Postdiagenetic Changes in Kerogen Properties and Type by Bacterial Oxidation and Dehydrogenation
Abstract
:1. Introduction
2. Results and Discussion
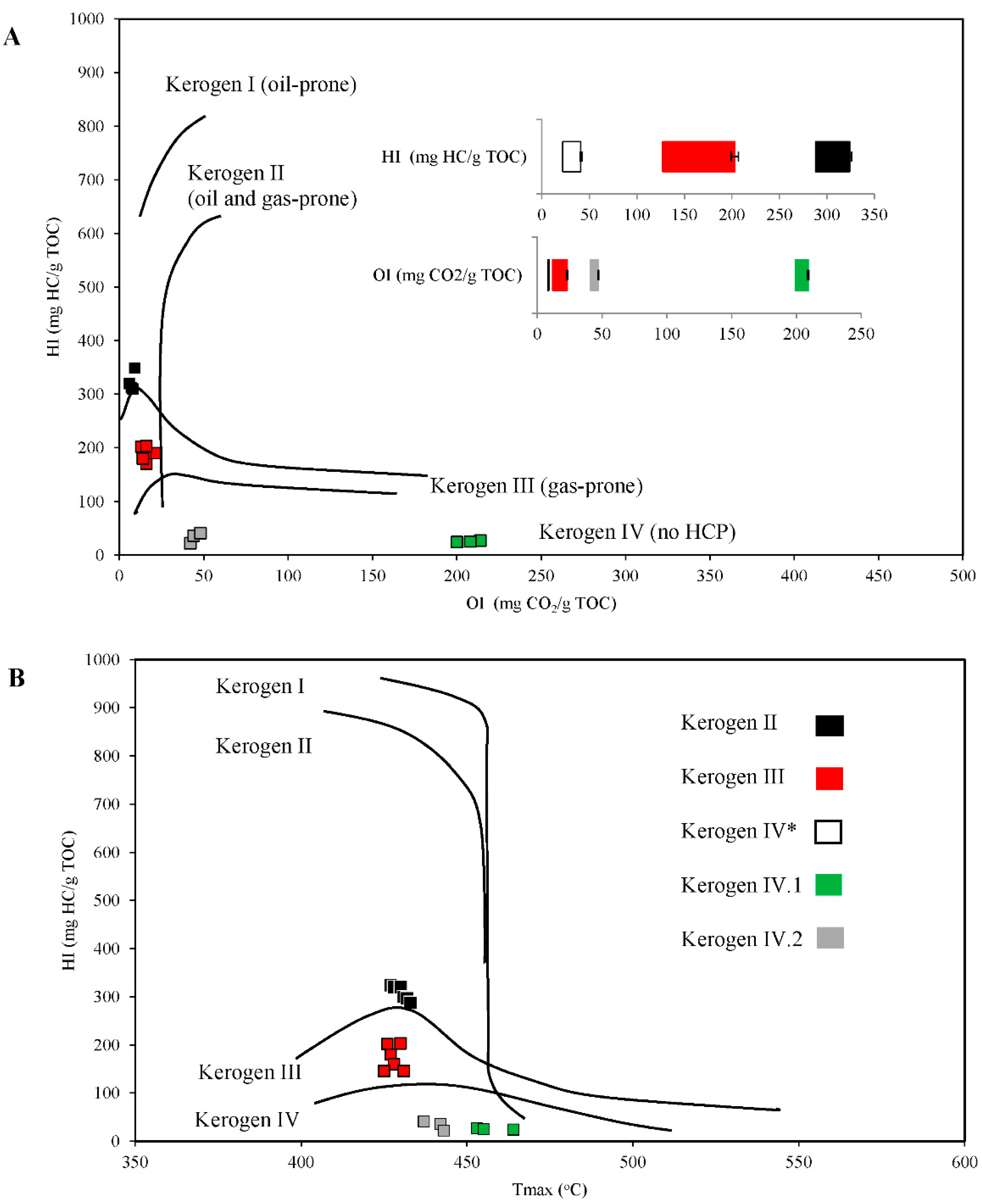
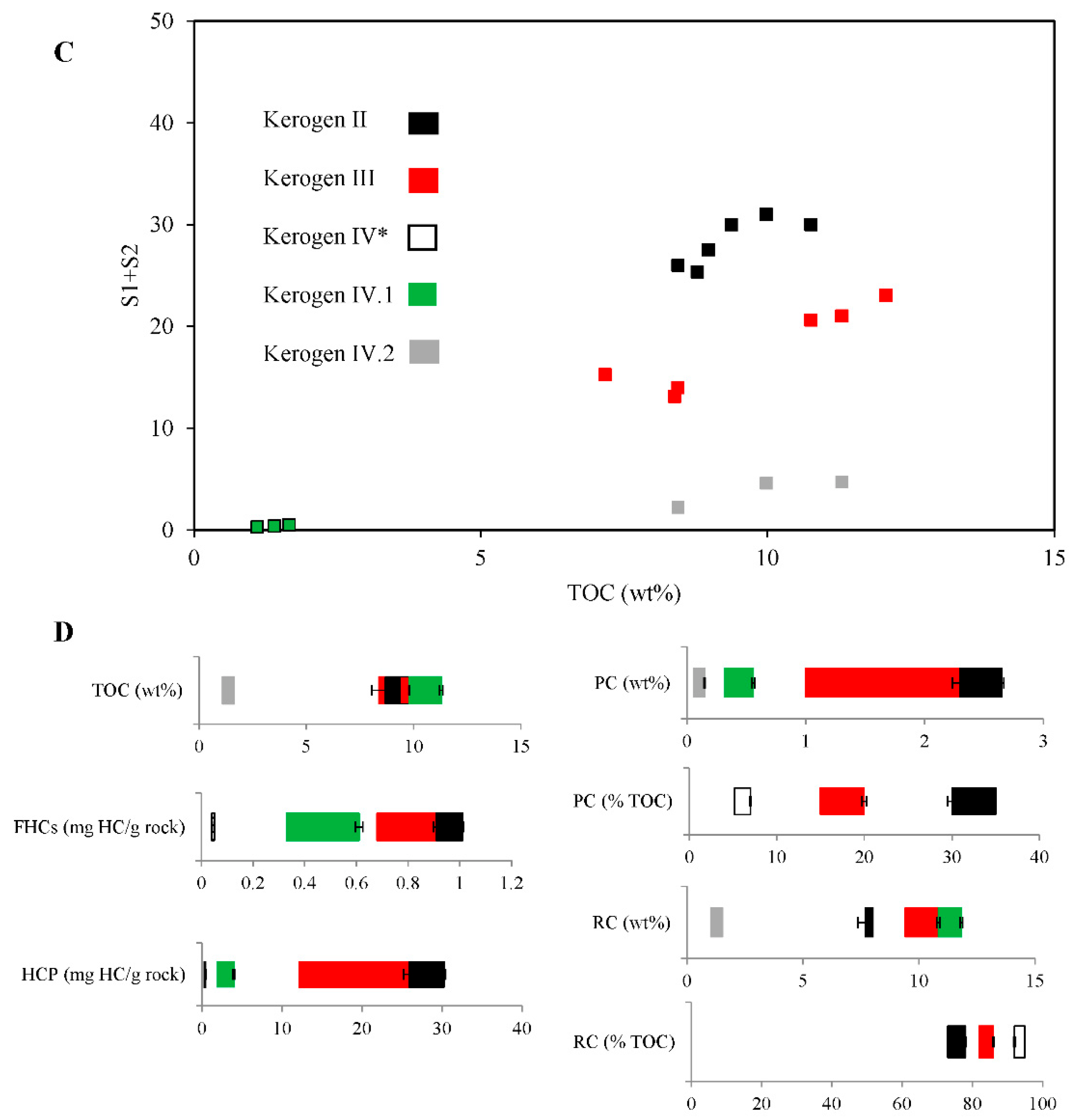
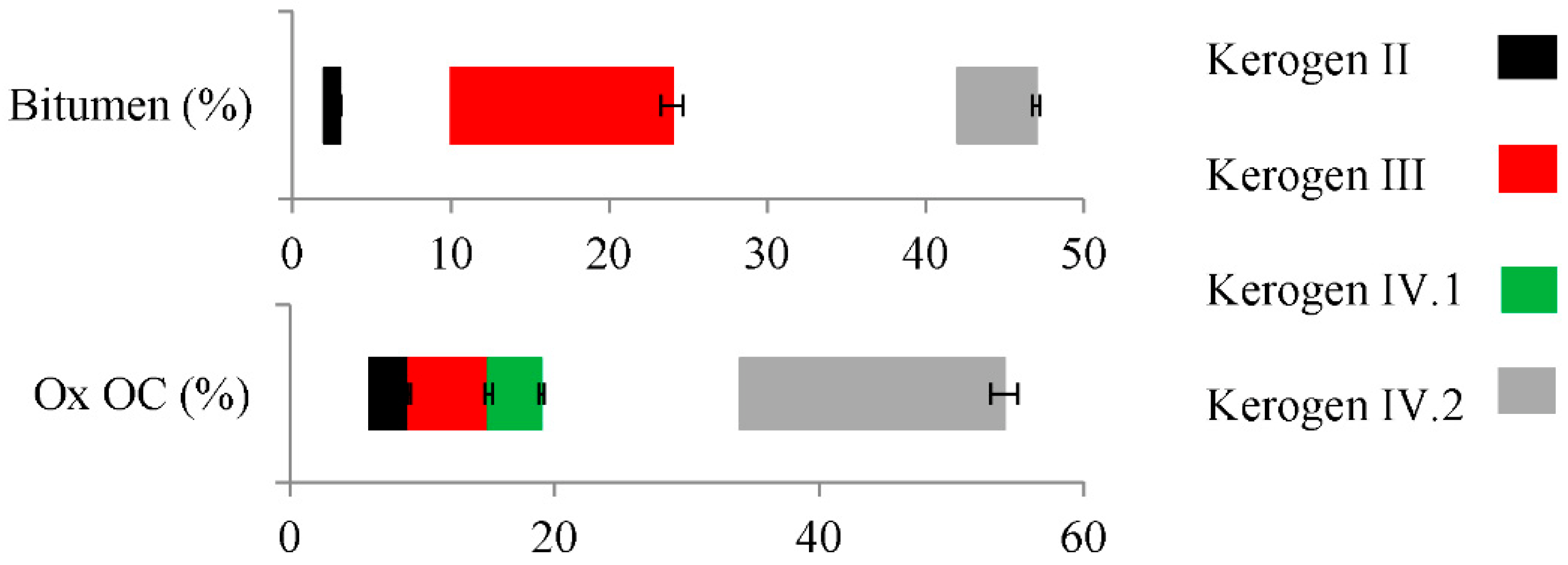
2.1. Properties and Type of Kerogen in Shale Rock on the Fore-Sudetic Monocline
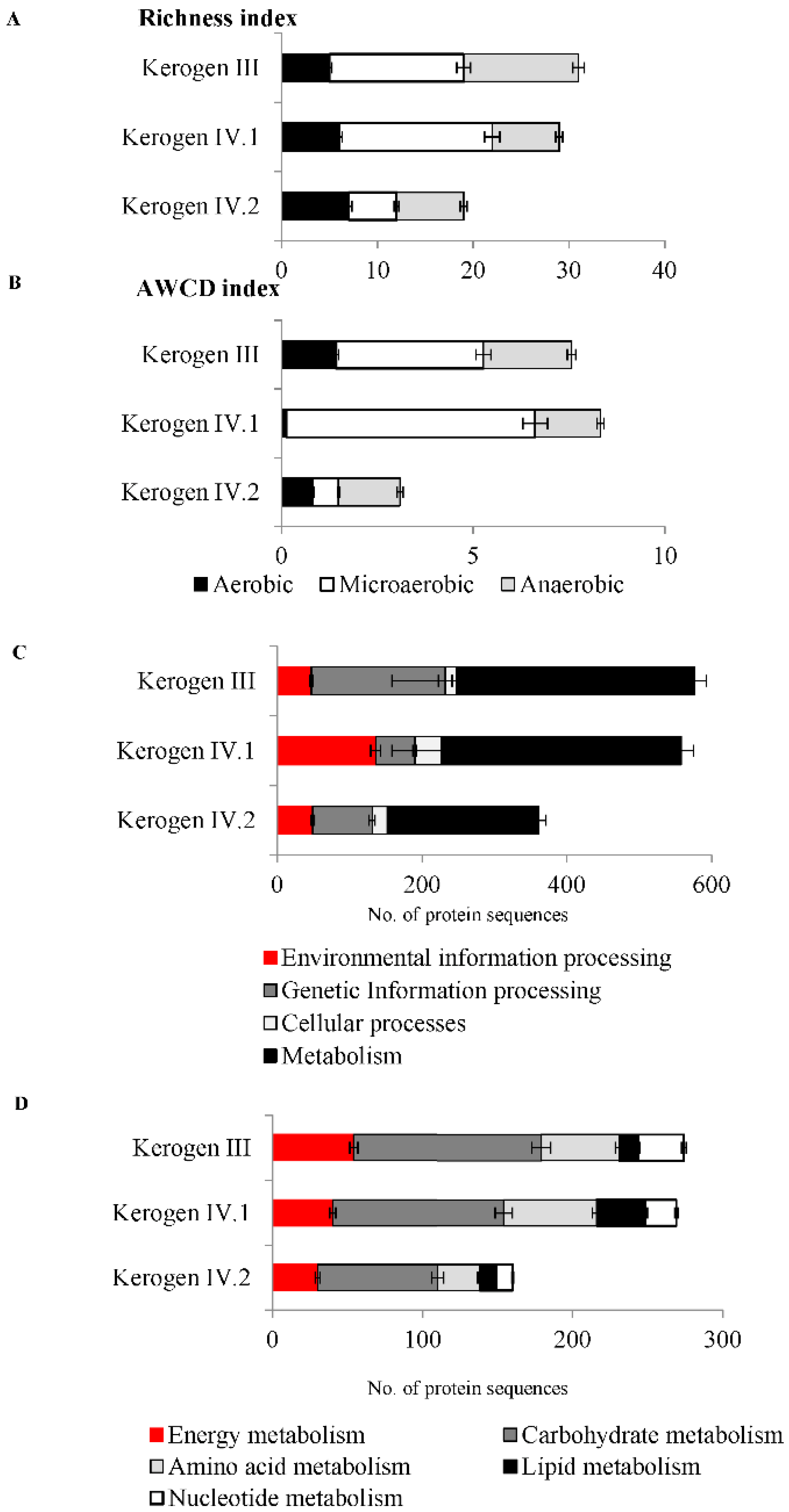
2.2. Detection of Microorganisms and Determination of Their Viability Based on Metabolic Activity
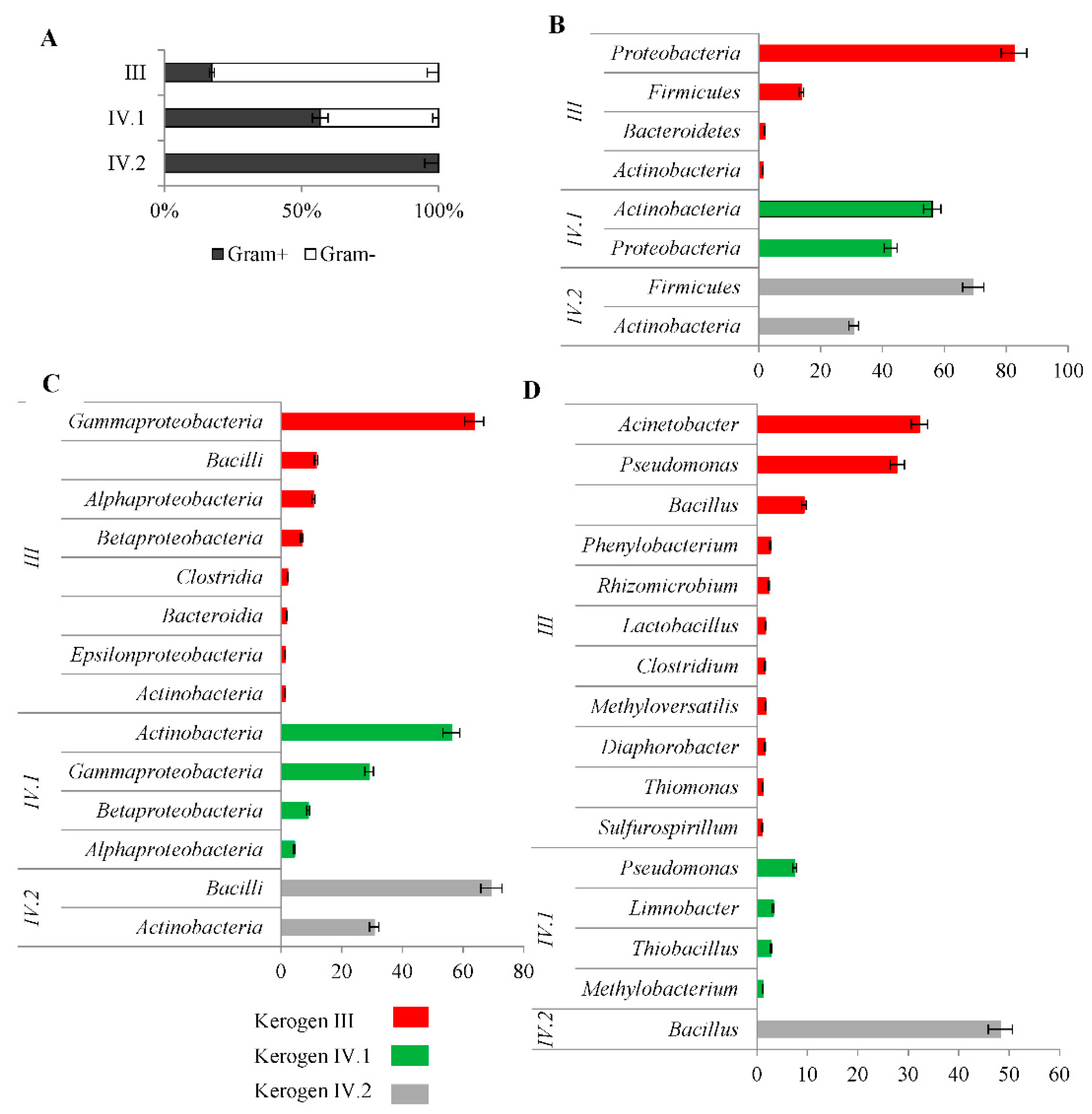
2.3. Bacterial Communities Inhabiting the Shale Rock
2.4. Bacterial Enzymes Potentially Involved in Kerogen Oxidation and Dehydrogenation
3. Materials and Methods
3.1. Description of the Sampling Site and Samples
3.2. Pyrolytic Analysis
3.3. Extraction of Organic Compounds
3.4. Analysis of Extractable Fossil Organic Matter (FOM)
3.5. Selection, Identification, and Classification of Organic Compounds
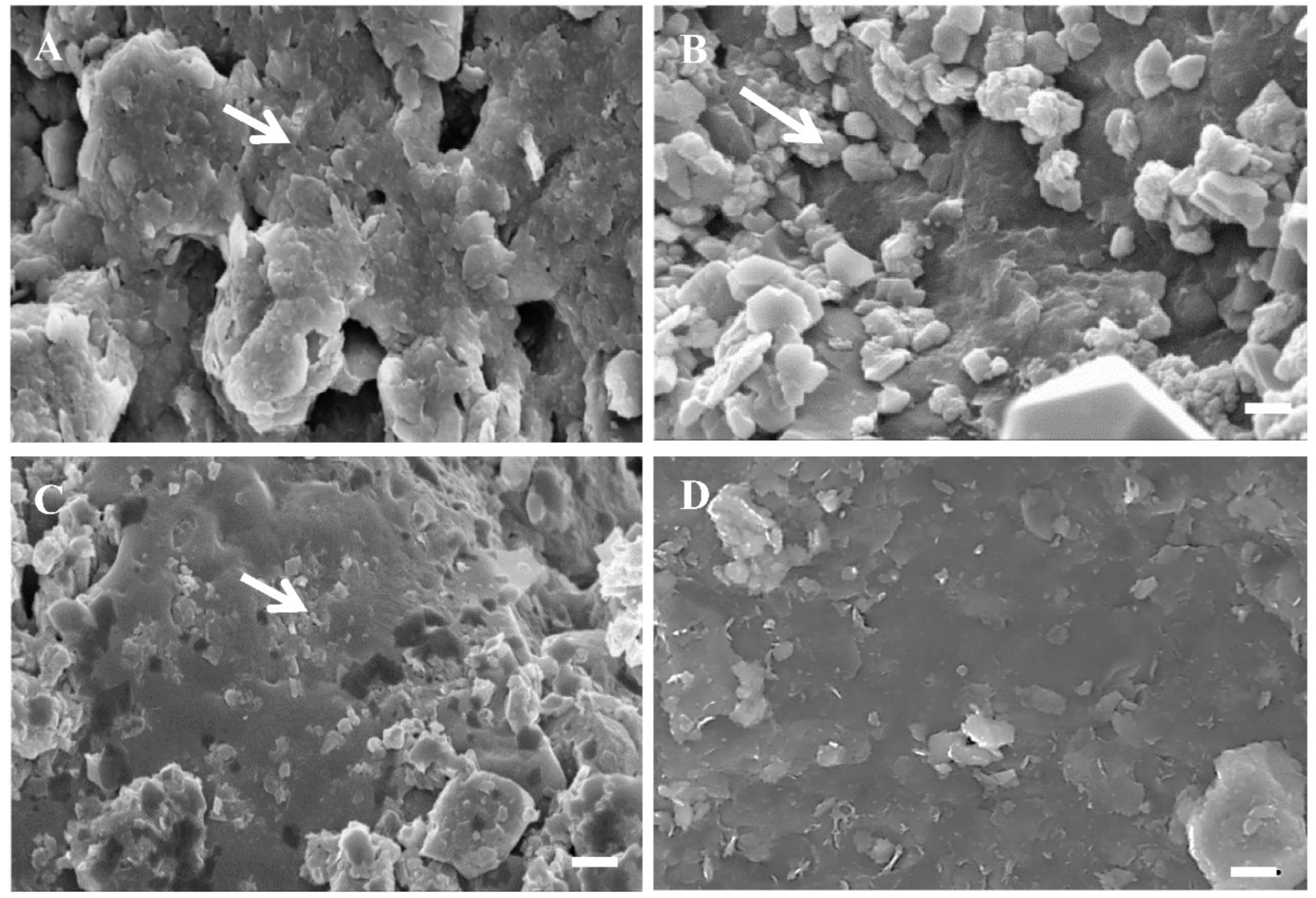
3.6. Microscopic Observations
3.7. Carbon Metabolism Analyses
3.8. DNA Isolation
3.9. DNA Amplification, Sequencing, and Bioinformatics Analysis
3.10. Isolation of Proteins
3.11. Identification of Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Tissot, B.P.; Welte, D.H. Petroleum Formation and Occurrence; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 1984. [Google Scholar]
- Waples, D.W. Geochemistry in Petroleum Exploration; International Human Resources Development Corporation: Boston, MA, USA, 1985. [Google Scholar]
