1. Introduction
Extra virgin olive oil (EVOO) has attracted particular attention due to its high nutritional value. Mainly consisting of triacylglycerols, which represent more than 98% of the oil weight, and small quantities of free fatty acids (less than 0.8% in EVOOs [
1]), it is considered one of the most valuable edible fats worldwide, with several health benefits being derived by its consumption. Olive oil consumption has been associated with the prevention of cardiovascular disease, diabetes, cancer, age-related cognitive decline, and lower incidence of metabolic syndrome [
2]. In fact, bioactive compounds included in the minor fraction of 2%
w/
w together with monounsaturated fatty acids (MUFAs), mainly referring to oleic acid, which is the most predominant type, present an increasing potential for health protection by decreasing low-density lipoprotein levels [
3,
4]. In this ambit, acknowledging EVOO’s high nutritional value and the health benefits derived from its consumption, European Union (EU) has proceeded to the establishment of relative regulation frameworks and health claims. Specifically, regarding fatty acids, the EU health claim of 2006 acknowledges cis-MUFAs (e.g., oleic acid) and cis- polyunsaturated fatty acids (PUFAs) (e.g., linoleic acid and alpha-linolenic acid) to the maintenance of normal blood cholesterol levels [
5]. In the same aspect, the regulation (EU) 432/2012 stated that foods with concentration of alpha-linolenic acid above 0.6 g/100 g are considered as foods with high content in Ω-3 fatty acids, whereas foods with concentration of linoleic acid above 1.5 g/100 g helps maintain normal blood cholesterol levels [
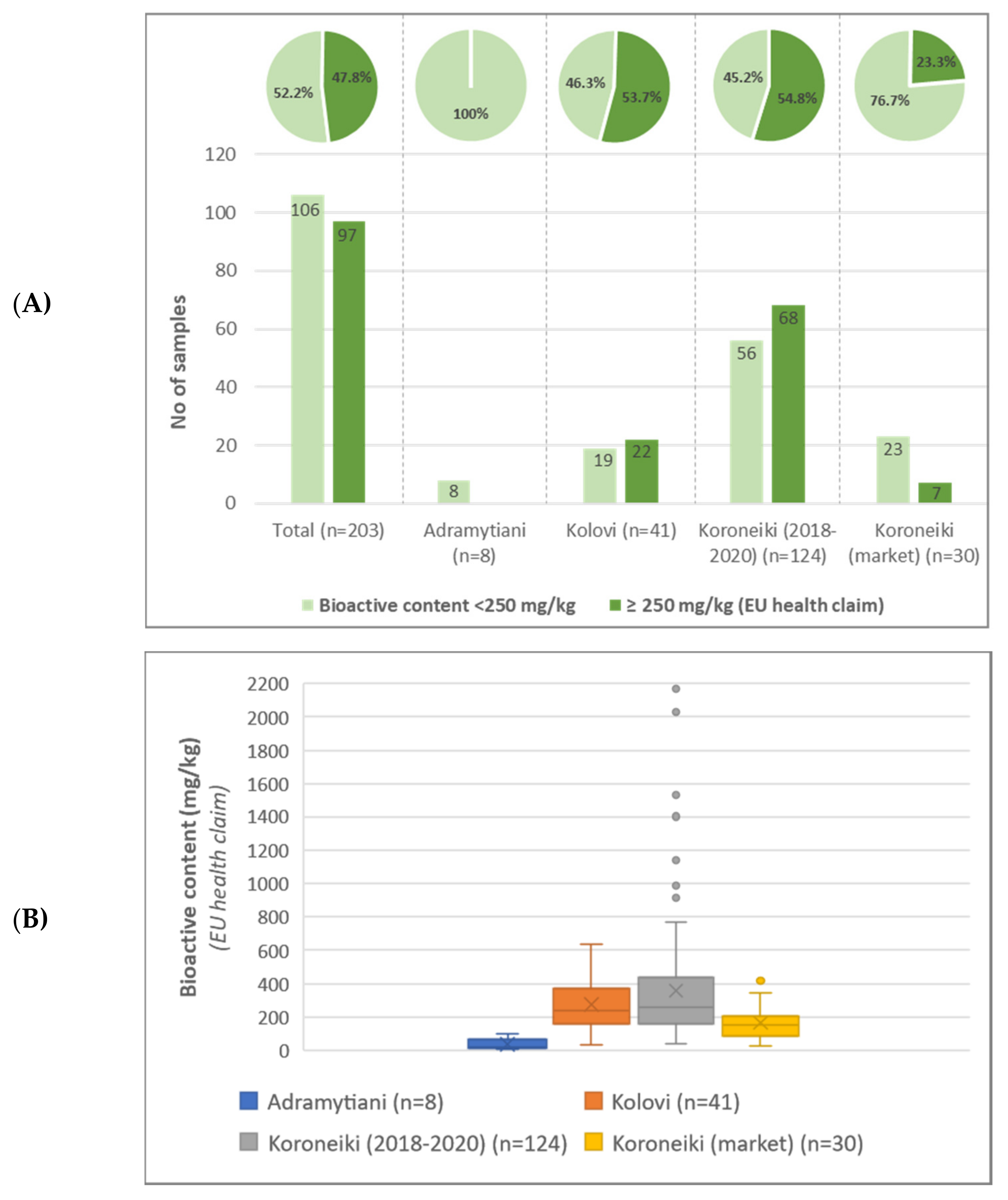
6]. The same regulation also highlights the health effects related to polyphenols, directly associated with European Food Safety Authority (EFSA) preceded substantiation about the benefits of EVOOs consumption [
7], setting the limit of “at least 5 mg of hydroxytyrosol and its derivatives (e.g., oleuropein complex and tyrosol) per 20 g of olive oil” [
6]. Different methodologies have been developed to assure that olive oil meets regulatory standards and to allow the use of health claims related to fatty acids and bioactive content. Regarding olive oil quality assurance, the methodologies applied by all control authorities are determined by the protocols established from regulatory bodies (e.g., European Commission Regulation) [
1].
