Expression and Molecular Modification of Chitin Deacetylase from Streptomyces bacillaris
Abstract
1. Introduction
2. Results
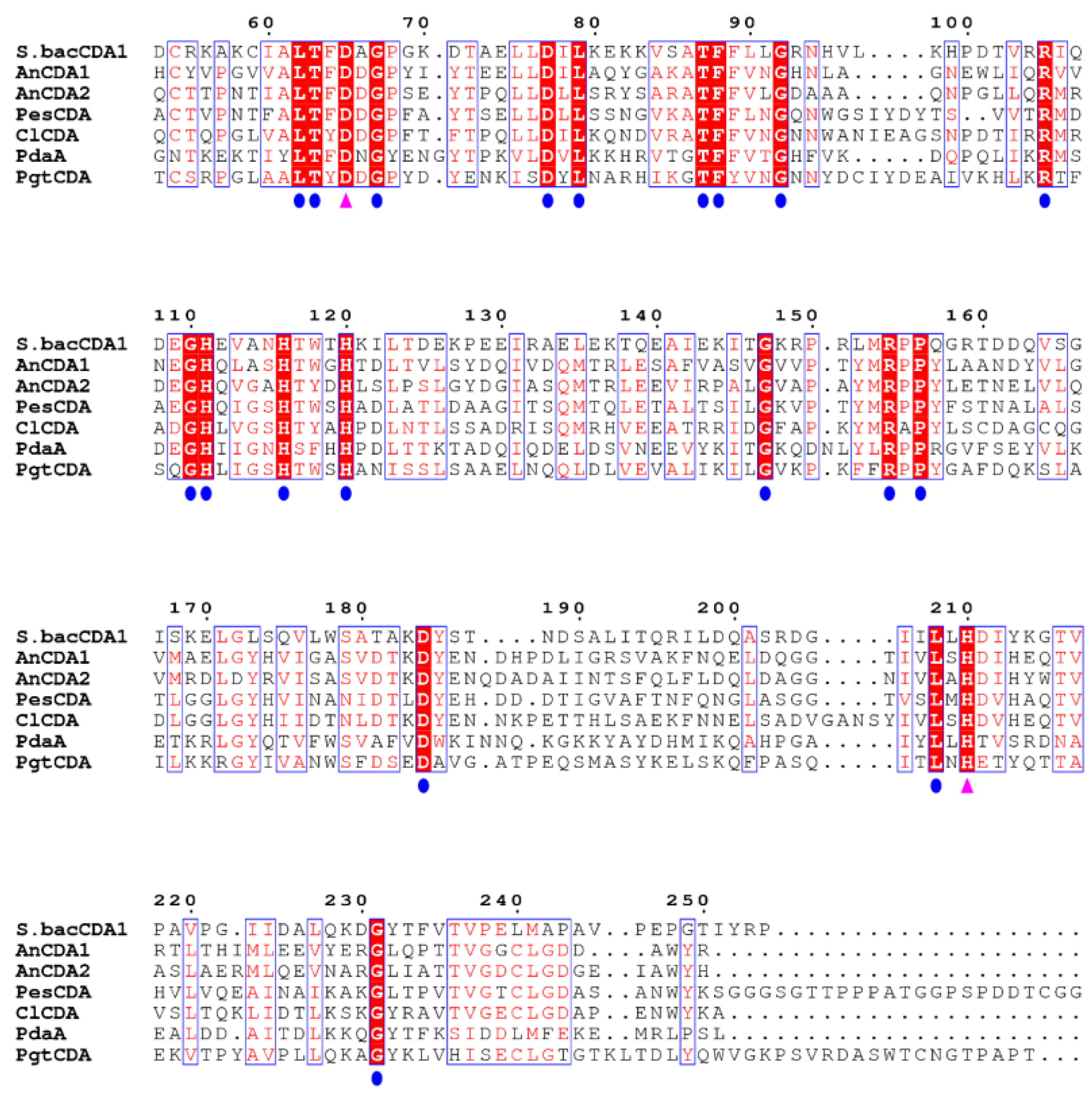
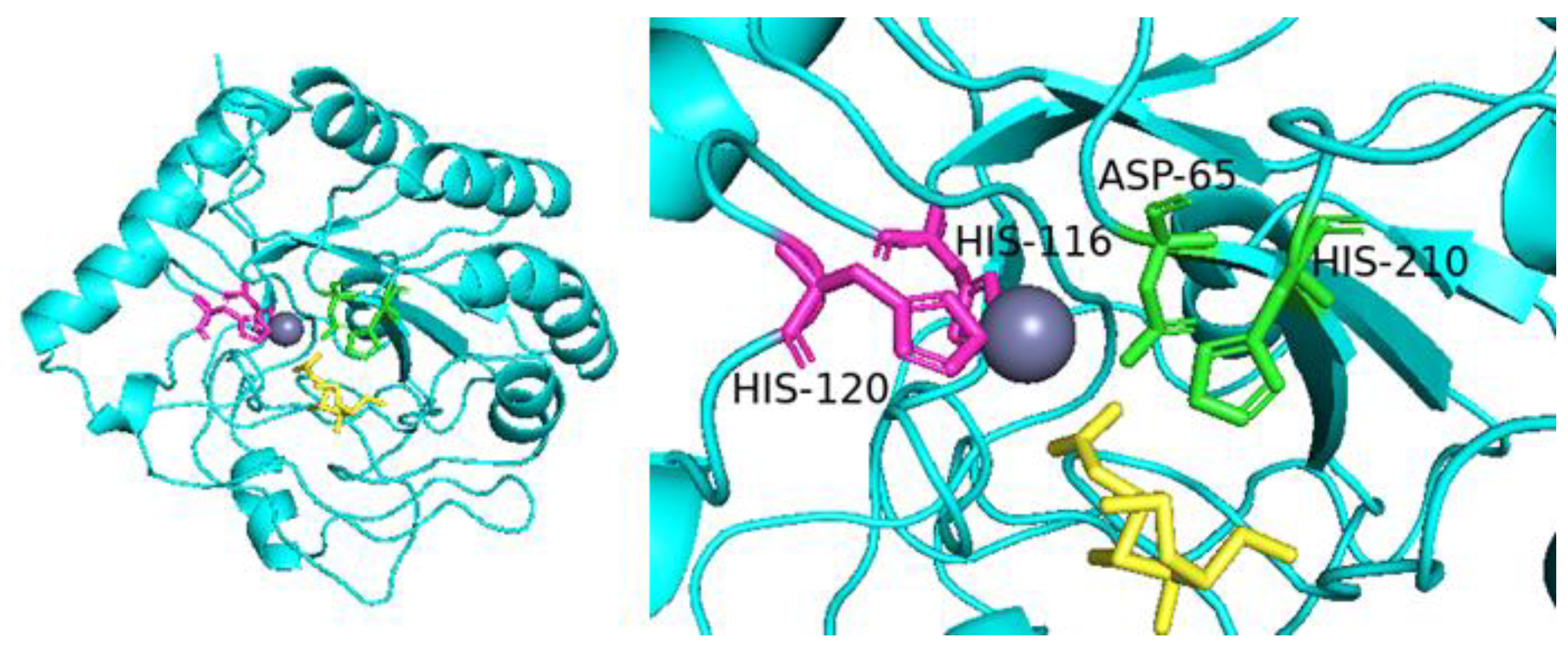
2.1. Bioinformatics Analysis of SbCDA
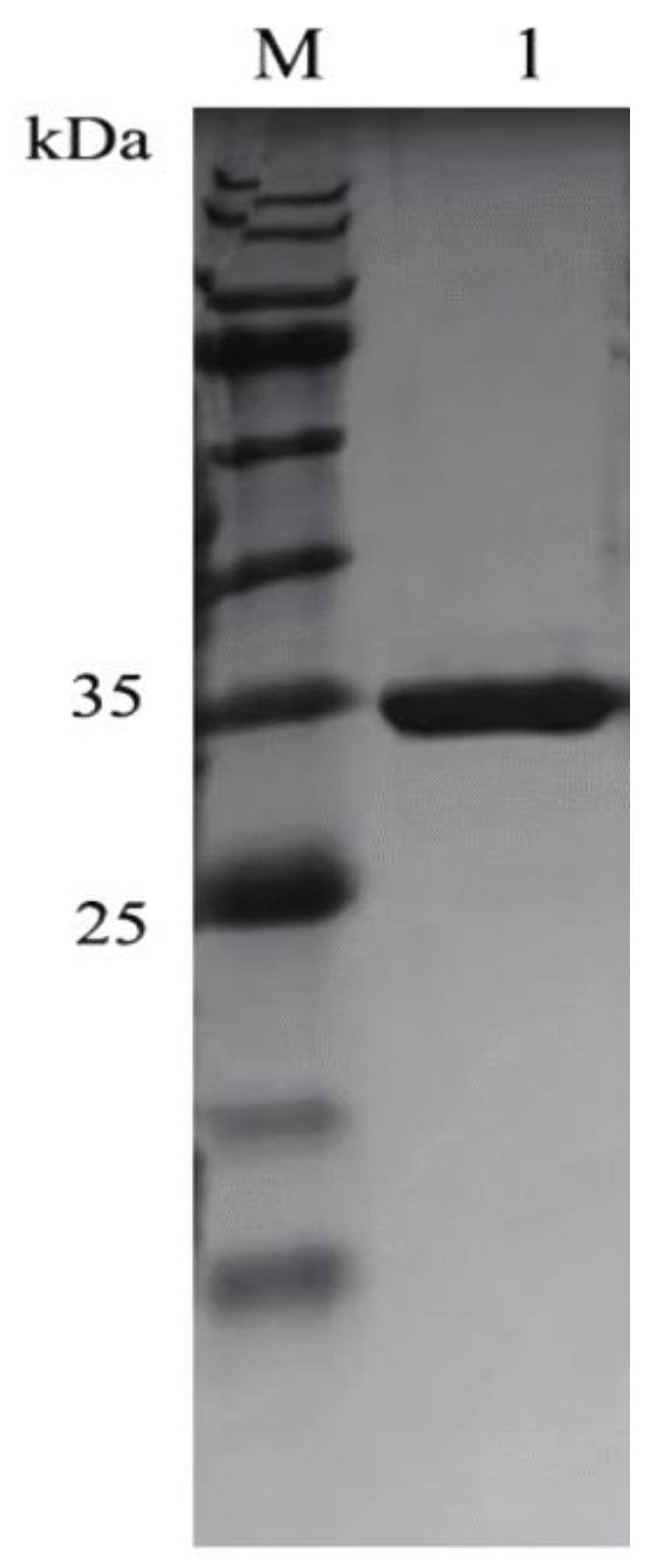
2.2. Expression and Purification of SbCDA
2.3. Characterization of SbCDA
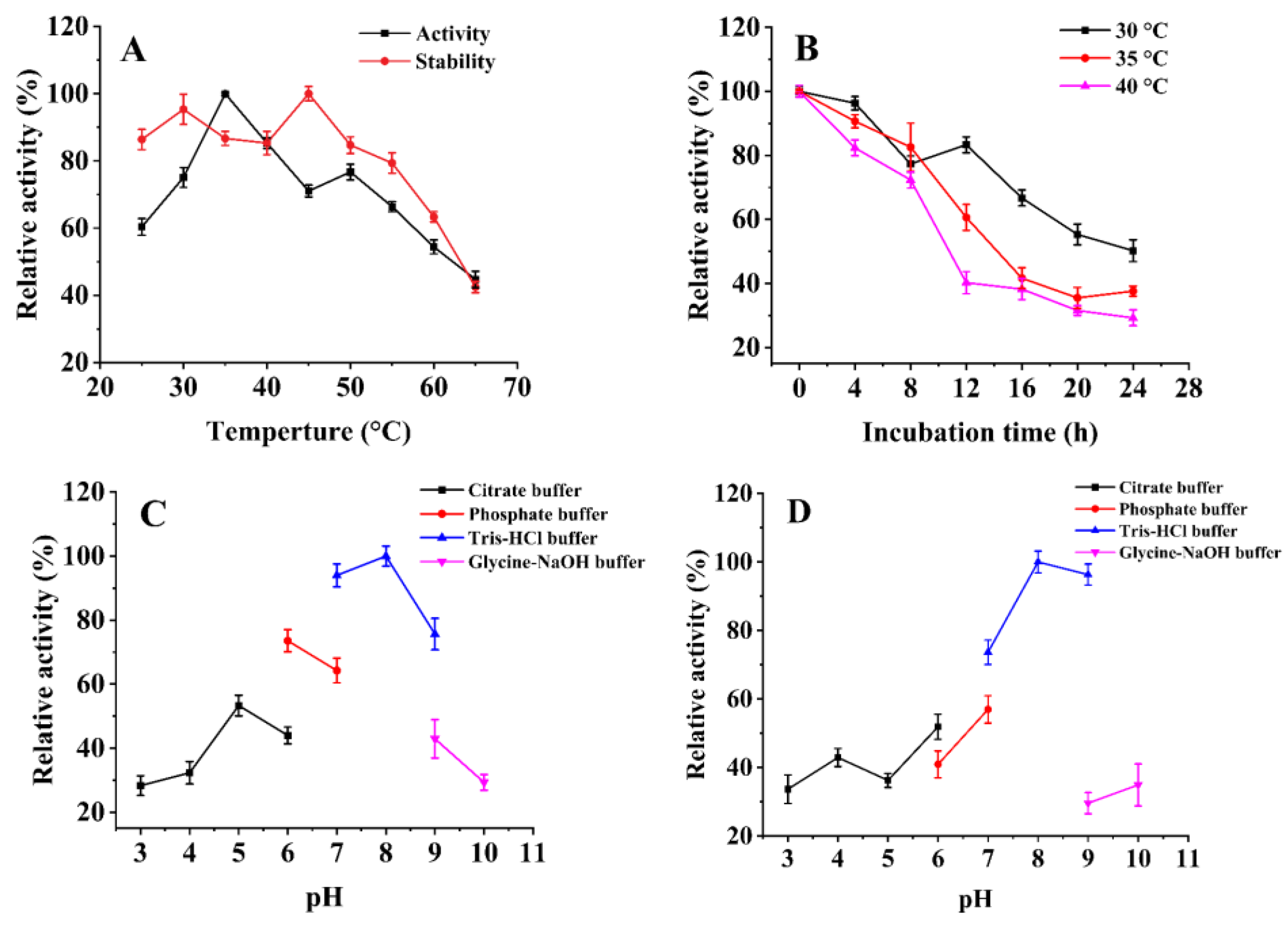
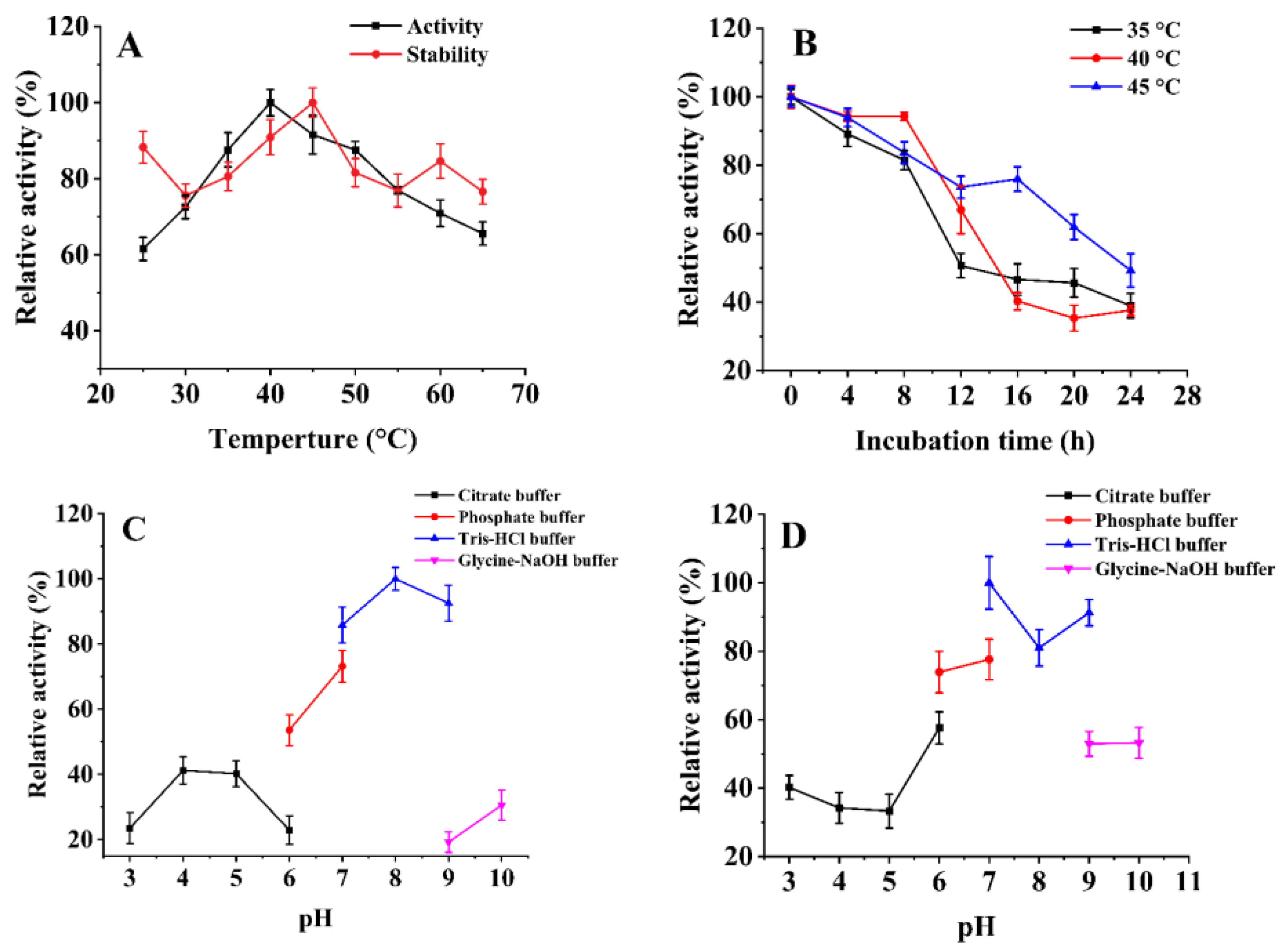
2.3.1. Effects of Temperature and pH on the Activity and Stability of SbCDA
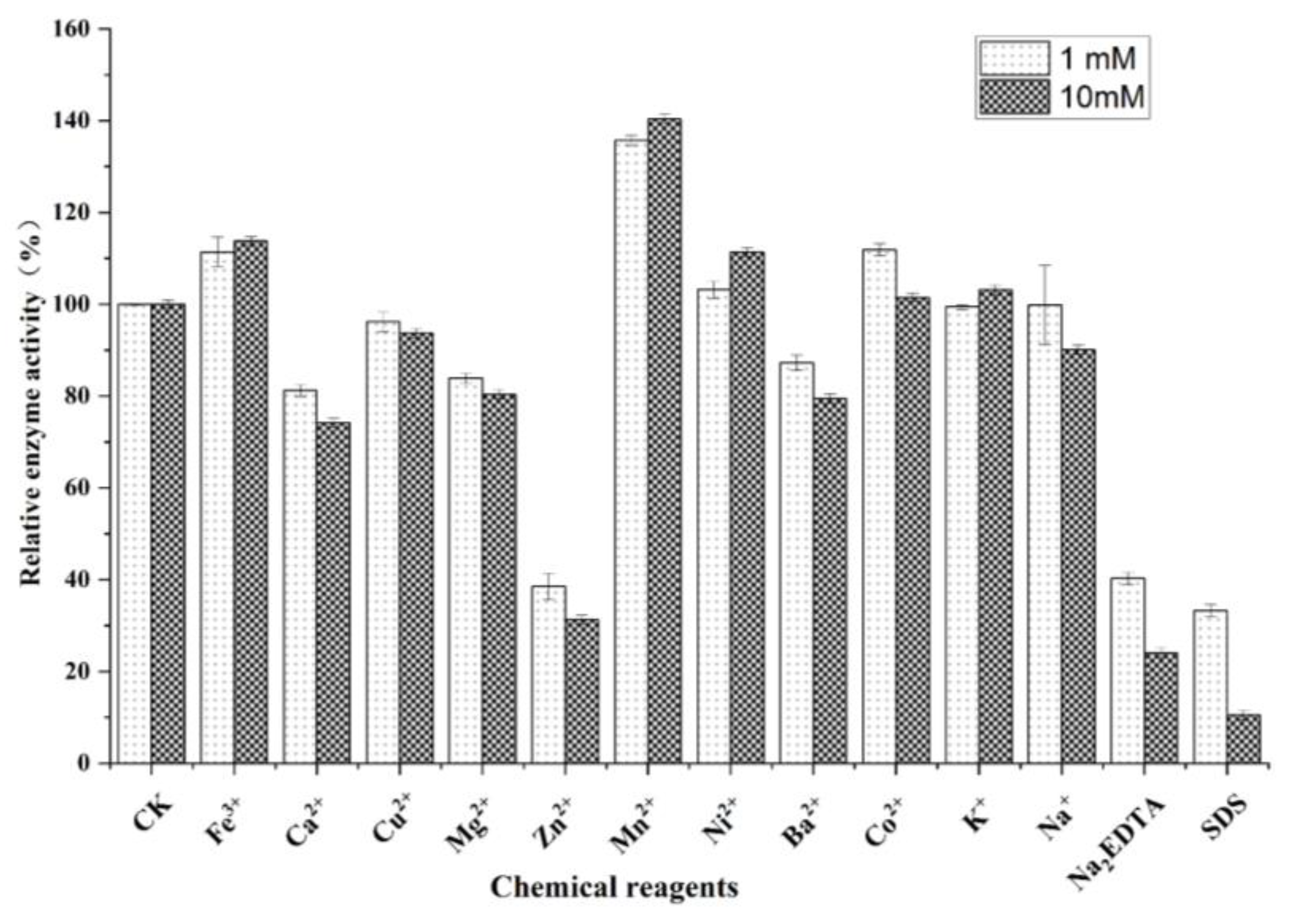
2.3.2. Effects of Chemical Reagents on SbCDA Enzyme Activity
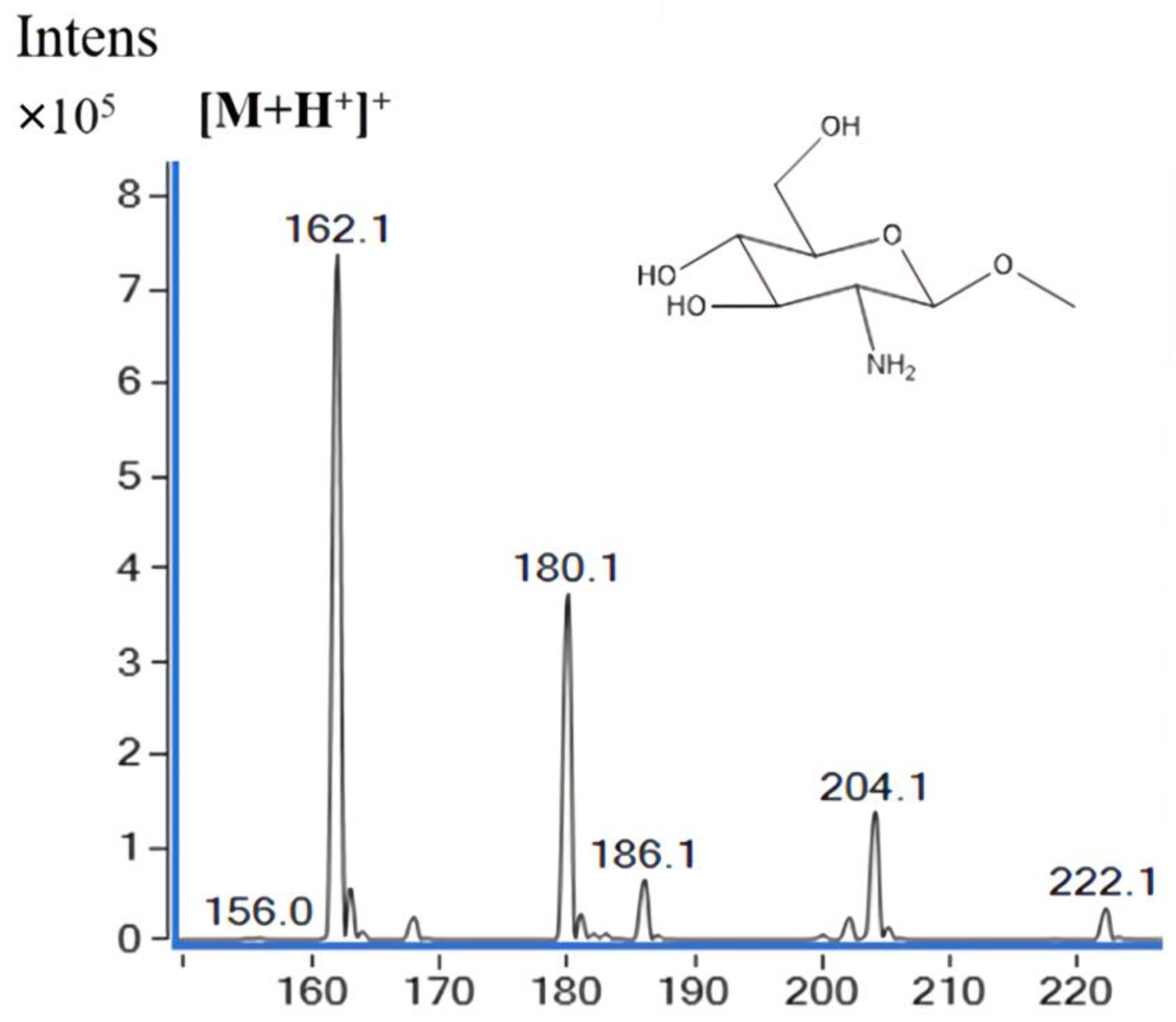
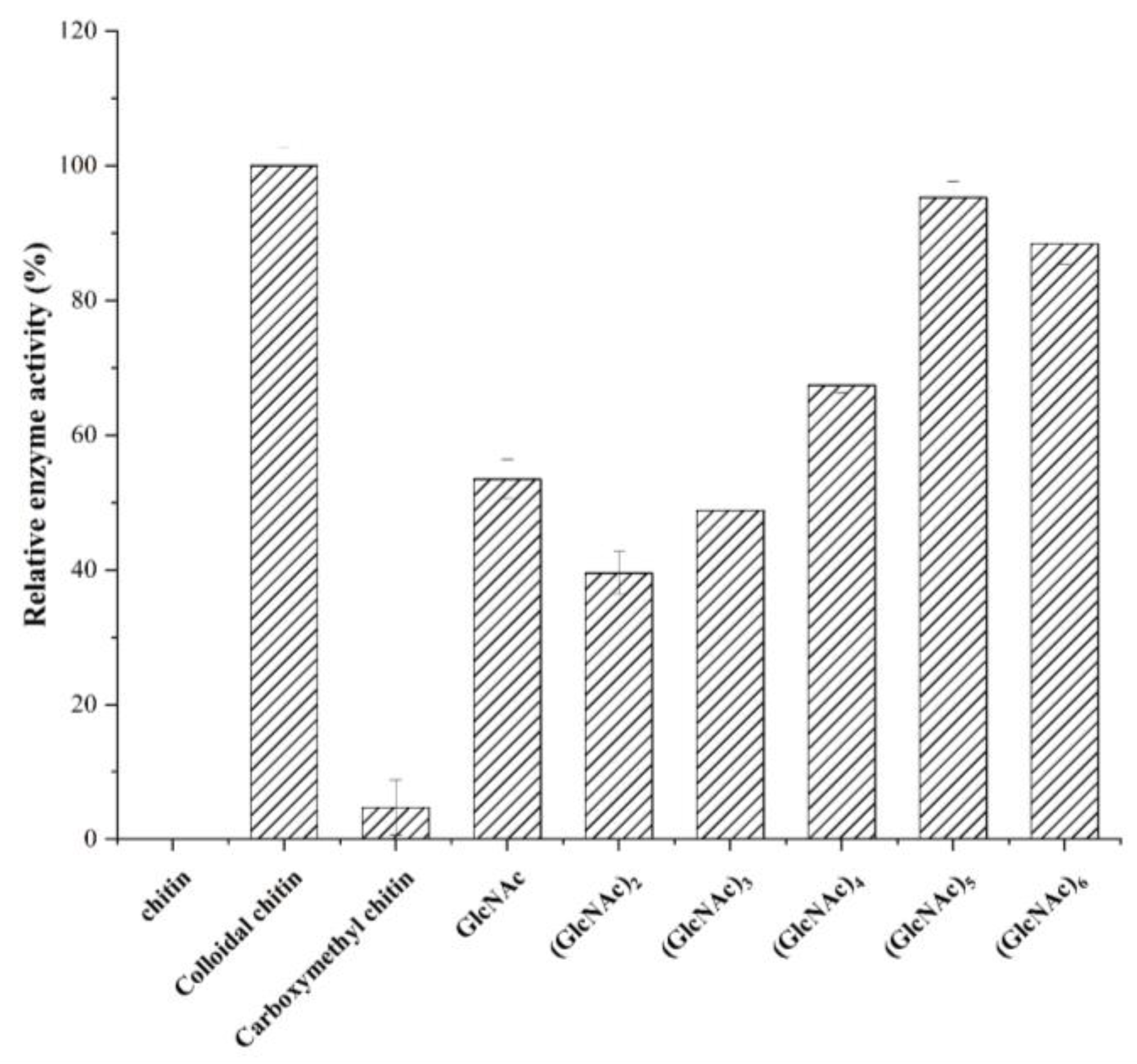
2.4. Substrate Preference of SbCDA
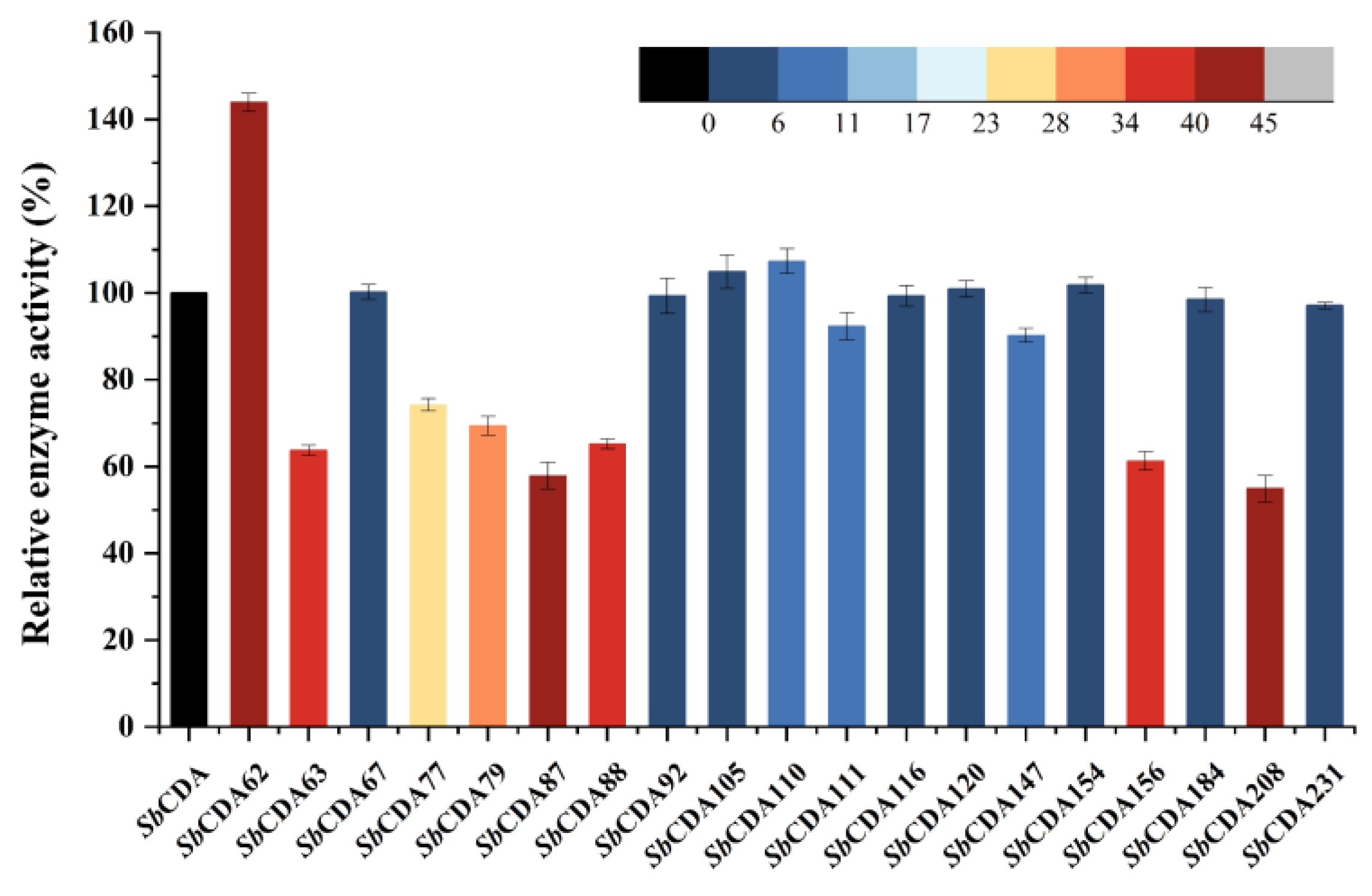
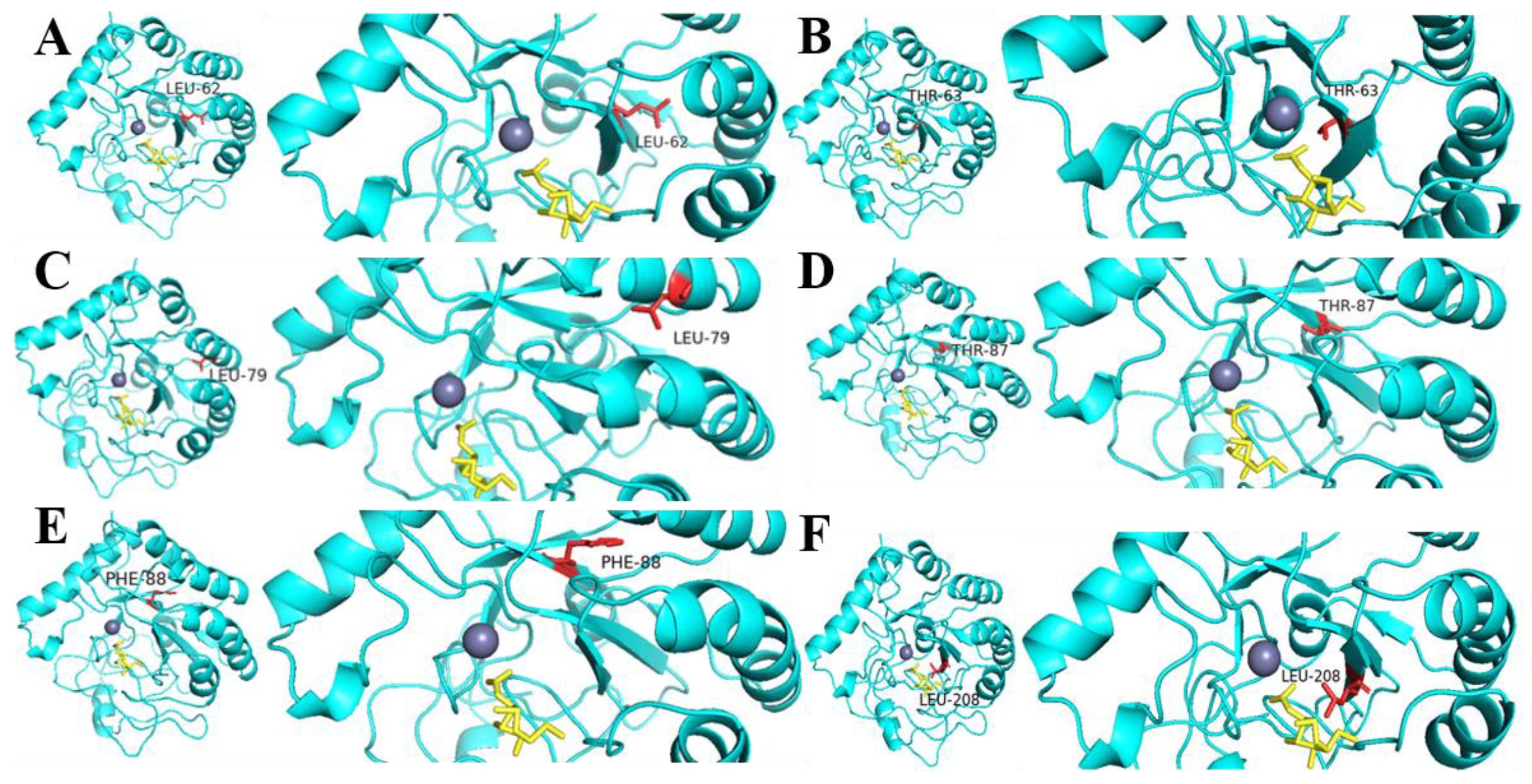
2.5. Comparison of Enzyme Activity of Mutants and Analysis of Homologous CDAs
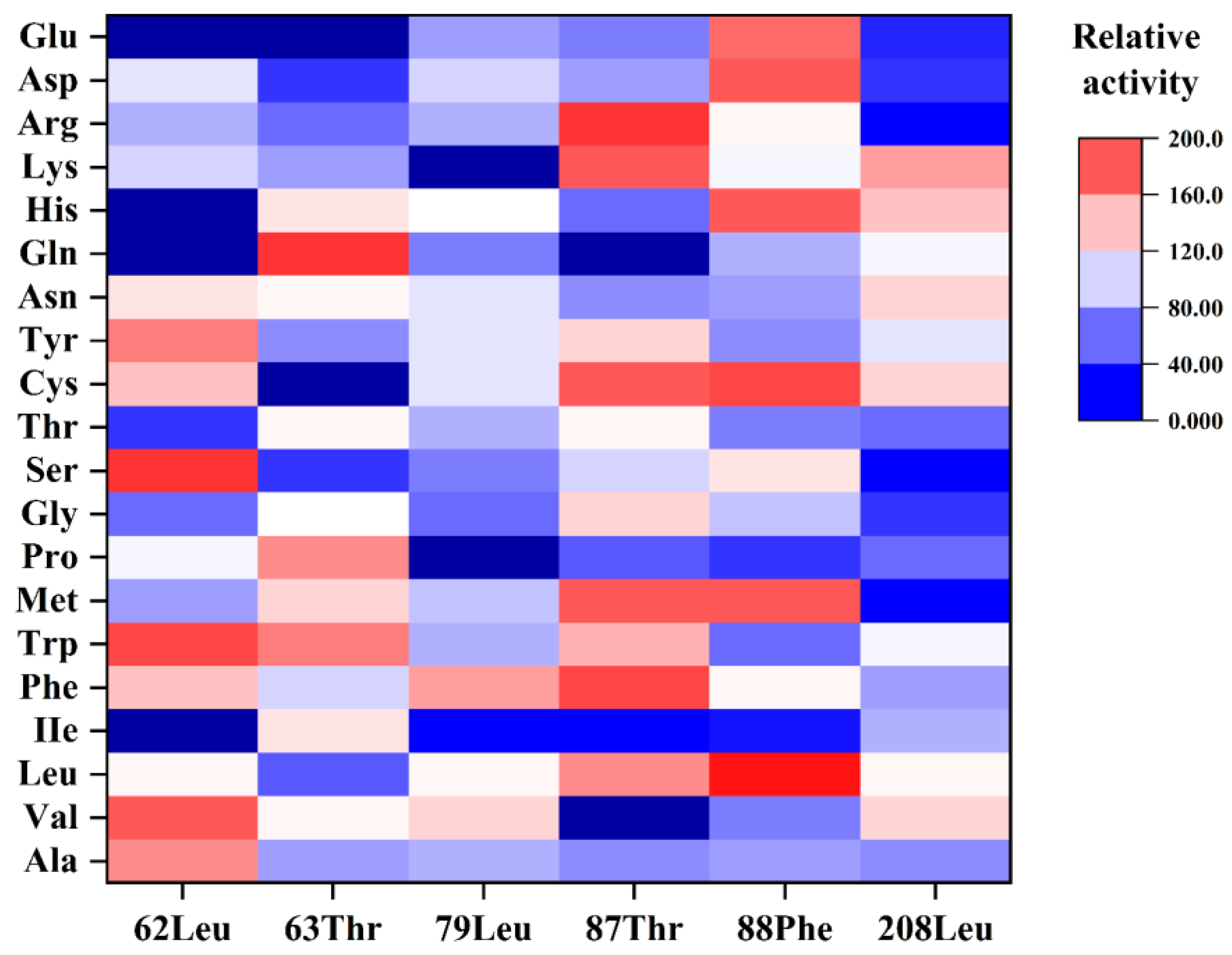
2.6. SbCDA Saturation Mutations at Six Amino Acid Sites
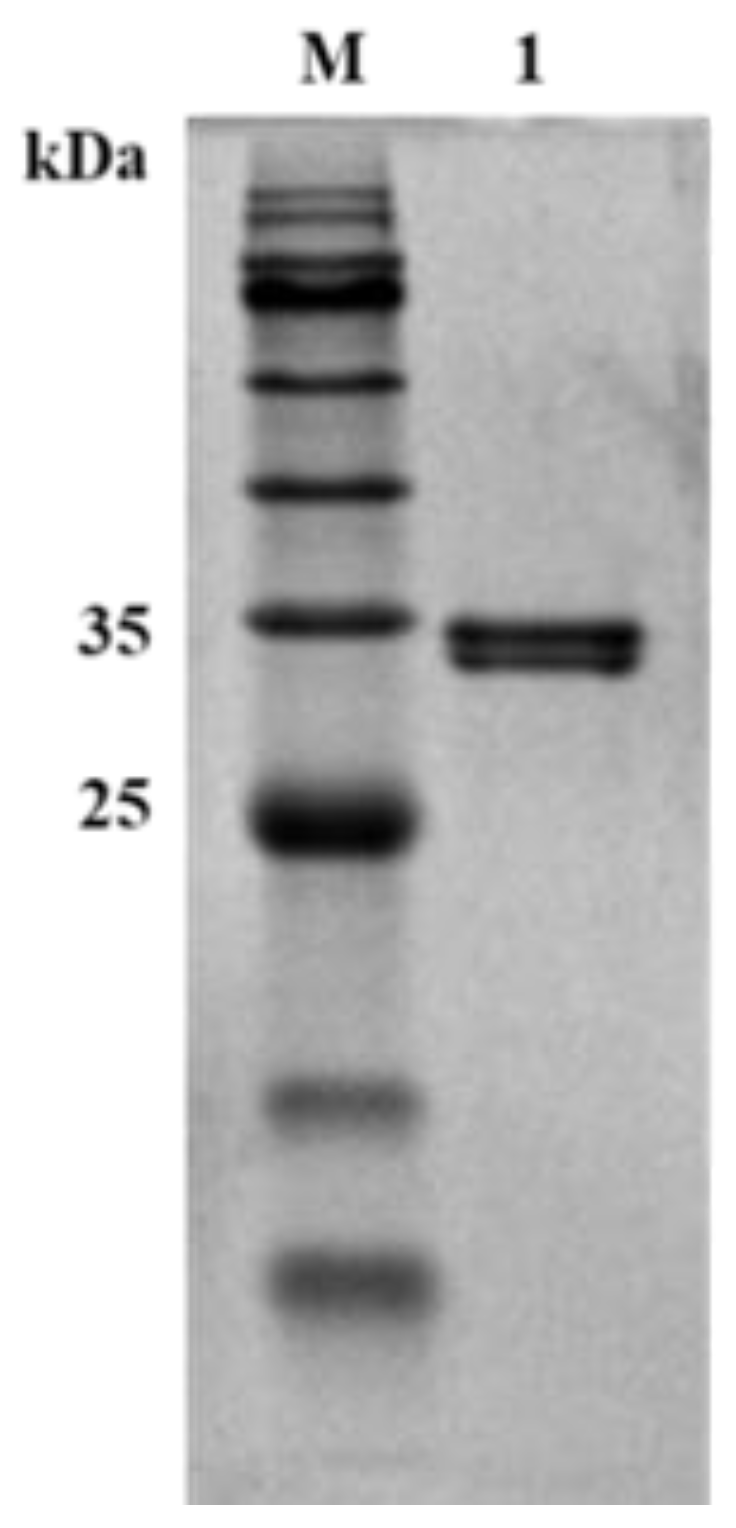
2.7. Characterization of Mutant SbCDA63G
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Strains and Culture Conditions
4.3. Bioinformatics Analysis, Homology Modeling, and Molecular Docking of CDAs
4.4. Construction of the SbCDA Expression Plasmid and Site-Directed Mutation
4.5. Expression and Purification of SbCDA and the Mutant Enzymes
4.6. Chitin Deacetylase Activity Assay
4.7. Biochemical Characterization of SbCDA and the Mutant Enzymes
4.8. Effects of Metal Ions and Chemical Reagents on SbCDA Activity
4.9. Substrate Preference of SbCDA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Appendix A
Appendix B
- >SbCDAMPKKMSVLGGGLAAALVATLTLALTGCSMETTAPASARRDAAPDAKGSFGRADCRKAKCIALTFDAGPGKDTAELLDILKEKKVSATFFLLGRNHVLKHPDTVRRIQDEGHEVANHTWTHKILTDEKPEEIRAELEKTQEAIEKITGKRPRLMRPPQGRTDDQVSGISKELGLSQVLWSATAKDYSTNDSALITQRILDQASRDGIILLHDIYKGTVPAVPGIIDALQKDGYTFVTVPELMAPAVPEPGTIYRP
References
- Younes, I.; Rinaudo, M. Chitin and Chitosan Preparation from Marine Sources. Structure, Properties and Applications. Mar. Drugs 2015, 13, 1133–1174. [Google Scholar] [CrossRef]