- Van Krevelen, D.W. Graphical-statistical method for the study of structure and reaction processes of coal. Fuel 1950, 29, 269–284. [Google Scholar]
- Sawłowicz, Z. Organic matter and its significance for the genesis of the copper-bearing shales (Kupferschiefer) from the Fore-Sudetic monocline (Poland). In Bitumens in ore Deposits; Parnell, J., Kucha, H., Landais, P., Eds.; Springer: Berlin, Germany, 1993; pp. 431–446. [Google Scholar]
- Sawłowicz, Z.; Gize, A.P.; Rospondek, M. Organic matter from Zechstein copper deposits (Kupferschiefer) in Poland. In Organic Matter and Mineralisation; Glikson, M., Mastalerz, M., Eds.; Springer: Dordrecht, The Netherlands; Berlin/Heidelberg, Germany, 2000; pp. 220–242. [Google Scholar]
- Lafargue, E.; Marquis, F.; Pillot, D. Rock-Eval 6 applications in hydrocarbon exploration, production, and soil contamination studies. Oil. Gas Sci. Technol. 1998, 53, 421–437. [Google Scholar] [CrossRef] [Green Version]
- Leythaeuser, D. Effects of weathering on organic matter in shales. Geochim. Cosmochim. Acta 1973, 37, 113–120. [Google Scholar] [CrossRef]
- Clayton, J.L.; Swetland, P.J. Subaerial weathering of sedimentary organic matter. Geochim. Cosmochim. Acta 1978, 42, 305–312. [Google Scholar] [CrossRef]
- Clayton, J.L.; King, J.D. Effects of weathering on biological marker and aromatic hydrocarbon composition of organic matter in Phosphoria shale outcrop. Geochim. Cosmochim. Acta 1987, 51, 2153–2157. [Google Scholar] [CrossRef]