Under the perspective of authenticity assessment and quality verification, more holistic approaches are needed in order to address challenging authenticity issues, such as variety or geographical origin identification of EVOO. High resolution mass spectrometry (HRMS)-metabolomics and optical spectroscopy are two cutting-edge methodologies that make full use of EVOOs chemical profile, allowing their thorough investigation. HRMS-metabolomics has demonstrated its potential on the field of EVOOs analysis with both targeted (focusing on a specific group of compounds-metabolites, identified and quantified) and untargeted approaches (investigating the whole profile) being reported so far [
8,
9,
10,
11,
12]. HRMS-workflows, often hyphenated with chromatographic techniques (e.g., liquid chromatography, LC), increase depth of coverage, with compounds of different chemical classes being detected in a single acquisition run. Hence, the methodology developed and applied by the authors [
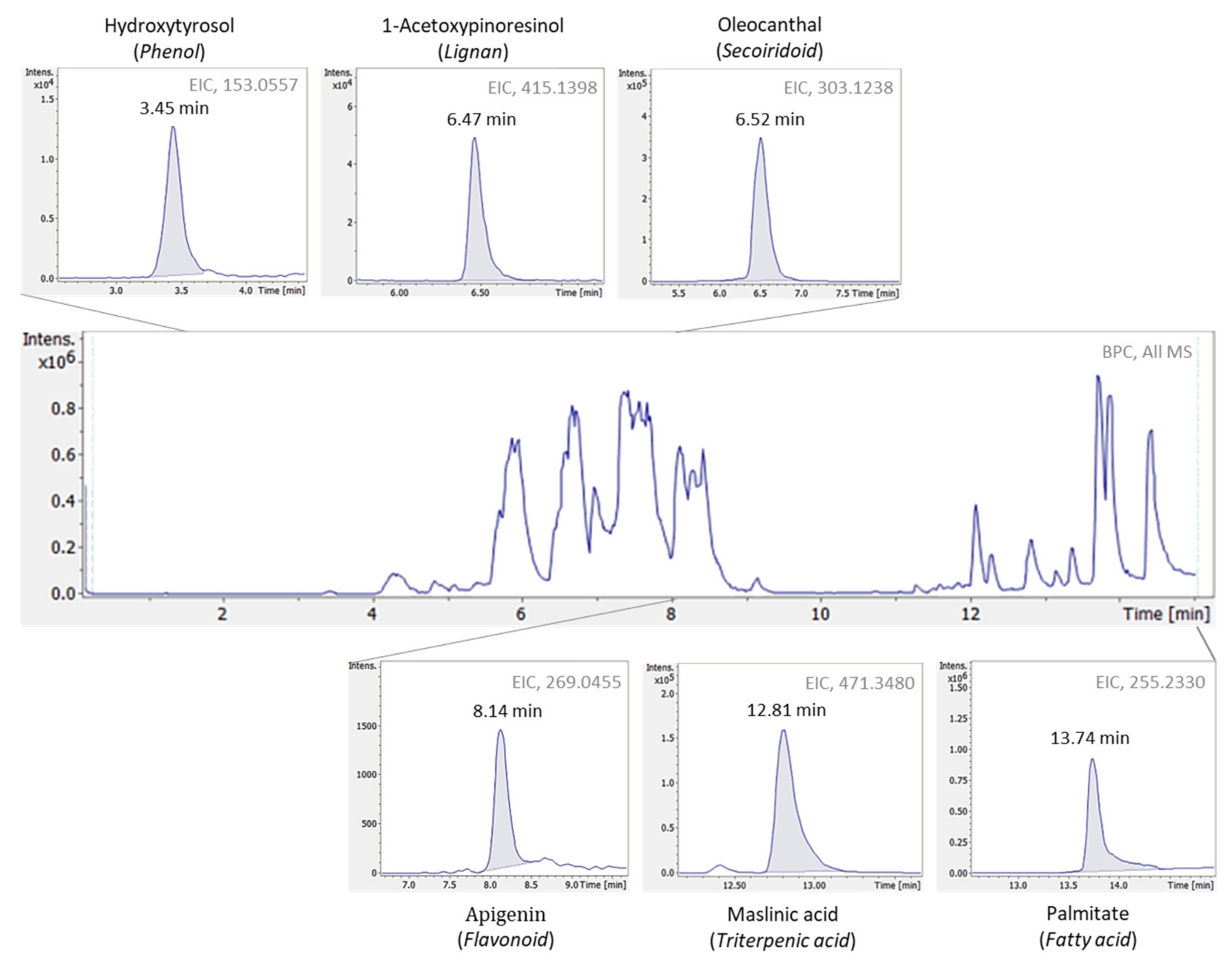
8] regarding EVOOs’ bioactive content determination, allows the detection of a large number of compounds separated according to the different chemical groups (i.e., phenols, lignans, terpenoids, flavonoids, fatty acids etc.). As a result, a holistic overview on EVOOs profile is implemented, which is crucial in metabolomics-based studies. Moreover, taking advantage of HRMS data dependent acquisition (DDA) mode, high-quality spectra are provided, thus enabling the accurate identification and structure annotation of even unknown compounds. To this purpose, the followed data treatment workflow comprises all information retrieved through LC-HRMS analysis (i.e., retention time, mass accuracy, isotopic pattern and MS2 fragmentation) aiming for tentative identification.
Optical spectroscopy has been introduced as a powerful analytical tool for EVOO authenticity determination, providing rapid and accurate results. Spectroscopic techniques such as absorption, fluorescence, Raman and FT-IR are non-invasive methods, relatively cheap, and user and environmentally friendly, as they have the advantage of the absence of reagents and solvents and require a small amount of sample. Over the last several years, the application of optical spectroscopic techniques combined with machine learning methods has been used to deal with various analytical problems of interest in the olive oil authenticity sector. Specifically, optical spectroscopic techniques were used to distinguish the EVOO samples based on their geographical origin [
13]. In addition, they have been used for detection of adulteration in extra virgin olive oil [
14] and for the verification of its quality parameters [
15,
16]. Moreover, these techniques have also been shown to be efficient tools for the identification of EVOOs based on their variety [
17,
18].
Both spectrometry and optical spectroscopy techniques derive data which contain a large amount of information. In order to eliminate noise and redundant information, machine learning analysis is considered a powerful and essential tool. Machine Learning methods, and in this case classification methods, give insight and intuition into the points of interest (i.e., identification of variety, geographical or botanical origin). Both targeted and untargeted techniques have been previously combined with machine learning methods, such as PLS-DA, SVMs, Neural Networks etc. Indicatively, various agricultural products such as honey [
19,
20] and wine [
21,
22,
23,
24,
25,
26] have been investigated, considering a wide range of issues and authentication purposes such as their quality, variety and geographical and botanical origin.
