Towards the Optimization of Microwave-Assisted Extraction and the Assessment of Chemical Profile, Antioxidant and Antimicrobial Activity of Wine Lees Extracts
Abstract
:1. Introduction
2. Results and Discussion
2.1. Extraction Solvent and Temperature
2.2. Implementation of DOE Models for MAE Optimization
2.2.1. Screening Design (23 Full Factorial Design)
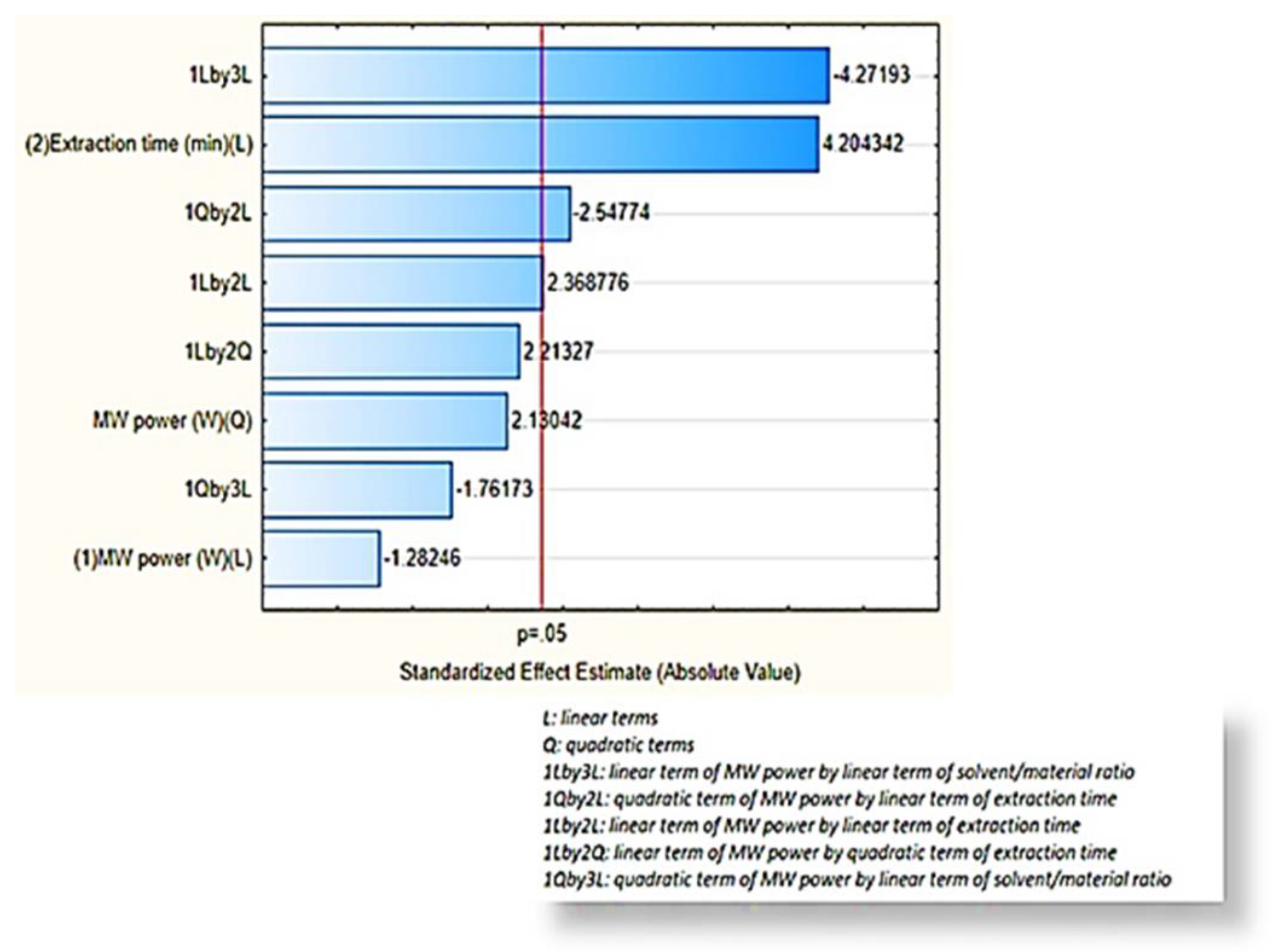
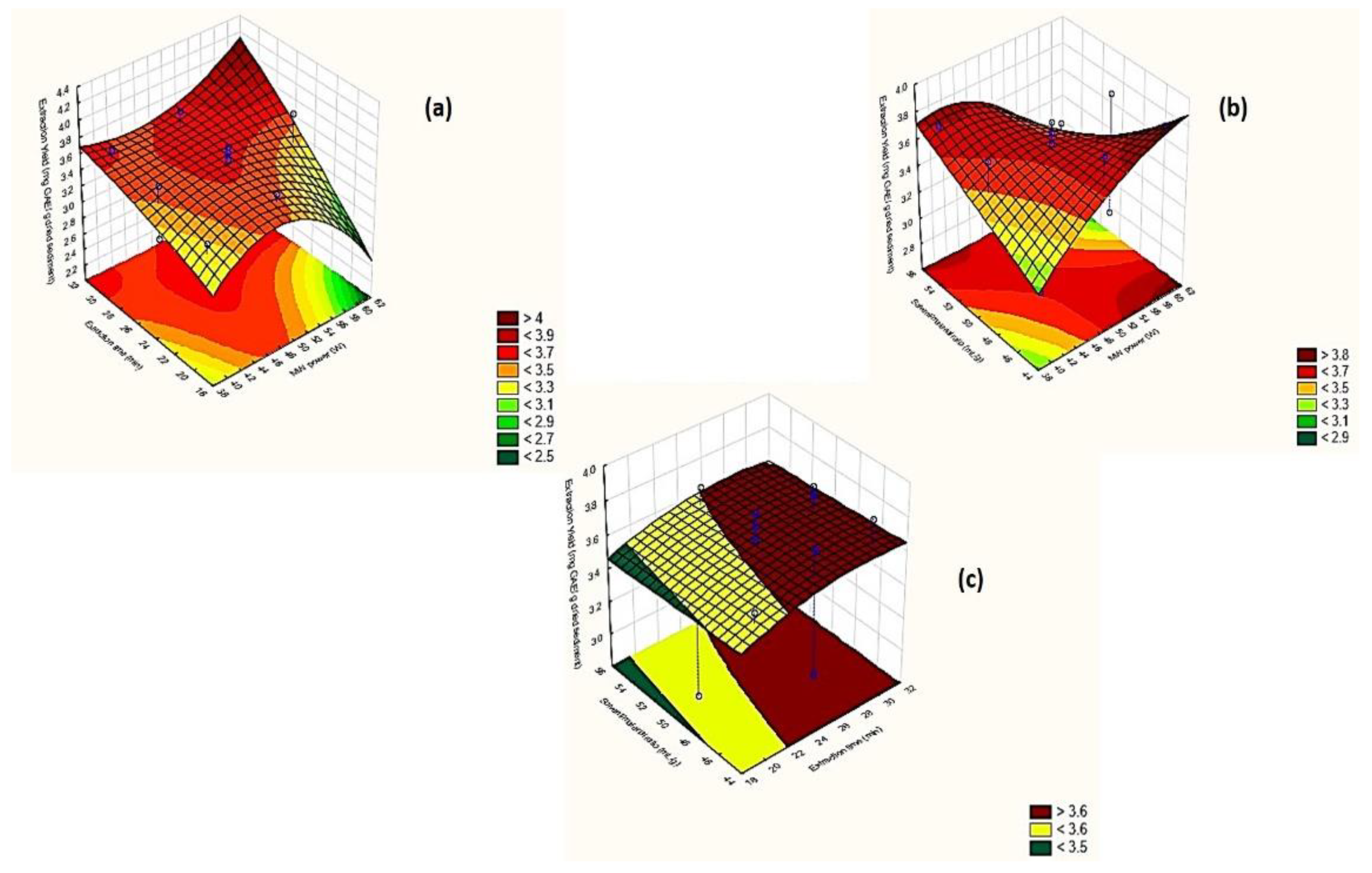
2.2.2. Response Surface Methodology (RSM)-Box–Behnken Design
2.2.3. Assessment of the Effects of the Extraction Factors under Optimization
MW Power
Extraction Time
Solvent/material Ratio
Optimal Extraction Conditions of MAE
2.3. Analysis of Wine Lees Samples by Spectrophotometric Methods
2.4. FT-IR Spectra Interpretation of Wine Lees Extracts
2.5. Antimicrobial Assay
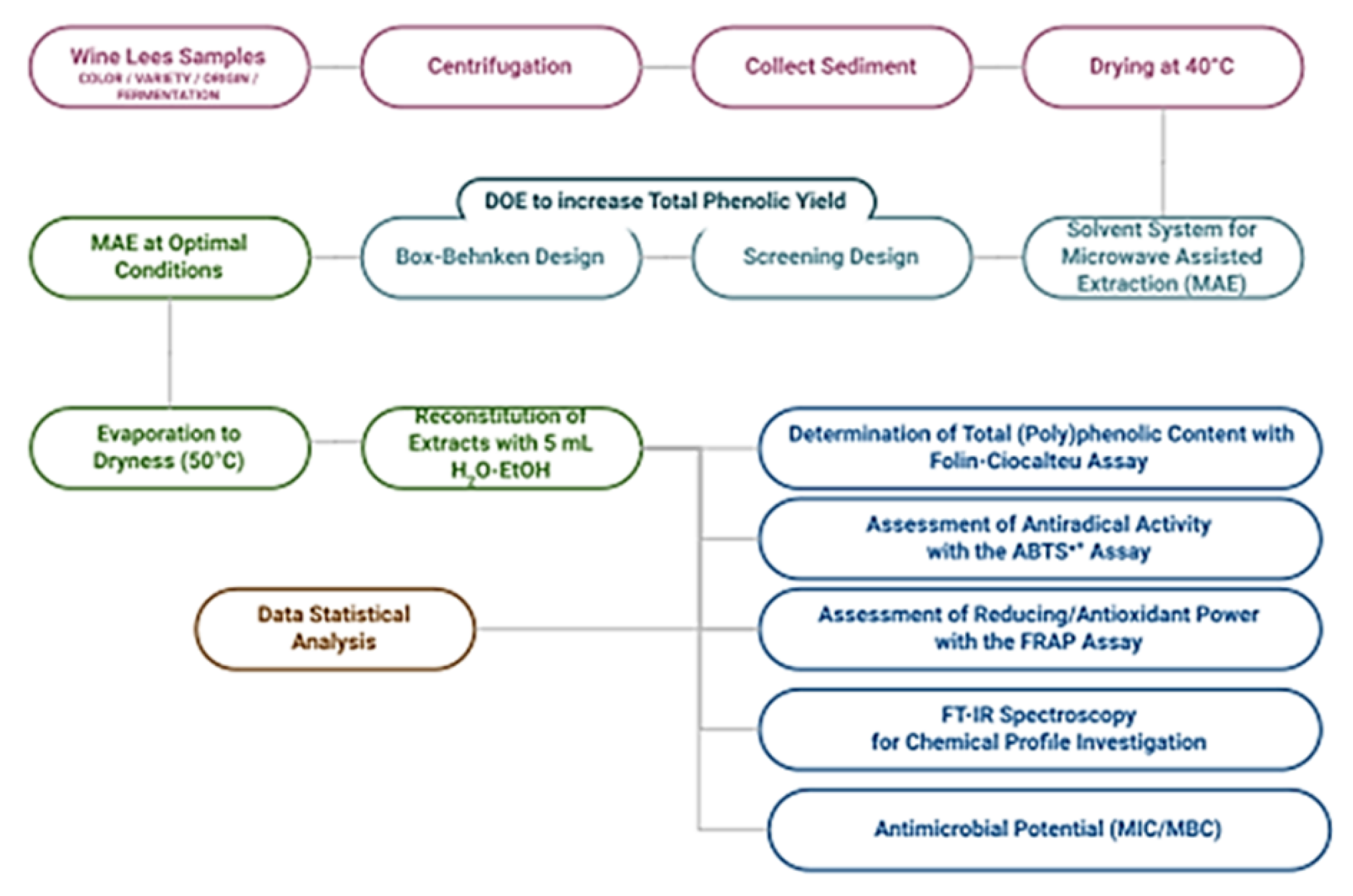
3. Materials and Methods
3.1. Reagents and Standards
3.2. Wine Sees Sample-Set and Sample Preparation
3.3. Microwave-Assisted Extraction (MAE) Instrumentation and Process
3.4. Experimental Design (DOE) Models
3.5. Spectrophotometric Analyses
3.5.1. Total Phenolic Content (TPC)
3.5.2. Scavenging Activity on 2,2′-Azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) Radical (ABTS●+)
3.5.3. Ferric Reducing/Antioxidant Power Assay (FRAP)
3.6. Fourier Transform Infrared Spectroscopy (FTIR)
3.7. Antimicrobial Activity
3.7.1. Tested Microorganisms
3.7.2. In Vitro Assays for Determination of Antibacterial Activity—Microdilution Method
3.8. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ilyas, T.; Chowdhary, P.; Chaurasia, D.; Gnansounou, E.; Pandey, A.; Chaturvedi, P. Sustainable Green Processing of Grape Pomace for the Production of Value-Added Products: An Overview. Environ. Technol. Innov. 2021, 23, 101592. [Google Scholar] [CrossRef]
- Nanni, A.; Parisi, M.; Colonna, M. Wine By-Products as Raw Materials for the Production of Biopolymers and of Natural Reinforcing Fillers: A Critical Review. Polymers 2021, 13, 381. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Wu, D.; Zhang, Q.-A.; Sun, D.-W. Ultrasound-Assisted Extraction of Phenolics from Wine Lees: Modeling, Optimization and Stability of Extracts during Storage. Ultrason. Sonochemistry 2014, 21, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Serradilla, J.A.; de Castro, M.D.L. Role of Lees in Wine Production: A Review. Food Chem. 2008, 111, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Troilo, M.; Difonzo, G.; Paradiso, V.M.; Summo, C.; Caponio, F. Bioactive Compounds from Vine Shoots, Grape Stalks, and Wine Lees: Their Potential Use in Agro-Food Chains. Foods 2021, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Romero-Díez, R.; Rodríguez-Rojo, S.; Cocero, M.J.; Duarte, C.M.M.; Matias, A.A.; Bronze, M.R. Phenolic Characterization of Aging Wine. Food Chem. 2018, 259, 188–195. [Google Scholar] [CrossRef] [Green Version]
- Landeka, I.; Dora, M.; Guberović, I.; Petras, M.; Brnčić, R.S.; Đikić, D. Polyphenols from Wine Lees as a Novel Functional Bioactive Compound in the Protection Against Oxidative Stress and Hyperlipidaemia. Food Technol. Biotechnol. 2017, 55, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Ilieva, F.; Petrov, K.; Veličkovska, S.K.; Gunova, N.; Dimovska, V.; Rocha, J.M.F.; Esatbeyoglu, T. Influence of Autochthonous and Commercial Yeast Strains on Fermentation and Quality of Wines Produced from Vranec and Cabernet Sauvignon Grape Varieties from Tikveš Wine-Growing Region, Republic of North Macedonia. Appl. Sci. 2021, 11, 6135. [Google Scholar] [CrossRef]
- Sancho-Galán, P.; Amores-Arrocha, A.; Jiménez-Cantizano, A.; Ferreiro-González, M.; Palacios, V.; Barbero, G.F. Ultrasound-assisted extraction of anthocyanins and total phenolic compounds in Vitis vinifera L. ‘Tempranillo’ winemaking lees. Vitis: J. Grapevine Res. 2019, 58, 39–47. [Google Scholar]