- Littke, R.; Klussmann, U.; Krooss, B.; Leythaeuser, D. Quantification of loss of calcite, pyrite, and organic matter due to weathering of Toarcian black shales and effects on kerogen and bitumen characteristics. Geochim. Cosmochim. Acta 1989, 55, 3369–3378. [Google Scholar] [CrossRef]
- Petsch, S.T.; Berner, R.A.; Eglinton, T.I. A field study of the chemical weathering of ancient sedimentary organic matter. Org. Geochem. 2000, 31, 475–487. [Google Scholar] [CrossRef]
- Petsch, S.T.; Edwards, K.J.; Eglinton, T.I. Microbial transformations of organic matter in black shales and implications for global biogeochemical cycles. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2005, 219, 157–170. [Google Scholar] [CrossRef]
- Berlendis, S.; Beyssac, O.; Derenne, S.; Benzerara, K.; Anquetil, C.; Guillaumet, M.; Estève, I.; Capelle, B. Comparative mineralogy, organic geochemistry and microbial diversity of the Autun black shale and Graissessac coal (France). Int. J. Coal Geol. 2014, 132, 147–157. [Google Scholar] [CrossRef]
- Seifert, A.G.; Trumbore, S.; Xu, X.; Zhang, D.; Kothe, E.; Gleixner, G. Variable effects of labile carbon on the carbon use of different microbial groups in black slate degradation. Geochim. Cosmochim. Acta 2012, 75, 2557–2570. [Google Scholar] [CrossRef] [Green Version]
- Wengel, M.; Kothe, E.; Schmidt, C.M.; Heide, K.; Gleixner, G. Degradation of organic matter from black shales and charcoal by the wood-rotting fungus Schizophyllum commune and release of DOC and heavy metals in the aqueous phase. Sci. Total Environ. 2006, 367, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Stasiuk, R.; Włodarczyk, A.; Karcz, P.; Janas, M.; Skłodowska, A.; Matlakowska, R. Bioweathering of fossil organic matter and organic carbon mobilization from subsurface organic-rich Kupferschiefer black shale—Long-term laboratory studies. Environ. Microbiol. Rep. 2017, 10, 1758–2229. [Google Scholar] [CrossRef]