In the present collaborative study, both targeted and untargeted approaches have been established to evaluate olive oil quality and assure its authentication in terms of variety identification. Greek EVOOs have been analyzed and assessed to meet the established health claims, thus highlighting their nutritional value, while thorough study of their profile has also been performed. Regarding the latter, both HRMS and optical spectroscopy techniques were implemented to gain perspective and investigate different chemical classes. All information retrieved was utilized to its fullest, contributing to the creation of predictive models, according to the parameter of analysis (i.e., fatty acids content, metabolomics, visible absorption, Raman and fluorescence). For the construction of the predictive models, the most widely used classification methods were employed, namely the Logistic Regression, Random Forest, SVMs and KNN. For each different technique (FAMEs, HRMS, visible absorption, Raman and fluorescence), the best-performing classification model was chosen based on the cross-validation procedure. Each model was validated and evaluated for its ability to correctly discriminate between three emblematic Greek olive oil varieties (Koroneiki, Kolovi and Adramytiani), performing a comparative study through the different analytical platforms. Finally, the constructed models were further evaluated with 30 commercially available EVOO samples in order to verify their variety.
3. Materials and Methods
3.1. Chemicals and Reagents
In fatty acids analysis, compounds were identified by the use of two commercial standards (i.e., FAME Mix C24-C22 and FAME Mix C4-C24) purchased from Supelco (Sigma, Saint Louis, MO, USA), while analytical grade methanol, heptane and potassium hydroxide were purchased from Carlo Erba Reagents.
All standards and reagents used in HRMS analysis were of high-purity grade (>95%): p-coumaric acid, eriodictyol, pinoresinol and syringaldehyde were acquired from Sigma-Aldrich (Stenheim, Germany). Hydroxytyrosol and luteolin were purchased from Santa Cruz-Biotechnology (Santa Cruz, CA, USA), while apigenin, naringenin, tyrosol and vanillin were purchased from Alfa Aesar (Karlsruche, Germany). Ligstroside aglycone, oleacein, oleocanthal, oleocanthalic acid, oleomissional and oleuropein aglycone were obtained from Prof. P. Magiatis laboratory (Laboratory of Pharmacognosy and Natural Products Chemistry, Faculty of Pharmacy, University of Athens, Athens, Greece). The standards had been previously isolated from olive oil extracts and their structure and purity grade were evaluated by NMR analysis [
34]. Methanol (MeOH) (LC-MS grade) was purchased from Merck (Darmstadt, Germany), whereas 2-propanol (LC-MS grade) was purchased from Fisher Scientific (Geel, Belgium). Sodium hydroxide monohydrate for trace analysis ≥99.9995% and ammonium acetate ≥99.0% were purchased from Fluka (Buchs, Switzerland). Distilled water was provided by a Milli-Q purification apparatus (Millipore Direct-Q UV, Bedford, MA, USA). Finally, regenerated cellulose syringe filters (RC, pore size 0.2 μm, diameter 15 mm) were purchased from Phenomenex (Torrance, CA, USA). Stock standard solutions of individual compounds (1000 mg L
−1) were prepared in MeOH and stored at −20 °C in dark glass bottles. Working mix solutions at six different concentrations levels (0.5, 1, 2, 5, 10 and 20 mg L
−1) were prepared by gradient dilution of the stock solutions in MeOH/H
2O 80:20,
v/
v and analyzed each time at the beginning of each sequence.
3.2. EVOOs Samples and Sample Preparation Protocols
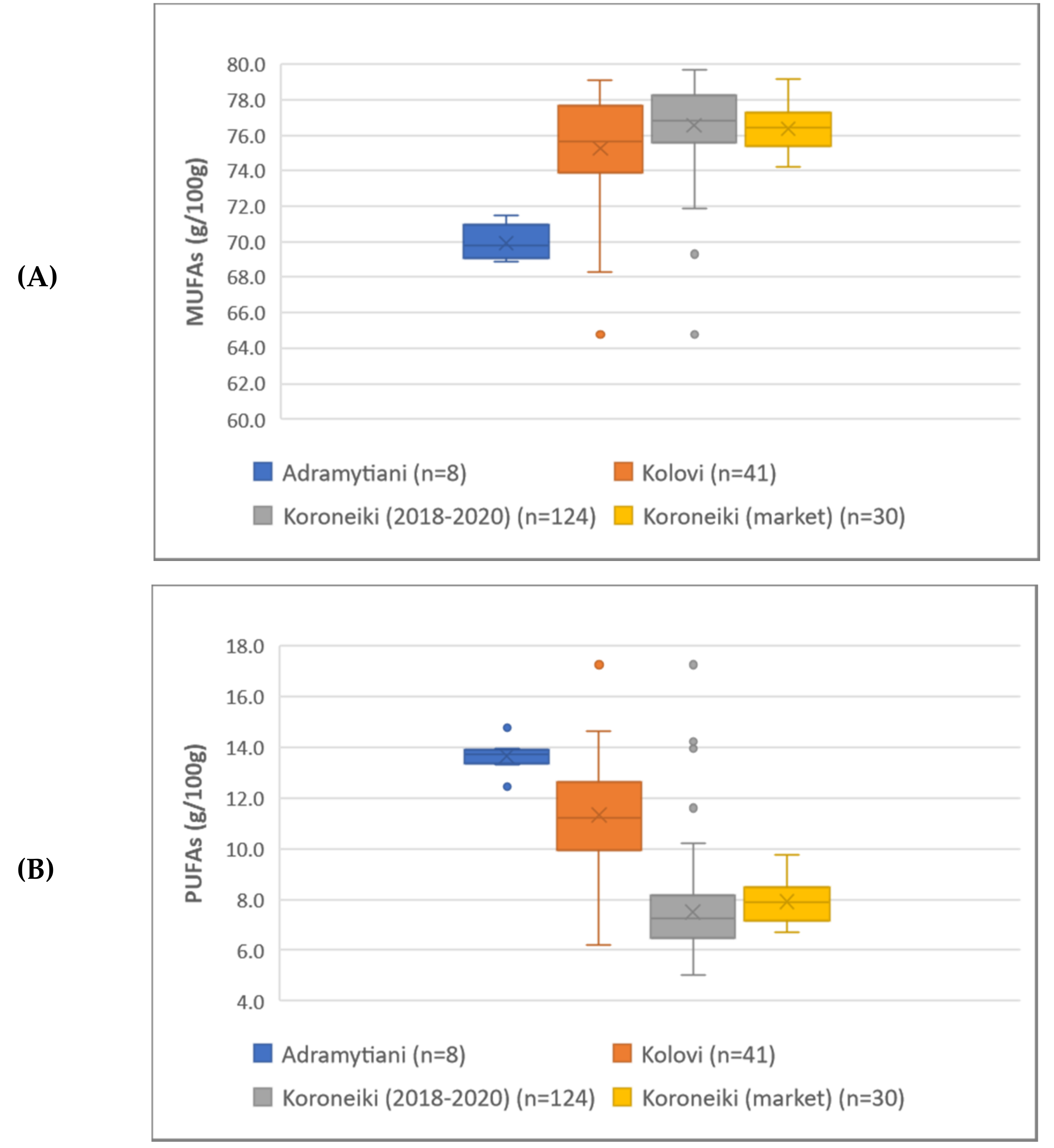
A total of 173 monovarietal EVOOs produced from three cultivars (Koroneiki, Kolovi and Andramytiani) were collected from the three major olive-cultivated regions in Greece, namely, Crete, Peloponnese and Lesvos in descending order, during the harvesting periods of 2018–2020. Koroneiki variety is the cultivar more widely cultivated in Greece (found in all 3 regions mentioned), representing around 60% of the total Greek olive-growing land [
35]. The widely cultivated olive variety, i.e., Koroneiki, allowed the selection of a large number of samples (154-market samples included). On the other side, Kolovi and Adramytiani cultivars are mainly cultivated in North Aegean region (the majority of which in Lesvos) with Kolovi representing the majority of cultivation in this geographical area. Therefore, during EVOO sampling a significant number of 41 Kolovi samples were selected, while only 8 monovarietal Adramytiani samples available were acquired. It also needs to be clarified at this point that only monovarietal samples were included in our study in order to study the influence of variety in EVOOs classification, which consequently reduced the number of Adramytiani samples included in the study Nevertheless, the limited number of Adramytiani samples has been addressed but also reported in previous studies [