- Socas-Rodríguez, B.; Torres-Cornejo, M.V.; Álvarez-Rivera, G.; Mendiola, J.A. Deep Eutectic Solvents for the Extraction of Bioactive Compounds from Natural Sources and Agricultural By-Products. Appl. Sci. 2021, 11, 4897. [Google Scholar] [CrossRef]
- Kandylis, P.; Dimitrellou, D.; Moschakis, T. Recent Applications of Grapes and Their Derivatives in Dairy Products. Trends Food Sci. Technol. 2021, 114, 696–711. [Google Scholar] [CrossRef]
- Silva, A.; Silva, V.; Igrejas, G.; Gaivão, I.; Aires, A.; Klibi, N.; Dapkevicius, M.L.E.; Valentão, P.; Falco, V.; Poeta, P. Valorization of Winemaking By-Products as a Novel Source of Antibacterial Properties: New Strategies to Fight Antibiotic Resistance. Molecules 2021, 26, 2331. [Google Scholar] [CrossRef] [PubMed]
- Zervakis, G.I.; Koutrotsios, G. Solid-State Fermentation of Plant Residues and Agro-Industrial Wastes for the Production of Medicinal Mushrooms. In Medicinal Plants and Fungi: Recent Advances in Research and Development; Agrawal, D.C., Tsay, H.-S., Shyur, L.-F., Wu, Y.-C., Wang, S.-Y., Eds.; Medicinal and Aromatic Plants of the World; Springer: Singapore, 2017; pp. 365–396. [Google Scholar]
- Pattnaik, M.; Pandey, P.; Martin, G.J.O.; Mishra, H.N.; Ashokkumar, M. Innovative Technologies for Extraction and Microencapsulation of Bioactives from Plant-Based Food Waste and Their Applications in Functional Food Development. Foods 2021, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Tsiaka, T.; Sinanoglou, V.J.; Zoumpoulakis, P. Chapter 8-Extracting Bioactive Compounds From Natural Sources Using Green High-Energy Approaches: Trends and Opportunities in Lab- and Large-Scale Applications. In Ingredients Extraction by Physicochemical Methods in Food; Grumezescu, A.M., Holban, A.M., Eds.; Handbook of Food Bioengineering; Academic Press: Cambridge, MA, USA, 2017; pp. 307–365. [Google Scholar]
- Pagano, I.; Campone, L.; Celano, R.; Piccinelli, A.L.; Rastrelli, L. Green Non-Conventional Techniques for the Extraction of Polyphenols from Agricultural Food by-Products: A Review. J. Chromatogr. A 2021, 1651, 462295. [Google Scholar] [CrossRef]
- Garrido, T.; Nikolaidis, M.J.; Leceta, I.; Urdanpilleta, M.; Guerrero, P.; la Caba, K.; Kilmartin, P.A. Optimizing the extraction process of natural antioxidants from chardonnay grape marc using microwave-assisted extraction. Waste Manag. 2019, 88, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha, C.B.; Noreña, C.P.Z. Microwave-Assisted Extraction and Ultrasound-Assisted Extraction of Bioactive Compounds from Grape Pomace. Int. J. Food Eng. 2020, 16, 20190191. [Google Scholar] [CrossRef]
- Moret, S.; Conchione, C.; Srbinovska, A.; Lucci, P. Microwave-Based Technique for Fast and Reliable Extraction of Organic Contaminants from Food, with a Special Focus on Hydrocarbon Contaminants. Foods 2019, 8, 503. [Google Scholar] [CrossRef] [Green Version]
- Kaderides, K.; Papaoikonomou, L.; Serafim, M.; Goula, A.M. Microwave-assisted extraction of phenolics from pomegranate peels: Optimization, kinetics, and comparison with ultrasounds extraction. Chem. Eng. Process 2019, 137, 1–11. [Google Scholar] [CrossRef]
- Ren, B.; Chen, C.; Li, C.; Fu, X.; You, L.; Liu, R.H. Optimization of microwave-assisted extraction of Sargassum thunbergii polysaccharides and its antioxidant and hypoglycemic activities. Carbohydr. Polym. 2017, 73, 192–201. [Google Scholar] [CrossRef]
- Putnik, P.; Kovačević, D.B.; Penić, M.; Fegeš, M.; Dragović-Uzelac, V. Microwave-Assisted Extraction (MAE) of Dalmatian Sage Leaves for the Optimal Yield of Polyphenols: HPLC-DAD Identification and Quantification. Food Anal. Methods 2016, 9, 2385–2394. [Google Scholar] [CrossRef]
- Álvarez, A.; Poejo, J.; Matias, A.A.; Duarte, C.M.M.; Cocero, M.J.; Mato, R.B. Microwave Pretreatment to Improve Extraction Efficiency and Polyphenol Extract Richness from Grape Pomace. Effect on Antioxidant Bioactivity. Food Bioprod. Process. 2017, 106, 162–170. [Google Scholar] [CrossRef]