- Włodarczyk, A.; Lirski, M.; Fogtman, A.; Koblowska, M.; Bidziński, G.; Matlakowska, R. The oxidative metabolism of fossil hydrocarbons and sulfide minerals by the lithobiontic microbial community inhabiting deep subterrestrial Kupferschiefer black shale. Front Microbiol. 2018, 9, 972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prince, R.C.; Amande, T.J.; McGenity, T.J. Prokaryotic Hydrocarbon Degraders. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; McGenicty, T.J., Ed.; Springer Nature: Cham, Switzerland, 2019. [Google Scholar]
- Slater, J.H.; Bull, A.T.; Hardman, D.J. Microbial dehalogenation of halogenated alkanoic acids, alcohols and alkanes. Adv. Microbial. Physiol. 1996, 38, 133–176. [Google Scholar]
- Chan, S.I.; Chen, K.H.; Yu, S.S.; Chen, C.L.; Kuo, S.S. Toward delineating the structure and function of the particulate methane monooxygenase from methanotrophic bacteria. Biochemistry 2004, 43, 4421–4430. [Google Scholar] [CrossRef]
- Fetzner, S. Bacterial degradation of pyridine, indole, quinoline, and their derivatives under different redox conditions. Appl. Microbiol. Biotechnol. 1998, 49, 237–250. [Google Scholar] [CrossRef]
- Chaudhry, G.R.; Chapalamadugu, S. Biodegradation of halogenated organic compounds. Microbiol. Rev. 1991, 55, 59–79. [Google Scholar] [CrossRef]