8]. Samples were packaged in dark-brown glass bottles and stored at 4 °C until analysis. Furthermore, during the same period, a total of 30 branded monovarietal (Koroneiki cultivar) EVOOs were sampled from the market.
The fatty acid composition was determined according to the official method of the Commission Regulation (EEC) No 2568/91 [
1]. Briefly, the fatty acid methyl esters were prepared by vigorous shaking of 0.1 g olive oil in 3 mL heptane, with 0.2 mL of 2 mg L
−1 methanolic potassium hydroxide in screwcap vials. After 30 min, 1 μL of the upper phase of the vial was injected into the gas chromatograph system for analysis.
To study the metabolomic profile of olive oil, a liquid-liquid extraction with MeOH/H
2O (80:20,
v/
v) used as extraction solvent was implemented, previously reported in the literature [
36]. Syringaldehyde was used as internal standard (IS) at 1.30 mg L
−1 [
9]. Olive oil extracts were subjected to a 2-fold dilution with MeOH/H
2O 80:20,
v/
v before analysis. Procedural blank was also prepared and analyzed to detect potential contamination. At the beginning of each instrumental sequence a six-points analytical standards’ curve was analyzed (from 0.5 to 20 mg L
−1) to check system suitability. Quality control (QC) samples were also prepared and analyzed to ensure analytical performance. The QC samples were prepared by mixing same-volume aliquots of 15 EVOOs, derived from the 3 different varieties (5 EVOOs of each variety). A QC sample was injected at the beginning of each sequence (six times for conditioning) and also at regular intervals (every 10 injections) to monitor potential instrumental drifts. The intensity of three exact mass retention time (EMRT) pairs (
m/
z 153.0557_3.5 min,
m/
z 271.0612_7.0 min, and
m/
z 319.1187_5.6 min, corresponding to hydroxytyrosol, naringenin and oleacein respectively) were monitored, and their ratios to IS peak intensity were calculated to evaluate system stability, recording in all cases ratios within QC limits (±2 s) (
Figure S1).
For the optical measurements no sample pretreatment was necessary.
3.3. Instrumental Analysis
3.3.1. Fatty Acids Determination
The fatty acids methyl esters (FAMEs) profile, expressed as % m/m using the peak area, was determined using an Agilent 7890A Chromatograph (Agilent Technologies, Santa Clara, SA, USA) coupled to a flame ionization detector (FID). The separation of fatty acid methyl esters was accomplished with a TR-FAME column, 50 m × 0.22 mm, i.d. 0.25 μm film thickness (Thermo Fisher Scientific, Waltham, MA, USA). The injector was set at 250 °C and the detector at 260 °C. The oven temperature was initially retained at 160 °C for 2 min, then raised with a rate of 1 °C min−1 up to 165 °C and kept for 30 min, then raised again with a rate of 3 °C min−1 up to 200 °C and kept for 8 min, resulting to a total run time of 57 min. Helium was used as carrier gas at a flow rate of 3 mL min−1. Each sample was injected twice, in split mode (50:1) with an injection volume of 1 μL, using an autosampler. FAMEs analysis was performed at Hellenic Food Authority (EFET).
3.3.2. HRMS-Metabolomics
Metabolomics analysis was carried out using an ultra-high performance liquid chromatography system (UHPLC) with an HPG-3400 pump (Dionex Ultimate 3000 RSLC, Thermo Fischer Scientific, Dreieich, Germany) coupled to a quadrupole time-of-flight mass spectrometer (QTOF) (Maxis Impact, Bruker Daltonics, Bremen, Germany). Samples were analyzed in reversed phase liquid chromatography (RPLC) using an Acclaim C18 column (2.1 × 100 mm, 2.2 μm) for the chromatographic separation obtained from Thermo Fischer Scientific (Dreieich, Germany) and preceded by a guard column of the same packaging material, thermostatted at 30 °C. The mobile phases consisted of water/methanol 90/10 (solvent A) and methanol (solvent B), both acidified with 5 mM ammonium acetate. A gradient elution program was adopted starting with 1% B (flow rate of 0.2 mL min−1) for 1 min, increased to 39% in 2 min and then to 99.9% (flow rate of 0.4 mL min−1) for another 11 min. At this point, 99.9% of B is being kept constant for 2 min (flow rate of 0.48 mL min−1) and then initial conditions are restored within 0.1 min, for the next 3 min; then the flow rate decreases to 0.2 mL min−1. Injection volume was set to 5 μL. The QTOF-MS system was equipped with an electrospray ionization interface (ESI), operating in negative mode, with the following operation parameters: capillary voltage 3500 V; end plate offset 500 V; nebulizer pressure 2 bar; drying gas 8 L min−1 and gas temperature 200 °C. The QTOF-MS system was operated in data independent acquisition mode (broadband collision-induced dissociation, bbCID), as well as in data-dependent acquisition mode (AutoMS/MS), and recorded spectra over the range of m/z 50–1000, with a scan rate of 2 Hz. A QTOF-MS external calibration was performed daily with the manufacturer’s solution. The typical resolving power of the instrument was ranged (FWHM) between 36,000 and 40,000 at m/z 226.1593, 430.9137, and 702.8636. HRMS-metabolomics was performed at National and Kapodistrian University of Athens.