- Bagade, S.B.; Patil, M. Recent Advances in Microwave Assisted Extraction of Bioactive Compounds from Complex Herbal Samples: A Review. Crit. Rev. Anal. Chem. 2021, 51, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.N.; Zhang, J.J.; Li, Y.; Meng, X.; Li, H.B. Microwave-Assisted Extraction of Phenolic Compounds from Melastoma Sanguineum Fruit: Optimization and Identification. Molecules 2018, 23, 2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drevelegka, I.; Goula, A.M. Recovery of grape pomace phenolic compounds through optimized extraction and adsorption processes. Chem. Eng. Process. 2020, 149, 107845. [Google Scholar] [CrossRef]
- Salehi, B.; Vlaisavljevic, S.; Adetunji, C.O.; Adetunji, J.B.; Kregiel, D.; Antolak, H.; Pawlikowska, E.; Uprety, Y.; Mileski, K.S.; Devkota, H.P.; et al. Plants of the Genus Vitis: Phenolic Compounds, Anticancer Properties and Clinical Relevance. Trends Food Sci. Technol. 2019, 91, 362–379. [Google Scholar] [CrossRef]
- Mitrevska, K.; Grigorakis, S.; Loupassaki, S.; Calokerinos, A.C. Antioxidant Activity and Polyphenolic Content of North Macedonian Wines. Appl. Sci. 2020, 10, 2010. [Google Scholar] [CrossRef] [Green Version]
- Makris, D.P.; Psarra, E.; Kallithraka, S.; Kefalas, P. The Effect of Polyphenolic Composition as Related to Antioxidant Capacity in White Wines. Food Res. Int. 2003, 36, 805–814. [Google Scholar] [CrossRef]
- Guendez, R.; Kallithraka, S.; Makris, D.P.; Kefalas, P. Determination of Low Molecular Weight Polyphenolic Constituents in Grape (Vitis vinifera Sp.) Seed Extracts: Correlation with Antiradical Activity. Food Chem. 2005, 89, 1–9. [Google Scholar] [CrossRef]
- Rockenbach, I.I.; Gonzaga, L.V.; Rizelio, V.M.; de Souza Schmidt Gonçalves, A.E.; Genovese, M.I.; Fett, R. Phenolic Compounds and Antioxidant Activity of Seed and Skin Extracts of Red Grape (Vitis vinifera and Vitis labrusca) Pomace from Brazilian Winemaking. Food Res. Int. 2011, 44, 897–901. [Google Scholar] [CrossRef]
- Lingua, M.S.; Fabani, M.P.; Wunderlin, D.A.; Baroni, M.V. From Grape to Wine: Changes in Phenolic Composition and Its Influence on Antioxidant Activity. Food Chem. 2016, 208, 228–238. [Google Scholar] [CrossRef]
- Romero-Díez, R.; Matos, M.; Rodrigues, L.; Bronze, M.R.; Rodríguez-Rojo, S.; Cocero, M.J.; Matias, A.A. Microwave and Ultrasound Pre-Treatments to Enhance Anthocyanins Extraction from Different Wine Lees. Food Chem. 2019, 272, 258–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzachristas, A.; Pasvanka, K.; Liouni, M.; Tataridis, P.; Proestos, C. Effect of Hippophae Rhamnoides, L. Leaves Treatment on the Radical Scavenging Activity, Reducing Power, Total Phenol Content and Sensory Profile of Dry White Wines Vinified with and without the Use of Sulphur Dioxide. Appl. Sci. 2018, 10, 3444. [Google Scholar] [CrossRef]
- Karvela, E.; Makris, D.P.; Karathanos, V.T. Implementation of Response Surface Methodology to Assess the Antiradical Behaviour in Mixtures of Ascorbic Acid and α-Tocopherol with Grape (Vitis vinifera) Stem Extracts. Food Chem. 2012, 132, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Kallithraka, S.; Mohdaly, A.A.-A.; Makris, D.P.; Kefalas, P. Determination of Major Anthocyanin Pigments in Hellenic Native Grape Varieties (Vitis vinifera Sp.): Association with Antiradical Activity. J. Food Compos. Anal. 2005, 18, 375–386. [Google Scholar] [CrossRef]
- Majkić, T.M.; Torović, L.D.; Lesjak, M.M.; Četojević-Simin, D.D.; Beara, I.N. Activity Profiling of Serbian and Some Other European Merlot Wines in Inflammation and Oxidation Processes. Food Res. Int. 2019, 121, 151–160. [Google Scholar] [CrossRef]
- Trošt, K.; Klančnik, A.; Mozetič Vodopivec, B.; Sternad Lemut, M.; Jug Novšak, K.; Raspor, P.; Smole Možina, S. Polyphenol, Antioxidant and Antimicrobial Potential of Six Different White and Red Wine Grape Processing Leftovers. J. Sci Food Agric. 2016, 96, 4809–4820. [Google Scholar] [CrossRef]