- Muraki, T.; Taki, M.; Hasegawa, Y.; Iwaki, H.; Lau, P.C.K. Prokaryotic homologs of the eukaryotic 3-hydroxyanthranilate3, 4-dioxygenase and 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase in the 2-nitrobenzoate degradation pathway of Pseudomonas fluorescens strain KU-7. Appl. Environ. Microbiol. 2003, 69, 1564–1572. [Google Scholar] [CrossRef] [Green Version]
- Harwood, C.S.; Parales, R.E. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996, 50, 553–590. [Google Scholar] [CrossRef]
- Haigler, B.E.; Spain, J.C. Degradation of p-chlorotoluene by a mutant of Pseudomonas sp. strain JS6. Appl. Environ. Microbiol. 1989, 55, 372–379. [Google Scholar] [CrossRef] [Green Version]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef] [PubMed]
- Ferrandez, A.; Minambres, B.; Garcia, B.; Olivera, E.R.; Luengo, J.M.; Garcia, J.L.; Diaz, E. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 1998, 273, 25974–25986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, P.S.; Goradia, B.; Saxena, A. Bacterial laccase: Recent update on production, properties and industrial applications. 3 Biotech 2017, 7, 323. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Llorens, J.M.; Patrauchan, M.A.; Stewart., G.R.; Davies., J.E.; Eltis., L.D.; Mohn, W.W. Phenylacetate Catabolism in Rhodococcus sp. Strain RHA1: A Central Pathway for Degradation of Aromatic Compounds. J. Bacteriol. 2005, 187, 4497–4504. [Google Scholar] [CrossRef] [Green Version]