- Ma, X.; Gözaydın, G.; Yang, H.; Ning, W.; Han, X.; Poon, N.Y.; Liang, H.; Yan, N.; Zhou, K. Upcycling chitin-containing waste into organonitrogen chemicals via an integrated process. Proc. Natl. Acad. Sci. USA 2020, 117, 7719–7728. [Google Scholar] [CrossRef]
- Tharanathan, R.N.; Kittur, F.S. Chitin—The Undisputed Biomolecule of Great Potential. Crit. Rev. Food Sci. Nutr. 2003, 43, 61–87. [Google Scholar] [CrossRef]
- Yan, N.; Chen, X. Sustainability: Don’t waste seafood waste. Nature 2015, 524, 155–157. [Google Scholar] [CrossRef]
- Liaqat, F.; Eltem, R. Chitooligosaccharides and their biological activities: A comprehensive review. Carbohydr. Polym. 2018, 184, 243–259. [Google Scholar] [CrossRef]
- de Assis, C.F.; Araújo, N.K.; Pagnoncelli, M.G.B.; da Silva Pedrini, M.R.; de Macedo, G.R.; dos Santos, E.S. Chitooligosaccharides enzymatic production by Metarhizium anisopliae. Bioprocess. Biosyst. Eng. 2010, 33, 893–899. [Google Scholar] [CrossRef]
- Nawrotek, K.; Tylman, M.; Decherchi, P.; Marqueste, T.; Rudnicka, K.; Gatkowska, J.; Wieczorek, M. Assessment of degradation and biocompatibility of electrodeposited chitosan and chitosan–carbon nanotube tubular implants. J. Biomed. Mater. Res. Part A 2016, 104, 2701–2711. [Google Scholar] [CrossRef]
- Hu, Z.; Lu, S.; Cheng, Y.; Kong, S.; Li, S.; Li, C.; Yang, L. Investigation of the Effects of Molecular Parameters on the Hemostatic Properties of Chitosan. Molecules 2018, 23, 3147. [Google Scholar] [CrossRef]
- Sahariah, P.; Másson, M. Antimicrobial Chitosan and Chitosan Derivatives: A Review of the Structure–Activity Relationship. Biomacromolecules 2017, 18, 3846–3868. [Google Scholar] [CrossRef]
- Chaiwong, N.; Leelapornpisid, P.; Jantanasakulwong, K.; Rachtanapun, P.; Seesuriyachan, P.; Sakdatorn, V.; Leksawasdi, N.; Phimolsiripol, Y. Antioxidant and Moisturizing Properties of Carboxymethyl Chitosan with Different Molecular Weights. Polymers 2020, 12, 1445. [Google Scholar] [CrossRef]