- Doshi, P.; Adsule, P.; Banerjee, K.; Oulkar, D. Phenolic Compounds, Antioxidant Activity and Insulinotropic Effect of Extracts Prepared from Grape (Vitis vinifera L.) Byproducts. J. Food Sci Technol 2015, 52, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Smith, B.C. Distinguishing Structural Isomers: Mono- and Disubstituted Benzene Rings. Spectroscopy 2016, 31, 36–39. [Google Scholar]
- Murugesh, S.; Vino, P. Phytochemical Constituents, Antioxidant Activity and FT-IR Analysis of Pisonia Grandis Leaf Extracts. Phyto 2017, 9, 933–938. [Google Scholar] [CrossRef] [Green Version]
- Baciu, A.; Ranga, F.; Fetea, F.; Zavoi, S.; Socaciu, C. Fingerprinting Food Supplements and Their Botanical Ingredients by Coupled UV/Vis/FTIR Spectrometry. Bull. Univ. Agric. Sci. Vet. Med. Cluj-Napoca Food Sci. Technol. 2013, 70, 8–15. [Google Scholar] [CrossRef] [Green Version]
- Andreou, V.; Strati, I.F.; Fotakis, C.; Liouni, M.; Zoumpoulakis, P.; Sinanoglou, V.J. Herbal Distillates: A New Era of Grape Marc Distillates with Enriched Antioxidant Profile. Food Chem. 2018, 253, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Leopold, L.; Diehl, H.; Socaciu, C. Quantification of Glucose, Fructose and Sucrose in Apple Juices Using ATR-MIR Spectroscopy Coupled with Chemometry. UASVMCN-AGR 1970, 66, 350–357. [Google Scholar] [CrossRef]
- Grassi, S.; Amigo, J.M.; Lyndgaard, C.B.; Foschino, R.; Casiraghi, E. Beer Fermentation: Monitoring of Process Parameters by FT-NIR and Multivariate Data Analysis. Food Chem. 2014, 155, 279–286. [Google Scholar] [CrossRef]
- Sinanoglou, V.J.; Zoumpoulakis, P.; Fotakis, C.; Kalogeropoulos, N.; Sakellari, A.; Karavoltsos, S.; Strati, I.F. On the Characterization and Correlation of Compositional, Antioxidant and Colour Profile of Common and Balsamic Vinegars. Antioxidants 2018, 7, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banc, R.; Loghin, F.; Miere, D.; Fetea, F.; Socaciu, C. Romanian Wines Quality and Authenticity Using FT-MIR Spectroscopy Coupled with Multivariate Data Analysis. Not. Bot. Horti Agrobot. Cluj-Napoca 2014, 42, 556–564. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, D.A.; Salvador, A.A.; Smânia, A.; Smânia, E.F.A.; Maraschin, M.; Ferreira, S.R.S. Antimicrobial Activity and Composition Profile of Grape (Vitis vinifera) Pomace Extracts Obtained by Supercritical Fluids. J. Biotechnol. 2013, 164, 423–432. [Google Scholar] [CrossRef]
- Moschona, A.; Liakopoulou-Kyriakides, M. Encapsulation of Biological Active Phenolic Compounds Extracted from Wine Wastes in Alginate-Chitosan Microbeads. J. Microencapsul. 2018, 35, 229–240. [Google Scholar] [CrossRef]
- Merkytė, V.; Longo, E.; Windisch, G.; Boselli, E. Phenolic Compounds as Markers of Wine Quality and Authenticity. Foods 2020, 9, 1785. [Google Scholar] [CrossRef]
- Gutiérrez-Escobar, R.; Aliaño-González, M.J.; Cantos-Villar, E. Wine Polyphenol Content and Its Influence on Wine Quality and Properties: A Review. Molecules 2021, 26, 718. [Google Scholar] [CrossRef]
- Tsiaka, T.; Fotakis, C.; Lantzouraki, D.Z.; Tsiantas, K.; Siapi, E.; Sinanoglou, V.J.; Zoumpoulakis, P. Expanding the Role of Sub-Exploited DOE-High Energy Extraction and Metabolomic Profiling towards Agro-Byproduct Valorization: The Case of Carotenoid-Rich Apricot Pulp. Molecules 2020, 25, 2702. [Google Scholar] [CrossRef]
- Lantzouraki, D.Z.; Tsiaka, T.; Soteriou, N.; Asimomiti, G.; Spanidi, E.; Natskoulis, P.; Gardikis, K.; Sinanoglou, V.J.; Zoumpoulakis, P. Antioxidant Profiles of Vitis Vinifera L. and Salvia Triloba L. Leaves Using High-Energy Extraction Methodologies. J. AOAC Int. 2020, 103, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Stegger, M.; Andersen, P.S.; Kearns, A.; Pichon, B.; Holmes, M.A.; Edwards, G.; Laurent, F.; Teale, C.; Skov, R.; Larsen, A.R. Rapid Detection, Differentiation and Typing of Methicillin-Resistant Staphylococcus Aureus Harbouring Either MecA or the New MecA Homologue MecALGA251. Clin. Microbiol. Infect. 2012, 18, 395–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilker, M. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2006; Volume 26, pp. M7–A7.