3.3.3. Optical Spectroscopy
Absorption spectra were recorded with a Shimadzu UV-1900 spectrophotometer (Shimadzu Corporation, Kyoto, Japan), which covers the spectral range from 190 to 1100 nm, as reported in a previous work by the authors [
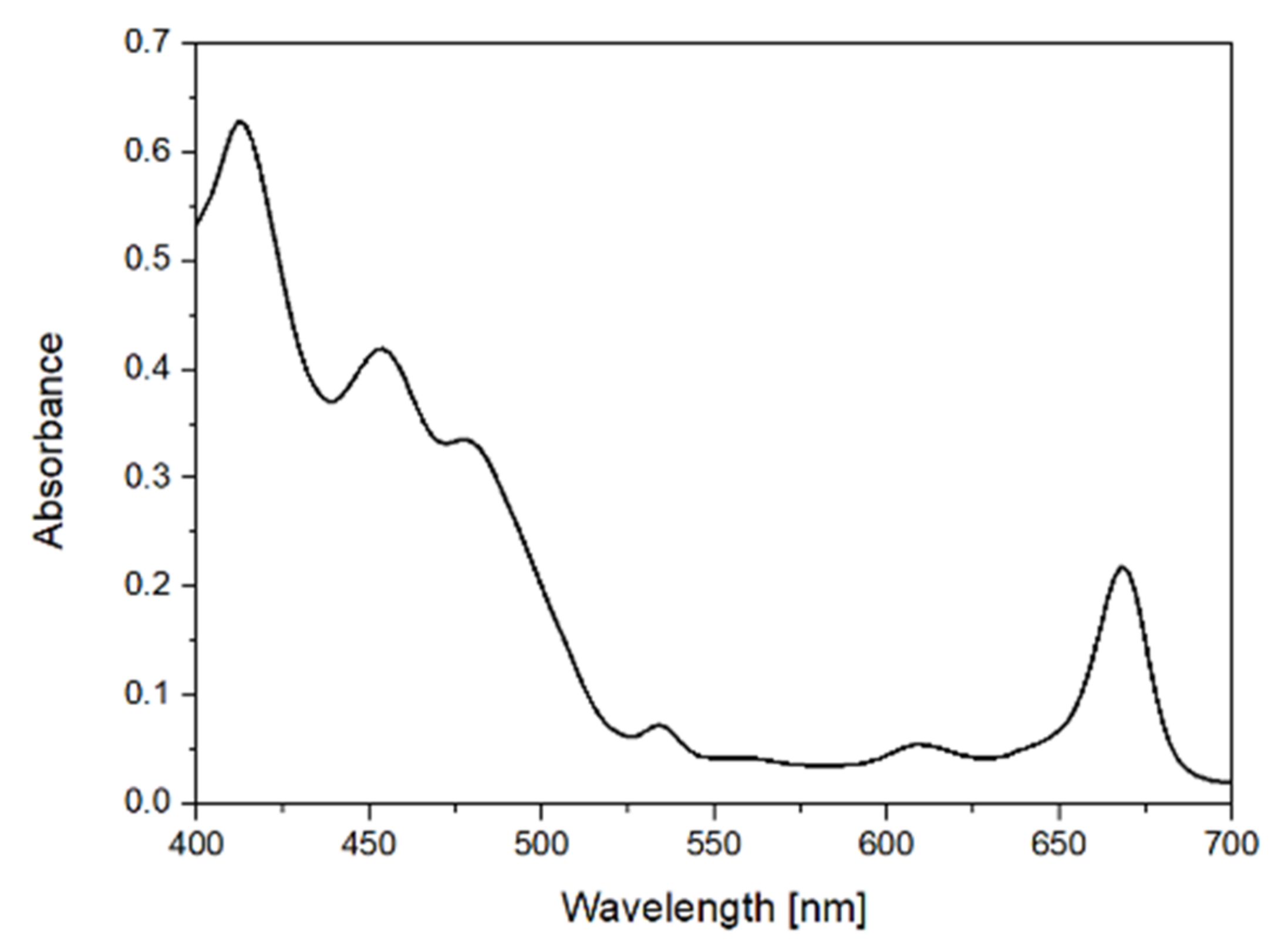
13]. For the visible region in the range of 400 nm to 700 nm, a quartz cuvette with a 2 nm pathlength was used. The corresponding spectral resolution for the absorption spectroscopy was 2 nm.
The fluorescence spectra were collected with Horiba FluoroMax-3 (HORIBA Ltd., Tokyo, Japan). For the measurements, a quartz cuvette, with a 10 mm pathlength was used and placed in the front face geometry (at 35° to the incident beam—in order to avoid inner filter effects) without any prior pre-treatment. Synchronous fluorescence spectra (SFS) were recorded by performing simultaneously scanning of the excitation and emission monochromators. In this case the emission wavelength (λemission) was following the excitation wavelength (λexcitation) at a constant distance (Δλ) so that λemission= λexcitation + Δλ. For each sample seven different SFS were collected with wavelength intervals (Δλ) of 30, 60, 90, 120, 150, 180 and 210 nm and the excitation wavelength ranged from 270 nm to 400 nm, in 2 nm step. The integration time was set at 0.25 s and the excitation and emission bandwidths were 1 and 2 nm, respectively.