- Lai, W.-F.; Zhao, S.; Chiou, J. Antibacterial and clusteroluminogenic hypromellose-graft-chitosan-based polyelectrolyte complex films with high functional flexibility for food packaging. Carbohydr. Polym. 2021, 271, 118447. [Google Scholar] [CrossRef]
- Bhatnagar, A.; Sillanpää, M. Applications of chitin- and chitosan-derivatives for the detoxification of water and wastewater—A short review. Adv. Colloid Interface Sci. 2009, 152, 26–38. [Google Scholar] [CrossRef]
- Khan, A.; Wang, B.; Ni, Y. Chitosan-Nanocellulose Composites for Regenerative Medicine Applications. Curr. Med. Chem. 2020, 27, 4584–4592. [Google Scholar] [CrossRef]
- Shahrajabian, M.H.; Chaski, C.; Polyzos, N.; Tzortzakis, N.; Petropoulos, S.A. Sustainable Agriculture Systems in Vegetable Production Using Chitin and Chitosan as Plant Biostimulants. Biomolecules 2021, 11, 819. [Google Scholar] [CrossRef]
- No, H.K.; Meyers, S.P. Preparation and Characterization of Chitin and Chitosan—A Review. J. Aquat. Food Prod. Technol. 1995, 4, 27–52. [Google Scholar] [CrossRef]
- Kumari, S.; Rath, P.; Sri Hari Kumar, A.; Tiwari, T.N. Extraction and characterization of chitin and chitosan from fishery waste by chemical method. Environ. Technol. Innov. 2015, 3, 77–85. [Google Scholar] [CrossRef]
- Kou, S.; Peters, L.M.; Mucalo, M.R. Chitosan: A review of sources and preparation methods. Int. J. Biol. Macromol. 2021, 169, 85–94. [Google Scholar] [CrossRef]
- Chen, X.; Yang, H.; Zhong, Z.; Yan, N. Base-catalysed, one-step mechanochemical conversion of chitin and shrimp shells into low molecular weight chitosan. Green Chem. 2017, 19, 2783–2792. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- John, M.; Röhrig, H.; Schmidt, J.; Wieneke, U.; Schell, J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. USA 1993, 90, 625–629. [Google Scholar] [CrossRef]
- Aragunde, H.; Biarnés, X.; Planas, A. Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases. Int. J. Mol. Sci. 2018, 19, 412. [Google Scholar] [CrossRef]