- Tsukatani, T.; Suenaga, H.; Shiga, M.; Noguchi, K.; Ishiyama, M.; Ezoe, T.; Matsumoto, K. Comparison of the WST-8 Colorimetric Method and the CLSI Broth Microdilution Method for Susceptibility Testing against Drug-Resistant Bacteria. J. Microbiol. Methods 2012, 90, 160–166. [Google Scholar] [CrossRef]
- Ghasemi, A.; Zahediasl, S. Normality Tests for Statistical Analysis: A Guide for Non-Statisticians. Int. J. Endocrinol. Metab. B 2012, 10, 486–489. [Google Scholar] [CrossRef] [Green Version]
| Extraction Solvent (v/v) | Extraction Yield (mg of GAE g−1 Dry Sediment) (± stdev), n = 3 1 |
|---|---|
| Ethanol | 1.60 (± 0.21) b |
| Water | 0.46 (± 0.21) c |
| Ethanol:water 1:1 | 2.034 (± 0.064) a |
| Extraction Factors | Optimal Conditions |
|---|---|
| MW power (W) | 54 |
| Extraction time (minutes) | 35 |
| Solvent/material ratio (mL g−1) | 60 |
| Extraction yield (mg of GAE g−1 dry sediment) (±stdev), n = 3 1 | 3.58 (±0.17) |
| A/A. 2 | Average mg of GAE g−1 of Dry Sediment (±S.D.), n = 3 1 | Average mg of TE g−1 of Dry Sediment (±stdev), n = 3 1 | Average mg of Fe (II) g−1 of Dry Sediment (±stdev), n = 3 1 |
|---|---|---|---|
| 1 | 3.57(±0.40) k | 33.7(±1.8) i,j | 15.9(±1.9) r |
| 2 | 5.81(±0.69) h,i,j,k | 20.7(±2.7) l,m | 46.7(±3.7) m,n,o |
| 3 | 4.82(±0.95) i,j,k | 15.77(±0.80) m,n | 52.86(±0.58) l,m |
| 4 | 4.28(±0.31) j,k | 12.67(±0.90) n | 35.4(±1.4) p,q |
| 5 | 7.65(±0.84) g,h,i,j | 40.6(±2.9) g,h,i | 76.1(±5.2) k |
| 6 | 11.5(±2.1) e,f,g | 48.4(±1.3) f | 103.8(±5.0) i |
| 7 | 7.48(±0.88) h,i,j,k | 48.2(±2.3) f | 44.7(±2.1) m,n,o,p |
| 8 | 4.09(±0.65) j,k | 30.7(±1.4) j,k | 32.7(±2.1) q |
| 9 | 5.61(±0.72) h,i,j,k | 20.6(±3.0) l,m | 57.7(±1.9) l |
| 10 | 8.72(±0.81) f,g,h,i | 24.6(±4.0) k,l | 76.0(±3.9) k |
| 11 | 5.1(±1.3) i,j,k | 25.4(±1.3) k,l | 34.43(±0.39) q |
| 12 | 6.9(±1.2) h,i,j,k | 40.8(±1.5) g,h,i | 39.5(±2.7) o,p,q |
| 13 | 6.01(±0.25) h,i,j,k | 43.3(±2.4) f,g,h | 41.63(±0.58) n,o,p,q |
| 14 | 6.73(±0.84) h,i,j,k | 42.41(±0.90) f,g,h | 49.5(±2.7) l,m,n |
| 15 | 13.22(±0.79) d,e | 67.3(±3.8) e | 117.5(±1.7) h |
| 16 | 17.2(±1.2) c | 72.2(±2.3) d,e | 151.1(±2.3) f |
| 17 | 16.9(±2.0) c,d | 76.5(±3.5) d | 178.9(±3.1) e |
| 18 | 12.6(±1.6) e,f | 36.4(±1.7) h,i,j | 89.3(±0.97) j |
| 19 | 7.10(±0.85) h,i,j,k | 25.2(±2.5) k,l | 58.85(±0.39) l |
| 20 | 22.7(±1.2) a,b | 120.4(±2.2) a | 224.8(±6.4) b |
| 21 | 25.3(±1.5) a | 120.4(±3.6) a | 243.8(±1.4) a |
| 22 | 24.0(±2.8) a | 113.5(±3.5) a,b | 234.2(±3.7) a,b |
| 23 | 24.2(±1.5) a | 100.9(±2.5) c | 192.5(±1.5) d |
| 24 | 26.0(±1.1) a | 114.08(±0.30) a,b | 203.4(±4.6) c |
| 25 | 18.6(±1.1) c | 111.5(±0.70) b | 139.8(±3.1) g |
| 26 | 19.0(±1.6) b,c | 94.2(±1.2) c | 231.4(±3.7) b |
| 27 | 8.0(±1.4) g,h,i,j | 43.9(±4.0) f,g | 70.3(±1.7) k |
| 28 | 9.25(±0.63) f,g,h | 41.4(±1.9) f,g,h | 78.0(±5.6) k |
| A/A | Citric Acid | Lactic Acid | Tartaric Acid | Glycerol | Fructose | Glucose | Saccharose |
|---|---|---|---|---|---|---|---|
| 1 | N.D. | 5.9 | 15.5 | 57.0 | 131.5 | 213.9 | 3.0 |
| 2 | 6.9 | 9.0 | 17.2 | 53.5 | 1.5 | N.D. | 8.5 |
| 3 | 4.4 | 4.5 | 15.3 | 50.5 | 22.5 | 12.5 | 3.0 |
| 4 | 7.7 | 9.1 | 4.1 | 57.5 | 12.0 | 14.5 | N.D. |
| 5 | N.D. | 5.3 | 16.5 | 60.5 | 193.9 | 113.5 | N.D. |
| 6 | 4.2 | 9.6 | 14.8 | 73.4 | 167.4 | 71.9 | 1.0 |
| 7 | 6.5 | 17.2 | 16.8 | 52.5 | N.D. | N.D. | 8.5 |
| 8 | 11.5 | 11.8 | 16.0 | 45.0 | N.D. | N.D. | 6.0 |
| 9 | 5.8 | 5.1 | 18.2 | 32.0 | 2.0 | 4.0 | 11.0 |
| 10 | 16.0 | 8.0 | 49.9 | 81.6 | N.D. | 8.3 | 21.6 |
| 11 | 15.9 | 14.3 | 0.1 | 43.0 | N.D. | 10.5 | 0.5 |
| 12 | 4.3 | 9.5 | 18.2 | 35.5 | 4.5 | 3.5 | 6.5 |
| 13 | 3.4 | 10.7 | 19.4 | 43.0 | 6.5 | 3.0 | 8.0 |
| 14 | 8.1 | 9.2 | 17.0 | 41.5 | 1.5 | 7.0 | 10.5 |
| 15 | 9.3 | 13.1 | 14.3 | 68.0 | 11.5 | 6.5 | 7.0 |
| 16 | 3.7 | 6.8 | 20.2 | 66.0 | 6.0 | 11.0 | 4.0 |
| 17 | 4.2 | 9.7 | 18.1 | 62.9 | 12.5 | 8.0 | 9.5 |
| 18 | N.D. | 4.8 | 17.3 | 61.5 | 192.5 | 141.0 | N.D. |
| 19 | N.D. | 2.0 | 20.5 | 56.0 | 178.9 | 134.4 | N.D. |
| 20 | 6.8 | 8.9 | 17.0 | 56.0 | 3.5 | 1.0 | 8.0 |
| 21 | 6.7 | 8.2 | 14.6 | 63.0 | 1.0 | 4.5 | 3.5 |
| 22 | 5.4 | 9.6 | 15.4 | 70.0 | 3.5 | 3.5 | 6.5 |