- Cerniglia, C.E.; Sutherland, J.B. Degradation of polycyclic aromatic HCs by fungi. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., McGenity, T.J., van der Meer, J.R., De Lorenzo, V., Eds.; Springer: Berlin, Germany, 2010; pp. 2080–2110. [Google Scholar]
- Johannes, C.; Majcherczyk, A. Natural mediators in the oxidation of polycyclic aromatic hydrocarbons by laccase mediator systems. Appl. Environ. Microbiol. 2000, 6, 524–528. [Google Scholar] [CrossRef] [Green Version]
- Silverstein, R.M.; Webster, F.X.; Kiemle, D.J.; Bryce, D.L. Spectrometric Identification of Organic Compounds; John Wiley and Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Gomez, E.; Garland, J.; Conti, M. Reproducibility in the response of soil bacterial community-level physiological profiles from a land use intensification gradient. Appl. Soil Ecol. 2004, 26, 21–30. [Google Scholar] [CrossRef]
- Garland, J. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol. Ecol. 1997, 24, 289–300. [Google Scholar] [CrossRef]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community; sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Ram, R.J.; VerBerkmoes, N.C.; Thelen, M.P.; Tyson, G.W.; Baker, B.J.; Blake, R.C., II; Shah, M.; Hettich, R.L.; Banfield, J.F. Community Proteomics of a Natural Microbial Biofilm. Science 2005, 308, 1915–1920. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Enzyme | Kerogen III | Kerogen IV.1 | Kerogen IV.2 |
|---|---|---|---|
| Dioxygenases | |||
| Catechol 1,2-dioxygenase | - | gi|498129511 | - |
| 2-Nitropropane dioxygenase | - | gi|490244293 gi|496990711 gi|490758414 | - |
| Monooxygenases | |||
| Methane monooxygenase protein B | gi|223717934 | - | - |