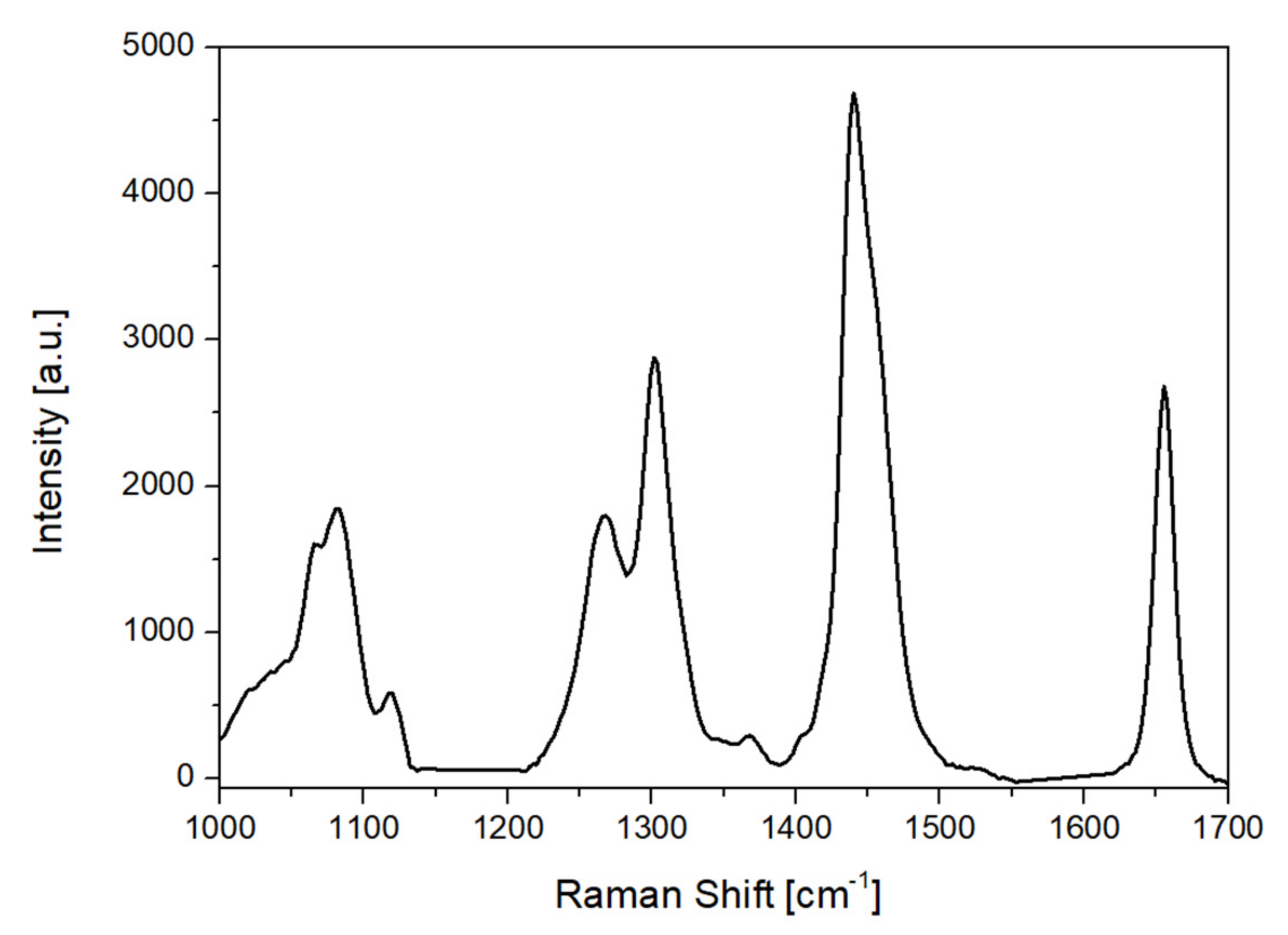
The Raman spectra were carried out with a mobile Raman spectrometer (HE 785, Horiba Jobin Yvon, Rue de Lille, France). The light source was a semiconductor diode laser, emitting at 785 nm. The instrumentation and the collection parameters have been described in previous work [
13]. Optical spectroscopy measurements were performed at IESL-FORTH.
3.4. Data Treatment
Data acquired from HRMS analysis were processed using target, suspect and un-target screening workflows. DataAnalysis 5.2 and TASQ 2.1 software (Bruker Daltonics) were used to the target and suspect approaches, while data treatment in the un-targeted approach was performed using MetaboScape® 4.0 software (Bruker Daltonics).
The target database used in the screening workflow comprised of 70 bioactive compounds found in natural products and in olive oil in particular, classified under the chemical class of phenols (i.e., flavonoids, lignans, phenolic acids, secoiridoids or simple phenols). The database included information of compounds’ molecular formulas, pseudomolecular ions ([M-H]
−), retention time (min), MS
2 fragments (qualifier ions) as well as their elemental formulas. Compounds’ identification was performed setting specific matching criteria: mass accuracy (mass error ≤ 2 mDa), isotopic fitting (mSigma ≤ 50), retention time (±0.2 min in relevance to standard’s analysis), MS
2 qualifier ions, minimum peak area threshold at 800 and minimum intensity threshold at 200, as reported in previous study by the authors [
8]. Quantification of the analytes was performed through an external standard calibration curve [
8].
“Suspect” compounds, often occurring in olive oil, but with no reference standard available, were successfully identified based on the aforementioned criteria, described in the targeted approach. Quantification was performed according to the in-house semi-quantitation methodology, previously reported by the authors [
9], which relies on compounds chemical similarity. In total 9 “suspect” compounds were identified in olive oil samples and further quantified, namely 10-hydroxy decarboxymethyl oleuropein aglycone, 10-hydroxy-10-methyl oleuropein aglycone, 10-hydroxyoleuropein aglycone, 1-acetoxypinoresinol, elenolic acid, hydroxylated form of elenolic acid, hydroxytyrosol acetate, methyl oleuropein aglycone and syringaresinol.