| 23 | 9.0 | 7.9 | 11.1 | 44.5 | N.D. | 3.0 | 13.0 |
| 24 | 7.5 | 11.4 | 15.9 | 66.0 | 5.5 | 6.0 | 4.5 |
| 25 | 3.8 | 7.3 | 14.3 | 50.0 | 1.0 | 6.0 | 5.0 |
| 26 | 5.3 | 7.9 | 22.9 | 61.5 | 6.0 | 11.5 | 10.0 |
| 27 | 0.2 | 16.2 | 24.4 | 74.5 | 13.0 | N.D. | 5.0 |
| 28 | 4.7 | 13.8 | 17.3 | 54.0 | 8.0 | 3.0 | 8.0 |
| A/A | MIC/MBC Values | E.c. | S.t. | En.cl. | P.a | L.m | M.f. | B.c. | S.a. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MIC | 1.13 ± 0.11 a,b,k | 0.63 ± 0.06 a,b,c,g | 0.67 ± 0.11 a,b | 0.6 ± 0.00 a,b,c,d | 0.53 ± 0.11 a,b,d | 0.56 ± 0.06 a,b,c,e | 0.67 ± 0.11 a,b,c,e | 0.63 ± 0.06 a,b,c,e |
| MBC | 2.27 ± 0.23 a,b,f,i | 1.30 ± 0.17 a,f | 1.13 ± 0.11 a,b | 1.2 ± 0.00 a,b,j,e | 1.10 ± 0.17 a,e | 1.30 ± 0.17 a,b,c,d | 1.13 ± 0.11 a,b,f | 1.23 ± 0.06 a,d | |
| 2 | MIC | 6.63 ± 0.11 a | 3.40 ± 0.09 a | 3.35 ± 0.00 a | 3.23 ± 0.2 a | 3.30 ± 0.1 a | 3.27 ± 0.14 a | 6.47 ± 0.4 a | 3.23 ± 0.2 a |
| MBC | 13.33 ± 0.3 a | 6.47 ± 0.4 a | 6.63 ± 0.11 a | 6.7 ± 0.00 a,b | 6.57 ± 0.23 a | 6.47 ± 0.4 a | 13.33 ± 0.3 a | 6.47 ± 0.4 a | |
| 3 | MIC | 0.3 ± 0.00 a,h | 0.53 ± 0.11a,c | 0.33 ± 0.06 a,b,d,e | 0.6 ± 0.00 a,b,c | 0.67 ± 0.11 a,b,c,e | 0.27 ± 0.06 a,b | 0.37 ± 0.11 a | 0.33 ± 0.06 a |
| MBC | 0.67 ± 0.11 a,b | 1.13 ± 0.11 a | 0.73 ± 0.23 a,b,d | 1.13 ± 0.11 a,b,e | 1.2 ± 0.00 a,b,c,e | 0.53 ± 0.11 a,c,d | 0.6 ± 0.00 a,b | 0.67 ± 0.11 a,b | |
| 4 | MIC | 3.23 ± 0.11 a | 1.6 ± 0.00 a,b | 1.53 ± 0.11 a | 1.63 ± 0.06 a,b | 1.67 ± 0.11 a | 3.20 ± 0.17 a | 0.87 ± 0.11 a | 3.20 ± 0.17 a |
| MBC | 6.40 ± 0.35 a | 3.7 ± 0.23 a | 3.20 ± 0.17 a | 3.37 ± 0.11 a | 3.3 ± 0.00 a,b | 6.40 ± 0.35 a | 1.53 ± 0.11 a,b,c,f | 6.57 ± 0.06 a,b | |
| 5 | MIC | 0.93 ± 0.06 a,b | 0.37 ± 0.14 a,e | 0.53 ± 0.14 a,b | 0.42 ± 0.06 a,f | 0.47 ± 0.03 a,b,d | 0.50 ± 0.09 a,b,f | 0.45 ± 0.00 a | 0.47 ± 0.03 a,e |
| MBC | 1.87 ± 0.11 a,b,i | 0.9 ± 0.11 a,b,f,g | 0.83 ± 0.11 a,b | 0.77 ± 0.23 a,b | 0.83 ± 0.06 a,b,c,g | 0.93 ± 0.06 a,b,c | 0.83 ± 0.11 a,b,f | 0.93 ± 0.06 a,b,c | |
| 6 | MIC | 0.87 ± 0.11 a,b | 0.93 ± 0.23 a,b,c,e,g | 0.83 ± 0.06 a,b,d | 0.8 ± 0.00 a,b,c,i | 0.7 ± 0.17 a,b | 0.73 ± 0.11 a | 0.33 ± 0.11 a | 0.73 ± 0.11 a,c,g |
| MBC | 1.57 ± 0.06 a,b,i | 1.6 ± 0.00 a,b,f | 1.73 ± 0.23 a,d | 1.53 ± 0.11 a,b | 1.56 ± 0.06 a | 1.6 ± 0.00 a,b,c,d | 0.87 ± 0.11 a,b,c,f | 1.47 ± 0.23 a,e | |
| 7 | MIC | 0.93 ± 0.06 a,b | 0.47 ± 0.03 a | 0.5 ± 0.1 a,b,d | 0.45 ± 0.00 a,d | 0.40 ± 0.1 a,f | 0.43 ± 0.03 a,f,e | 0.45 ± 0.00 a | 0.48 ± 0.06 a |
| MBC | 1.73 ± 0.11 a,i | 0.93 ± 0.06 a,b,d | 0.87 ± 0.06 a,b | 0.93 ± 0.06 a,b,j | 0.77 ± 0.23 a,b,c | 0.97 ± 0.11 a,b,c | 0.9 ± 0.00 a,b,d | 0.93 ± 0.06 a,b,c | |
| 8 | MIC | 1.9 ± 0.08 a,b | 1.73 ± 0.2 a | 0.95 ± ±0.04 a | 1.85 ± 0.00 a,b | 1.77 ± 0.14 a,b | 1.85 ± 0.00 a | 1.90 ± 0.09 a | 0.95 ± 0.04 a,b,e,i |
| MBC | 3.7 ± 0.00 a | 3.63 ± 0.11a | 1.90 ± 0.09 a,d | 3.47 ± 0.4 a | 3.63 ± 0.11 a | 3.7 ± 0.00 a | 3.63 ± 0.11 a | 1.85 ± 0.00 a,b,d | |
| 9 | MIC | 2.53 ± 0.5 a | 2.7 ± 0.17 a | 1.43 ± 0.06 a | 1.27 ± 0.2 a,b | 2.8 ± 0.00 a | 2.73 ± 0.11 a | 0.38 ± 0.01 a | 1.33 ± 0.11 a |
| MBC | 5.80 ± 0.17 a | 5.6 ± 0.11 a | 2.87 ± 0.11 a | 2.70 ± 0.17 a | 5.47 ± 0.4 a | 5.63 ± 0.11 a | 0.83 ± 0.15 a,b | 2.53 ± 0.46 a,b | |
| 10 | MIC | 0.5 ± 0.00 a,b | 0.15 ± 0.04 a,g | 0.27 ± 0.02 a,b | 0.14 ± 0.01 a,b | 0.15 ± 0.04 a,b | 0.27 ± 0.02 a,b | 0.17 ± 0.07 a,b,c | 0.12 ± 0.17 a,c,k |
| MBC | 1 ± 0.00 a,b,c,f,i | 0.34 ± 0.11 a,b,g | 0.58 ± 0.10 a,b | 0.32 ± 0.1 a | 0.34 ± 0.13 a,c,f | 0.68 ± 0.27 a,b,c | 0.26 ± 0.00 a,b | 0.29 ± 0.05 a,c | |
| 11 | MIC | 0.68 ± 0.01 a,b,j | 1.4 ± 0.09 a | 0.72 ± 0.07 a,b,c | 1.35 ± 0.00 a,b | 1.40 ± 0.1 a,b | 0.70 ± 0.04 a,b,e | 0.67 ± 0.00 a,b,d | 1.40 ± 0.09 a,i |
| MBC | 1.23 ± 0.20 a,b | 2.47 ± 0.4 a | 1.40 ± 0.08 a,b,d | 2.63 ± 0.11 a | 2.7 ± 0.00 a,b | 1.30 ± 0.09 a,b | 1.23 ± 0.2 a,b | 2.63 ± 0.11 a | |
| 12 | MIC | 0.33 ± 0.06 a,b,k | 0.53 ± 0.11 a | 0.37 ± 0.11 a,b,c | 0.67 ± 0.11 a,b | 0.63 ± 0.06 a,e | 0.7 ± 0.06 a,b | 0.23 ± 0.11 a,e | 0.27 ± 0.06 a,g |
| MBC | 0.73 ± 0.23 a,b | 1.13 ± 0.11 a,c | 0.6 ± 0.00 a,b | 1.2 ± 0.00 a,b,e | 1.10 ± 0.17 a | 0.70 ± 0.17 a,d | 0.6 ± 0.00 a,b | 0.73 ± 0.23 a,e | |