| Particulate methane monooxygenase B-subunit | - | gi|306921967 | - |
| FAD-binding monooxygenase | - | gi|517722695 | - |
| Quinol monooxygenase | gi|501096931 | - | - |
| Other oxidizing enzymes | |||
| Carboxymuconolactone decarboxylase | gi|488115986 | - | - |
| Dienelactone hydrolase | - | gi|490775052 | - |
| Haloacid dehalogenase | gi|503054342 gi|503823017 | gi|544647509 | - |
| Phenylacetic acid degradation protein | gi|523636522 | - | - |
| Isoquinoline 1-oxidoreductase subunit alpha | gi|522028532 | - | - |
| YfiH family protein (laccase)/polyphenol oxidase | gi|523636522 | - | - |
| Alcohol dehydrogenases | |||
| Alcohol dehydrogenase | - | gi|446065336 gi|481050357 gi|490834704 gi|491126456 gi|517727322 gi|489375334 gi|489392338 gi|490768969 gi|498009882 gi|499606054 | WP_004631200.1 WP_011286788.1 |
| Alkyl alcohol (butanol) dehydrogenase | gi|283480587 | - | gi|283480587 |
| (R,R)-butanediol dehydrogenase | - | gi|494048403 | - |
| Cyclohexanol dehydrogenase | - | gi|9965291 | - |
| Lanthanide-dependent methanol dehydrogenase | - | - | gi|177826798, gi|495339880 |
| Methanol dehydrogenase | gi|492375028 | - | - |
| Aldehyde dehydrogenases | |||
| Acetaldehyde dehydrogenase | - | gi|494200545 | - |
| Aldehyde dehydrogenase | gi|430004500 gi|742727449 gi|501570423 gi|499520282 | gi|518406973 gi|489377479 gi|490789497 gi|491118422 gi|491126465 gi|493682336 gi|500262322 gi|518406973 gi|635596374 | WP_0195771801 BAO81531.1 |
| Phenylacetaldehyde dehydrogenase | CAA67780.1 | ||
| Enzymes participating in degradation of fatty acids | |||