- Zhao, Y.; Park, R.-D.; Muzzarelli, R.A.A. Chitin Deacetylases: Properties and Applications. Mar. Drugs 2010, 8, 24–46. [Google Scholar] [CrossRef]
- ElMekawy, A.; Hegab, M.H.; El-Baz, A.; Hudson, M.S. Kinetic Properties and Role of Bacterial Chitin Deacetylase in the Bioconversion of Chitin to Chitosan. Recent Pat. Biotechnol. 2013, 7, 234–241. [Google Scholar] [CrossRef]
- Raval, R.; Simsa, R.; Raval, K. Expression studies of Bacillus licheniformis chitin deacetylase in E. coli Rosetta cells. Int. J. Biol. Macromol. 2017, 104, 1692–1696. [Google Scholar] [CrossRef]
- Blair, D.E.; van Aalten, D.M.F. Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine. FEBS Lett. 2004, 570, 13–19. [Google Scholar] [CrossRef]
- Andreou, A.; Giastas, P.; Christoforides, E.; Eliopoulos, E.E. Structural and Evolutionary Insights within the Polysaccharide Deacetylase Gene Family of Bacillus anthracis and Bacillus cereus. Genes 2018, 9, 386. [Google Scholar] [CrossRef]
- Urch, J.E.; Hurtado-Guerrero, R.; Brosson, D.; Liu, Z.; Eijsink, V.G.H.; Texier, C.; van Aalten, D.M.F. Structural and functional characterization of a putative polysaccharide deacetylase of the human parasite Encephalitozoon cuniculi. Protein Sci. 2009, 18, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Karthik, N.; Binod, P.; Pandey, A. SSF production, purification and characterization of chitin deacetylase from Aspergillus flavus. Biocatal. Biotransform. 2018, 36, 296–306. [Google Scholar] [CrossRef]
- Xie, M.; Zhao, X.; Lü, Y.; Jin, C. Chitin deacetylases Cod4 and Cod7 are involved in polar growth of Aspergillus fumigatus. MicrobiologyOpen 2020, 9, e00943. [Google Scholar] [CrossRef]
- Liu, Z.; Gay, L.M.; Tuveng, T.R.; Agger, J.W.; Westereng, B.; Mathiesen, G.; Horn, S.J.; Vaaje-Kolstad, G.; van Aalten, D.M.F.; Eijsink, V.G.H. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017, 7, 1746. [Google Scholar] [CrossRef]
- Liu, L.; Zhou, Y.; Qu, M.; Qiu, Y.; Guo, X.; Zhang, Y.; Liu, T.; Yang, J.; Yang, Q. Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family. J. Biol. Chem. 2019, 294, 5774–5783. [Google Scholar] [CrossRef]