| 13 | MIC | 0.53 ± 0.06 a,b,d | 0.33 ± 0.14 a,c | 0.25 ± 0.00 a | 0.33 ± 0.14 a,b,d | 0.32 ± 0.11 a,b | 0.25 ± 0.00 a | 0.27 ± 0.03 a | 0.31 ± 0.11 a |
| MBC | 1 ± 0.00 a,b,i | 0.67 ± 0.3 a,f | 0.60 ± 0.17 a,b | 0.5 ± 0.00 a | 0.57 ± 0.11 a,b,e | 0.63 ± 0.23 a,b | 0.5 ± 0.00 a | 0.67 ± 0.3 a | |
| 14 | MIC | 0.77 ± 0.11 a,b,h | 0.41 ± 0.11 a | 0.33 ± 0.3 a,b,d | 0.35 ± 0.00 a,f,c | 0.40 ± 0.1 a,f | 0.80 ± 0.17 a,e | 0.20 ± 0.04 a,b,d,e | 0.42 ± 0.11 a,b |
| MBC | 1.27 ± 0.23 a,b | 0.7 ± 0.26 a,f | 0.7 ± 0.00 a,b | 0.77 ± 0.11 a,b | 0.7 ± 0.26 a,c | 1.13 ± 0.11 a,b,d | 0.42 ± 0.11 a,d | 0.7 ± 0.00 a | |
| 15 | MIC | 1.07 ± 0.06 a,b,d | 0.70 ± 0.26 a,c | 0.28 ± 0.11 a,b,d | 1.13 ± 0.06 a,b,d | 0.70 ± 0.25 a | 2.13 ± 0.11 a | 0.30 ± 0.4 a,b,e,g | 0.28 ± 0.014 a |
| MBC | 2.13 ± 0.11 a,b,f | 1.23 ± 0.23 a,b | 0.7 ± 0.3 a,b,c,f | 0.63 ± 0.14 a,b,e | 1.1 ± 0.00 a | 4.27 ± 0.23 a | 0.70 ± 0.26 a,b | 0.57 ± 0.03 a,c | |
| 16 | MIC | 0.9 ± 0.1 a,b | 0.43 ± 0.06 a | 0.47 ± 0.11 a,b,f | 0.4 ± 0.00 a,i | 0.3 ± 0.17 a,b,e | 0.43 ± 0.06 a,f,e | 0.47 ± 0.11 a | 0.4 ± 0.00 a,b |
| MBC | 1.63 ± 0.11 a | 0.87 ± 0.11 a,b,c,d | 0.8 ± 0.09 a,b,d | 0.73 ± 0.11 a,b,c,j | 0.83 ± 0.06 a,b,c | 0.8 ± 0.00 a,c | 0.87 ± 0.11 a,b,f | 0.70 ± 0.17 a | |
| 17 | MIC | 0.83 ± 0.14 a,b,h | 0.23 ± 0.06 a | 0.40 ± 0.04 a,e,f | 0.21 ± 0.03 a,b | 0.19 ± 0.01 a,d | 0.18 ± 0.02 a,c | 0.23 ± 0.06 a | 0.40 ± 0.04 a,b |
| MBC | 1.33 ± 0.3 a,b | 0.45 ± 0.12 a,c | 0.80 ± 0.09 a,c,d | 0.34 ± 0.07 a,b | 0.38 ± 0.00 a,g | 0.40 ± 0.04 a,d | 0.42 ± 0.07 a,d | 0.80 ± 0.09 a,b,e | |
| 18 | MIC | 1.13 ± 0.06 a,b,k | 0.70 ± 0.2 a,c,b,e,g | 0.67 ± 0.2 a,b.d | 0.70 ± 0.3 a,b,c,d,f | 0.60 ± 0.1 a | 1.1 ± 0.06 a,b,e | 0.70 ± 0.3 a,b,e,g | 0.55 ± 0.00 a,k |
| MBC | 2.13 ± 0.11 a,b | 1.07 ± 0.06 a,b | 1.23 ± 0.23 a,b,d | 1.17 ± 0.11 a,b,j | 1.07 ± 0.06 a | 2.30 ± 0.17 a,b | 1.23 ± 0.23 a,b | 1.03 ± 0.11 a | |
| 19 | MIC | 0.67 ± 0.30 a,l,j | 0.33 ± 0.14 a | 0.25 ± 0.00 a | 0.28 ± 0.06 a,b,f | 0.32 ± 0.11 a,b,c | 0.5 ± 0.00 a,b,f | 0.27 ± 0.11 a | 0.37 ± 0.2 a,e |
| MBC | 1.17 ± 0.30 a,b,f,i | 0.67 ± 0.3 a,f | 0.5 ± 0.00 a,b | 0.57 ± 0.11 a,b | 0.60 ± 0.17 a,b | 1.16 ± 0.28 a,c | 0.67 ± 0.28 a,b,f | 0.43 ± 0.11 a,e | |
| 20 | MIC | 0.08 ± 0.01 a,b | 0.075 ± 0.00 a,g | 0.075 ± 0.00 a,b,f | 0.07 ± 0.01 a | 0.17 ± 0.03 a,b | 0.20 ± 0.08 a,e | 0.15 ± 0.00 a,b | 0.13 ± 0.03 a |
| MBC | 0.17 ± 0.03 a,b | 0.20 ± 0.08 a,b,d | 0.15 ± 0.00 a,b | 0.17 ± 0.03 a,b | 0.33 ± 0.06 a,c,e | 0.3 ± 0.00 a,b,c | 0.37 ± 0.11 a,c,f | 0.27 ± 0.06 a,b,e | |
| 21 | MIC | 0.2 ± 0.00 a,b,j,k | 0.13 ± 0.06 a,e | 0.27 ± 0.11 a | 0.1 ± 0.00 a,b | 0.17 ± 0.11 a,b | 0.20 ± 0.17 a,e | 0.13 ± 0.06 a,b | 0.27 ± 0.11 a,g |
| MBC | 0.4 ± 0.00 a,b,d | 0.17 ± 0.06 a,b,f | 0.47 ± 0.11 a,b | 0.2 ± 0.00 a,b,c | 0.27 ± 0.11 a,b,c | 0.33 ± 0.23 a,b | 0.30 ± 0.17 a,b,c | 0.50 ± 0.17 a | |
| 22 | MIC | 0.27 ± 0.1 a,b,d,h | 0.1 ± 0.00 a,c | 0.13 ± 0.06 a | 0.17 ± 0.11 a,d | 0.13 ± 0.1 a | 0.27 ± 0.11 a,b | 0.23 ± 0.06 a,e | 0.1 ± 0.00 a,b |
| MBC | 0.33 ± 0.11 a,b | 0.27 ± 0.11 a,b,d | 0.30 ± 0.17 a,b | 0.2 ± 0.00 a,b,c | 0.17 ± 0.06 a,b,c | 0.47 ± 0.11 a,b | 0.50 ± 0.17 a | 0.2 ± 0.00 a,b,e | |
| 23 | MIC | 0.8 ± 0.00 a,b,d,h,k | 0.33 ± 0.11 a,c | 0.73 ± 0.11 a,b,e | 0.77 ± 0.06 a,b,c,d,f | 0.33 ± 0.11 a,b,e | 0.8 ± 0.00 a,e | 0.27 ± 0.11 a | 0.37 ± 0.06 a |
| MBC | 1.53 ± 0.11 a,b,f | 0.87 ± 0.11 a,b,c | 1.53 ± 0.11 a,d | 1.40 ± 0.34 a,b,j | 0.87 ± 0.11 a,b,f,e | 1.56 ± 0.06 a,b,c | 0.4 ± 0.00 a,f | 0.87 ± 0.11 a,b,e | |
| 24 | MIC | 0.87 ± 0.11 a,b,k | 0.47 ± 0.11 a | 0.43 ± 0.06 a | 0.4 ± 0.00 a,i | 0.33 ± 0.11 a,b | 0.73 ± 0.11 a | 0.27 ± 0.11 a | 0.23 ± 0.06 a,c |
| MBC | 1.40 ± 0.35 a,b,f | 0.87 ± 0.11 a,b,d | 0.73 ± 0.11 a,b,d | 0.8 ± 0.00 a,b | 0.87 ± 0.11 a,b,f,e | 1.6 ± 0.00 a,b,c,d | 0.53 ± 0.23 a,b | 0.47 ± 0.11 a | |
| 25 | MIC | 0.18 ± 0.06 a,b,j | 0.08 ± 0.01 a,b,g | 0.08 ± 0.01 a,f | 0.075 ± 0.00 a,b,c | 0.075 ± 0.00 a,b,f | 0.07 ± 0.01 a,b,c,f | 0.075 ± 0.00 a | 0.08 ± 0.01 a,b |
| MBC | 0.3 ± 0.00 a,i | 0.18 ± 0.06 a,c | 0.13 ± 0.03 a,b | 0.15 ± 0.00 a,b | 0.20 ± 0.08 a,b,c | 0.17 ± 0.03 a,b,c | 0.15 ± 0.00 a,b | 0.13 ± 0.03 a,b,e | |