| Acyl-CoA synthesis | |||
| Long-chain-fatty-acid-CoA ligase | WP_003869413.1 WP_007704349.1 | gi|371548208| gb|EHN76535.1| gi|493470804| | | - |
| Beta-oxidation | |||
| Acyl-CoA oxidase | sp|Q9Y7B1.1 | - | - |
| Enoyl-CoA hydratase | WP_016918268.1 WP_029919295.1 | - | WP_016918268.1 |
| 3-hydroxyacyl-CoA dehydrogenase | WP_014612827.1 | gi|504425725 | - |
| Medium-chain acyl-CoA dehydrogenase | - | gi|501277187 | - |
| Other enzymes | |||
| Acetyl-CoA C-acetyltransferase | AAG30258.1 | - | WP_011531005.1 |
| Glutaryl-CoA dehydrogenase (ETF) | - | - | WP_018763741.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilamowska, A.; Koblowska, M.; Matlakowska, R. Postdiagenetic Changes in Kerogen Properties and Type by Bacterial Oxidation and Dehydrogenation. Molecules 2022, 27, 2408. https://doi.org/10.3390/molecules27082408
Wilamowska A, Koblowska M, Matlakowska R. Postdiagenetic Changes in Kerogen Properties and Type by Bacterial Oxidation and Dehydrogenation. Molecules. 2022; 27(8):2408. https://doi.org/10.3390/molecules27082408
Chicago/Turabian StyleWilamowska, Agnieszka, Marta Koblowska, and Renata Matlakowska. 2022. "Postdiagenetic Changes in Kerogen Properties and Type by Bacterial Oxidation and Dehydrogenation" Molecules 27, no. 8: 2408. https://doi.org/10.3390/molecules27082408
APA StyleWilamowska, A., Koblowska, M., & Matlakowska, R. (2022). Postdiagenetic Changes in Kerogen Properties and Type by Bacterial Oxidation and Dehydrogenation. Molecules, 27(8), 2408. https://doi.org/10.3390/molecules27082408