- Sarmiento, K.P.; Panes, V.A.; Santos, M.D. Molecular cloning and expression of chitin deacetylase 1 gene from the gills of Penaeus monodon (black tiger shrimp). Fish Shellfish Immunol. 2016, 55, 484–489. [Google Scholar] [CrossRef]
- Fan, B.; Li, Y.; Huang, Y.; Zhang, M.; Liu, Z.; Fan, W.; Zhao, Y. Cloning and expression of chitin deacetylase 1 from Macrobrachium nipponense, and the effects of dietary protein on growth, body composition and digestive enzymes. Aquacult. Nutr. 2018, 24, 1664–1678. [Google Scholar] [CrossRef]
- Cord-Landwehr, S.; Melcher, R.L.J.; Kolkenbrock, S.; Moerschbacher, B.M. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 2016, 6, 38018. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ohnishi-Kameyama, M.; Hayashi, K.; Mori, Y. Cloning and expression of chitin deacetylase gene from a deuteromycete, Colletotrichum lindemuthianum. J. Biosci. Bioeng. 1999, 87, 418–423. [Google Scholar] [CrossRef]
- Naqvi, S.; Cord-Landwehr, S.; Singh, R.; Bernard, F.; Kolkenbrock, S.; El Gueddari Nour, E.; Moerschbacher Bruno, M. A Recombinant Fungal Chitin Deacetylase Produces Fully Defined Chitosan Oligomers with Novel Patterns of Acetylation. Appl. Environ. Microbiol. 2016, 82, 6645–6655. [Google Scholar] [CrossRef]
- Wang, G.; Guo, Z.; Zhang, X.; Wu, H.; Bai, X.; Zhang, H.; Hu, R.; Han, S.; Pang, Y.; Gao, Z.a.; et al. Heterologous expression of pediocin/papA in Bacillus subtilis. Microb. Cell Fact. 2022, 21, 104. [Google Scholar] [CrossRef]
- Murashima, K.; Chen, C.-L.; Kosugi, A.; Tamaru, Y.; Doi Roy, H.; Wong, S.-L. Heterologous Production of Clostridium cellulovorans engB, Using Protease-Deficient Bacillus subtilis, and Preparation of Active Recombinant Cellulosomes. J. Bacteriol. 2002, 184, 76–81. [Google Scholar] [CrossRef]
- Tuveng, T.R.; Rothweiler, U.; Udatha, G.; Vaaje-Kolstad, G.; Smalas, A.; Eijsink, V.G.H. Structure and function of a CE4 deacetylase isolated from a marine environment. PLoS ONE 2017, 12, 0187544. [Google Scholar] [CrossRef]