| 26 | MIC | 0.67 ± 0.3 a,b,k | 0.33 ± 0.14 a,c | 0.27 ± 0.03 a | 0.25 ± 0.00 a,c | 0.32 ± 0.11 a,b,c | 0.67 ± 0.28 a,b | 0.25 ± 0.00 a,g | 0.33 ± 0.14 a |
| MBC | 1.17 ± 0.3 a,b,d | 0.67 ± 0.3a,f | 0.47 ± 0.06a,b | 0.57 ± 0.11a,b | 0.67 ± 0.28 a,b,e | 1 ± 0.00a,b,d | 0.5 ± 0.00a | 0.67 ± 0.3a | |
| 27 | MIC | 1.33 ± 0.3 a,d,h,k | 0.33 ± 0.11 a,c | 0.37 ± 0.06 a,b,c,d | 0.43 ± 0.06 a,d,f | 0.4 ± 0.00 a,f | 0.47 ± 0.11 a,c | 0.50 ± 0.17 a | 0.37 ± 0.06 a |
| MBC | 2.67 ± 0.6 a,b | 0.87 ± 0.11 a,b,f | 0.77 ± 0.06 a | 0.70 ± 0.17 a,e,j | 0.73 ± 0.11 a,c | 0.87 ± 0.11 a,b,e | 0.8 ± 0.00 a,b,c | 0.87 ± 0.11 a,b,e | |
| 28 | MIC | 0.5 ± 0.00 a,b | 0.16 ± 0.03 a | 0.14 ± 0.01 a,b,f | 0.16 ± 0.03 a,b,c | 0.13 ± 0.03 a,d | 0.14 ± 0.00 a,b,f | 0.17 ± 0.06 a,b,c | 0.15 ± 0.02 a,e |
| MBC | 1 ± 0.00 a,b,i | 0.28 ± 0.01 a | 0.30 ± 0.03 a,b,d | 0.35 ± 0.13 a,b | 0.25 ± 0.05 a,b | 0.28 ± 0.00 a,b,e | 0.29 ± 0.11 a,b | 0.27 ± 0.017 a,b,e | |
| Streptomycin | MIC | 0.10 ± 0.04 a,d | 0.10 ± 0.03 a,b | 0.025 ± 0.02 a,b,e | 0.15 ± 0.04 a,f | 0.10 ± 0.05 a,e | 0.05 ± 0.06 a | 0.025 ± 0.01 a | 0.10 ± 0.05 a |
| MBC | 0.20 ± 0.01 a | 0.20 ± 0.06 a,b | 0.05 ± 0.01 a,b | 0.30 ± 0.02 a,b | 0.20 ± 0.01 a,b | 0.1 ± 0.00 a,b | 0.05 ± 0.02 a,b | 0.20 ± 0.02 a,b | |
| Ampicillin | MIC | 0.15 ± 0.02 a,d | 0.10 ± 0.03 a,b | 0.10 ± 0.05 a,b,e | 0.15 ± 0.03 a,d,f,i | 0.30 ± 0.03 a,e | 0.10 ± 0.04 a | 0.10 ± 0.04 a | 0.10 ± 0.05 a |
| MBC | 0.20 ± 0.01 a | 0.20 ± 0.06 a,b | 0.15 ± 0. 04 a,b | 0.30 ± 0.01 a | 0.50 ± 0.02 a,b | 0.15 ± 0.01 a,b | 0.15 ± 0.03 a,b | 0.15 ± 0.01 a,b |
| A/A | Variety | Variety Code | Color 1 | Origin 2 | Fermentation Stage 3 |
|---|---|---|---|---|---|
| 1 | Kidonitsa | 1 | 1 | 2 | 1 |
| 2 | Kidonitsa | 1 | 1 | 2 | 2 |
| 3 | Savvatiano | 2 | 1 | 1 | 2 |
| 4 | Savvatiano | 2 | 1 | 1 | 2 |
| 5 | Savvatiano | 2 | 1 | 1 | 1 |
| 6 | Savvatiano | 2 | 1 | 1 | 1 |
| 7 | Savvatiano | 2 | 1 | 1 | 2 |
| 8 | Savvatiano | 2 | 1 | 1 | 2 |
| 9 | Chardonnay | 3 | 1 | 1 | 2 |
| 10 | Chardonnay | 3 | 1 | 1 | 2 |
| 11 | Chardonnay | 3 | 1 | 2 | 1 |
| 12 | Moschofilero | 4 | 1 | 2 | 2 |
| 13 | Moschofilero | 4 | 1 | 2 | 2 |
| 14 | Moschofilero | 4 | 1 | 2 | 2 |
| 15 | Moschofilero | 4 | 1 | 2 | 2 |
| 16 | Grenache rouge 1 | 5 | 2 | 1 | 1 |
| 17 | Grenache rouge 2 | 5 | 2 | 1 | 1 |
| 18 | Merlot | 6 | 2 | 1 | 1 |
| 19 | Merlot | 6 | 2 | 1 | 1 |
| 20 | Merlot | 6 | 2 | 2 | 2 |
| 21 | Cabernet | 7 | 2 | 2 | 2 |
| 22 | Cabernet | 7 | 2 | 2 | 2 |
| 23 | Agiorgitiko | 8 | 2 | 2 | 2 |
| 24 | Agiorgitiko | 8 | 2 | 2 | 1 |
| 25 | Agiorgitiko | 8 | 2 | 2 | 2 |
| 26 | Agiorgitiko | 8 | 2 | 2 | 2 |
| 27 | Agiorgitiko | 8 | 2 | 2 | 2 |
| 28 | Agiorgitiko | 8 | 2 | 2 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tagkouli, D.; Tsiaka, T.; Kritsi, E.; Soković, M.; Sinanoglou, V.J.; Lantzouraki, D.Z.; Zoumpoulakis, P. Towards the Optimization of Microwave-Assisted Extraction and the Assessment of Chemical Profile, Antioxidant and Antimicrobial Activity of Wine Lees Extracts. Molecules 2022, 27, 2189. https://doi.org/10.3390/molecules27072189
Tagkouli D, Tsiaka T, Kritsi E, Soković M, Sinanoglou VJ, Lantzouraki DZ, Zoumpoulakis P. Towards the Optimization of Microwave-Assisted Extraction and the Assessment of Chemical Profile, Antioxidant and Antimicrobial Activity of Wine Lees Extracts. Molecules. 2022; 27(7):2189. https://doi.org/10.3390/molecules27072189
Chicago/Turabian StyleTagkouli, Dimitra, Thalia Tsiaka, Eftichia Kritsi, Marina Soković, Vassilia J. Sinanoglou, Dimitra Z. Lantzouraki, and Panagiotis Zoumpoulakis. 2022. "Towards the Optimization of Microwave-Assisted Extraction and the Assessment of Chemical Profile, Antioxidant and Antimicrobial Activity of Wine Lees Extracts" Molecules 27, no. 7: 2189. https://doi.org/10.3390/molecules27072189
APA StyleTagkouli, D., Tsiaka, T., Kritsi, E., Soković, M., Sinanoglou, V. J., Lantzouraki, D. Z., & Zoumpoulakis, P. (2022). Towards the Optimization of Microwave-Assisted Extraction and the Assessment of Chemical Profile, Antioxidant and Antimicrobial Activity of Wine Lees Extracts. Molecules, 27(7), 2189. https://doi.org/10.3390/molecules27072189










