Comprehensive Metabolite Profiling of Four Different Beans Fermented by Aspergillus oryzae
Abstract
1. Introduction
2. Results and Discussion
2.1. Metabolite Profiling of Four Different Beans before Fermentation
2.2. Metabolite Changes of the Four Different Beans during Aspergillus-Oryzae-Mediated Fermentation
2.2.1. Multivariate Statistical Analyses and Heatmap Analyses of the Four Beans during Fermentation
2.2.2. Metabolite Pathway Analysis of the Four Beans Fermented with A. oryzae over Time
2.3. Antioxidant Activity, Total Flavonoid and Phenolic Contents
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Microbial Cultures and Bean Fermentation
3.3. Sample Preparation
3.4. Gas Chromatography Time-of-Flight Mass Spectrometry Analysis
3.5. Liquid Chromatography-Linear Trap Quadrupole-Orbitrap-Tandem Mass Spectrometry Analysis
3.6. Data Processing and Multivariate Statistical Analysis
3.7. Antioxidant Activity and Total Flavonoid and Phenolic Contents
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seo, H.S.; Lee, S.; Singh, D.; Shin, H.W.; Cho, S.A.; Lee, C.H. Untargeted metabolite profiling for koji-fermentative bioprocess unravels the effects of varying substrate types and microbial inocula. Food Chem. 2018, 266, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Sadh, P.K.; Saharan, P.; Duhan, J.S. Bio-augmentation of antioxidants and phenolic content of Lablab purpureus by solid state fermentation with GRAS filamentous fungi. Resour. Technol. 2017, 3, 285–292. [Google Scholar] [CrossRef]
- De Almeida Costa, G.E.; Da Silva Queiroz-Monici, K.; Pissini Machado Reis, S.M.; De Oliveira, A.C. Chemical composition, dietary fibre and resistant starch contents of raw and cooked pea, common bean, chickpea and lentil legumes. Food Chem. 2006, 94, 327–330. [Google Scholar] [CrossRef]
- Kwon, Y.S.; Lee, S.; Lee, S.H.; Kim, H.J.; Lee, C.H. Comparative evaluation of six traditional fermented soybean products in East Asia: A metabolomics approach. Metabolites 2019, 9, 183. [Google Scholar] [CrossRef]
- Song, D.H.; Chun, B.H.; Lee, S.; Son, S.Y.; Reddy, C.K.; Mun, H.I.; Jeon, C.O.; Lee, C.H. Comprehensive metabolite profiling and microbial communities of Doenjang (fermented soy paste) and Ganjang (fermented soy sauce): A comparative study. Foods 2021, 10, 641. [Google Scholar] [CrossRef] [PubMed]
- Do Prado, F.G.; Pagnoncelli, M.G.B.; de Melo Pereira, G.V.; Karp, S.G.; Soccol, C.R. Fermented soy products and their potential health benefits: A review. Microorganisms 2022, 10, 1606. [Google Scholar] [CrossRef]
- Juan, M.Y.; Wu, C.H.; Chou, C.C. Fermentation with Bacillus spp. as a bioprocess to enhance anthocyanin content, the angiotensin converting enzyme inhibitory effect, and the reducing activity of black soybeans. Food Microbiol. 2010, 27, 918–923. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Q.; Zhu, Y.; Lam, H.-M.; Cai, Z.; Guo, D. Comparative metabolic profiling reveals secondary metabolites correlated with soybean salt tolerance. J. Agric. Food Chem. 2008, 56, 11132–11138. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.; Singh, D.; Oh, J.Y.; Jeon, E.J.; Ryu, H.S.K.; Lee, D.W.; Kim, B.S.; Lee, C.H. Comparative evaluation of microbial diversity and metabolite profiles in Doenjang, a fermented soybean paste, during the two different industrial manufacturing processes. Food Chem. 2017, 221, 1578–1586. [Google Scholar] [CrossRef]
- Lin, C.H.; Wei, Y.T.; Chou, C.C. Enhanced antioxidative activity of soybean koji prepared with various filamentous fungi. Food Microbiol. 2006, 23, 628–633. [Google Scholar] [CrossRef]
- Singh, D.; Lee, C.H. Intraspecies volatile interactions affect growth rates and exometabolomes in Aspergillus oryzae KCCM 60345. J. Microbiol. Biotechnol. 2018, 28, 199–209. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, X.; Yuan, X.; Shi, J.; Zhang, C.; Yan, N.; Jing, C. Comparison of phenolic and flavonoid compound profiles and antioxidant and α-glucosidase inhibition properties of cultivated soybean (Glycine max) and wild soybean (Glycine soja). Plants 2021, 10, 813. [Google Scholar] [CrossRef] [PubMed]
- Colletti, A.; Attrovio, A.; Boffa, L.; Mantegna, S.; Cravotto, G. Valorisation of by-products from soybean (Glycine max (L.) Merr.) processing. Molecules 2020, 25, 2129. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Lee, S.; Van, K.; Kim, T.H.; Jeong, S.C.; Choi, I.Y.; Kim, D.S.; Lee, Y.S.; Park, D.; Ma, J.; et al. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci. USA 2010, 107, 22032–22037. [Google Scholar] [CrossRef]
- Li, Y.H.; Li, W.; Zhang, C.; Yang, L.; Chang, R.Z.; Gaut, B.S.; Qiu, L.J. Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single-nucleotide polymorphism loci. New Phytol. 2010, 188, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Jing, C.; Wen, Z.; Zou, P.; Yuan, Y.; Jing, W.; Li, Y.; Zhang, C. Consumption of black legumes Glycine soja and Glycine max lowers serum lipids and alters the gut microbiome profile in mice fed a high-fat diet. J. Agric. Food Chem. 2018, 66, 7367–7375. [Google Scholar] [CrossRef] [PubMed]
- Kofsky, J.; Zhang, H.; Song, B.H. The untapped genetic reservoir: The past, current, and future applications of the wild soybean (Glycine soja). Front. Plant Sci. 2018, 9, 949. [Google Scholar] [CrossRef]
- Beninger, C.W.; Hosfield, G.L. Antioxidant activity of extracts, condensed tannin fractions, and pure flavonoids from Phaseolus vulgaris L. seed coat color genotypes. J. Agric. Food Chem. 2003, 51, 7879–7883. [Google Scholar] [CrossRef]
- Hanapiah, N.F.H.; Sinniah, U.R.; Yusoff, M.M. Seed quality of lablab bean (Lablab purpureus) as influenced by seed maturity and drying methods. Agronomy 2022, 12, 363. [Google Scholar] [CrossRef]
- Ha, T.J.; Lee, B.W.; Park, K.H.; Jeong, S.H.; Kim, H.T.; Ko, J.M.; Baek, I.Y.; Lee, J.H. Rapid characterisation and comparison of saponin profiles in the seeds of Korean leguminous species using ultra performance liquid chromatography with photodiode array detector and electrospray ionisation/mass spectrometry (UPLC-PDA-ESI/MS) analysis. Food Chem. 2014, 146, 270–277. [Google Scholar] [CrossRef]
- Kim, Y.-W.; Lim, S.-J.; Kim, M.-A.; Choi, W.-C.; Yoon, H.-T. Determination and isolation of leaf isoflavone in hyacinth bean. KOREAN J. Crop Sci. 2001, 46, 449–452. [Google Scholar]
- Yang, Q.Q.; Gan, R.Y.; Ge, Y.Y.; Zhang, D.; Corke, H. Polyphenols in common beans (Phaseolus vulgaris L.): Chemistry, analysis, and factors affecting composition. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1518–1539. [Google Scholar] [CrossRef] [PubMed]
- Gil, H.J.; Lee, S.; Singh, D.; Lee, C.H. Varying inocula permutations (Aspergillus oryzae and Bacillus amyloliquefaciens) affect enzyme activities and metabolite levels in koji. J. Microbiol. Biotechnol. 2018, 28, 1971–1981. [Google Scholar] [CrossRef] [PubMed]
- Suganuma, T.; Fujita, K.; Kitahara, K. Some distinguishable properties between acid-stable and neutral types of α-amylases from acid-producing koji. J. Biosci. Bioeng. 2007, 104, 353–362. [Google Scholar] [CrossRef]
- Soung, S.H.; Lee, S.; Lee, S.H.; Kim, H.J.; Lee, N.R.; Lee, C.H. Metabolomic-based comparison of traditional and industrial Doenjang samples with antioxidative activities. Foods 2021, 10, 1377. [Google Scholar] [CrossRef]
- Hyeon, H.; Min, C.W.; Moon, K.; Cha, J.; Gupta, R.; Park, S.U.; Kim, S.T.; Kim, J.K. Metabolic profiling-based evaluation of the fermentative behavior of Aspergillus oryzae and Bacillus subtilis for soybean residues treated at different temperatures. Foods 2020, 9, 1–17. [Google Scholar] [CrossRef]
- Lee, H.; Lee, S.; Kyung, S.; Ryu, J.; Kang, S.; Park, M.; Lee, C. Metabolite Profiling and Anti-aging activity of rice koji fermented with Aspergillus oryzae and Aspergillus cristatus: A comparative study. Metabolites 2021, 11, 524. [Google Scholar] [CrossRef]
- Chancharoonpong, C.; Hsieh, P.-C.; Sheu, S.-C. Enzyme production and growth of Aspergillus oryzae S. on soybean koji fermentation. APCBEE Procedia 2012, 2, 57–61. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, S.; Lee, S.H.; Kim, H.J.; Singh, D.; Lee, C.H. Integrated metabolomics and volatolomics for comparative evaluation of fermented soy products. Foods 2021, 10, 2516. [Google Scholar] [CrossRef]
- Suh, D.H.; Jung, E.S.; Park, H.M.; Kim, S.H.; Lee, S.; Jo, Y.H.; Lee, M.K.; Jung, G.; Do, S.G.; Lee, C.H. Comparison of metabolites variation and antiobesity effects of fermented versus nonfermented mixtures of Cudrania tricuspidata, Lonicera caerulea, and soybean according to fermentation in Vitro and in Vivo. PLoS ONE 2016, 11, e0149022. [Google Scholar] [CrossRef]
- Mora, L.d.O.; Antunes, L.M.G.; Francescato, H.D.C.; Bianchi, M.d.L.P. The effects of oral glutamine on cisplatin-induced nephrotoxicity in rats. Pharmacol. Res. 2003, 47, 517–522. [Google Scholar] [CrossRef]
- Nemati, A.; Alipanah-Moghadam, R.; Molazadeh, L.; Baghi, A.N. The effect of glutamine supplementation on oxidative stress and matrix metalloproteinase 2 and 9 after exhaustive exercise. Drug Des. Devel. Ther. 2019, 13, 4215–4223. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Seo, M.H.; Oh, D.K.; Lee, C.H. Targeted metabolomics for Aspergillus oryzae-mediated biotransformation of soybean isoflavones, showing variations in primary metabolites. Biosci. Biotechnol. Biochem. 2014, 78, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.H.; Green-Johnson, J.M.; Buckley, N.D.; Lin, Q.Y. Bioactivity of soy-based fermented foods: A review. Biotechnol. Adv. 2019, 37, 223–238. [Google Scholar] [CrossRef]
- Lee, S.; Oh, D.G.; Singh, D.; Lee, J.S.; Lee, S.; Lee, C.H. Exploring the metabolomic diversity of plant species across spatial (leaf and stem) components and phylogenic groups. BMC Plant Biol. 2020, 20, 39. [Google Scholar] [CrossRef]
- Won, J.Y.; Son, S.Y.; Lee, S.; Singh, D.; Lee, S.; Lee, J.S.; Lee, C.H. Strategy for screening of antioxidant compounds from two ulmaceae species based on liquid chromatography-mass spectrometry. Molecules 2018, 23, 1830. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, D.E.; Singh, D.; Lee, C.H. Metabolomics reveal optimal grain preprocessing (milling) toward rice koji fermentation. J. Agric. Food Chem. 2018, 66, 2694–2703. [Google Scholar] [CrossRef]
- Lee, D.E.; Lee, S.; Jang, E.S.; Shin, H.W.; Moon, B.S.; Lee, C.H. Metabolomic profiles of Aspergillus oryzae and Bacillus amyloliquefaciens during rice koji fermentation. Molecules 2016, 21, 773. [Google Scholar] [CrossRef]
- Hodgson, S.; Griffin, T.J.; Reilly, C.; Harvey, S.; Witthuhn, B.A.; Sandri, B.J.; Kunisaki, K.M.; Wendt, C.H. Plasma sphingolipids in HIV-associated chronic obstructive pulmonary disease. BMJ Open Respir. Res. 2017, 4, e000180. [Google Scholar] [CrossRef]
- Pelosi, A.C.; Fernandes, A.M.A.P.; Maciel, L.F.; Silva, A.A.R.; Mendes, G.C.; Bueno, L.F.; Silva, L.M.F.; Bredariol, R.F.; Santana, M.G.; Porcari, A.M.; et al. Liquid chromatography coupled to high-resolution mass spectrometry metabolomics: A useful tool for investigating tumor secretome based on a three-dimensional co-culture model. PLoS ONE 2022, 17, e0274623. [Google Scholar] [CrossRef]
- Kim, M.J.; Son, S.Y.; Jeon, S.G.; Kim, J.G.; Lee, C.H. Metabolite profiling of Dioscorea (yam) leaves to identify bioactive compounds reveals their potential as renewable resources. Plants 2021, 10, 1751. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kumar, S.; Kumar, B. LC-MS Identification of proanthocyanidins in bark and fruit of six Terminalia species. Nat. Prod. Commun. 2018, 13, 555–560. [Google Scholar] [CrossRef]
- Leng, Z.; Zhong, B.; Wu, H.; Liu, Z.; Rauf, A.; Bawazeer, S.; Suleria, H.A.R. Identification of phenolic compounds in Australian-grown bell peppers by liquid chromatography coupled with electrospray ionization-quadrupole-time-of-flight-mass spectrometry and estimation of their antioxidant potential. ACS Omega 2022, 7, 4563–4576. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.P.; Yang, H.; Wang, Y.L.; Chen, X.X.; Zhang, K.; Wang, Y.L.; Sun, Y.F.; Huang, J.; Yang, L.; Wang, J.H. Determination of flavonoids compounds of three species and different harvesting periods in Crataegi folium based on LC-MS/MS. Molecules 2021, 26, 1602. [Google Scholar] [CrossRef] [PubMed]
- Li, M.J.; Jiang, G.F.; Wang, W. Metabolite changes in orange dead leaf butterfly Kallima inachus during ontogeny and diapause. Metabolites 2022, 12, 804. [Google Scholar] [CrossRef]
- Yang, T.; Hui, R.; Nouws, J.; Sauler, M.; Zeng, T.; Wu, Q. Untargeted metabolomics analysis of esophageal squamous cell cancer progression. J. Transl. Med. 2022, 20, 127. [Google Scholar] [CrossRef]
 (GM): G. max,
(GM): G. max,  (GS): G. soja,
(GS): G. soja,  (PV): P. vulgaris,
(PV): P. vulgaris,  (LP): L. purpureus).
(LP): L. purpureus).
 (GM): G. max,
(GM): G. max,  (GS): G. soja,
(GS): G. soja,  (PV): P. vulgaris,
(PV): P. vulgaris,  (LP): L. purpureus).
(LP): L. purpureus).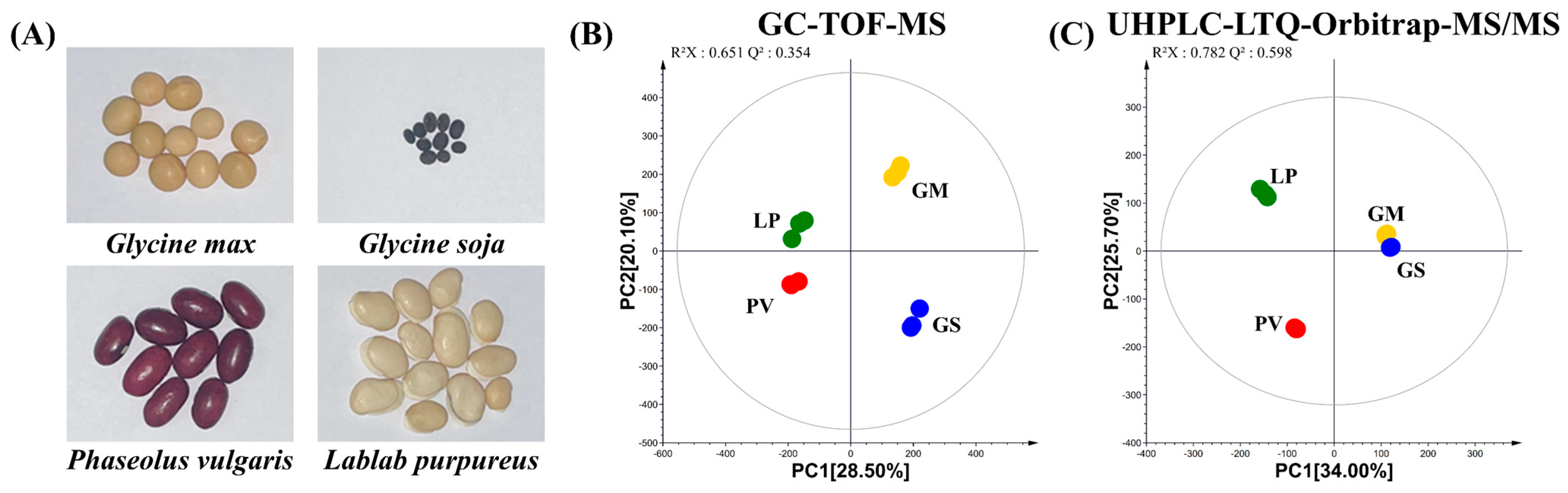



 (GM): G. max,
(GM): G. max,  (GS): G. soja,
(GS): G. soja,  (PV): P. vulgaris,
(PV): P. vulgaris,  (LP): L. purpureus, 0D: 0 day of fermentation, 1D: 1 day after fermentation, 2D: 2 days after fermentation, 3D: 3 days after fermentation.
(LP): L. purpureus, 0D: 0 day of fermentation, 1D: 1 day after fermentation, 2D: 2 days after fermentation, 3D: 3 days after fermentation.
 (GM): G. max,
(GM): G. max,  (GS): G. soja,
(GS): G. soja,  (PV): P. vulgaris,
(PV): P. vulgaris,  (LP): L. purpureus, 0D: 0 day of fermentation, 1D: 1 day after fermentation, 2D: 2 days after fermentation, 3D: 3 days after fermentation.
(LP): L. purpureus, 0D: 0 day of fermentation, 1D: 1 day after fermentation, 2D: 2 days after fermentation, 3D: 3 days after fermentation.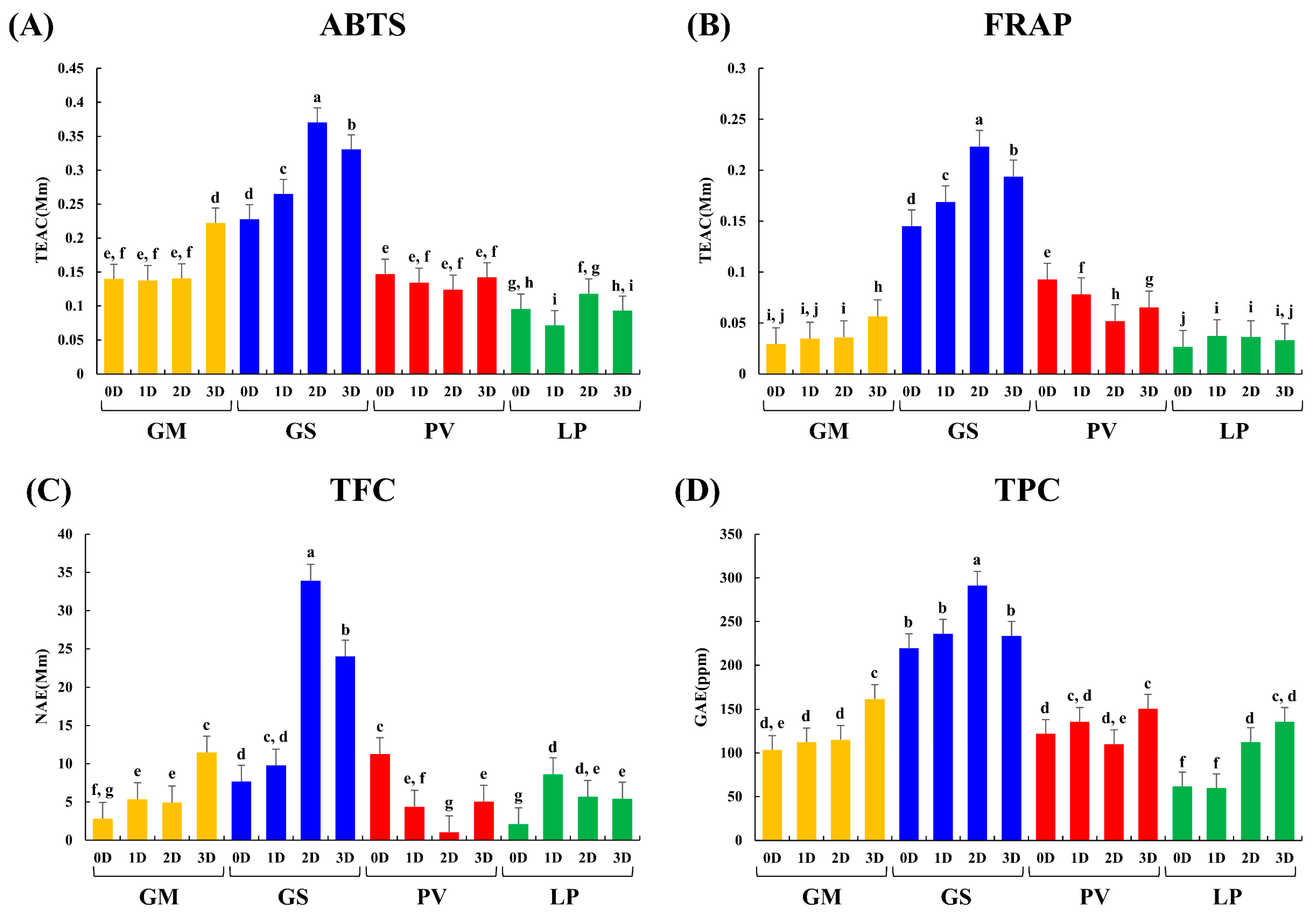
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.H.; Lee, N.-R.; Lee, C.H. Comprehensive Metabolite Profiling of Four Different Beans Fermented by Aspergillus oryzae. Molecules 2022, 27, 7917. https://doi.org/10.3390/molecules27227917
Lee YH, Lee N-R, Lee CH. Comprehensive Metabolite Profiling of Four Different Beans Fermented by Aspergillus oryzae. Molecules. 2022; 27(22):7917. https://doi.org/10.3390/molecules27227917
Chicago/Turabian StyleLee, Yeon Hee, Na-Rae Lee, and Choong Hwan Lee. 2022. "Comprehensive Metabolite Profiling of Four Different Beans Fermented by Aspergillus oryzae" Molecules 27, no. 22: 7917. https://doi.org/10.3390/molecules27227917
APA StyleLee, Y. H., Lee, N.-R., & Lee, C. H. (2022). Comprehensive Metabolite Profiling of Four Different Beans Fermented by Aspergillus oryzae. Molecules, 27(22), 7917. https://doi.org/10.3390/molecules27227917





