NMR-Based Approaches in the Study of Foods
Abstract
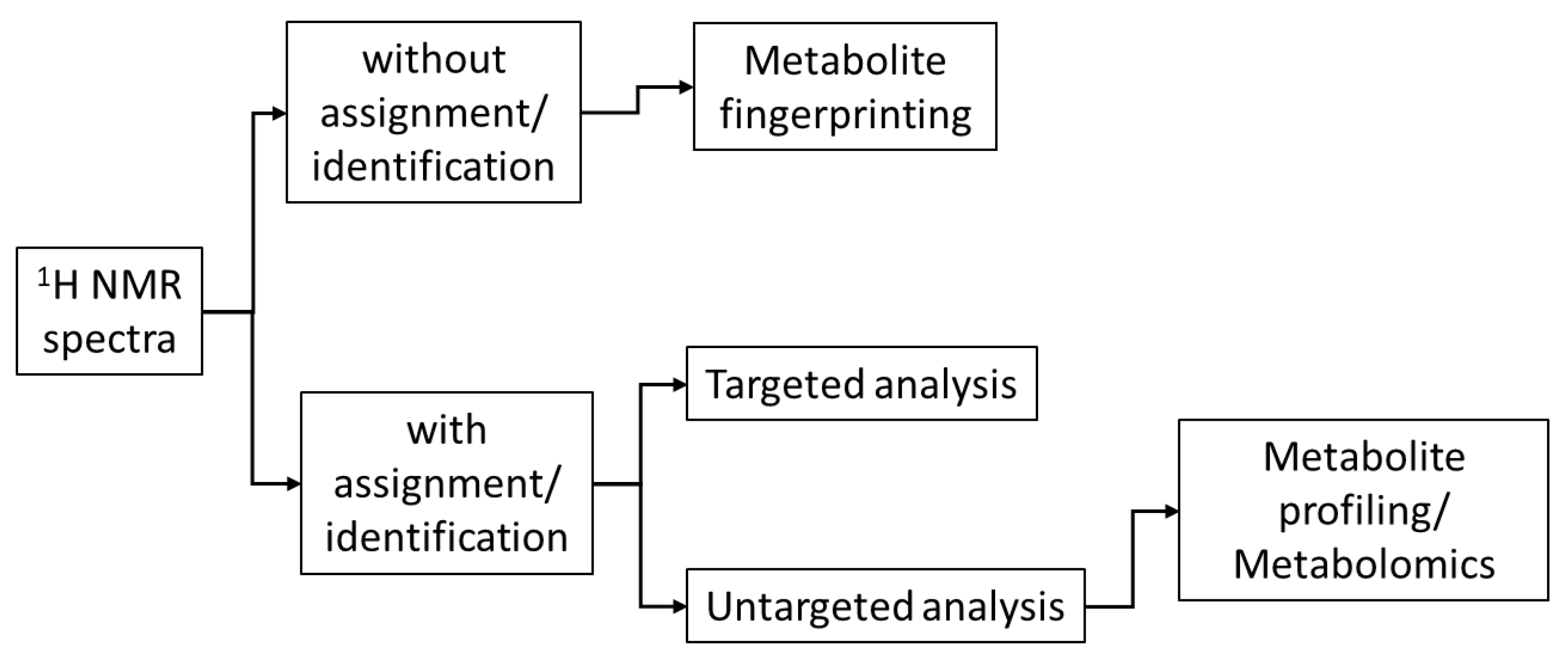
1. Introduction
- -
- High-resolution NMR of liquid samples [6] is used to define the chemical structure of isolated compounds or compounds in complex mixtures. It can be applied also to solid and semisolid food samples after extraction of soluble components;
- -
- High-resolution magic-angle spinning (HR-MAS) NMR methodology has been developed to analyze mobile molecular components in all types of matrixes and it is particularly useful in analyzing intact semisolid samples without any extraction. The rotation of the sample around an axis forming an angle of 54.7° (magic angle) with respect to the applied magnetic field B0 drastically reduces the broadening of signals due to magnetic susceptibility variations and makes it possible to obtain high-resolution spectra of all mobile molecular components;
- -
- Cross-polarization magic-angle spinning (CP-MAS) is the most frequently used methodology for the NMR analysis of solids. It combines three techniques: fast magic-angle rotation of the sample, high-power decoupling of protons, and cross-polarization (transferring part of the hydrogen magnetization to a heteronucleus to improve its sensitivity) [10]. Generally, in food samples, CP-MAS is applied to study the insoluble fraction (fibers, proteins, etc.), observing 13C as the heteronucleus. Moreover, 31P and 2H solid-state NMR spectroscopic techniques were largely employed to examine the interaction among lipids and many bioactive compounds from foods [11,12,13,14];
- -
- Low-field 1H NMR relaxometry [15] is a suitable tool to study the most abundant components of intact foodstuffs by measurement of relaxation parameters (longitudinal relaxation time, T1, water self-diffusion (Dw), T2) and amplitudes of the NMR signal. Information on food microstructure such as water compartments and diffusion can be obtained by detecting proton signals dominated by H2O contained in foodstuffs. Sample pretreatment is not required, and the method can be easily used for quality control applications;
- -
- MRI allows one to obtain detailed morphological images of internal sections of intact tissues [16]. Furthermore, using specific data acquisition protocols, it is able to provide multiple pieces of complementary information also relating to the composition and dynamics of the molecules that form the investigated tissue.