- Grifoll-Romero, L.; Sainz-Polo, M.A.; Albesa-Jové, D.; Guerin, M.E.; Biarnés, X.; Planas, A. Structure-function relationships underlying the dual N-acetylmuramic and N-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC. J. Biol. Chem. 2019, 294, 19066–19080. [Google Scholar] [CrossRef]
- Blair, D.E.; Schüttelkopf, A.W.; MacRae, J.I.; van Aalten, D.M.F. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15429–15434. [Google Scholar] [CrossRef]
- Giastas, P.; Andreou, A.; Papakyriakou, A.; Koutsioulis, D.; Balomenou, S.; Tzartos, S.J.; Bouriotis, V.; Eliopoulos, E.E. Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors. Biochemistry 2018, 57, 753–763. [Google Scholar] [CrossRef]
- Fadouloglou, V.E.; Balomenou, S.; Aivaliotis, M.; Kotsifaki, D.; Arnaouteli, S.; Tomatsidou, A.; Efstathiou, G.; Kountourakis, N.; Miliara, S.; Griniezaki, M.; et al. Unusual α-Carbon Hydroxylation of Proline Promotes Active-Site Maturation. J. Am. Chem. Soc. 2017, 139, 5330–5337. [Google Scholar] [CrossRef]
- Wang, Y.; Song, J.-Z.; Yang, Q.; Liu, Z.-H.; Huang, X.-M.; Chen, Y. Cloning of a Heat-Stable Chitin Deacetylase Gene from Aspergillus nidulans and its Functional Expression in Escherichia coli. Appl. Biochem. Biotechnol. 2010, 162, 843–854. [Google Scholar] [CrossRef]
- Alfonso, C.; Nuero, O.M.; Santamaría, F.; Reyes, F. Purification of a heat-stable chitin deacetylase from Aspergillus nidulans and its role in cell wall degradation. Curr. Microbiol. 1995, 30, 49–54. [Google Scholar] [CrossRef]
- Blair, D.E.; Hekmat, O.; Schüttelkopf, A.W.; Shrestha, B.; Tokuyasu, K.; Withers, S.G.; van Aalten, D.M.F. Structure and Mechanism of Chitin Deacetylase from the Fungal Pathogen Colletotrichum lindemuthianum. Biochemistry 2006, 45, 9416–9426. [Google Scholar] [CrossRef]
- Yamada, M.; Kurano, M.; Inatomi, S.; Taguchi, G.; Okazaki, M.; Shimosaka, M. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol. Lett. 2008, 289, 130–137. [Google Scholar] [CrossRef]
