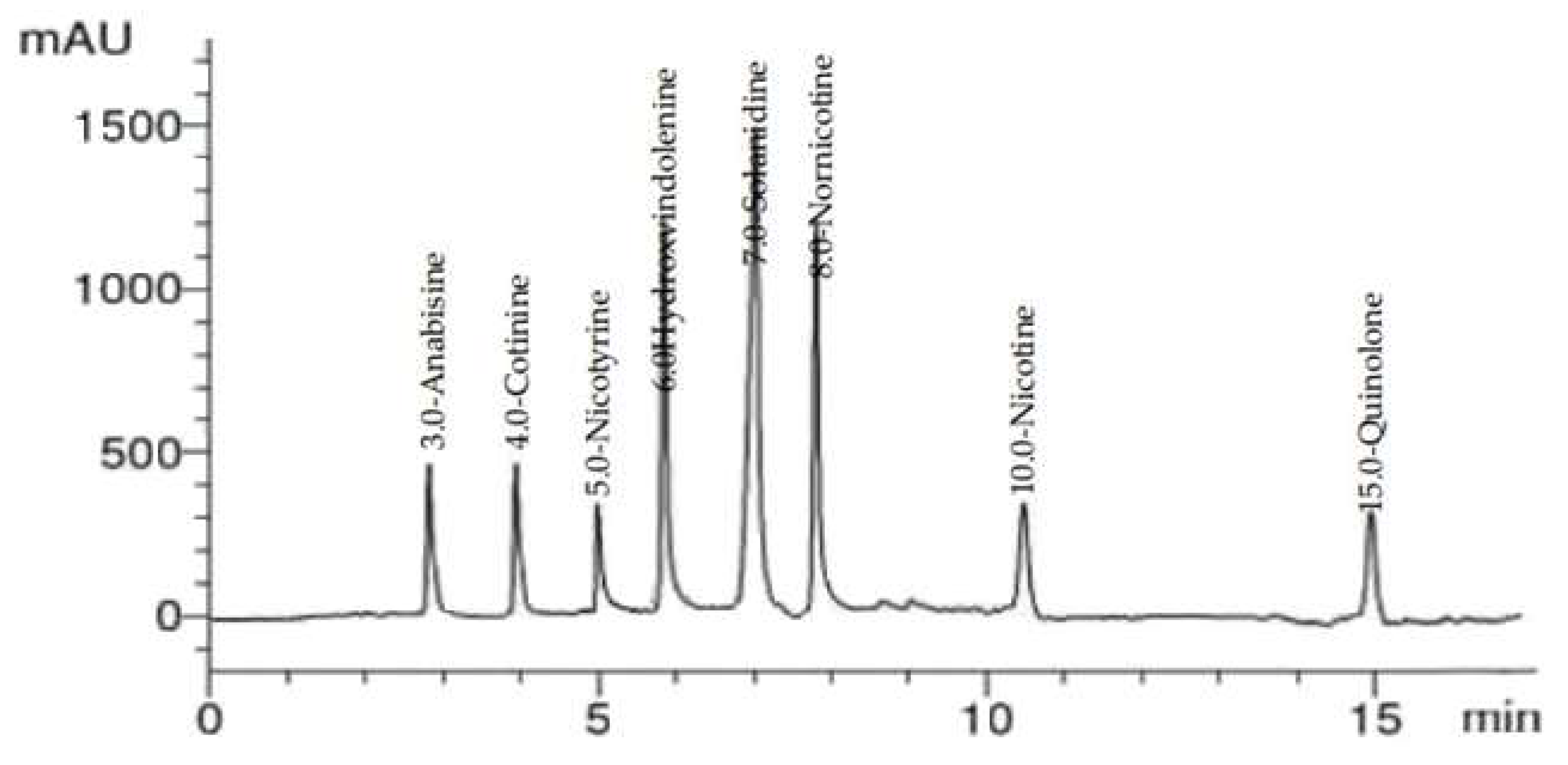
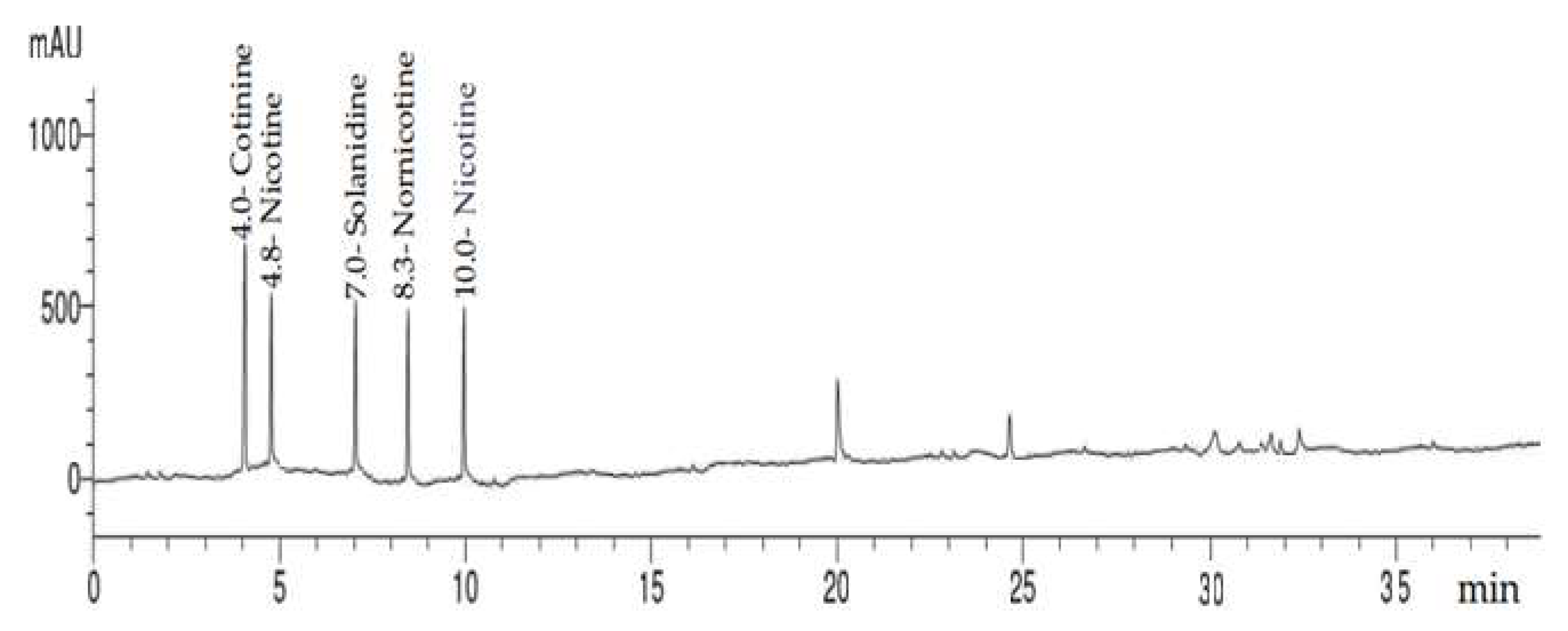
2.2. HPLC Analysis of Alkaloid Constituents
2.3. Characterization of ZnO-Nanoparticles
2.3.1. UV Analysis
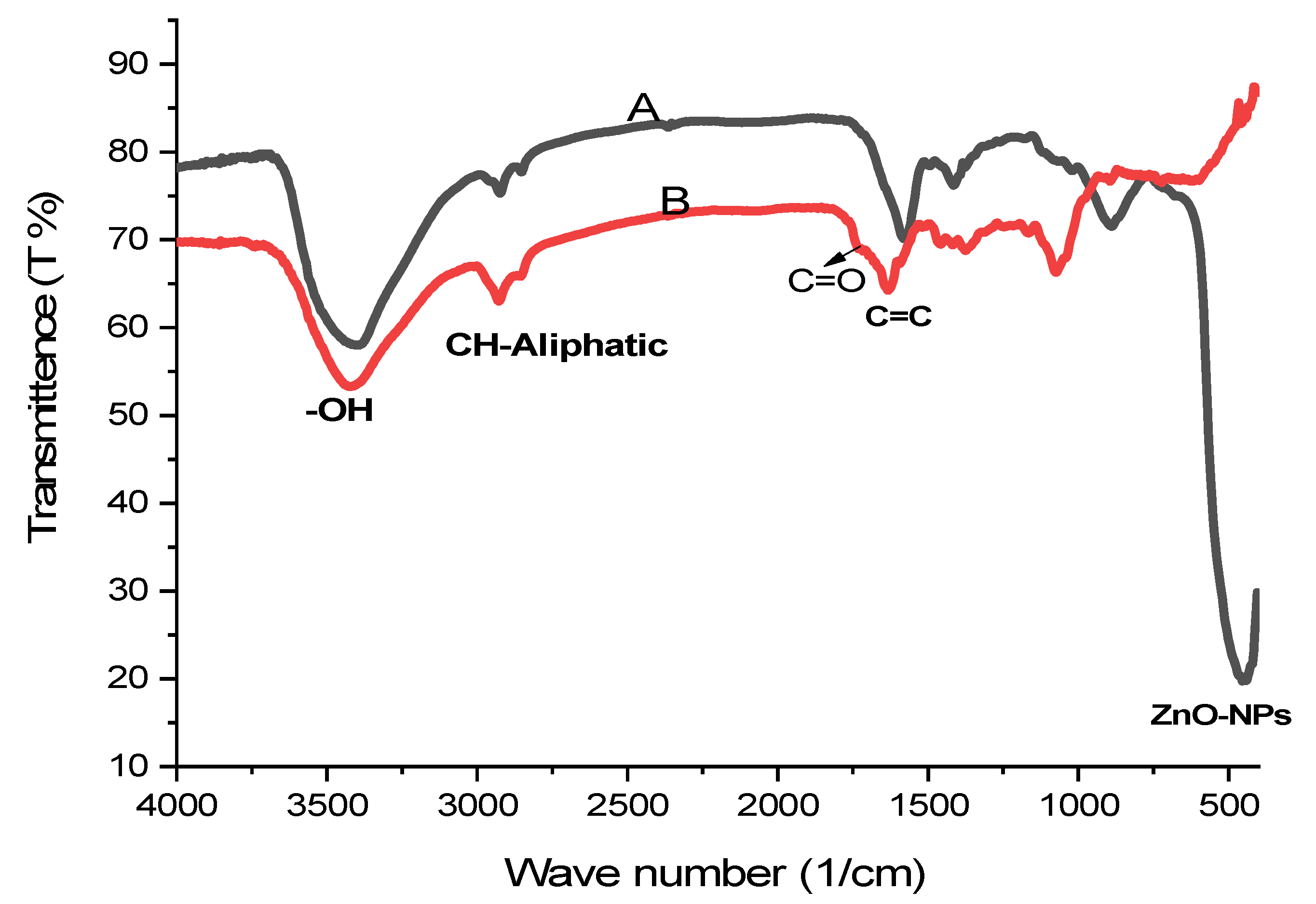
2.3.2. FT-IR Analysis of ZnO-NPs and C. diurnum L. Leaf Extract
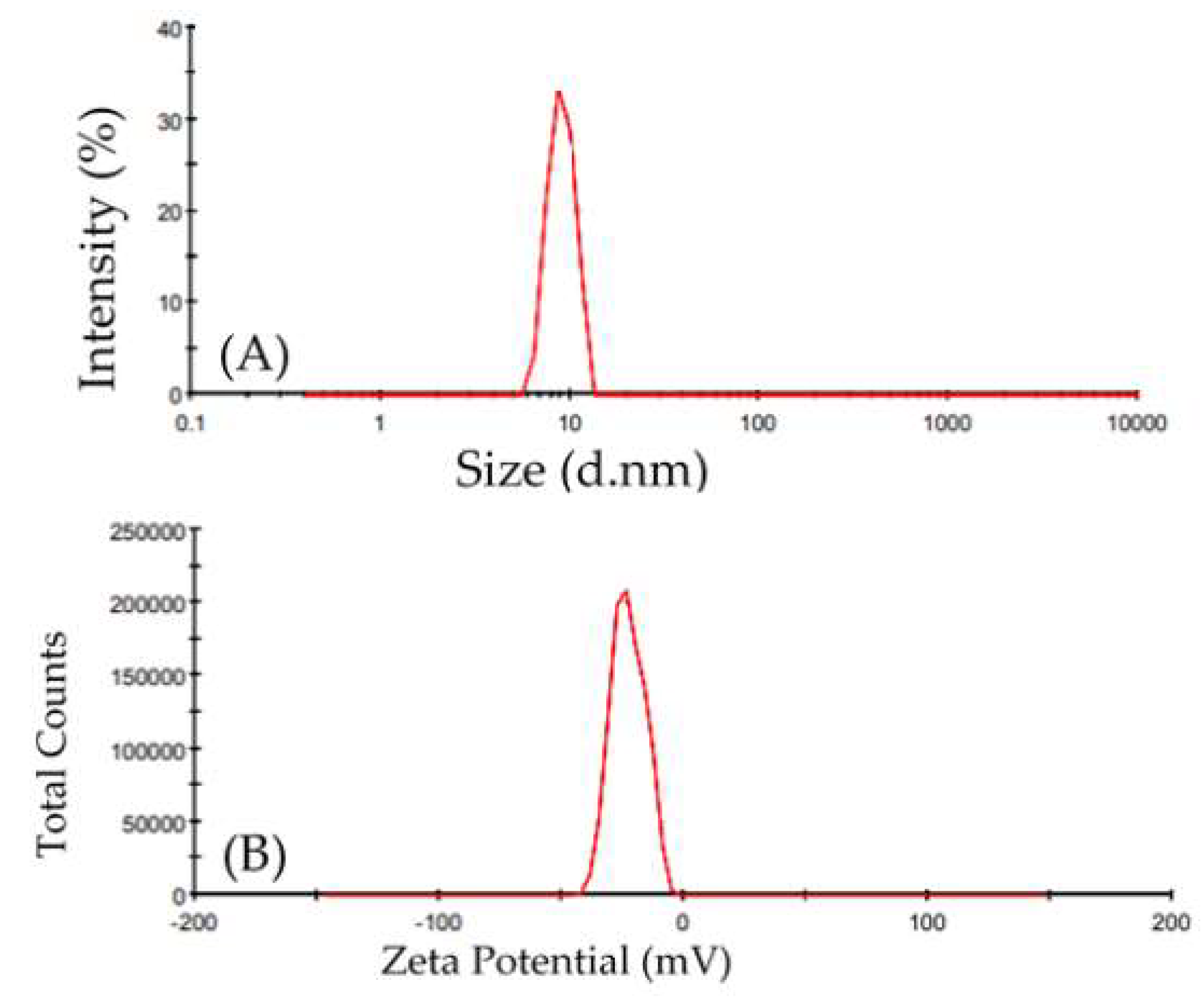
2.3.3. Light Scattering Dynamics and Zeta Potential
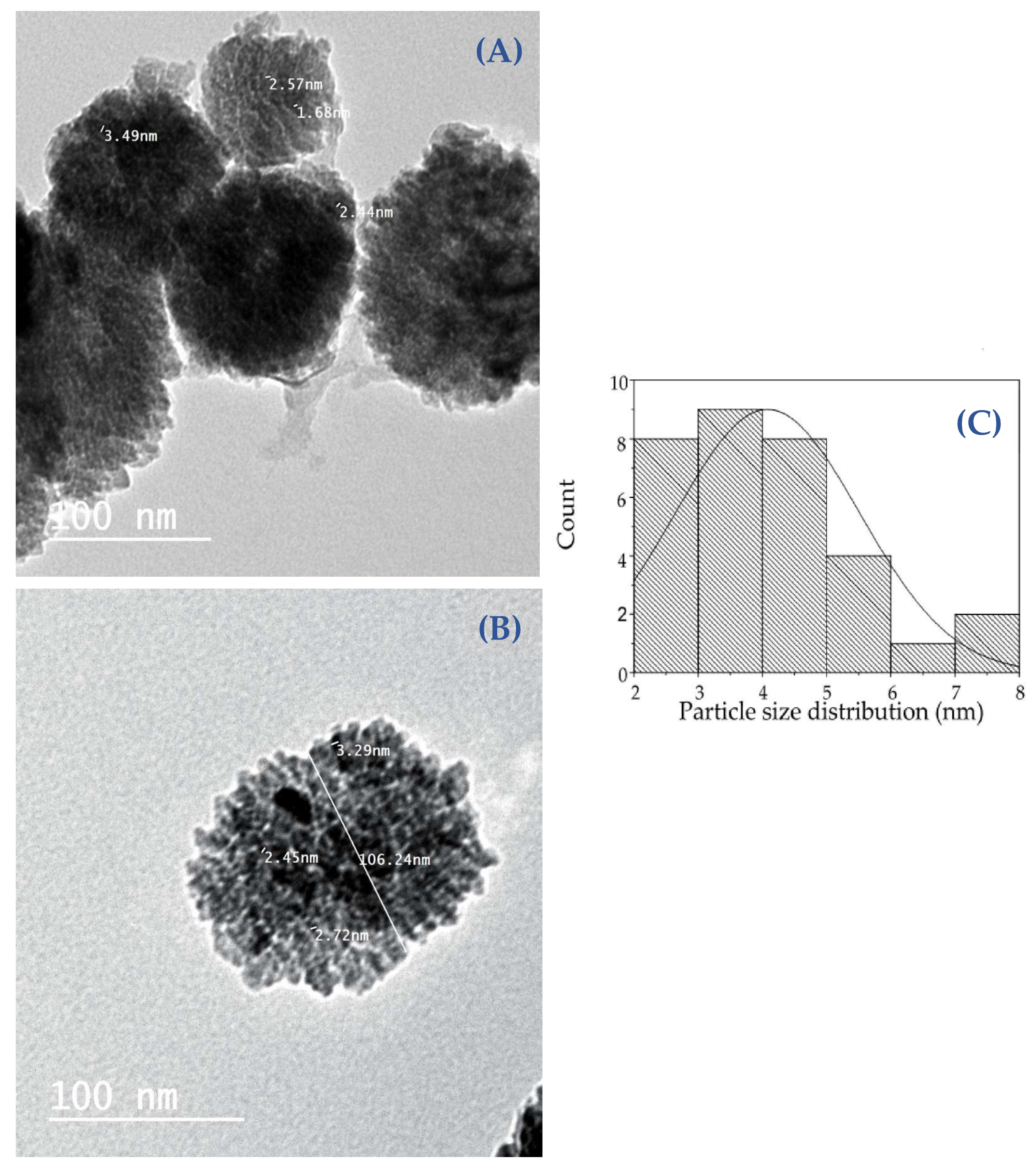
2.3.4. Transmission Electron Microscope (TEM) and Scanning Electron Microscopy (SEM) Analysis
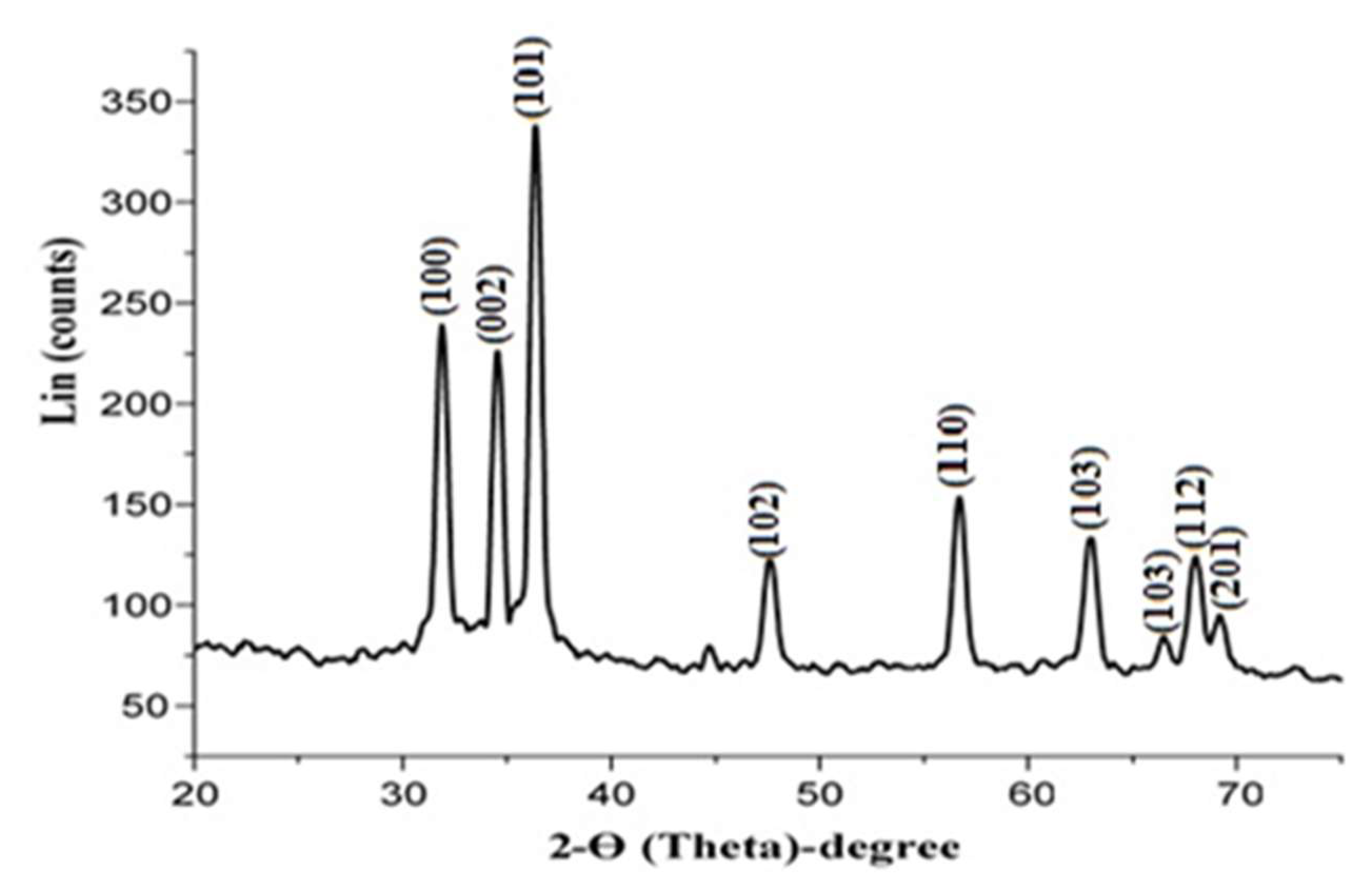
2.3.5. X-ray Diffraction (XRD) Analysis
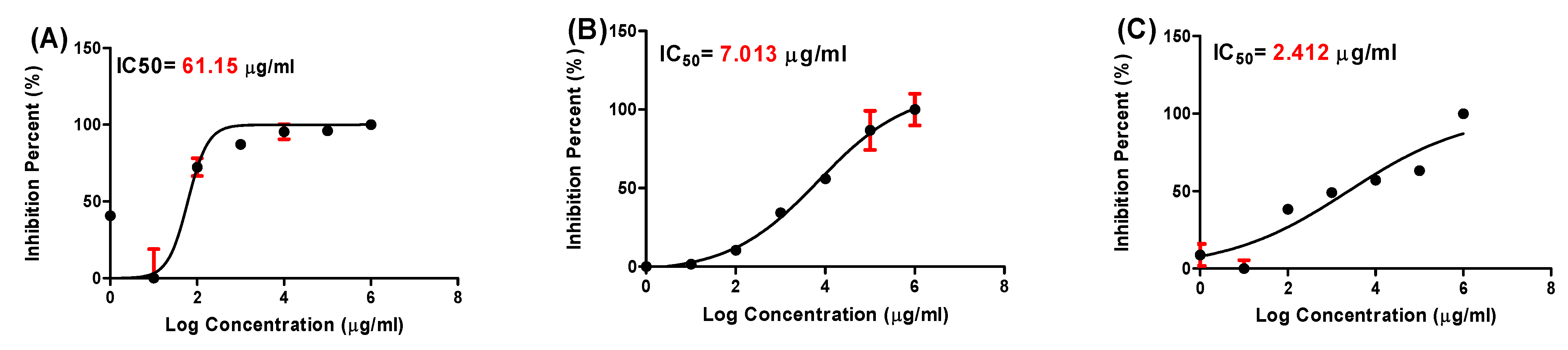
2.4. Antiviral Activity
2.5. In Silico Assessment and Molecular Docking Studies