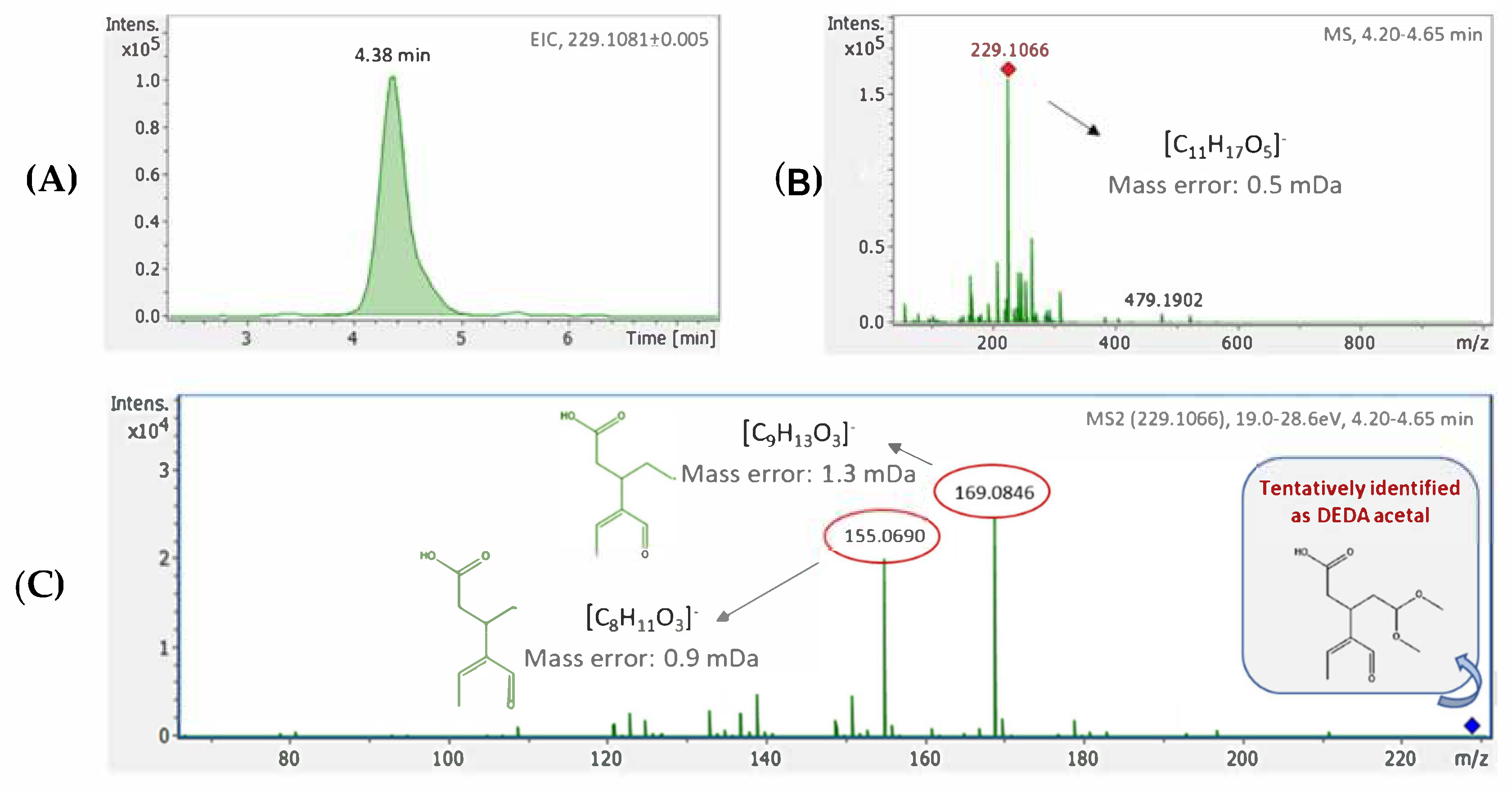
Regarding the un-targeted approach, a complete data pre-processing workflow, including peak-picking, mass calibration and time alignment was performed using Time aligned Region complete eXtraction (T-ReX) algorithm, embedded in Bruker’s MetaboScape software. In particular T-ReX 3D was used, compatible with LC-HRMS data acquired, which extracted features according to 3 particular parameters (i.e., retention time, mass to charge ratio (m/z) and intensity), resulting to the creation of a peak-list. In the peak-picking step, intensity threshold was set at 1000 counts and minimum peak length of 5 spectra was selected. Mass recalibration was automatically performed for each sample, using the same calibrant solution with that of external calibration (i.e., sodium acetate) at 0.1–0.25-time range (min). Retention time and mass range were also defined and set the same with analysis conditions (i.e., retention time range: 0.25–15 min, mass range: 50–1000 m/z). Primary ion of [M-H]− was selected for negative acquisition mode. Procedural blank subtraction was also performed in the software interface to subtract false positive peaks and avoid potential contamination. Only features of 3-fold higher intensity compared to the respective intensity of the blank were taken into account and included in the final peak-list.
3.5. Machine Learning Analysis
For the machine learning analysis a Python’s open-source library for Machine Learning, scikit-learn was used [
37].
3.5.1. Methods
Feature Selectiontitle
Prior to data classification, feature selection was performed. Feature Selection is the process of selecting a subset of features that is relevant to the target (i.e., geographical origin, variety identification etc.). It provides simplified models as well as a more intuitive interpretation of the model results. Among different feature selection techniques provided by scikit-learn, model-based feature selection was applied. Specifically, some learning algorithms perform feature selection as part of their overall operation [
38,
39]. For this scope, Random Forest was used as a feature selector. For this procedure the “SelectFromModel” function of scikit-learn was used.
Feature Selection was performed on the spectroscopic data as well as the HRMS data, which due to their nature might contain redundant information. For the FAMEs data, no feature selection was performed, as the data contain targeted information.
Classification
The classification procedure was performed to the data of the different techniques (FAMES, HRMS, Raman, Fluorescence, Visible). Specifically, four classification methods were examined: K Nearest Neighbors (KNN) [
40], Support Vector Machines (SVM) [
41], Logistic Regression (LR) [
42] and Random Forests (RF) [
43].
KNN assumes that the samples of each class are “close” to each other. Having the train data and their corresponding labels, each new sample is classified according to the majority class of its k-nearest neighbors.
Random forest is an ensemble model that fits multiple decision trees on different sub-samples of the initial train data. For a new sample, each tree in the Random Forest outputs a class prediction. The model finally predicts the most voted class. Moreover, the Random Forest model outputs the importance of each feature. The most important features are the ones that contribute the most to the classification results. Random Forest could also be used as a feature selector prior to the classification procedure, by selecting the most important features according to a threshold.