- Zhao, Y.; Kim, Y.J.; Oh, K.T.; Nguyen, V.N.; Park, R.D. Production and Characterization of Extracellular Chitin Deacetylase from Absidia corymbifera DY-9. J. Korean Soc. Appl. Biol. Chem. 2010, 53, 119–126. [Google Scholar] [CrossRef]
- Kafetzopoulos, D.; Martinou, A.; Bouriotis, V. Bioconversion of Chitin to Chitosan—Purification and Characterization of Chitin Deacetylase from Mucor-Rouxii. Proc. Natl. Acad. Sci. USA 1993, 90, 2564–2568. [Google Scholar] [CrossRef]
- Pareek, N.; Vivekanand, V.; Agarwal, P.; Saroj, S.; Singh, R.P. Bioconversion to chitosan: A two stage process employing chitin deacetylase from Penicillium oxalicum SAEM-51. Carbohydr. Polym. 2013, 96, 417–425. [Google Scholar] [CrossRef]
- Jayakumar, R.; Prabaharan, M.; Nair, S.V.; Tokura, S.; Tamura, H.; Selvamurugan, N. Novel carboxymethyl derivatives of chitin and chitosan materials and their biomedical applications. Prog. Mater Sci. 2010, 55, 675–709. [Google Scholar] [CrossRef]
- Bhat, P.; Pawaskar, G.-M.; Raval, R.; Cord-Landwehr, S.; Moerschbacher, B.; Raval, K. Expression of Bacillus licheniformis chitin deacetylase in E. coli pLysS: Sustainable production, purification and characterisation. Int. J. Biol. Macromol. 2019, 131, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, L.-X.; Wang, X.; Roseman, S. The Chitin Catabolic Cascade in the Marine Bacterium Vibrio Cholerae: Characterization of a Unique Chitin Oligosaccharide Deacetylase. Glycobiology 2007, 17, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Hou, X.; Lu, J.; Wang, M.; Wang, Y.; Huang, Y.; Liu, Q.; Liu, S.; Fang, Y. Enzymatic modification of native chitin and chitin oligosaccharides by an alkaline chitin deacetylase from Microbacterium esteraromaticum MCDA02. Int. J. Biol. Macromol. 2022, 203, 671–678. [Google Scholar] [CrossRef]
- Tsigos, I.; Bouriotis, V. Purification and Characterization of Chitin Deacetylase from Colletotrichum lindemuthianum (∗). J. Biol. Chem. 1995, 270, 26286–26291. [Google Scholar] [CrossRef]