2. First Starting Point: Food
2.1. Kiwifruit: Chemical Profile and Morphological Aspects
2.2. Kiwifruit Ripeness
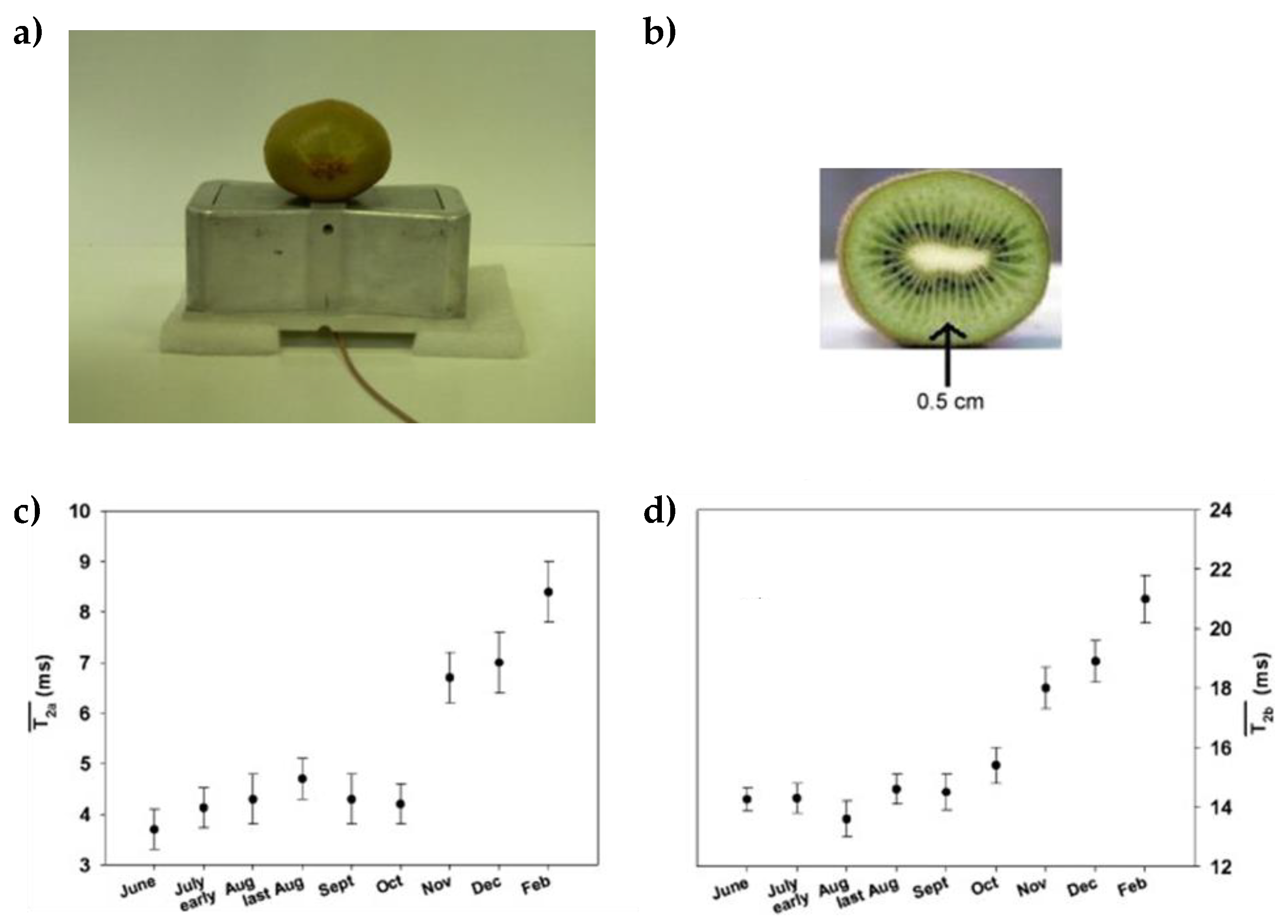
2.2.1. Chemical Profile over the Time (Ripeness and Postharvest Period)
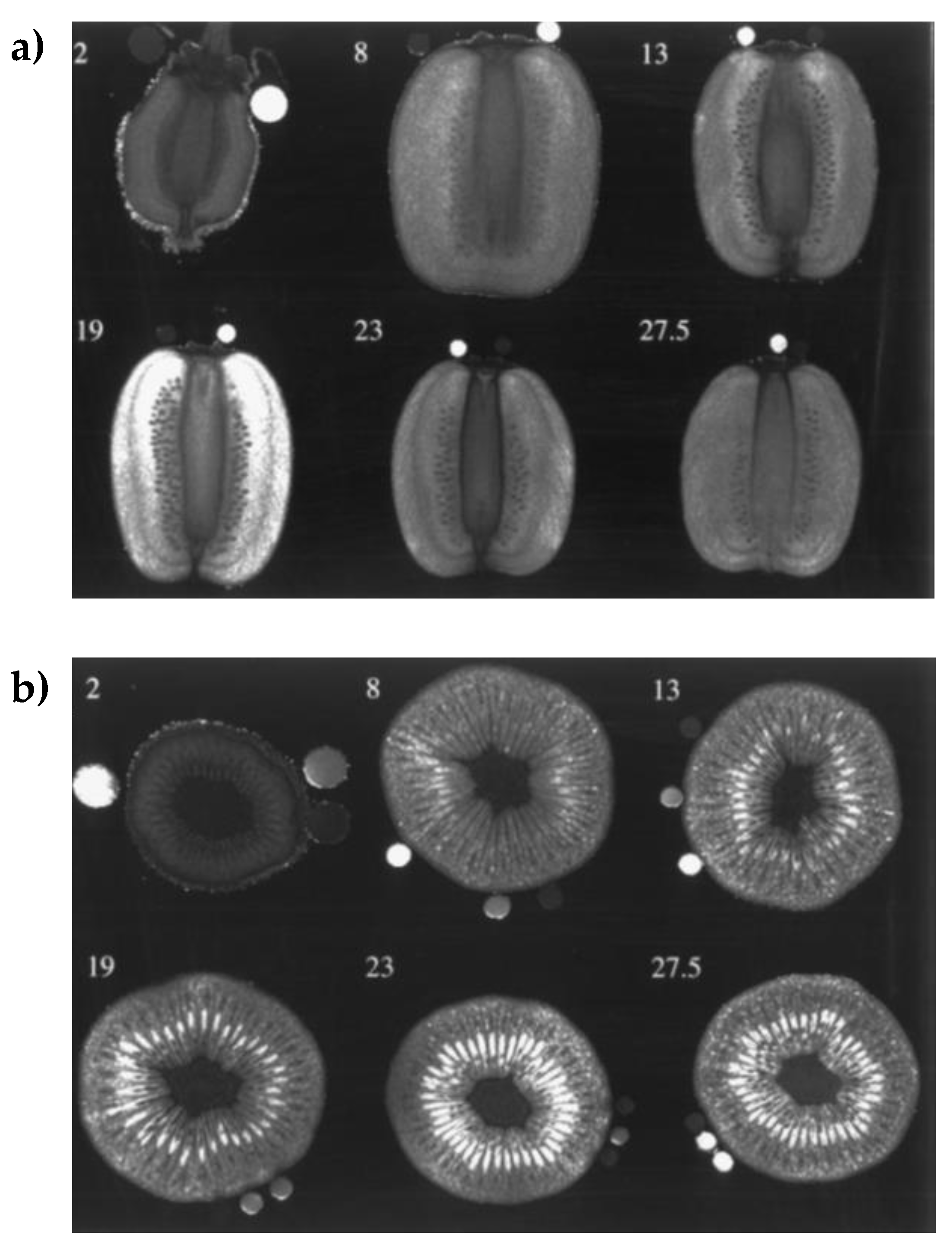
2.2.2. Morphological Changes over the Time
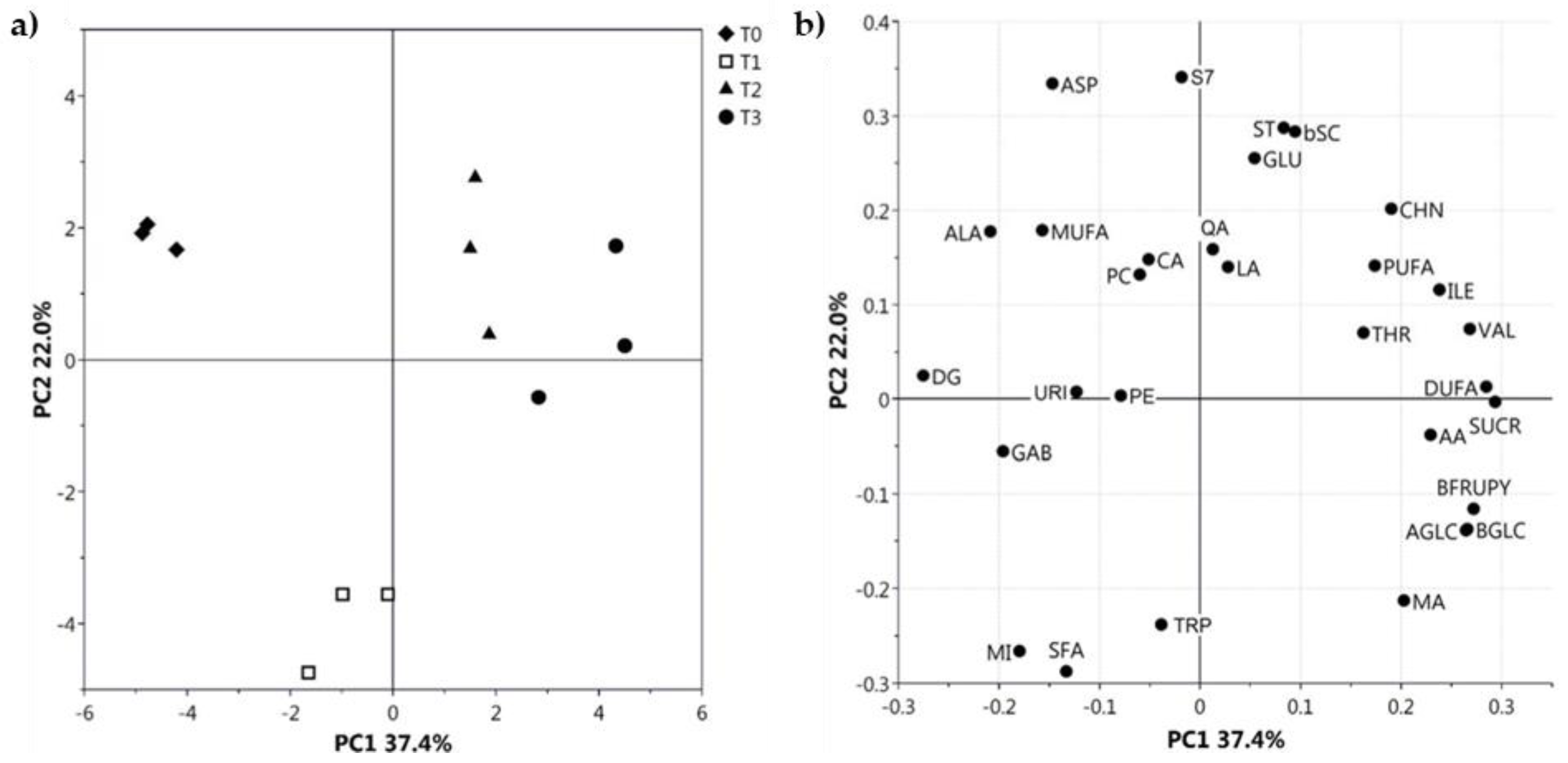
2.3. Comparison among Cultivars
2.3.1. MRI for Morphological and Physico-Chemical Aspects
2.3.2. Comparison among Healthy and Non-Healthy Kiwifruit
3. Second Starting Point: Common Food-Related Challenges
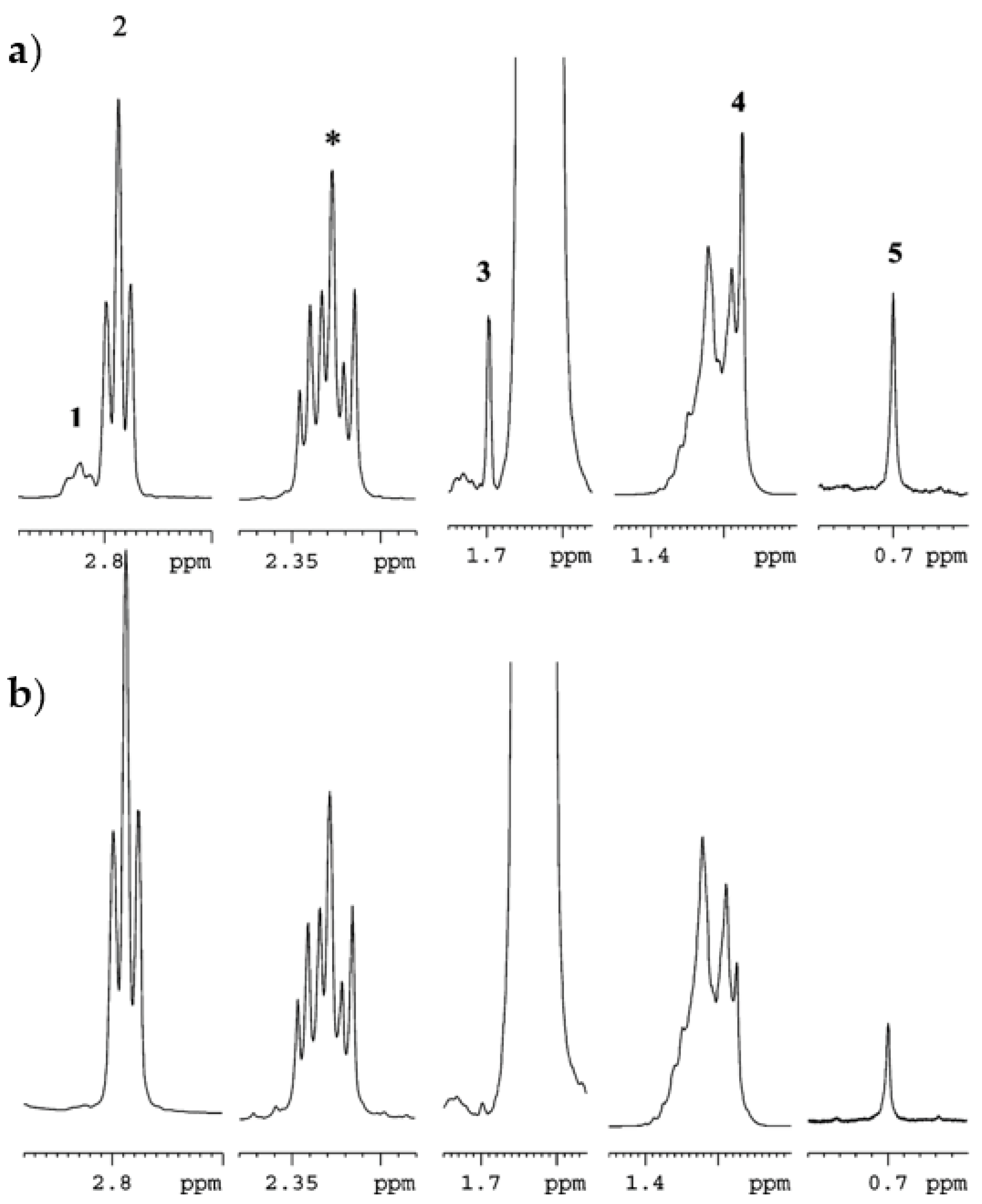
3.1. Olive Oils: Addition of Hazelnut Oils to Olive Oils
3.2. Honey: Fraudulent Designation of Botanical Origin and Addition of Sugars
3.2.1. Addition of Sugars in Honey Samples
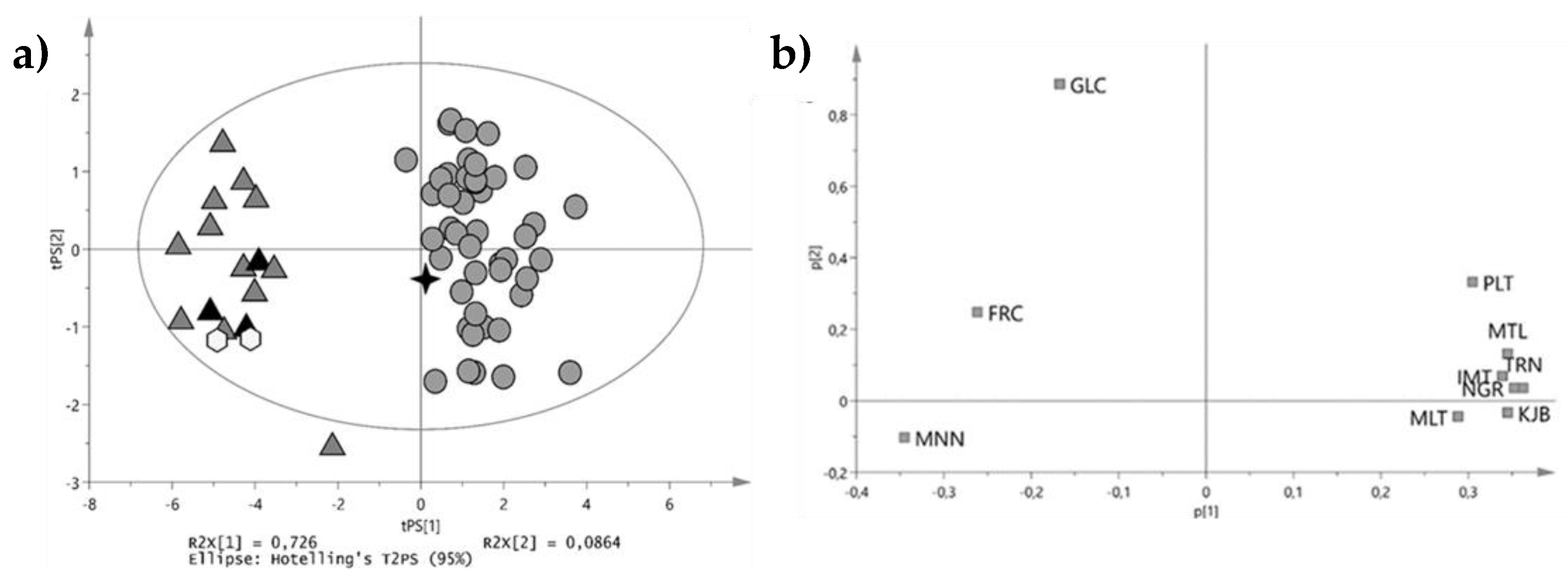
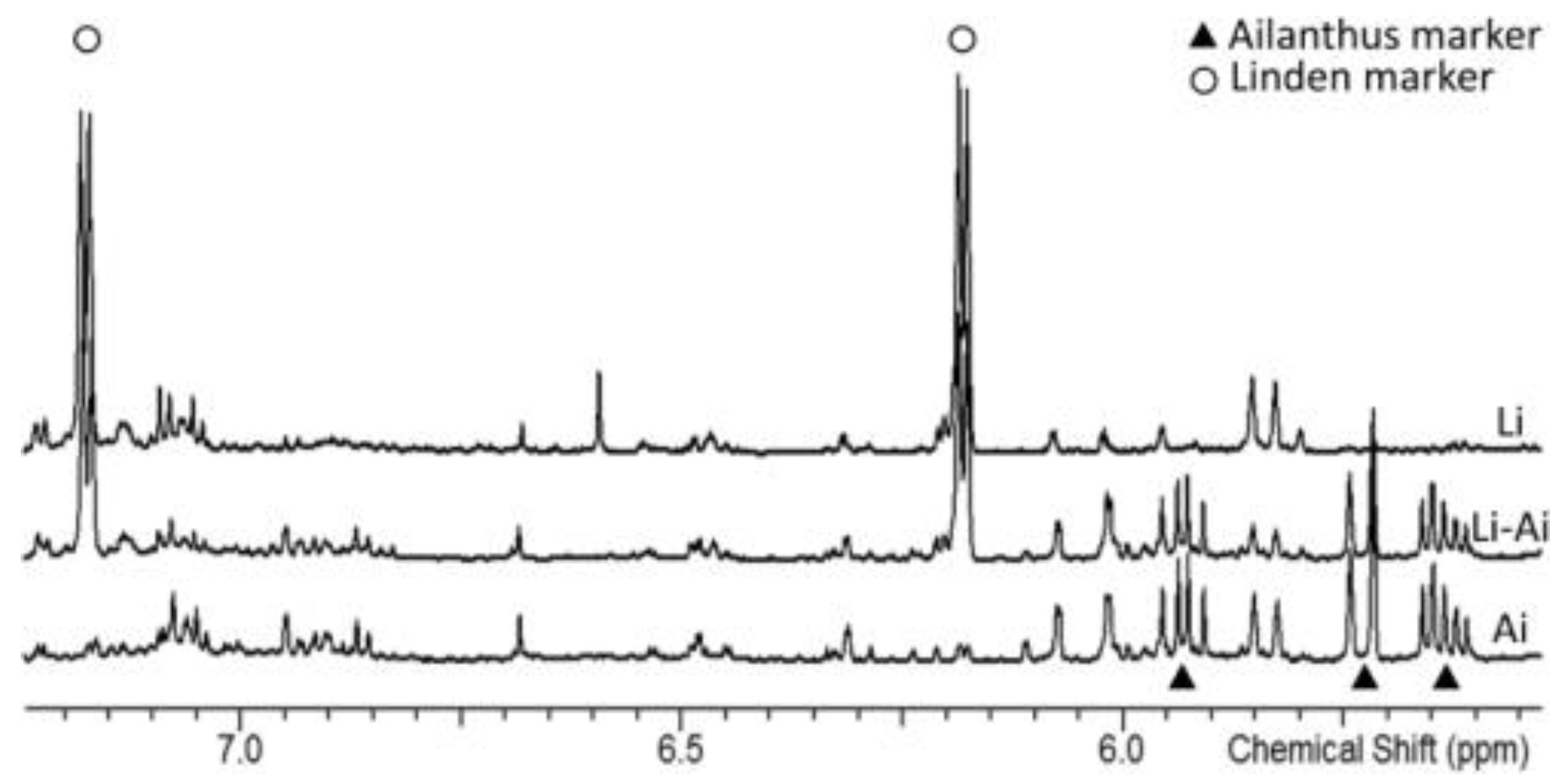
3.2.2. Fraudulent Designation of Botanical Origin
- Linden honey:
- -
- 4-(1-hydroxy-1-methylethyl)cyclohexa-1,3-diencarboxyl methyl ester
- -
- 4-(1-hydroxy-1-methylethyl)cyclohexa-1,3-dienecarboxylic acid
- -
- 4-(1-methylethenyl)cyclohexa-1,3-diencarboxylic acid
- -
- 4-(1-hydroxy-1-methylethyl) benzoic acid
- -
- (E)-2,6-dimethyl-3,7-octadiene-2,6-diol;
- Chestnut honey:
- -
- deoxyvasicinone
- -
- γ-lactam derivative of 3-(2′-pyrrolidinyl)-kinurenic acid
- -
- γ-LACT-3-PKA
- -
- 2-quinolone
- -
- 4-quinolone;
- Acacia honey:
- -
- pinocembrin
- -
- chrysin
- -
- alpinone
- -
- (Z, E)-abscissic acid;
- Orange honey:
- -
- caffeine
- -
- (E)-2,6-dimethylotta-2,7-dien-1,6-diol (8-hydroxylinalol);
- Eucalyptus honey:
- -
- dehydrovomifoliol
- -
- 3-oxo-α-ionone;
- Strawberry tree honey:
- -
- unedone homogenistic acid.
3.3. Coffee
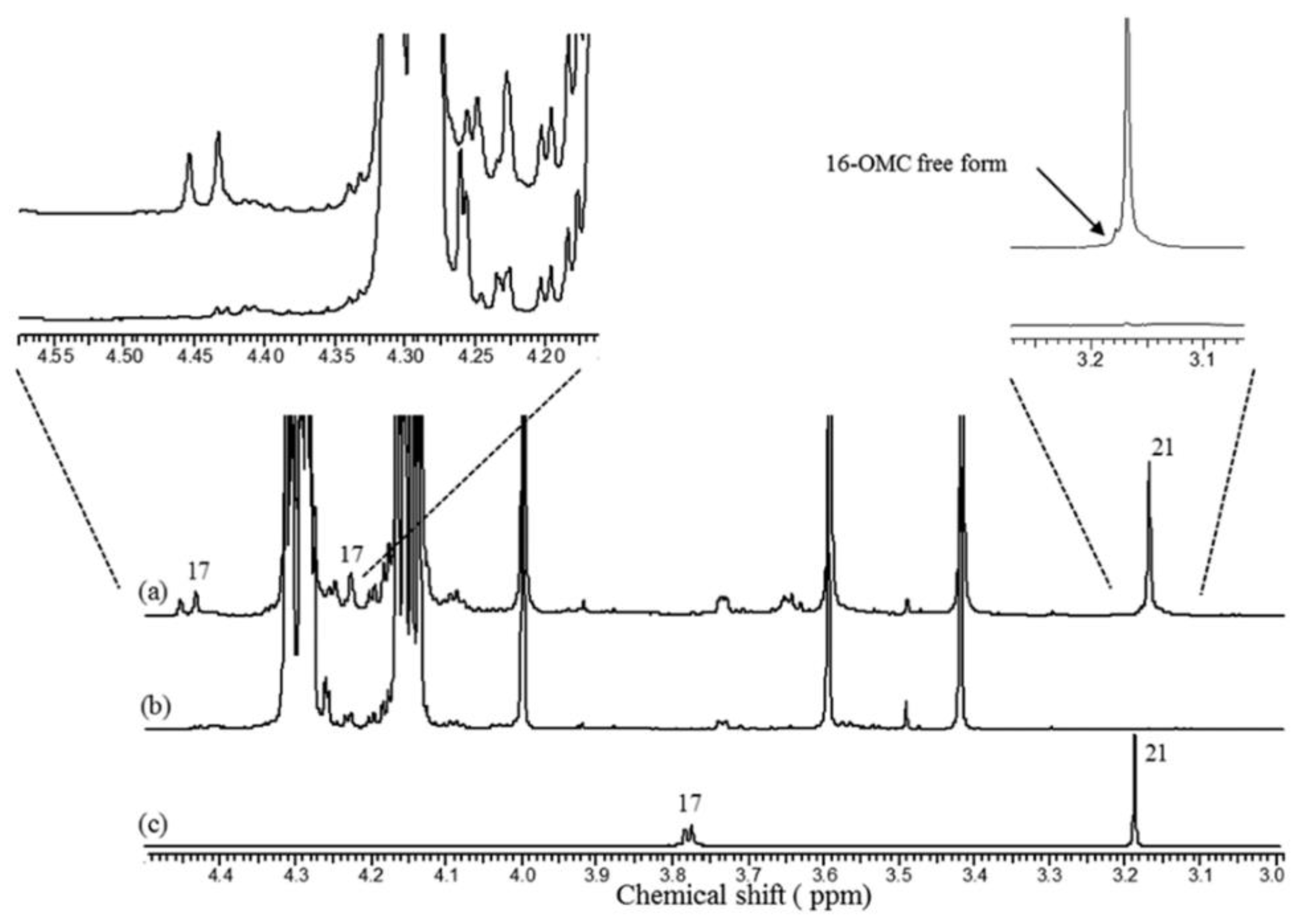
3.3.1. Fraudulent Addition of Robusta to Arabica Coffee
3.3.2. Geographical Origin
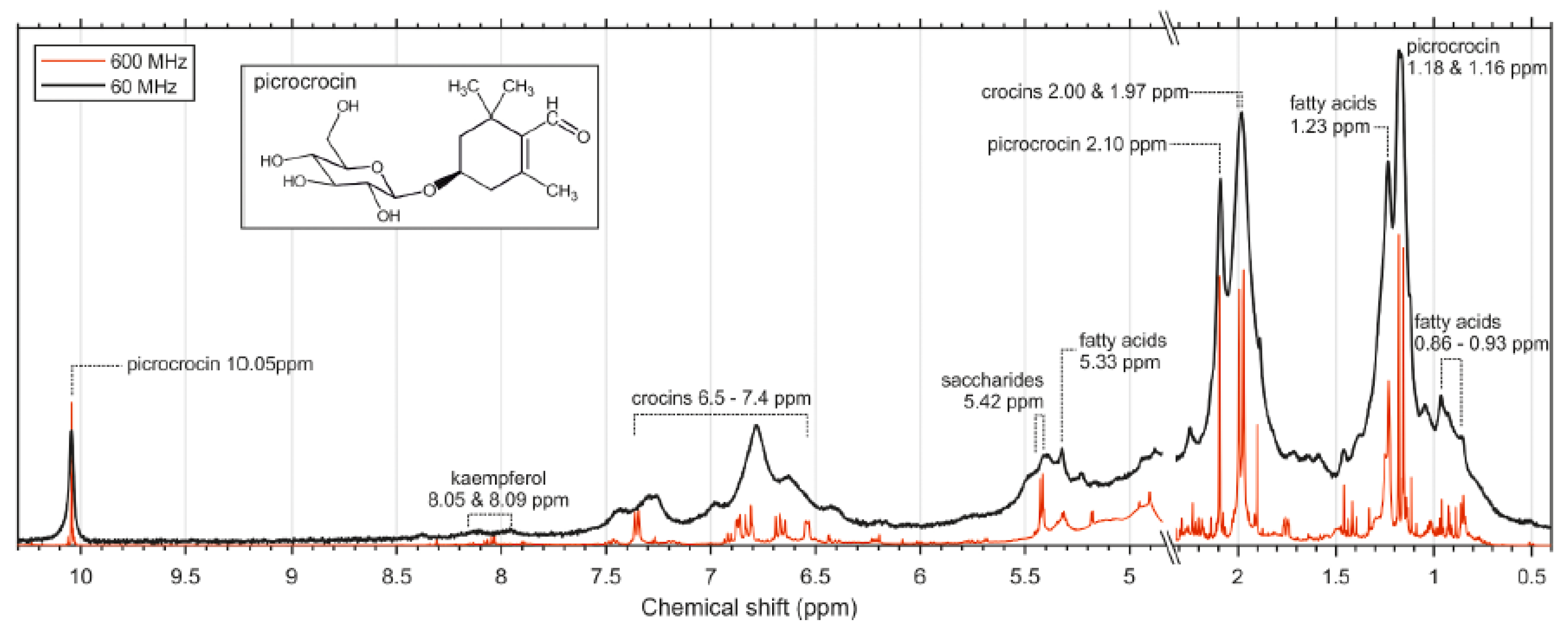
3.4. Saffron
3.5. Milk
4. Third Starting Point: NMR Methodology
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mannina, L.; Sobolev, A.P.; Aru, V.; Bellomaria, A.; Bertocchi, F.; Botta, B.; Cagliani, L.R.; Caligiani, A.; Capozzi, F.; Çela, D. NMR Methodologies in Food Analysis; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2017; ISBN 9781536122824. [Google Scholar]
- Solovyev, P.A.; Fauhl-Hassek, C.; Riedl, J.; Esslinger, S.; Bontempo, L.; Camin, F. NMR spectroscopy in wine authentication: An official control perspective. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2040–2062. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.J.; Musharraf, S.G.; Choudhary, M.I.; Rahman, A. Application of analytical methods in authentication and adulteration of honey. Food Chem. 2017, 217, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.D.C.; Fonseca, F.A.; Lião, L.M.; Alcantara, G.B.; Barison, A. High-resolution magic angle spinning nuclear magnetic resonance in foodstuff analysis. TrAC-Trends Anal. Chem. 2015, 73, 10–18. [Google Scholar] [CrossRef]
- Kirtil, E.; Cikrikci, S.; McCarthy, M.J.; Oztop, M.H. Recent advances in time domain NMR & MRI sensors and their food applications. Curr. Opin. Food Sci. 2017, 17, 9–15. [Google Scholar]
- Mannina, L.; Sobolev, A.P.; Viel, S. Liquid state 1H high field NMR in food analysis. Prog. Nucl. Magn. Reson. Spectrosc. 2012, 66, 1–39. [Google Scholar] [CrossRef]
- Hatzakis, E. Nuclear Magnetic Resonance (NMR) Spectroscopy in Food Science: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 189–220. [Google Scholar] [CrossRef]
- Sobolev, A.P.; Thomas, F.; Donarski, J.; Ingallina, C.; Circi, S.; Cesare Marincola, F.; Capitani, D.; Mannina, L. Use of NMR applications to tackle future food fraud issues. Trends Food Sci. Technol. 2019, 91, 347–353. [Google Scholar] [CrossRef]
- Marcone, M.F.; Wang, S.; Albabish, W.; Nie, S.; Somnarain, D.; Hill, A. Diverse food-based applications of nuclear magnetic resonance (NMR) technology. Food Res. Int. 2013, 51, 729–747. [Google Scholar] [CrossRef]
- Mao, J.; Cao, X.; Olk, D.C.; Chu, W.; Schmidt-Rohr, K. Advanced solid-state NMR spectroscopy of natural organic matter. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 100, 17–51. [Google Scholar] [CrossRef]
- Khodov, I.A.; Musabirova, G.S.; Klochkov, V.V.; Karataeva, F.K.; Huster, D.; Scheidt, H.A. Structural details on the interaction of fenamates with lipid membranes. J. Mol. Liq. 2022, 367, 120502. [Google Scholar] [CrossRef]
- Saad, A.; Bousquet, J.; Fernandez-Castro, N.; Loquet, A.; Géan, J. New Insights into Wine Taste: Impact of Dietary Lipids on Sensory Perceptions of Grape Tannins. J. Agric. Food Chem. 2021, 69, 3165–3174. [Google Scholar] [CrossRef] [PubMed]
- Huster, D.; Arnold, K.; Gawrisch, K. Influence of docosahexaenoic acid and cholesterol on lateral lipid organization in phospholipid mixtures. Biochemistry 1998, 37, 17299–17308. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chu, S.; Hagerman, A.E.; Lorigan, G.A. Probing the interaction of polyphenols with lipid bilayers by solid-state nmr spectroscopy. J. Agric. Food Chem. 2011, 59, 6783–6789. [Google Scholar] [CrossRef]
- van Duynhoven, J.; Voda, A.; Witek, M.; van As, H. Time-Domain NMR Applied to Food Products. Annu. Rep. NMR Spectrosc. 2010, 69, 145–197. [Google Scholar]
- Hills, B.P.; Godward, J.; Manning, C.E.; Biechlin, J.L.; Wright, K.M. Microstructural characterization of starch systems by NMR relaxation and Q-space microscopy. Magn. Reson. Imaging 1998, 16, 557–564. [Google Scholar] [CrossRef]
- Capitani, D.; Mannina, L.; Proietti, N.; Sobolev, A.P.; Tomassini, A.; Miccheli, A.; Di Cocco, M.E.; Capuani, G.; De Salvador, R.; Delfini, M. Monitoring of metabolic profiling and water status of Hayward kiwifruits by nuclear magnetic resonance. Talanta 2010, 82, 1826–1838. [Google Scholar] [CrossRef]
- Salzano, A.M.; Renzone, G.; Sobolev, A.P.; Carbone, V.; Petriccione, M.; Capitani, D.; Vitale, M.; Novi, G.; Zambrano, N.; Pasquariello, M.S.; et al. Unveiling kiwifruit metabolite and protein changes in the course of postharvest cold storage. Front. Plant Sci. 2019, 10, 71. [Google Scholar] [CrossRef]
- Panarese, V.; Laghi, L.; Pisi, A.; Tylewicz, U.; Rosa, M.D.; Rocculi, P. Effect of osmotic dehydration on Actinidia deliciosa kiwifruit: A combined NMR and ultrastructural study. Food Chem. 2012, 132, 1706–1712. [Google Scholar] [CrossRef]
- Clark, C.J.; Drummond, L.N.; MacFall, J.S. Quantitative NMR imaging of kiwifruit (Actinidia deliciosa) during growth and ripening. J. Sci. Food Agric. 1998, 78, 349–358. [Google Scholar] [CrossRef]
- Capitani, D.; Mannina, L.; Proietti, N.; Sobolev, A.P.; Tomassini, A.; Miccheli, A.; Di Cocco, M.E.; Capuani, G.; De Salvador, F.R.; Delfini, M. Metabolic profiling and outer pericarp water state in zespri, CI.GI, and hayward kiwifruits. J. Agric. Food Chem. 2013, 61, 1727–1740. [Google Scholar] [CrossRef]
- Burdon, J.; Clark, C. Effect of postharvest water loss on “hayward” kiwifruit water status. Postharvest Biol. Technol. 2001, 22, 215–225. [Google Scholar] [CrossRef]
- Taglienti, A.; Ritota, M.; Cozzolino, S.; Sequi, P.; Valentini, M.; Conte, L.; Terlizzi, M. MRI characterization of new kiwifruit selections. Proc. Acta Hortic. 2011. [Google Scholar] [CrossRef]
- Bonora, S.; Francioso, O.; Tugnoli, V.; Prodi, A.; Di Foggia, M.; Righi, V.; Nlpoti, P.; Filippini, G.; Pisi, A. Structural characteristics of “hayward” kiwifruits from elephantiasis-affected plants studied by drift, ft-raman, nmr, and sem techniques. J. Agric. Food Chem. 2009, 57, 4827–4832. [Google Scholar] [CrossRef] [PubMed]
- Maestrello, V.; Solovyev, P.; Bontempo, L.; Mannina, L.; Camin, F. Nuclear magnetic resonance spectroscopy in extra virgin olive oil authentication. Compr. Rev. Food Sci. Food Saf. 2022. [Google Scholar] [CrossRef] [PubMed]
- Mannina, L.; D’Imperio, M.; Capitani, D.; Rezzi, S.; Guillou, C.; Mavromoustakos, T.; Vilchez, M.D.M.; Fernández, A.H.; Thomas, F.; Aparicio, R. 1H NMR-based protocol for the detection of adulterations of refined olive oil with refined hazelnut oil. J. Agric. Food Chem. 2009, 57, 11550–11556. [Google Scholar] [CrossRef] [PubMed]
- Parker, T.; Limer, E.; Watson, A.D.; Defernez, M.; Williamson, D.; Kemsley, E.K. 60 MHz 1H NMR spectroscopy for the analysis of edible oils. TrAC-Trends Anal. Chem. 2014, 57, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Gouilleux, B.; Charrier, B.; Akoka, S.; Felpin, F.X.; Rodriguez-Zubiri, M.; Giraudeau, P. Ultrafast 2D NMR on a benchtop spectrometer: Applications and perspectives. TrAC-Trends Anal. Chem. 2016, 83, 65–75. [Google Scholar] [CrossRef]
- Gouilleux, B.; Marchand, J.; Charrier, B.; Remaud, G.S.; Giraudeau, P. High-throughput authentication of edible oils with benchtop Ultrafast 2D NMR. Food Chem. 2018, 244, 153–158. [Google Scholar] [CrossRef] [PubMed]
- FAO. Codex Alimentarius Commission Revised Codex Standard for Honey Codex Stan 12-1981, Rev. 1 (1987), Rev. 2 (2001); FAO (Food and Agriculture Organization of the United Nations): Rome, Italy, 1981; pp. 1–7. [Google Scholar]
- Schwarzinger, S.; Brauer, F.; Rösch, P.; Kämpf, B. Large-Scale Screening of Food Products for Quality and Authenticity. New Food Magazine 2015. [Google Scholar]
- Schievano, E.; Sbrizza, M.; Zuccato, V.; Piana, L.; Tessari, M. NMR carbohydrate profile in tracing acacia honey authenticity. Food Chem. 2020, 309, 125788. [Google Scholar] [CrossRef]
- Schievano, E.; Mammi, S.; Menegazzo, I. Nuclear Magnetic Resonance as a Method to Predict the Geographical and Entomological Origin of Pot-Honey. In Pot-Honey a Leg. Stingless Bees; Springer Science & Business Media: New York, NY, USA, 2013; pp. 429–445. [Google Scholar]
- Schievano, E.; Morelato, E.; Facchin, C.; Mammi, S. Characterization of markers of botanical origin and other compounds extracted from unifloral honeys. J. Agric. Food Chem. 2013, 61, 1747–1755. [Google Scholar] [CrossRef]
- Schievano, E.; Finotello, C.; Uddin, J.; Mammi, S.; Piana, L. Objective Definition of Monofloral and Polyfloral Honeys Based on NMR Metabolomic Profiling. J. Agric. Food Chem. 2016, 64, 3645–3652. [Google Scholar] [CrossRef] [PubMed]
- Schievano, E.; Stocchero, M.; Morelato, E.; Facchin, C.; Mammi, S. An NMR-based metabolomic approach to identify the botanical origin of honey. Metabolomics 2012, 8, 679–690. [Google Scholar] [CrossRef]
- Analysis of Coffee and Coffee Products—Determination of 16-O-Methyl Cafestol Content of Roasted Coffee—HPLC-Method. Available online: https://www.en-standard.eu/din-10779-analysis-of-coffee-and-coffee-products-determination-of-16-o-methyl-cafestol-content-of-roasted-coffee-hplc-method/ (accessed on 6 October 2022).
- Monakhova, Y.B.; Ruge, W.; Kuballa, T.; Ilse, M.; Winkelmann, O.; Diehl, B.; Thomas, F.; Lachenmeier, D.W. Rapid approach to identify the presence of Arabica and Robusta species in coffee using 1H NMR spectroscopy. Food Chem. 2015, 182, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Schievano, E.; Finotello, C.; De Angelis, E.; Mammi, S.; Navarini, L. Rapid authentication of coffee blends and quantification of 16-O-methylcafestol in roasted coffee beans by nuclear magnetic resonance. J. Agric. Food Chem. 2014, 62, 12309–12314. [Google Scholar] [CrossRef]
- Gunning, Y.; Defernez, M.; Watson, A.D.; Beadman, N.; Colquhoun, I.J.; Le Gall, G.; Philo, M.; Garwood, H.; Williamson, D.; Davis, A.P.; et al. 16-O-methylcafestol is present in ground roast Arabica coffees: Implications for authenticity testing. Food Chem. 2018, 248, 52–60. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.R.; Cogliati, C. NMR based geographical characterization of roasted coffee. Talanta 2012, 88, 420–426. [Google Scholar] [CrossRef]
- Cagliani, L.R.; Pellegrino, G.; Giugno, G.; Consonni, R. Quantification of Coffea arabica and Coffea canephora var. robusta in roasted and ground coffee blends. Talanta 2013, 106, 169–173. [Google Scholar] [CrossRef]
- Arana, V.A.; Medina, J.; Alarcon, R.; Moreno, E.; Heintz, L.; Schäfer, H.; Wist, J. Coffee’s country of origin determined by NMR: The Colombian case. Food Chem. 2015, 175, 500–506. [Google Scholar] [CrossRef]
- Charlton, A.J.; Farrington, W.H.H.; Brereton, P. Application of 1H NMR and multivariate statistics for screening complex mixtures: Quality control and authenticity of instant coffee. J. Agric. Food Chem. 2002, 50, 3098–3103. [Google Scholar] [CrossRef]
- Cardone, L.; Castronuovo, D.; Perniola, M.; Cicco, N.; Candido, V. Saffron (Crocus sativus L.), the king of spices: An overview. Sci. Hortic. 2020, 272, 109560. [Google Scholar] [CrossRef]
- Koocheki, A.; Milani, E. Saffron adulteration. In Saffron; Woodhead Publishing: Sawston, UK, 2020. [Google Scholar]
- Petrakis, E.A.; Cagliani, L.R.; Polissiou, M.G.; Consonni, R. Evaluation of saffron (Crocus sativus L.) adulteration with plant adulterants by1H NMR metabolite fingerprinting. Food Chem. 2015, 173, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Dowlatabadi, R.; Farshidfar, F.; Zare, Z.; Pirali, M.; Rabiei, M.; Khoshayand, M.R.; Vogel, H.J. Detection of adulteration in Iranian saffron samples by 1H NMR spectroscopy and multivariate data analysis techniques. Metabolomics 2017, 13, 1–11. [Google Scholar] [CrossRef]
- Musio, B.; Todisco, S.; Antonicelli, M.; Garino, C.; Arlorio, M.; Mastrorilli, P.; Latronico, M.; Gallo, V. Non-Targeted NMR Method to Assess the Authenticity of Saffron and Trace the Agronomic Practices Applied for Its Production. Appl. Sci. 2022, 12, 2583. [Google Scholar] [CrossRef]
- Gunning, Y.; Davies, K.S.; Kemsley, E.K. Authentication of saffron using 60 MHz 1H NMR spectroscopy. Food Chem. 2023, 404, 134649. [Google Scholar] [CrossRef]
- Erich, S.; Schill, S.; Annweiler, E.; Waiblinger, H.U.; Kuballa, T.; Lachenmeier, D.W.; Monakhova, Y.B. Combined chemometric analysis of 1H NMR, 13C NMR and stable isotope data to differentiate organic and conventional milk. Food Chem. 2015, 188, 1–7. [Google Scholar] [CrossRef]
- Bruschetta, G.; Notti, A.; Lando, G.; Ferlazzo, A. A promising 31P NMR-multivariate analysis approach for the identification of milk phosphorylated metabolites and for rapid authentication of milk samples. Biochem. Biophys. Rep. 2021, 27, 101087. [Google Scholar] [CrossRef]
- Mazzei, P.; Piccolo, A. NMR-based metabolomics of water-buffalo milk after conventional or biological feeding. Chem. Biol. Technol. Agric. 2018, 5, 3. [Google Scholar] [CrossRef]
- Bergana, M.M.; Adams, K.M.; Harnly, J.; Moore, J.C.; Xie, Z. Non-targeted detection of milk powder adulteration by 1H NMR spectroscopy and conformity index analysis. J. Food Compos. Anal. 2019, 78, 49–58. [Google Scholar] [CrossRef]
- Rysova, L.; Legarova, V.; Pacakova, Z.; Hanus, O.; Nemeckova, I.; Klimesova, M.; Havlik, J. Detection of bovine milk adulteration in caprine milk with N-acetyl carbohydrate biomarkers by using 1H nuclear magnetic resonance spectroscopy. J. Dairy Sci. 2021, 104, 9583–9595. [Google Scholar] [CrossRef]
- Santos, P.M.; Pereira-Filho, E.R.; Colnago, L.A. Detection and quantification of milk adulteration using time domain nuclear magnetic resonance (TD-NMR). Microchem. J. 2016, 124, 15–19. [Google Scholar] [CrossRef]
- Novoa-Carballal, R.; Fernandez-Megia, E.; Jimenez, C.; Riguera, R. NMR methods for unravelling the spectra of complex mixtures. Nat. Prod. Rep. 2011, 28, 78–98. [Google Scholar] [CrossRef] [PubMed]
- Sandusky, P.; Raftery, D. Use of selective TOCSY NMR experiments for quantifying minor components in complex mixtures: Application to the metabonomics of amino acids in honey. Anal. Chem. 2005, 77, 2455–2463. [Google Scholar] [CrossRef] [PubMed]
- Gil, A.M.; Duarte, I.; Cabrita, E.; Goodfellow, B.J.; Spraul, M.; Kerssebaum, R. Exploratory applications of diffusion ordered spectroscopy to liquid foods: An aid towards spectral assignment. Anal. Chim. Acta 2004, 506, 215–223. [Google Scholar] [CrossRef]
- Bingol, K.; Zhang, F.; Bruschweiler-Li, L.; Brüschweiler, R. TOCCATA: A customized carbon total correlation spectroscopy NMR metabolomics database. Anal. Chem. 2012, 84, 9395–9401. [Google Scholar] [CrossRef][Green Version]
- Tulpan, D.; Léger, S.; Belliveau, L.; Culf, A.; Čuperlović-Culf, M. MetaboHunter: An automatic approach for identification of metabolites from 1H-NMR spectra of complex mixtures. BMC Bioinform. 2011, 12, 400. [Google Scholar] [CrossRef]
- Okaru, A.O.; Scharinger, A.; De Rezende, T.R.; Teipel, J.; Kuballa, T.; Walch, S.G.; Lachenmeier, D.W. Validation of a quantitative proton nuclear magnetic resonance spectroscopic screening method for coffee quality and authenticity (NMR coffee screener). Foods 2020, 9, 47. [Google Scholar] [CrossRef]
- Burton, I.W.; Martinez Farina, C.F.; Ragupathy, S.; Arunachalam, T.; Newmaster, S.; Berrué, F. Quantitative NMR Methodology for the Authentication of Roasted Coffee and Prediction of Blends. J. Agric. Food Chem. 2020, 68, 14643–14651. [Google Scholar] [CrossRef]
- Yang, W.B.; Shu, H.W.; Yi, T.C. QNMR as a Tool for Determination of Six Common Sugars in Foods. In Applications of NMR Spectroscopy: Volume 8; Bentham Science: Sharjah, United Arab Emirates, 2020; pp. 1–24. ISBN 9789811439971. [Google Scholar]
- Mandrone, M.; Marincich, L.; Chiocchio, I.; Petroli, A.; Gođevac, D.; Maresca, I.; Poli, F. NMR-based metabolomics for frauds detection and quality control of oregano samples. Food Control 2021, 127, 108141. [Google Scholar] [CrossRef]
- Jungen, M.; Schütz, B.; Schweiggert, R. Influence of species and processing techniques on phlorin in Citrus juices as quantified by 1H-NMR spectroscopy. LWT 2020, 134, 109949. [Google Scholar] [CrossRef]
- Jungen, M.; Lotz, P.; Patz, C.D.; Steingass, C.B.; Schweiggert, R. Coumarins, psoralens, and quantitative 1H-NMR spectroscopy for authentication of lemon (Citrus limon [L.] Burm.f.) and Persian lime (Citrus × latifolia [Yu.Tanaka] Tanaka) juices. Food Chem. 2021, 359, 129804. [Google Scholar] [CrossRef]
- Cagliani, L.R.; Maestri, G.; Consonni, R. Detection and evaluation of saccharide adulteration in Italian honey by NMR spectroscopy. Food Control 2022, 133, 108574. [Google Scholar] [CrossRef]
- Li, F.F.; Guo, C.C.; Zhang, Y.L.; Wang, N.; Shi, Y.; Wang, S.Q. A screening method based on 1D CSSF-TOCSY for the identification and quantification of 11 illegal adulterants in herbal medicines. Microchem. J. 2020, 153, 104495. [Google Scholar] [CrossRef]
- Teipel, J.C.; Hausler, T.; Sommerfeld, K.; Scharinger, A.; Walch, S.G.; Lachenmeier, D.W.; Kuballa, T. Application of 1h nuclear magnetic resonance spectroscopy as spirit drinks screener for quality and authenticity control. Foods 2020, 9, 1355. [Google Scholar] [CrossRef] [PubMed]
- Scettri, A.; Schievano, E. Quantification of polyols in sugar-free foodstuffs by qNMR. Food Chem. 2022, 390, 133125. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Zhao, C.; Zhu, H.; Tong, J.; Zhao, X.; Yang, H.; Guo, H. NMR detection of fatty acids content in walnut oil and compared with liquid chromatography. J. Food Meas. Charact. 2021, 15, 2716–2726. [Google Scholar] [CrossRef]
- Donarski, J.A.; Roberts, D.P.T.; Charlton, A.J. Quantitative NMR spectroscopy for the rapid measurement of methylglyoxal in manuka honey. Anal. Methods 2010, 2, 1479–1483. [Google Scholar] [CrossRef]
- Siciliano, C.; Bartella, L.; Mazzotti, F.; Aiello, D.; Napoli, A.; De Luca, P.; Temperini, A. 1H NMR quantification of cannabidiol (CBD) in industrial products derived from Cannabis sativa L. (hemp) seeds. In Proceedings of the IOP Conference Series: Materials Science and Engineering; IOP Publishing Ltd: Bristol, UK, 2019; Volume 572. [Google Scholar]
- Barthlott, I.; Scharinger, A.; Golombek, P.; Kuballa, T.; Lachenmeier, D.W. A quantitative1h nmr method for screening cannabinoids in cbd oils. Toxics 2021, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Araneda, J.F.; Chu, T.; Leclerc, M.C.; Riegel, S.D.; Spingarn, N. Quantitative analysis of cannabinoids using benchtop NMR instruments. Anal. Methods 2020, 12, 4853–4857. [Google Scholar] [CrossRef]
- Mannina, L.; Sobolev, A.P.; Capitani, D.; Iaffaldano, N.; Rosato, M.P.; Ragni, P.; Reale, A.; Sorrentino, E.; D’Amico, I.; Coppola, R. NMR metabolic profiling of organic and aqueous sea bass extracts: Implications in the discrimination of wild and cultured sea bass. Talanta 2008, 77, 433–444. [Google Scholar] [CrossRef]
- Ingallina, C.; Cerreto, A.; Mannina, L.; Circi, S.; Vista, S.; Capitani, D.; Spano, M.; Sobolev, A.P.; Marini, F. Extra-virgin olive oils from nine italian regions: An 1H NMR-chemometric characterization. Metabolites 2019, 9, 65. [Google Scholar] [CrossRef]
- Agiomyrgianaki, A.; Petrakis, P.V.; Dais, P. Influence of harvest year, cultivar and geographical origin on Greek extra virgin olive oils composition: A study by NMR spectroscopy and biometric analysis. Food Chem. 2012, 135, 2561–2568. [Google Scholar] [CrossRef] [PubMed]
- Papadia, P.; Del Coco, L.; Muzzalupo, I.; Rizzi, M.; Perri, E.; Cesari, G.; Simeone, V.; Mondelli, D.; