Comparison of the Effectiveness and Environmental Impact of Selected Methods for the Determination of Fatty Acids in Milk Samples
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimization of GC-FID Conditions
2.2. Comparison of Preparation Methods for FAMEs Determination
2.3. Assessment of the Method Greenness
3. Materials and Methods
3.1. Milk Sample
3.2. Chemicals and Reagents
3.3. Lipid Extraction and FAME Preparation
3.4. GC Analysis
3.5. Statistical Analysis
3.6. Calculation of Fatty Acid Contents
- mi—is the mas of FAME, i, in the reference mixture,
- ∑A—is the sum of all areas of all FAMEs of the reference mixture,
- Ai—is the area of FAME, i, in the reference mixture,
- ∑m—is the total of the masses of the various components, as FAMEs of the reference mixture.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Karrar, E.; Ahmed, I.A.M.; Huppertz, T.; Wei, W.; Jin, J.; Wang, X. Fatty acid composition and stereospecificity and sterol composition of milk fat from different species. Int. Dairy J. 2022, 128, 105313. [Google Scholar] [CrossRef]
- Nateghi, L.; Yousefi, M.; Zamani, E.; Gholamian, M.; Mohammadzadeh, M. The effect of different seasons on the milk quality. Eur. J. Exp. Biol. 2014, 4, 550–552. [Google Scholar]
- Ferrand, M.; Huquet, B.; Barbey, S.; Barillet, F.; Faucon, F.; Larroque, H.; Leray, O.; Trommenschlager, J.M.; Brochard, M. Determination of fatty acid profile in cow’s milk using mid-infrared spectrometry: Interest of applying a variable selection by genetic algorithms before a PLS regression. Chemom. Intell. Lab. Syst. 2011, 106, 183–189. [Google Scholar] [CrossRef]
- Liu, Z.; Moate, P.; Rochfort, S. A simplified protocol for fatty acid profiling of milk fat without lipid extraction. Int. Dairy J. 2019, 90, 68–71. [Google Scholar] [CrossRef]
- Fleming, A.; Schenkel, F.S.; Malchiodi, F.; Ali, R.A.; Mallard, B.; Sargolzaei, M.; Jamrozik, J.; Johnston, J.; Miglior, F. Genetic correlations of mid-infrared-predicted milk fatty acid groups with production traits. J. Dairy Sci. 2018, 101, 4295–4306. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Li, C.; Rochfort, S. Development of one-step sample preparation methods for fatty acid profiling of milk fat. Food Chem. 2020, 315, 126281. [Google Scholar] [CrossRef]
- Samková, E.; Kalač, P. Rapeseed supplements affect propitiously fatty acid composition of cow milk fat: A meta-analysis. Livest. Sci. 2021, 244, 104382. [Google Scholar] [CrossRef]
- Amores, G.; Virto, M. Total and free fatty acids analysis in milk and dairy fat. Separations 2019, 6, 14. [Google Scholar] [CrossRef]
- Folch, L.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipids from animal tissue. J. Biol. Chem. 1957, 726, 497–509. [Google Scholar] [CrossRef]
- Ilieva, Y.; Ivanova, S.; Penchev, P. Fatty-acid composition of buffalo milk under intensive and pasture farming. J. Cent. Eur. Agric. 2020, 21, 722–732. [Google Scholar] [CrossRef]
- de Melo Soares, É.S.; Ítavo, C.C.B.F.; Ítavo, L.C.V.; dos Santos, G.T.; Nazário, C.E.D.; Soares, I.S.S.; Cavalheiro, L.F. Comparison of analytical methods for the fatty acid profile in ewes’ milk. PLoS ONE 2022, 17, e0263071. [Google Scholar]
- Sajid, M.; Płotka-Wasylka, J. Green analytical chemistry matrics: A review. Talanta 2022, 238, 123046. [Google Scholar] [CrossRef] [PubMed]
- Płotka-Wasylka, J.; Mohamed, H.M.; Kurowska-Susdorf, A.; Dewani, R.; Fares, M.Y.; Andruch, V. Green analytical chemistry as an integral part of sustainable education development. Curr. Opin. Green Sustain. Chem. 2021, 31, 100508. [Google Scholar] [CrossRef]
- Visentainer, J.V.; Claus, T.; Santos, O.O., Jr.; Chiavelli, L.U.R.; Maruyama, S.A. Analytical aspects of the fames ionization detection in comparison with mass spectrometry with emphasis on fatty acids and their esters. In Advances in Gas Chromatography; IntechOpen: London, UK, 2014; pp. 39–56, Chapter 2. [Google Scholar]
- Span, C.S.; Beach, C.A.; Jones, A.J.; Dauenhauer, P.J. Increasing flame ionization detector (FID) sensitivity using post-column oxidation-methanation. Anal. Methods 2017, 9, 1928–1934. [Google Scholar]
- Bannon, C.D.; Craske, J.D.; Hilliker, A.E. Analysis of fatty acid methyl esters with high accuracy and reliability. V. Validation of theoretical relative response factors of unsaturated esters in the flame ionization detector. J. Am. Oil Chem. Soc. 1986, 63, 105–110. [Google Scholar] [CrossRef]
- Wang, F.; Chen, M.; Luo, R.; Huang, G.; Wu, X.; Zheng, N.; Zhang, Y.; Wang, J. Fatty acid profiles of milk from Holstein cows, Jersey cows, buffalos, yaks, humans, goats, camels, and donkeys based on gas chromatography-mass spectrometry. J. Dairy Sci. 2021, 105, 1687–1700. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Lock, A.L.; Garnsworthy, P.C. Technical Note: A rapid lipid separation method for determining fatty acid composition in milk. J. Dairy Sci. 2004, 87, 3785–3788. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Wang, P.; Yang, L.; Ma, Y.; Day, L. Quantification of fatty acids in human, cow, buffalo, goat, yak, and camel milk using an improved one-step GC-FID method. Food Anal. Methods 2017, 10, 2881–2891. [Google Scholar] [CrossRef]
- Liu, Z.; Ezernieks, V.; Rochfort, S.; Cocks, B. Comparison of methylation methods for fatty acid analysis of milk fat. Food Chem. 2018, 261, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Gałuszka, A.; Migaszewski, Z.M.; Konieczka, P.; Namieśnik, J. Analytical Eco-Scale for assessing the greenness of analytical procedures. TrAC–Trends Anal. Chem. 2012, 37, 61–72. [Google Scholar] [CrossRef]
- Ahmed, A.B.; Gamal, M.; Naguib, I.A.; Ali, H.M. Environmental impact of the reported chromatographic methods for the determination of the first FDA-Approved therapy for COVID-19 Patients. Remdesivir: A comparative study. Microchem. J. 2022, 176, 107242. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, D.; Fouad, M.M. Application of NEMI. Analytical Eco-Scale and GAPI tools for greenness assessment of three developed chromatographic methods for quantification of sulfadiazine and trimethoprim in bovine meat and chicken muscles: Comparison to greenness profile of reported HPLC methods. Microchem. J. 2020, 157, 104873. [Google Scholar]
- Płotka-Wasylka, J. A new tool for the evaluation of the analytical procedure: Green Analytical Procedure Index. Talanta 2018, 181, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Wojnowski, W.; Tobiszewska, M.; Pena-Pereira, F.; Psillakis, E. AGREEprep–Analytical greenness metric for sample preparation. Trends Anal. Chem. 2022, 149, 11655. [Google Scholar] [CrossRef]
- Supelco. Comparison of 37 Component FAME Standard on Four Capillary GC Columns; Bulletin 907; Supelco, Inc.: Bellefonte, PA, USA, 1996. [Google Scholar]
- ISO 14156:2001; Milk and Milk Products—Extraction Methods for Lipids and Liposoluble Compounds. International Organization for Standardization: London, UK, 2001.
- ISO 15885/IDF 184:2002; Milk Fat—Determination of the Fatty Acid Composition by Gas-Liquid Chromatography. International Organization for Standardization: London, UK, 2002.
- ISO 15884/IDF 182:2002; Milk Fat—Preparation of Fatty Acid Methyl Esters. International Organization for Standardization: London, UK, 2002.
- ISO 16958:2015; Milk; Milk Products, Infant Formula and Adult Nutritional’s—Determination of Fatty Acids Composition—Capillary Gas Chromatographic Method. International Organization for Standardization: London, UK, 2015.
- ISO 12966-4:2015; Animal and Vegetable Fats and Oils—Gas Chromatography of Fatty Acid Methyl Esters—Part 4: Determination by Capillary Gas Column. International Organization for Standardization: London, UK, 2015.
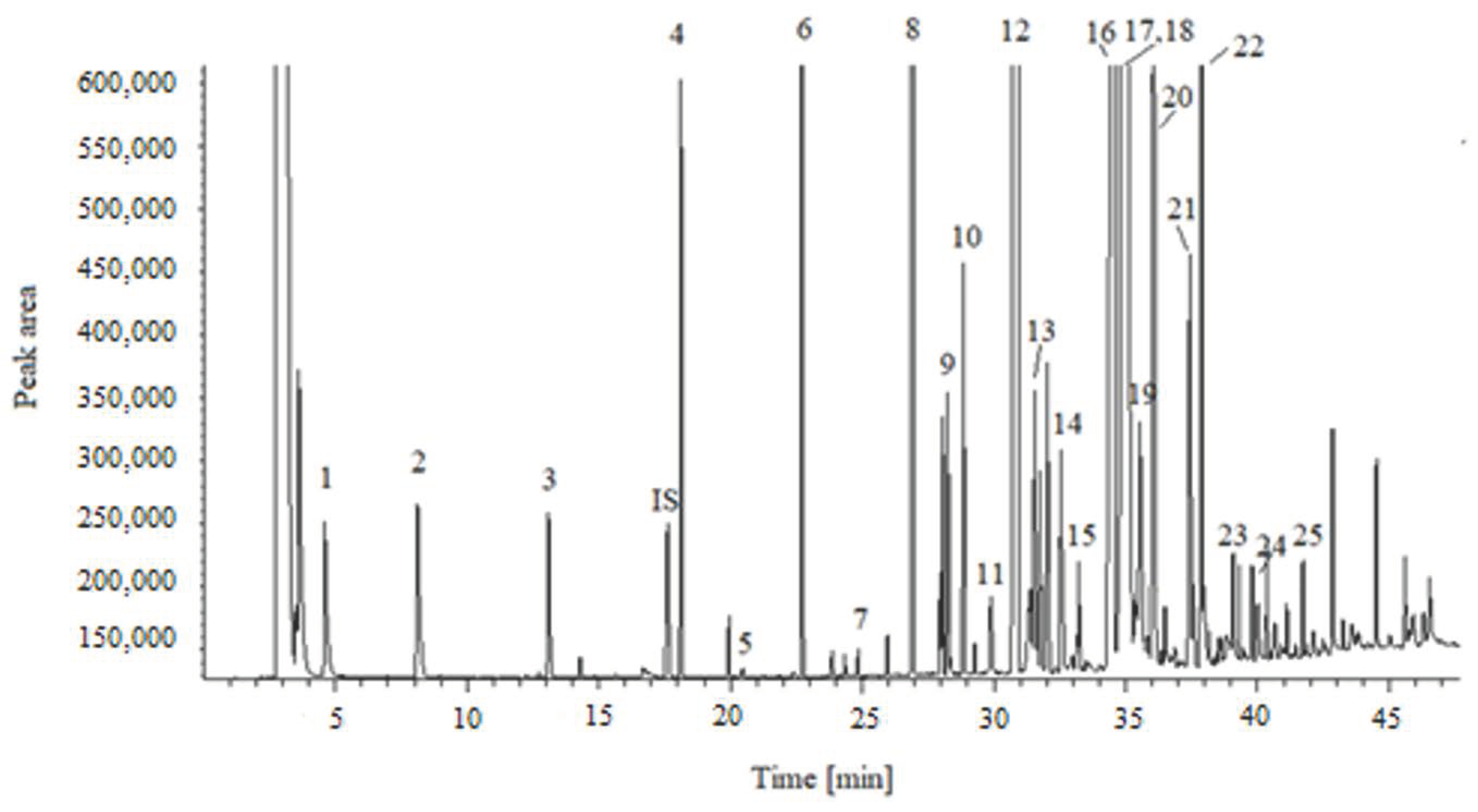
| Fatty Acid | ERF 1 | TRF 2 | EF 3 | Intra-Day RSD (%) | Inter-Day RSD (%) | |
|---|---|---|---|---|---|---|
| butyric acid | C4:0 | 2.3260 | 1.5742 | 1.4776 | 7.5 | 8.0 |
| caproic acid | C6:0 | 1.5308 | 1.3378 | 1.1443 | 6.6 | 2.4 |
| caprylic acid | C8:0 | 1.1036 | 1.2195 | 0.9050 | 4.1 | 1.8 |
| capric acid | C10:0 | 0.9640 | 1.2702 | 0.7589 | 2.5 | 1.7 |
| undecanoic acid | C11:0 | 0.9501 | 1.1486 | 0.8272 | 2.3 | 2.9 |
| lauric acid | C12:0 | 0.9433 | 1.1013 | 0.8566 | 0.6 | 1.3 |
| oleic acid | C13:0 | 0.9549 | 1.0831 | 0.8817 | 1.2 | 0.9 |
| myristic acid | C14:0 | 0.9678 | 1.0675 | 0.9066 | 1.6 | 0.8 |
| myristoleic acid | C14:1 | 0.9835 | 1.0587 | 0.9290 | 1.6 | 1.0 |
| pentadecylic acid | C15:0 | 0.9514 | 1.0540 | 0.9026 | 1.1 | 0.7 |
| ginkgolic acid | C15:1 | 0.9439 | 1.0457 | 0.9027 | 1.0 | 1.7 |
| palmitic acid | C16:0 | 0.9418 | 1.0422 | 0.9037 | 2.0 | 0.6 |
| palmitoleic acid | C16:1 | 0.9909 | 1.0345 | 0.9579 | 1.7 | 0.5 |
| heptadecanoic acid | C17:0 | 0.9418 | 1.0318 | 0.9127 | 1.9 | 1.1 |
| 10-heptadecenoic acid | C17:1 | 0.9019 | 1.0244 | 0.8804 | 1.7 | 4.5 |
| stearic acid | C18:0 | 0.9183 | 1.0225 | 0.8981 | 3.5 | 1.6 |
| elaidic acid + oleic acid | C18:1n9t + C18:1n9c | 0.9000 | 1.0155 | 0.8863 | 1.8 | 1.1 |
| linolealidic acid | C18:2n6c | 0.9510 | 1.0087 | 0.9428 | 8.8 | 1.4 |
| linoleic acid | C18:2n6t | 0.9112 | 1.0087 | 0.9033 | 1.7 | 6.0 |
| arachidic acid | C20:0 | 0.9813 | 1.0067 | 0.9748 | 1.4 | 6.6 |
| alpha-linolenic acid | C18:3n3 | 0.9060 | 1.0017 | 0.9044 | 8.9 | 1.6 |
| 11-eicosenoic acid | C20:1n9 | 0.9359 | 1.0005 | 0.9354 | 7.3 | 10.0 |
| behenic acid | C22:0 | 0.9326 | 0.9939 | 0.9384 | 2.9 | 2.7 |
| arachidonic acid | C20:4n6 | 1.0020 | 0.9819 | 1.0205 | 2.4 | 5.8 |
| Nr of FA | FA | Method A | Method B | Method C | Method D | Method E | Method F | Method G | Method G’ |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C4:0 | 1.19 ± 0.05 c | 1.46 ± 0.01 a | 1.41 ± 0.01 a | 1.83 ± 0.05 d | 1.43 ± 0.08 a | 1.57 ± 0.06 a | 3.97 ± 0.09 b | 3.87 ± 0.11 b |
| 2 | C6:0 | 1.69 ± 0.09 a | 1.79 ± 0.14 a | 1.68 ± 0.00 a | 2.24 ± 0.04 a | 1.83 ± 0.12 a | 1.90 ± 0.02 a | 2.05 ± 0.10 a | 2.15 ± 0.80 a |
| 3 | C8:0 | 1.12 ± 0.05 a | 1.07 ± 0.08 ac | 1.02 ± 0.04 a | 1.27 ± 0.03 a | 1.19 ± 0.07 a | 1.11 ± 0.02 a | 0.97 ± 0.07 ab | 0.95 ± 0.04 bc |
| 4 | C10:0 | 2.66 ± 0.05 a | 2.52 ± 0.09 a | 2.42 ± 0.06 ac | 2.72 ± 0.02 a | 2.79 ± 0.05 a | 2.84 ± 0.09 a | 2.06 ± 0.15 b | 2.16 ± 0.19 bc |
| 5 | C11:0 | 0.06 ± 0.00 a | 0.06 ± 0.00 a | 0.06 ± 0.00 a | 0.07 ± 0.00 b | 0.08 ± 0.00 bc | 0.08 ± 0.01 c | 0.05 ± 0.00 d | 0.03 ± 0.00 e |
| 6 | C12:0 | 3.31 ± 0.02 a | 3.13 ± 0.02 a | 3.06 ± 0.06 a | 3.19 ± 0.01 a | 3.38 ± 0.15 a | 3.35 ± 0.15 a | 2.53 ± 0.16 b | 2.46 ± 0.11 b |
| 7 | C13:0 | 0.13 ± 0.00 a | 0.12 ± 0.00 a | 0.12 ± 0.00 a | 0.12 ± 0.00 a | 0.13 ± 0.01 a | 0.14 ± 0.01 a | 0.10 ± 0.01 b | 0.09 ± 0.01 b |
| 8 | C14:0 | 11.99 ± 0.04 a | 11.57 ± 0.14 a | 11.46 ± 0.18 a | 11.26 ± 0.06 a | 11.93 ± 0.40 a | 11.50 ± 0.37 a | 9.77 ± 0.38 b | 9.54 ± 0.39 b |
| 9 | C14:1 | 1.61 ± 0.01 a | 1.54 ± 0.02 a | 1.52 ± 0.03 a | 1.57 ± 0.01 a | 1.58 ± 0.05 a | 1.59 ± 0.04 a | 1.06 ± 0.06 b | 1.09 ± 0.07 b |
| 10 | C15:0 | 1.30 ± 0.00 a | 1.27 ± 0.02 a | 1.27 ± 0.02 a | 1.25 ± 0.01 a | 1.30 ± 0.04 a | 1.25 ± 0.02 a | 1.13 ± 0.03 b | 1.11 ± 0.02 b |
| 11 | C15:1 | 0.26 ± 0.00 a | 0.26 ± 0.01 a | 0.26 ± 0.00 a | 0.25 ± 0.00 a | 0.26 ± 0.00 a | 0.25 ± 0.00 a | 0.23 ± 0.00 b | 0.21 ± 0.00 b |
| 12 | C16:0 | 31.34 ± 0.10 a | 32.11 ± 0.79 a | 31.87 ± 0.83 a | 30.49 ± 0.15 a | 31.58 ± 0.48 a | 31.71 ± 0.30 a | 29.73 ± 0.18 b | 29.52 ± 0.27 b |
| 13 | C16:1 | 2.26 ± 0.01 a | 2.00. ± 0.04 b | 2.01 ± 0.04 b | 2.01 ± 0.03 b | 2.08 ± 0.16 ab | 1.95 ± 0.04 b | 1.67 ± 0.03 c | 1.62 ± 0.03 c |
| 14 | C17:0 | 0.69 ± 0.01 a | 0.71 ± 0.02 a | 0.72 ± 0.01 a | 0.63 ± 0.00 ac | 0.70 ± 0.00 a | 0.71 ± 0.03 a | 0.61 ± 0.01 bc | 0.58 ± 0.01 b |
| 15 | C17:1 | 0.29 ± 0.00 ac | 0.27 ± 0.01 a | 0.28 ± 0.02 a | 0.29 ± 0.00 a | 0.28 ± 0.00 a | 0.27 ± 0.01 a | 0.33 ± 0.00 b | 0.31 ± 0.00 bc |
| 16 | C18:0 | 10.53 ± 0.09 a | 11.3 ± 0.39 a | 11.27 ± 0.38 a | 10.23 ± 0.00 a | 10.17 ± 0.10 a | 10.50 ± 0.15 a | 13.61 ± 0.67 b | 13.29 ± 0.35 b |
| 17 + 18 | C18:1n9t + C18:1n9c | 25.76 ± 0.31 a | 25.7 ± 0.63 a | 26.35 ± 0.42 a | 26.31 ± 0.09 a | 25.35 ± 0.44 a | 25.69 ± 0.72 a | 24.69 ± 0.85 a | 24.89 ± 0.47 a |
| 19 | C18:2n6c | 2.25 ± 0.05 ac | 2.22 ± 0.04 ac | 2.24 ± 0.01 ac | 2.27 ± 0.03 ac | 2.19 ± 0.02 a | 2.21 ± 0.01 ab | 2.52 ± 0.06 d | 2.31 ± 0.05 bc |
| 20 | C18:2n6t | 0.49 ± 0.01 a | 0.49 ± 0.01 a | 0.51 ± 0.01 a | 0.49 ± 0.00 a | 0.48 ± 0.01 a | 0.47 ± 0.03 a | 1.05 ± 0.08 b | 1.03 ± 0.04 b |
| 21 | C20:0 | 0.18 ± 0.00 a | 0.19 ± 0.01 a | 0.19 ± 0.01 ab | 0.16 ± 0.00 a | 0.18 ± 0.00 ab | 0.21 ± 0.00 bd | 0.26 ± 0.01 e | 0.23 ± 0.01 d |
| 22 | C18:3n3 | 0.46 ± 0.01 a | 0.44 ± 0.01 a | 0.45 ± 0.00 a | 0.47 ± 0.00 a | 0.44 ± 0.01 a | 0.45 ± 0.01 a | 0.45 ± 0.01 a | 0.39 ± 0.01 b |
| 23 | C20:1n9 | 0.53 ± 0.01 a | 0.54 ± 0.01 a | 0.54 ± 0.01 a | 0.55 ± 0.01 a | 0.52 ± 0.01 a | 0.54 ± 0.01 a | 0.76 ± 0.05 b | 0.72 ± 0.06 b |
| 24 | C22:0 | 0.13 ± 0.00 a | 0.13 ± 0.00 a | 0.13 ± 0.00 a | 0.13 ± 0.00 a | 0.13 ± 0.00 a | 0.13 ± 0.00 a | 0.20 ± 0.02 b | 0.19 ± 0.02 b |
| 25 | C20:4n6 | 0.18 ± 0.00 a | 0.18 ± 0.00 a | 0.18 ± 0.00 a | 0.19 ± 0.01 acd | 0.17 ± 0.00 a | 0.18 ± 0.00 ac | 0.21 ± 0.01 b | 0.20 ± 0.01 bd |
| Sums | |||||||||
| ƩSFA 1 | 65.92 a | 66.35 a | 65.66 a | 65.60 a | 66.80 a | 66.90 a | 67.04 a | 67.11 a | |
| ƩUFA 2 | 34.08 a | 33.65 a | 34.34 a | 34.40 a | 33.20 a | 33.10 a | 32.96 a | 32.89 a | |
| ƩMUFA 3 | 30.70 a | 30.31 a | 30.97 a | 30.98 a | 30.08 ac | 30.29 ac | 28.73 bc | 28.89 bc | |
| ƩPUFA 4 | 3.38 a | 3.33 a | 3.38 a | 3.42 a | 3.12 a | 3.13 a | 4.23 b | 3.99 b | |
| Method | Analytical Eco-Scale Score | GAPI Pictogram | AGREEprep Pictogram |
|---|---|---|---|
| A | 70 acceptable green analysis |  (1 green, 6 yellow, 8 red) |  |
| B | 72 acceptable green analysis |  (1 green, 6 yellow, 8 red) | 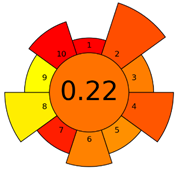 |
| C | 72 acceptable green analysis | 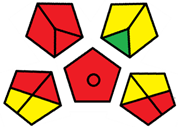 (1 green, 6 yellow, 8 red) | 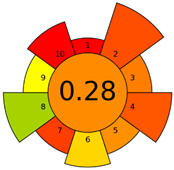 |
| D | 73 acceptable green analysis | 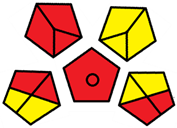 (0 green, 7 yellow, 8 red) | 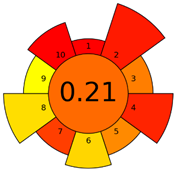 |
| E | 71 acceptable green analysis | 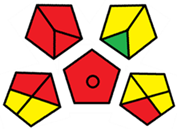 (1 green, 7 yellow, 7 red) | 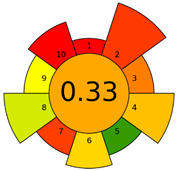 |
| F | 71 acceptable green analysis | 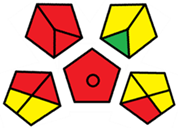 (1 green, 7 yellow, 7 red) | 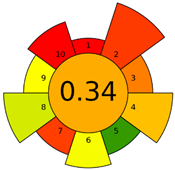 |
| G | 24 inadequate green analysis |  (0 green, 4 yellow, 11 red) | 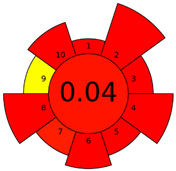 |
| G’ | 45 inadequate green analysis | 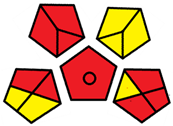 (0 green, 6 yellow, 9 red) | 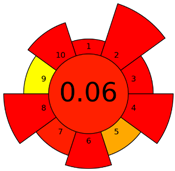 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narloch, I.; Wejnerowska, G. Comparison of the Effectiveness and Environmental Impact of Selected Methods for the Determination of Fatty Acids in Milk Samples. Molecules 2022, 27, 8242. https://doi.org/10.3390/molecules27238242
Narloch I, Wejnerowska G. Comparison of the Effectiveness and Environmental Impact of Selected Methods for the Determination of Fatty Acids in Milk Samples. Molecules. 2022; 27(23):8242. https://doi.org/10.3390/molecules27238242
Chicago/Turabian StyleNarloch, Izabela, and Grażyna Wejnerowska. 2022. "Comparison of the Effectiveness and Environmental Impact of Selected Methods for the Determination of Fatty Acids in Milk Samples" Molecules 27, no. 23: 8242. https://doi.org/10.3390/molecules27238242
APA StyleNarloch, I., & Wejnerowska, G. (2022). Comparison of the Effectiveness and Environmental Impact of Selected Methods for the Determination of Fatty Acids in Milk Samples. Molecules, 27(23), 8242. https://doi.org/10.3390/molecules27238242