- Chai, J.; Hang, J.; Zhang, C.; Yang, J.; Wang, S.; Liu, S.; Fang, Y. Purification and characterization of chitin deacetylase active on insoluble chitin from Nitratireductor aquimarinus MCDA3-3. Int. J. Biol. Macromol. 2020, 152, 922–929. [Google Scholar] [CrossRef]
- Porter, N.J.; Christianson, D.W. Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases. Curr. Opin. Struct. Biol. 2019, 59, 9–18. [Google Scholar] [CrossRef]
- Canon, N.; Schein, C.; Chen, X.; Pozzoli, M.; Pathy, V.; Dreskin, S. Alanine scans of IgE-binding to linear epitopes of Ara h 2 reveal critical amino acids. J. Allergy Clin. Immunol. 2021, 147, AB89. [Google Scholar] [CrossRef]
- Matthews, B.W.; Nicholson, H.; Becktel, W.J. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc. Natl. Acad. Sci. USA 1987, 84, 6663–6667. [Google Scholar] [CrossRef]
- Fernández-Lucas, J. New Insights on Enzyme Stabilization for Industrial Biocatalysis. ACS Sustain. Chem. Eng. 2021, 9, 15073–15074. [Google Scholar] [CrossRef]
- Folch, B.; Dehouck, Y.; Rooman, M. Thermo- and Mesostabilizing Protein Interactions Identified by Temperature-Dependent Statistical Potentials. Biophys. J. 2010, 98, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, L.; Wang, Q.; Sun, J.; Mao, X. Expression and Molecular Modification of Chitin Deacetylase from Streptomyces bacillaris. Molecules 2023, 28, 113. https://doi.org/10.3390/molecules28010113
Yin L, Wang Q, Sun J, Mao X. Expression and Molecular Modification of Chitin Deacetylase from Streptomyces bacillaris. Molecules. 2023; 28(1):113. https://doi.org/10.3390/molecules28010113
Chicago/Turabian StyleYin, Lili, Qi Wang, Jianan Sun, and Xiangzhao Mao. 2023. "Expression and Molecular Modification of Chitin Deacetylase from Streptomyces bacillaris" Molecules 28, no. 1: 113. https://doi.org/10.3390/molecules28010113
APA StyleYin, L., Wang, Q., Sun, J., & Mao, X. (2023). Expression and Molecular Modification of Chitin Deacetylase from Streptomyces bacillaris. Molecules, 28(1), 113. https://doi.org/10.3390/molecules28010113