Logistic Regression is a generalized linear model used for classification purposes. Specifically, it fits a logistic function (i.e., sigmoid function) to the training data, that takes values in [0, 1]. For each new sample, the model outputs the probability that it belongs to the positive class based on the fitted logistic function. If the probability is over 0.5 the model predicts that the sample belongs to the positive class, or else to the negative class. The multiclass case is handled according to a one-vs-rest scheme.
Support Vector Machines for binary classification tasks aim to find a hyperplane in the p-dimensional space (p features in the data) that discriminates the samples of different classes. The support vectors are defined as the closest samples to the hyperplane and specify its position and orientation. SVMs can handle data that are not linearly separable by mapping them into a high-dimensional feature space, where they become linearly separable (kernel trick) [
41]. The multiclass case is handled according to a one-vs-one scheme.
Both feature selection and classification methods require some user-defined parameters (hyperparameters). For example, KNN’s hyperparameter is the number of the nearest neighbors; the hyperparameters of SVM are the type of kernel (linear, RBF, polynomial, sigmoid) and the regularization parameter C. The hyperparameter tuning was performed during the cross-validation procedure. The different values of the hyperparameters of each method are shown in
Table S10.
3.5.2. Methodology
The data were randomly split into train (70%) and test set (30%) in a stratified manner such that the percentage of samples for each class is preserved as observed in the original data. The train set, total number 121 samples (87 Koroneiki, 29 Kolovi, 5 Adramytiani) was used for the model construction and the test set, total number 52 samples (37 Koroneiki, 12 Kolovi, 3 Adramytiani) was used for testing purposes.
In order to determine the model with the best hyperparameter set, stratified k-fold cross validation was performed in the train set. During the stratified k-fold cross validation the train samples are randomly partitioned into k equal sized folds. Each fold contains approximately the same percentage of samples of each class as in the train set. Each model is trained in the k-1 folds and the left-out fold is used for testing. The procedure is repeated k times, where every fold is used exactly one time as the test fold. The final estimation originates from the mean performance of all repetitions. In our case k was equal to 5.
After the model with the best hyperparameters was determined from cross-validation, it was trained to the entire train set. The model’s performance was then evaluated on the test set. The results were compared to the baseline (“DummyClassifier” of scikit-learn), which is a model that always predicts the most frequent class, in our case Koroneiki.
As metrics of evaluation the accuracy score, the sensitivity and specificity, as well as the confusion matrix were used. Accuracy is the percentage of correctly predicted samples. The sensitivity and specificity are separately measured for each class. For each class, sensitivity (True Positive Rate) is shown in the following equation:
Specifically, it refers to the proportion of samples that are correctly predicted to belong to the class (True positives-TP) among all samples that actually belong to the class (True Positives + False Negatives (TP + FN)). The specificity for each class refers to the proportion of samples that are correctly predicted that they do not belong to the class (True Negative-TN) among all samples that actually do not belong to the class (True Negative + False Positive (TN + FN)). It is shown in the following equation:
The highest value for sensitivity and specificity is 100%. The mean sensitivity (specificity) is the average sensitivity (specificity) of all classes. The confusion matrix is also presented for further intuition of the results. It is a c-by-c matrix, where c is the number of classes. Each row represents the true label and each column the predicted one. The diagonal elements represent the correct predictions, while the non-diagonal elements represent the misclassifications. The highest performance is achieved when the diagonal elements are equal to the number of samples per class and all the non-diagonal elements are zero.
3.5.3. Data Pre-Processing
In general, machine learning algorithms benefit from data pre-processing. In order to evaluate the data of each different technique, the most appropriate and required pre-processing methods were applied. Specifically, autoscale (z-score) was performed in the FAMEs data.
In HRMS data, extracted features were subjected to normalization by internal standard (i.e., syringaldehyde) to reduce variation between samples that may attributed to sample procedure, instrumental conditions, or data-processing factors.
For the Raman spectroscopic data, baseline subtraction was performed to correct the baseline offset caused by sample fluorescence on the Raman spectra, using the airPLS algorithm (Zhangfit) [
31]. SNV and autoscale pre-processing were furtherly applied for extracting the spectral information from Raman spectroscopic data. In a similar way, fluorescence spectroscopic data and the visible absorption data were processed with a combination of Savitzky-Golay [
44], SNV and autoscale pre-processing techniques.
To avoid information leakage from the test set, both for the FAMEs and spectroscopic data, the autoscale procedure was performed independently on each feature of the samples in the train set. The statistics were stored in order to be used on the test data.
4. Conclusions
In this work, a thorough study has been performed for the varietal identification of Greek EVOOs. Different analytical techniques have been implemented and compared, from more conventional ones such as gas chromatographic FAMEs’ determination, to more sophisticated ones, such as spectrometric (i.e., HRMS) and spectroscopic techniques (i.e., visible absorption, Raman and fluorescence spectroscopy). The analytical results were combined with Machine Learning algorithms for the classification of the samples to Koroneiki, Kolovi and Adramytiani varieties. For each technique, the predictive model with the best hyperparameter set was chosen. The models’ predictive power was evaluated on test data. The results indicated that the FAMES and HRMS analysis achieve the highest predictive ability. In parallel, the spectroscopic techniques exhibited high performance with a small number of misclassifications. The models were furtherly tested using commercially available market EVOOs for the verification of their variety. The results demonstrated that both targeted and untargeted techniques correctly predicted the variety of all the commercially available market EVOOs, except from the visible absorption spectroscopy, where only 3 out of the 30 market EVOOs were misclassified.
Based on these encouraging results, HRMS metabolomics as well as optical spectroscopic techniques combined with machine learning methods provide an alternative approach for the identification of olive oil variety, compared to the official methods already established (e.g., FAMEs). Through untargeted metabolomics a thorough investigation on EVOOs chemical profile may be accomplished, while that identification of compounds of interest is also enabled, exploiting HRMS full capabilities. Regarding optical techniques, they have prevailed in the field as they are facile, rapid and cost-effective compared to other analytical techniques. They also require no sample pre-treatment, while their predictive performance is comparable to the modern analytical procedures.
Finally, to the best of the authors’ knowledge, this is the first time where different techniques from the fields of standard analysis, spectrometry and spectroscopy are applied to the same EVOO samples, and the results are compared and evaluated by machine learning techniques. The findings of this work are valuable for providing a novel methodology for EVOO authentication that might help in the field of production, packaging and trading of EVOO.