2.5.1. Docking Studies of Experimental Ligands
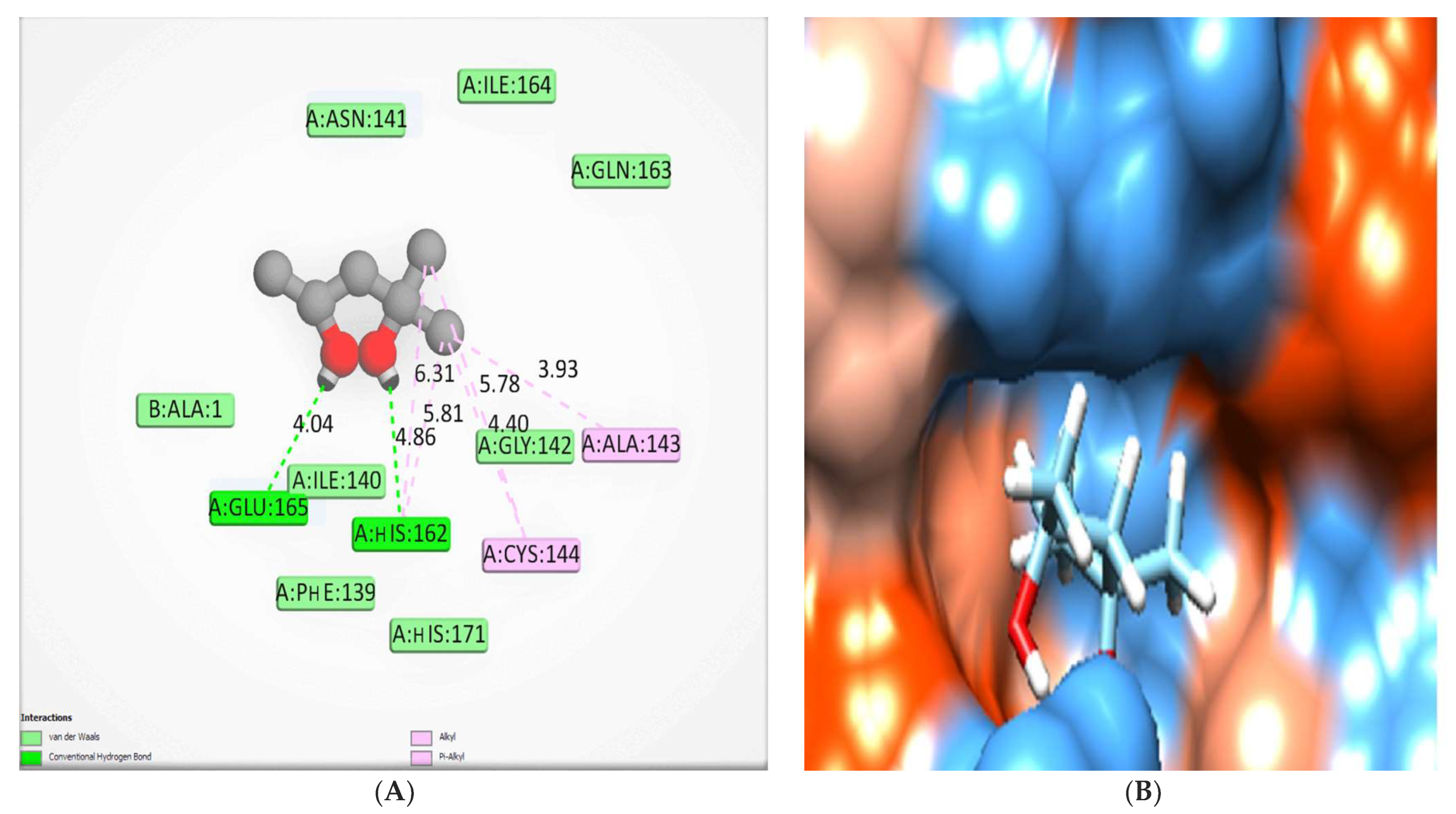
2.5.2. Docking Studies of Phenolic Compounds
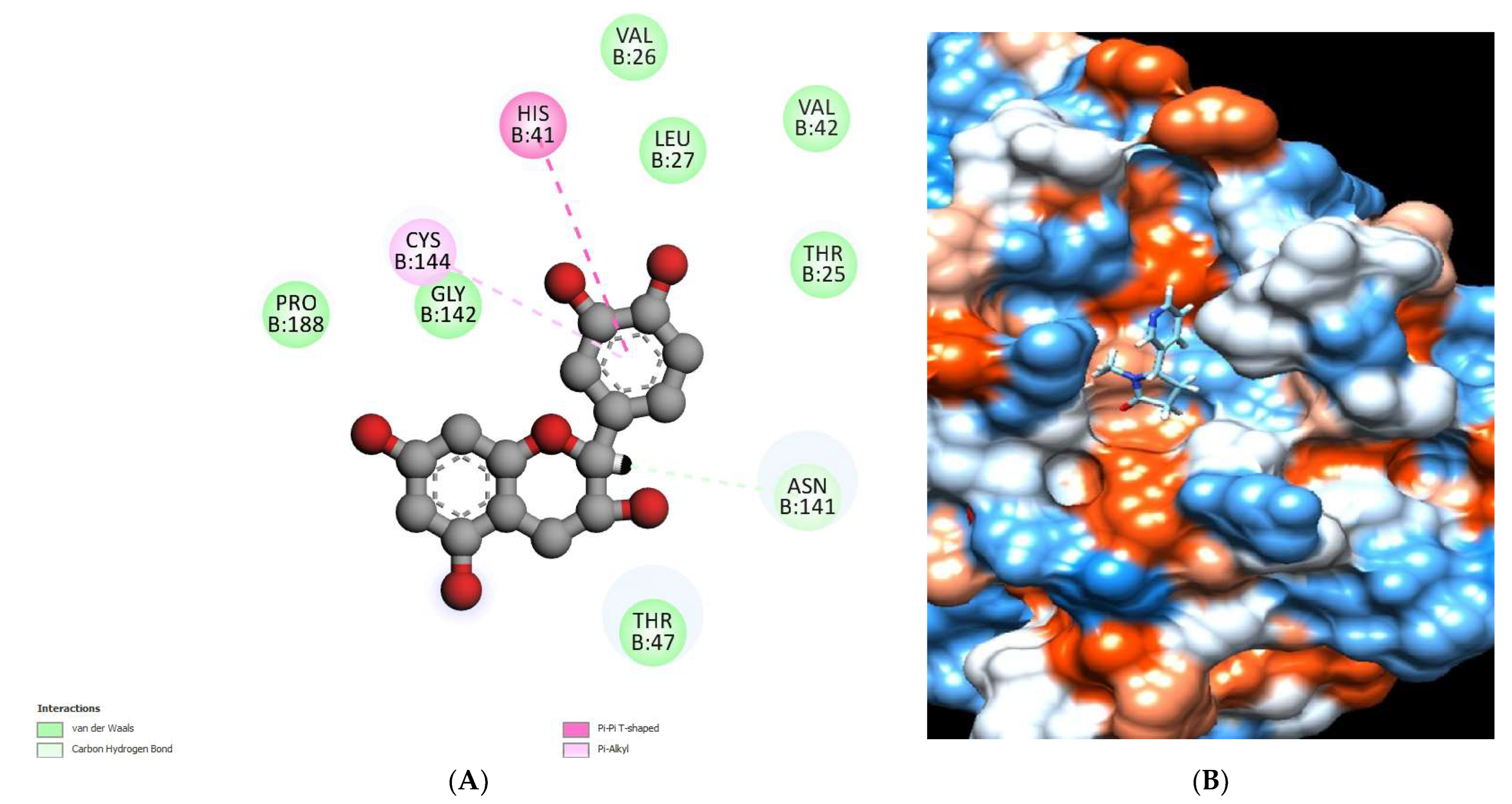
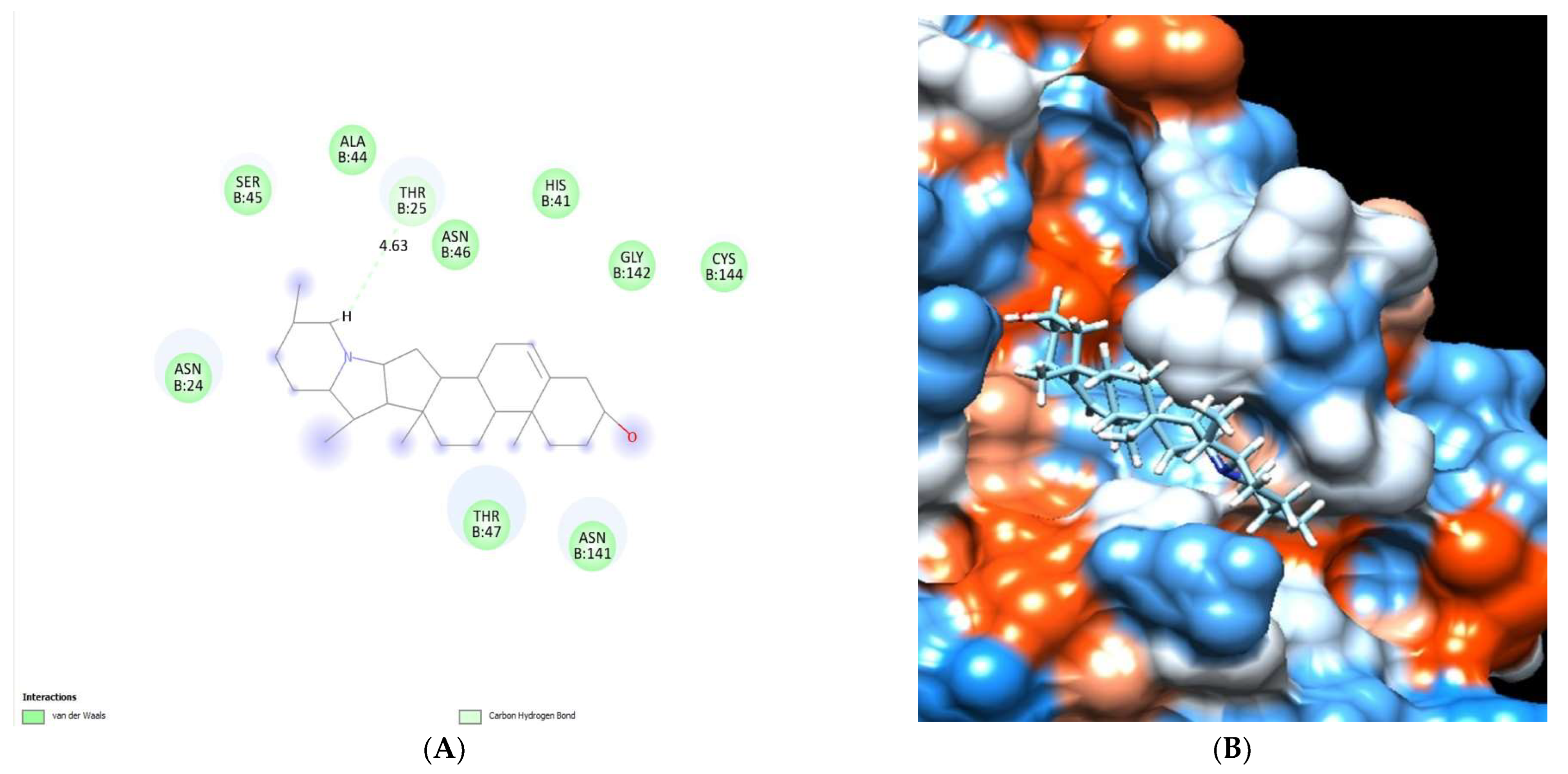
2.5.3. Docking Studies of Alkaloid Compounds
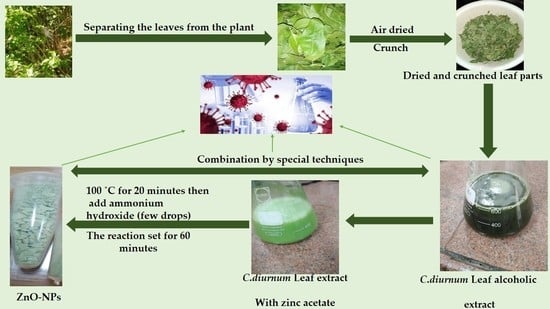
3. Material and Methods
3.1. Plant Material
3.2. Extraction
3.3. HPLC Analysis of Phenolic Constituents
3.4. HPLC Analysis of Alkaloid Constituents
3.5. Green Synthesis of Zinc Oxide Nanoparticles
3.6. Characterization of Zinc Oxide Nanoparticles
3.6.1. V-Vis Spectral Analysis
3.6.2. FT-IR Analysis
3.6.3. Zeta-Sizer Measurements
3.6.4. Transmission Electron Microscopy (TEM) and Scanning Electron Microscopy (SEM) Analysis
3.6.5. X-ray Diffraction (XRD)
3.7. Evaluation of the Antiviral Activity
3.7.1. Cytotoxicity Assessment
3.7.2. Antiviral Assessment
3.8. Mode of Action of Antiviral Activity
3.8.1. Adsorption Mechanism
3.8.2. Replication Mechanism
3.9. In Silico Assessment and Molecular Docking Studies
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Monajjemi, M.; Mollaamin, F.; Shojaei, S. An overview on Coronaviruses family from past to COVID-19: Introduce some inhibitors as antiviruses from Gillan’s plants. Biointerface Res. Appl. Chem. 2020, 3, 5575–5585. [Google Scholar]
- Anand, K.B.; Karade, S.; Sen, S.; Gupta, R.M. SARS-CoV-2: Camazotz’s curse. Med. J. Armed Forces India 2020, 76, 136–141. [Google Scholar] [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encycl. Virol. 2021, 428–440. [Google Scholar] [CrossRef]
- Pene, F.; Merlat, A.; Vabret, A.; Rozenberg, F.; Buzyn, A.; Dreyfus, F.; Lebon, P. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 2003, 37, 929–932. [Google Scholar] [CrossRef] [PubMed]
- Zarei, K.; Farahbakhsh, R.; Crespi, N.; Tyson, G. A first instagram dataset on covid-19. arXiv 2020, arXiv:2004.12226. [Google Scholar]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 128. [Google Scholar] [CrossRef]
- WHO. WHO Coronavirus Disease (COVID-19) Dashboard. 2022. Available online: https://covid19.who.int/?gclid=Cj0KCQiA4L2BBhCvARIsAO0SBdZbO3TKPgQ73OHj0Qvo8Eq_pVbDJcr0xKePgUsABuHCyikOhqQULDQaAtQyEALw_wcB (accessed on 25 August 2022).
- Attia, G.H.; Moemen, Y.S.; Youns, M.; Ibrahim, A.M.; Abdou, R.; El Raey, M.A. Antiviral zinc oxide nanoparticles mediated by hesperidin and in silico comparison study between antiviral phenolics as anti-SARS-CoV-2. Colloids Surf. B: Biointerfaces 2021, 203, 111724. [Google Scholar] [CrossRef]
- Elhawary, S.; Hala, E.H.; Mokhtar, F.A.; Mansour Sobeh, E.M.; Osman, S.; El-Raey, M. Green synthesis of silver nanoparticles using extract of Jasminum officinal l. leaves and evaluation of cytotoxic activity towards bladder (5637) and breast cancer (MCF-7) cell lines. Int. J. Nanomed. 2020, 15, 9771. [Google Scholar] [CrossRef]
- Melk, M.M.; El-Hawary, S.S.; Melek, F.R.; Saleh, D.O.; Ali, O.M.; El Raey, M.A.; Selim, N.M. Nano zinc oxide green-synthesized from plumbago auriculata lam. alcoholic extract. Plants 2021, 10, 2447. [Google Scholar] [CrossRef]
- Kusumoputro, S.; Tseng, S.; Tse, J.; Au, C.; Lau, C.; Wang, X.; Xia, T. Potential nanoparticle applications for prevention, diagnosis, and treatment of COVID-19. View 2020, 1, 20200105. [Google Scholar] [CrossRef]
- Melk, M.M.; El-Hawary, S.S.; Melek, F.R.; Saleh, D.O.; Ali, O.M.; El Raey, M.A.; Selim, N.M. Antiviral Activity of Zinc Oxide Nanoparticles Mediated by Plumbago indica L. Extract against Herpes Simplex Virus Type 1 (HSV-1). Int. J. Nanomed. 2021, 16, 8221. [Google Scholar] [CrossRef] [PubMed]
- Rónavári, A.; Kovács, D.; Igaz, N.; Vágvölgyi, C.; Boros, I.M.; Kónya, Z.; Pfeiffer, I.; Kiricsi, M. Biological activity of green-synthesized silver nanoparticles depends on the applied natural extracts: A comprehensive study. Int. J. Nanomed. 2017, 12, 871. [Google Scholar] [CrossRef] [PubMed]
- Alamdari, S.; Sasani Ghamsari, M.; Lee, C.; Han, W.; Park, H.H.; Tafreshi, M.J.; Afarideh, H.; Ara, M.H.M. Preparation and characterization of zinc oxide nanoparticles using leaf extract of Sambucus ebulus. Appl. Sci. 2020, 10, 3620. [Google Scholar] [CrossRef]
- l-Hawwary, S.S.; Abd Almaksoud, H.M.; Saber, F.R.; Elimam, H.; Sayed, A.M.; El Raey, M.A.; Abdelmohsen, U.R. Green-synthesized zinc oxide nanoparticles, anti-Alzheimer potential and the metabolic profiling of Sabal blackburniana grown in Egypt supported by molecular modelling. RSC Adv. 2021, 11, 18009–18025. [Google Scholar] [CrossRef]
- Alamdari, S.; Mirzaee, O.; Jahroodi, F.N.; Tafreshi, M.J.; Ghamsari, M.S.; Shik, S.S.; Ara, M.H.M.; Lee, K.; Park, H.H. Green synthesis of multifunctional ZnO/chitosan nanocomposite film using wild Mentha pulegium extract for packaging applications. Surf. Interfaces 2022, 34, 102349. [Google Scholar] [CrossRef]
- McClements, D.J.; Xiao, H. Is nano safe in foods? Establishing the factors impacting the gastrointestinal fate and toxicity of organic and inorganic food-grade nanoparticles. Npj Sci. Food 2017, 1, 6. [Google Scholar] [CrossRef]
- Swain, P.S.; Rao, S.B.; Rajendran, D.; Dominic, G.; Selvaraju, S. Nano zinc, an alternative to conventional zinc as animal feed supplement: A review. Anim. Nutr. 2016, 2, 134–141. [Google Scholar] [CrossRef]
- Begum, A.S.; Goyal, M. Phcog Mag.: Review Article Research and Medicinal Potential of the genus Cestrum (Solanaceae)—A Review. Pharmacogn. Rev. 2007, 1, 320–332. [Google Scholar]
- Nasr, S.M.; Elwan, N.M.; Abdel-Motagaly, M.; Abdel-Aziz AW, A.; Ghareeb, M. Phytochemical investigation and differential effects of Cestrum elegans isolated compounds as antimicrobial and virucidal against hepatitis A virus. Egypt. J. Chem. 2021, 64, 3729–3738. [Google Scholar]
- Bhattacharjee, I.; Ghosh, A.; Chandra, G. Antimicrobial activity of the essential oil of Cestrum diurnum (L.) (Solanales: Solanaceae). Afr. J. Biotechnol. 2005, 4, 371–374. [Google Scholar]
- Fouad, M.A.; Mohamed, K.M.; Kamel, M.S.; Matsunami, K.; Otsuka, H. Cesdiurins I–III, steroidal saponins from Cestrum diurnum L. J. Nat. Med. 2008, 62, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Sana, S. SADIQ, SANA Phytochemical and Biological Evaluation of Cestrum diurnum (Solanaceae). Doctoral Dissertation, Bahria University, Islamabad, Pakistan, 2020. [Google Scholar]
- Mohamed, K.M.; Fouad, M.A.; Matsunami, K.; Kamel, M.S.; Otsuka, H. A new norlignan glycoside from Cestrum diurnum L. Arkivoc 2007, 13, 63–70. [Google Scholar] [CrossRef]
- Khatun, A.; Rahman, M.; Nesa, M.; Looi, C.Y.; Wong, W.F.; Hazni, H.; bin Mahdzir, M.A.; Uddin, S.J.; Awang, K.; Shilpi, J.A. Analgesic, anti-inflammatory and NF-κB inhibitory activity of aerial parts of Cestrum diurnum. Clin. Phytosci. 2022, 8, 11. [Google Scholar] [CrossRef]
- Khatun, A.; Chowdhury, U.K.; Jahan, A.; Rahman, M. Cytotoxic and thrombolytic activity of the aerial part of Cestrum diurnum L. (Solanaceae). PharmacolOnline 2014, 1, 109–113. [Google Scholar]
- Lacaille-Dubois, M.A.; Wagner, H. A review of the biological and pharmacological activities of saponins. Phytomedicine 1996, 2, 363–386. [Google Scholar] [CrossRef]
- Badshah, S.L.; Faisal, S.; Muhammad, A.; Poulson, B.G.; Emwas, A.H.; Jaremko, M. Antiviral activities of flavonoids. Biomed. Pharmacother. 2021, 140, 111596. [Google Scholar] [CrossRef]
- MacRae, W.D.; Hudson, J.B.; Towers, G.H.N. The antiviral action of lignans. Planta Med. 1989, 55, 531–535. [Google Scholar] [CrossRef]
- Elkaeed, E.B.; Eissa, I.H.; Elkady, H.; Abdelalim, A.; Alqaisi, A.M.; Alsfouk, A.A.; Elwan, A.; Metwaly, A.M. A Multistage In Silico Study of Natural Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Int. J. Mol. Sci. 2022, 23, 8407. [Google Scholar] [CrossRef]
- Nasr, S.M.; Ghareeb, M.A.; Mohamed, M.A.; Elwan, N.M.; El-WanesAnter Abdel-Aziz, A.; Abdel-Aziz, M.S. High Performance Liquid Chromatography Fingerprint Analyses, In vitro Cytotoxicity, Antimicrobial and Antioxidant Activities of the Extracts of Two Cestrum Species Growing in Egypt. Pharmacogn. Res. 2018, 10, 173–180. [Google Scholar] [CrossRef]
- Doshi, G.M. Pharmacognostic quantification of flavonoids by high performance thin layer chromatography and in vitro cell line study on developed herbal formulation from Cestrum nocturnum plant extract. Int. J. Green Pharm. (IJGP) 2016, 10, 183–188. [Google Scholar]
- Nagels, L.; Van Dongen, W.; Parmentier, F. Cestric acid, a caffeic acid ester from Cestrum euanthes. Phytochemistry 1982, 21, 743–746. [Google Scholar] [CrossRef]
- Halim, A.F.; Collins, R.P.; Berigari, M.S. Alkaloids produced by Cestrum nocturnum and Cestrum diurnum. Planta Med. 1971, 20, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, R.M.; Webster, F.X.; Kiemle, D.J.; Bryce, D.L. Spectrometric Identification of Organic Compounds; John Wiley & Sons: Hoboken, NJ, USA, 2014; pp. 72–125. [Google Scholar]
- Rónavári, A.; Bélteky, P.; Boka, E.; Zakupszky, D.; Igaz, N.; Szerencsés, B.; Pfeiffer, I.; Kónya, Z.; Kiricsi, M. Polyvinyl-Pyrrolidone-Coated Silver Nanoparticles—The Colloidal, Chemical, and Biological Consequences of Steric Stabilization under Biorelevant Conditions. Int. J. Mol. Sci. 2021, 22, 8673. [Google Scholar] [CrossRef] [PubMed]
- Attia, H.G.; Albarqi, H.A.; Said, I.G.; Alqahtani, O.; Raey, M.A.E. Synergistic Effect between Amoxicillin and Zinc Oxide Nanoparticles Reduced by Oak Gall Extract against Helicobacter pylori. Molecules 2022, 27, 4559. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, A.A.; El Raey, M.A.; Abdelsalam, E.; Ibrahim, A.M.; Alqahtani, O.; Torky, Z.A.; Attia, H.G. The Biosynthesized Zinc Oxide Nanoparticles’ Antiviral Activity in Combination with Pelargonium zonale Extract against the Human Corona 229E Virus. Molecules 2022, 27, 8362. [Google Scholar] [CrossRef] [PubMed]
- AbouAitah, K.; Allayh, A.K.; Wojnarowicz, J.; Shaker, Y.M.; Swiderska-Sroda, A.; Lojkowski, W. Nanoformulation Composed of Ellagic Acid and Functionalized Zinc Oxide Nanoparticles Inactivates DNA and RNA Viruses. Pharmaceutics 2021, 13, 2174. [Google Scholar] [CrossRef] [PubMed]
- Adem, Ş.; Eyupoglu, V.; Sarfraz, I.; Rasul, A.; Zahoor, A.F.; Ali, M.; Abdalla, M.; Ibrahim, I.M.; Elfiky, A.A. Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19. Phytomedicine 2021, 85, 153310. [Google Scholar] [CrossRef] [PubMed]
- Bailly, F.; Cotelle, P. Anti-HIV activities of natural antioxidant caffeic acid derivatives: Toward an antiviral supplementation diet. Curr. Med. Chem. 2005, 12, 1811–1818. [Google Scholar] [CrossRef]
- Ikeda, K.; Tsujimoto, K.; Uozaki, M.; Nishide, M.; Suzuki, Y.; Koyama, A.H.; Yamasaki, H. Inhibition of multiplication of herpes simplex virus by caffeic acid. Int. J. Mol. Med. 2011, 28, 595–598. [Google Scholar]
- Wang, G.F.; Shi, L.P.; Ren, Y.D.; Liu, Q.F.; Liu, H.F.; Zhang, R.J.; Li, Z.; Zhu, F.; He, P.; Tang, W.; et al. Anti-hepatitis B virus activity of chlorogenic acid, quinic acid and caffeic acid in vivo and in vitro. Antivir. Res. 2009, 83, 186–190. [Google Scholar] [CrossRef]
- Wang, W.X.; Zhang, Y.R.; Luo, S.Y.; Zhang, Y.S.; Zhang, Y.; Tang, C. Chlorogenic acid, a natural product as potential inhibitor of COVID-19: Virtual screening experiment based on network pharmacology and molecular docking. Nat. Prod. Res. 2022, 36, 2580–2584. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Cao, Z.; Cao, L.; Ding, G.; Wang, Z.; Xiao, W. Antiviral activity of chlorogenic acid against influenza A (H1N1/H3N2) virus and its inhibition of neuraminidase. Sci. Rep. 2017, 7, 45723. [Google Scholar] [CrossRef] [PubMed]
- Gravina, H.D.; Tafuri, N.F.; Júnior, A.S.; Fietto, J.L.R.; Oliveira, T.T.; Diaz, M.A.N.; Almeida, M.R. In vitro assessment of the antiviral potential of trans-cinnamic acid, quercetin and morin against equid herpesvirus 1. Res. Vet. Sci. 2011, 91, e158–e162. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, Z.; Pan, P.; Lao, Z.; Xu, J.; Li, Z.; Li, G. Cinnamic acid inhibits Zika virus by inhibiting RdRp activity. Antivir. Res. 2021, 192, 105117. [Google Scholar] [CrossRef] [PubMed]
- Elebeedy, D.; Elkhatib, W.F.; Kandeil, A.; Ghanem, A.; Kutkat, O.; Alnajjar, R.; Saleh, M.A.; el Maksoud, A.I.A.; Badawy, I.; Al-Karmalawy, A.A. Anti-SARS-CoV-2 activities of tanshinone IIA, carnosic acid, rosmarinic acid, salvianolic acid, baicalein, and glycyrrhetinic acid between computational and in vitro insights. RSC Adv. 2021, 11, 29267–29286. [Google Scholar] [CrossRef]
- Ibrahim, M.A.; Mohamed, E.A.; Abdelrahman, A.H.; Allemailem, K.S.; Moustafa, M.F.; Shawky, A.M.; Mahzari, A.; Hakami, A.R.; Abdeljawaad, K.A.A.; Atia, M.A. Rutin and flavone analogs as prospective SARS-CoV-2 main protease inhibitors: In silico drug discovery study. J. Mol. Graph. Model. 2021, 105, 107904. [Google Scholar] [CrossRef]
- Orfali, R.; Rateb, M.E.; Hassan, H.M.; Alonazi, M.; Gomaa, M.R.; Mahrous, N.; Sayed, A.M. Sinapic Acid Suppresses SARS CoV-2 Replication by Targeting Its Envelope Protein. Antibiotics 2021, 10, 420. [Google Scholar] [CrossRef]
- Keivan, Z.; Teoh, B.T.; Sam, S.S.; Wong, P.F.; Mustafa, M.R.; AbuBakar, S. In vitro antiviral activity of fisetin, rutin and naringenin against dengue virus type-2. J. Med. Plants Res. 2011, 5, 5534–5539. [Google Scholar]
- You, H.L.; Huang, C.C.; Chen, C.J.; Chang, C.C.; Liao, P.L.; Huang, S.T. Anti-pandemic influenza A (H1N1) virus potential of catechin and gallic acid. J. Chin. Med. Assoc. 2018, 81, 458–468. [Google Scholar] [CrossRef]
- Kim, K.H.; Tsao, R.; Yang, R.; Cui, S.W. Phenolic acid profiles and antioxidant activities of wheat bran extracts and the effect of hydrolysis conditions. Food Chem. 2006, 95, 466–473. [Google Scholar] [CrossRef]
- Hayden, F.G.; Cote, K.M.; Douglas, R.G. Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob. Agents Chemother. 1980, 17, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Kuo, C.J.; Ko, T.P.; Hsu, M.F.; Tsui, Y.C.; Chang, S.C.; Yang, S.; Chen, S.; Chen, H.; Hsu, N.; et al. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J. Biol. Chem. 2009, 284, 7646–7655. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Suppl. 2), W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sharma, A.; Gupta, U. Molecular Docking studies on the Anti-fungal activity of Allium sativum (Garlic) against Mucormycosis (black fungus) by BIOVIA discovery studio visualizer 21.1. 0.0. Ann. Antivir. Antiretrovir. 2021, 5, 28–32. [Google Scholar]
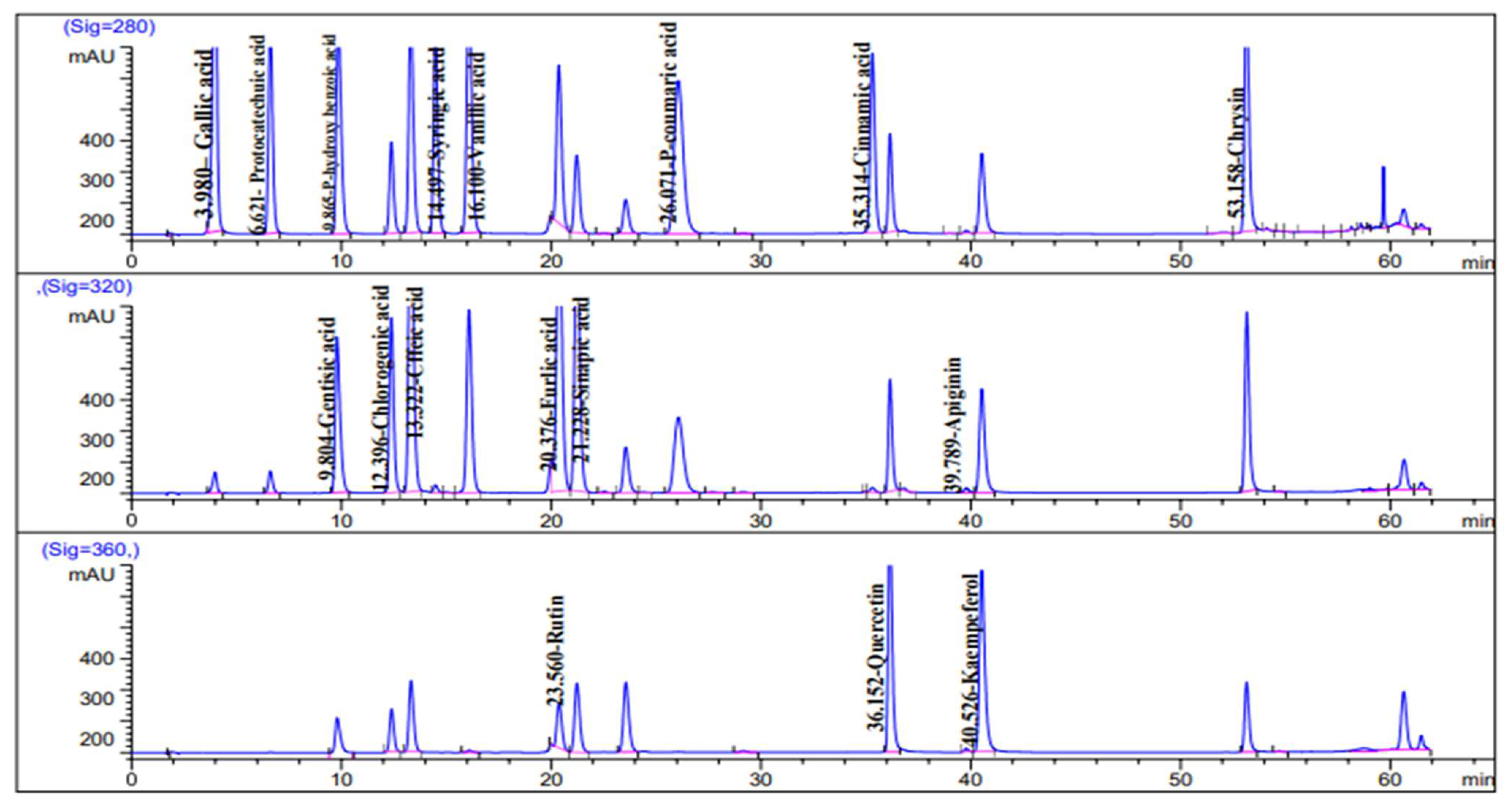
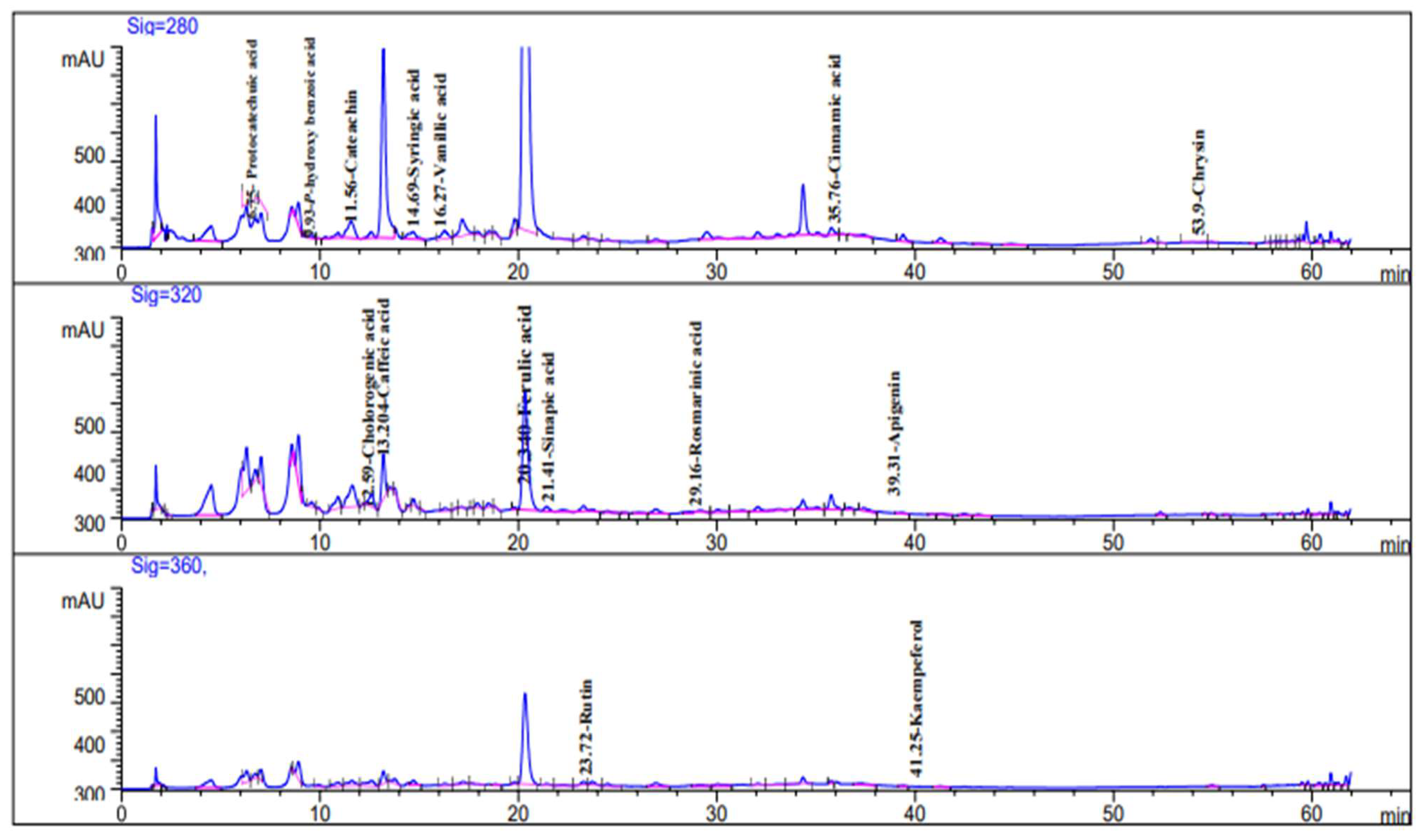

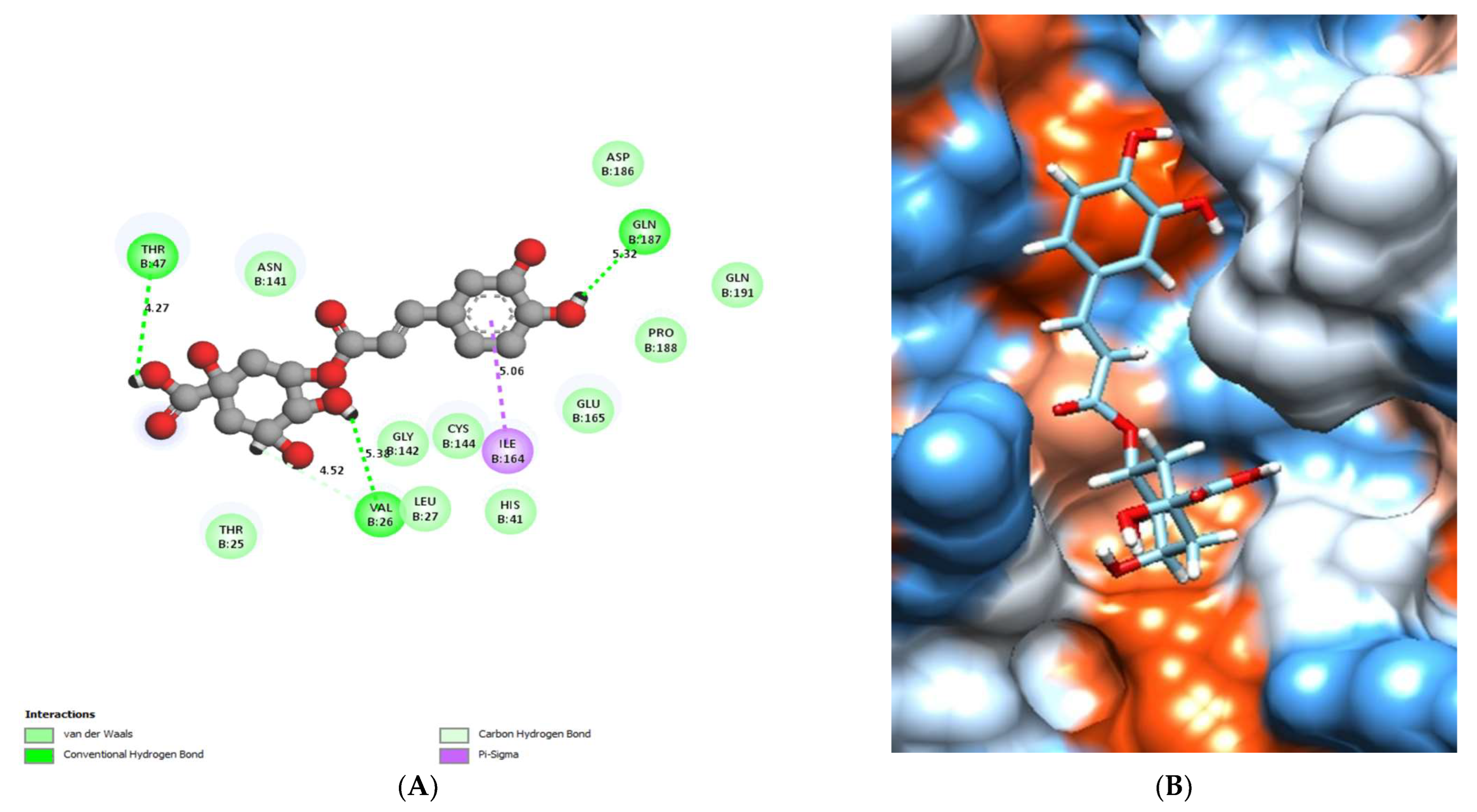

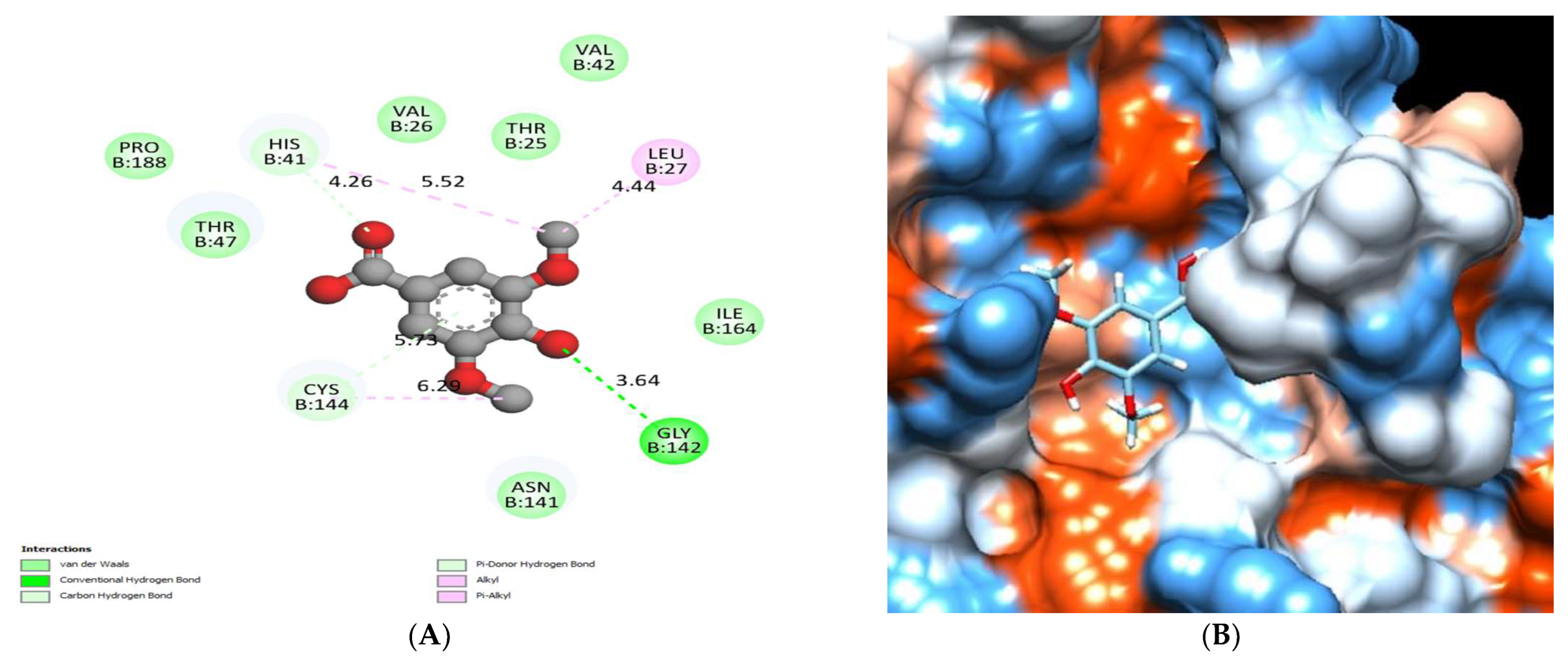
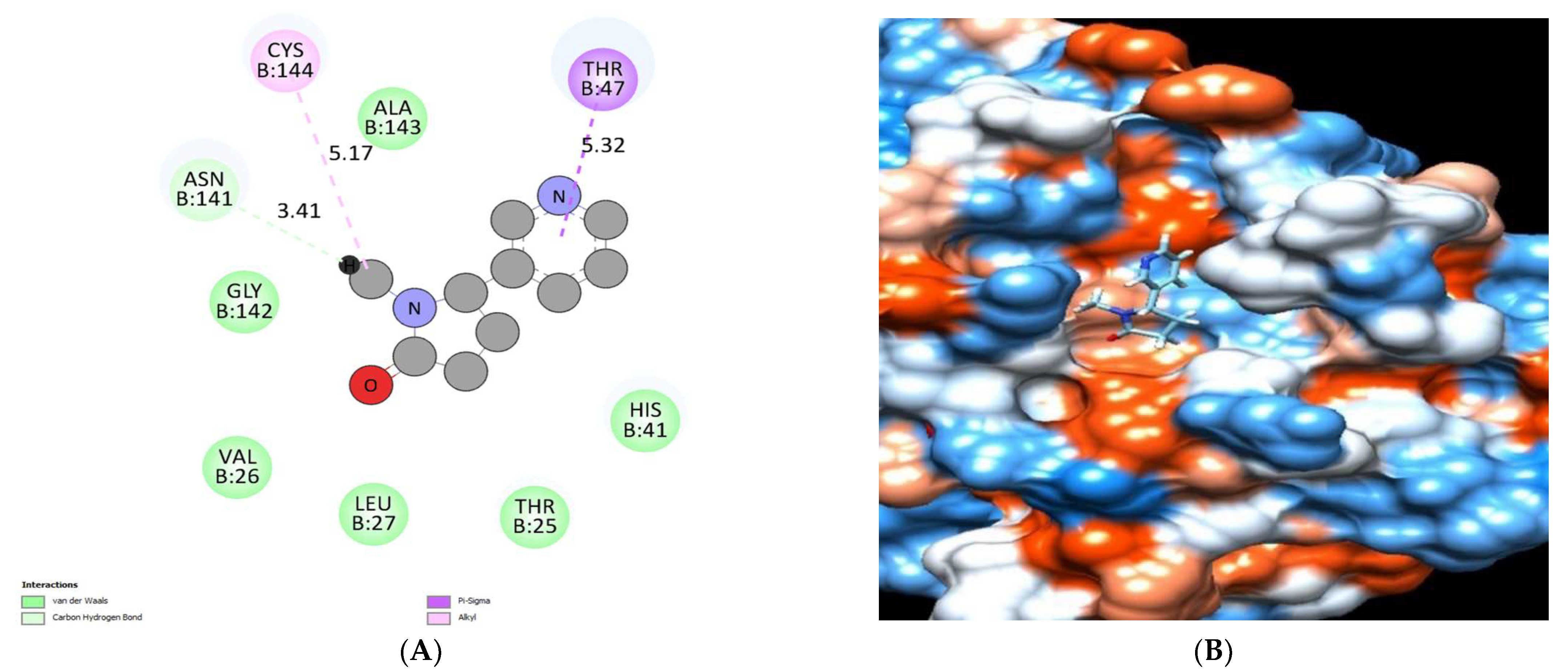
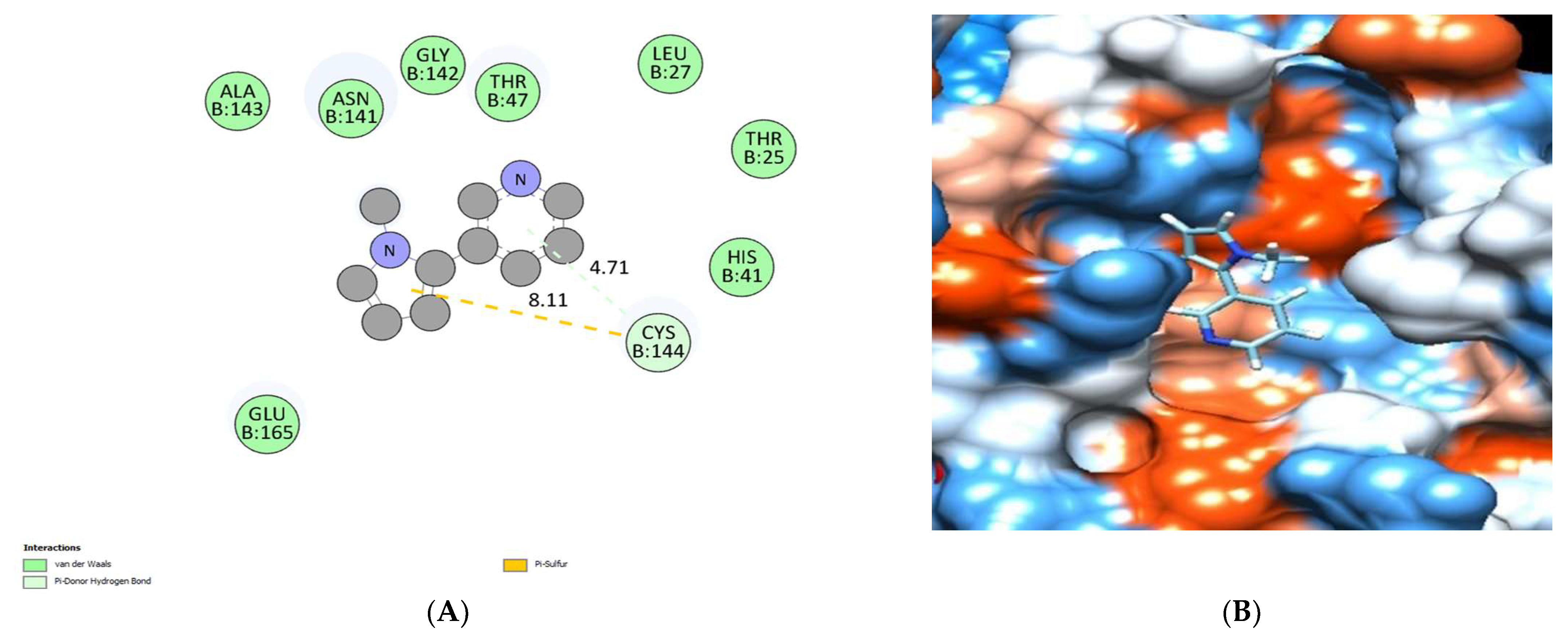
| No. | Compound | Rt * | RRT * | Area% | Conc (µg/g) |
|---|---|---|---|---|---|
| 1 | Protocatechuic acid | 6.751 | 0.58 | 1.649 | 91.02 |
| 2 | P-hydroxy benzoic acid | 9.935 | 0.86 | 0.1323 | 6.44 |
| 3 | Catechin | 11.566 | 1.0 | 10.656 | 1425.16 |
| 4 | Chlorogenic acid | 12.194 | 1.05 | 0.193 | 207.46 |
| 5 | Caffeic acid | 13.204 | 1.14 | 12.490 | 13.81 |
| 6 | Syringic acid | 14.698 | 1.27 | 5.0717 | 158.95 |
| 7 | Vanillic acid | 16.279 | 1.40 | 4.515 | 91.14 |
| 8 | Ferulic acid | 20.340 | 1.76 | 56.479 | 1004.68 |
| 9 | Sinapic acid | 21.419 | 1.85 | 1.954 | 38.16 |
| 10 | Rutin | 23.287 | 2.01 | 1.139 | 78.70 |
| 11 | Rosmarinic acid | 29.166 | 2.52 | 0.972 | 90.56 |
| 12 | Cinnamic acid | 35.768 | 3.09 | 3.051 | 25.74 |
| 13 | Apigenin | 39.371 | 2.97 | 0.808 | 16.64 |
| 14 | Kaempferol | 41.255 | 3.567 | 0.343 | 8.66 |
| 15 | Chrysin | 53.943 | 4.66 | 0.539 | 5.92 |
| No. | Compound | RT | RRT * | Area% | Conc (µg/g) |
|---|---|---|---|---|---|
| 1 | Cotinine | 4.0 | 1.0 | 25.70 | 10.66 |
| 2 | Nicotyrine | 4.8 | 1.2 | 20.39 | 9.74 |
| 3 | Solanidine | 7.0 | 1.75 | 16.69 | 9.65 |
| 4 | Nornicotine | 8.3 | 2.08 | 12.42 | 8.89 |
| 5 | Nicotine | 10.0 | 2.5 | 14.87 | 8.55 |
| Tested Samples | CC50 | IC50 | SI |
|---|---|---|---|
| Leaf extract | 237.68 | 61.15 | 3.89 |
| ZnO-NPs | 145.87 | 7.01 | 20.81 |
| Combination (leaf + ZnO-NPs) | 179.23 | 2.41 | 74.37 |
| Mechanism | Sample Concentration (µg/µL) | Virus Control Titer (PFU/mL) | Virus Titer before and Post Treatment (PFU/mL) | Viral Inhibition (%) |
|---|---|---|---|---|
| Replication | 100 | 7.0 × 103 | 1.9 × 103 | 72.8 |
| 50 | 3.1 × 103 | 55.7 | ||
| 25 | 5.7 × 103 | 18.5 | ||
| Adsorption | 100 | 7.0 × 103 | 5.8 × 103 | 17.2 |
| 50 | 6.7 × 103 | 4.3 | ||
| 25 | 7.0 × 103 | 0 | ||
| Virucidal | 100 | 7.0 × 103 | 3.0 × 103 | 57 |
| 50 | 4.9 × 103 | 30 | ||
| 25 | 5.8 × 103 | 17.2 |
| Compounds | Docking Score (kcal/mol) | Interactions | ||
|---|---|---|---|---|
| H. B. | Pi Interactions | Van der Waals | ||
| Crystal Ligand (MDP) | −5.66 | His162 and Glu165. | Ala143, Cys144 and His162. | Ala1, Phe139, Ile140, Gly142, Gln163 and His171. |
| Catechin | −6.52 | - | His41 and Cys144. | Thr25, Val26, Leu27, Val42, Asn141, Gly142 and Pro188. |
| Chlorogenic acid | −7.13 | Val 26, Thr47 and Gln187 | Ile164 | Thr25, Val 26, Leu27, His41, Gly142, Cys144, Glu165, Pro188 and Gln191. |
| Ferulic acid | −6.00 | Val 26. | Pro188. | Thr 25, Val 26, Leu27, His41, Thr 47, Asn141, Gly142, Cys144, Ile164 and Gln187. |
| Syringic acid | −6.25 | Gly142 | Leu27, His41 and Cys144. | Thr 25, Val 26, Thr 47, Ile164 and Pro188. |
| Cotinine | −6.12 | Asn141 | Thr47 and Cys144 | Thr 25, Val 26, Leu27, His41 Asn141, Gly142 and Ala143. |
| Nicotyrine | −5.75 | Cys144 | Cys144 | Thr 25, Leu27, His41, Thr 47, Asn141, Gly142, Ala143, Cys144 and Glu165. |
| Solanidine | −6.95 | Thr 25 | - | Asn24, Thr 25, His41, Ala44, Ser45, Asn46, Thr47 Asn141, Gly142 and Cys144. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alrabayah, I.N.; Elhawary, S.S.; Kandil, Z.A.; El-Kadder, E.M.A.; Moemen, Y.S.; Saleh, A.M.; El Raey, M.A. Green Synthesized Zinc Oxide Nanoparticles Based on Cestrum diurnum L. of Potential Antiviral Activity against Human Corona 229-E Virus. Molecules 2023, 28, 266. https://doi.org/10.3390/molecules28010266
Alrabayah IN, Elhawary SS, Kandil ZA, El-Kadder EMA, Moemen YS, Saleh AM, El Raey MA. Green Synthesized Zinc Oxide Nanoparticles Based on Cestrum diurnum L. of Potential Antiviral Activity against Human Corona 229-E Virus. Molecules. 2023; 28(1):266. https://doi.org/10.3390/molecules28010266
Chicago/Turabian StyleAlrabayah, Ibrahim N., Seham S. Elhawary, Zeinab A. Kandil, Essam M. Abd El-Kadder, Yasmine S. Moemen, Abdulrahman M. Saleh, and Mohamed A. El Raey. 2023. "Green Synthesized Zinc Oxide Nanoparticles Based on Cestrum diurnum L. of Potential Antiviral Activity against Human Corona 229-E Virus" Molecules 28, no. 1: 266. https://doi.org/10.3390/molecules28010266
APA StyleAlrabayah, I. N., Elhawary, S. S., Kandil, Z. A., El-Kadder, E. M. A., Moemen, Y. S., Saleh, A. M., & El Raey, M. A. (2023). Green Synthesized Zinc Oxide Nanoparticles Based on Cestrum diurnum L. of Potential Antiviral Activity against Human Corona 229-E Virus. Molecules, 28(1), 266. https://doi.org/10.3390/molecules28010266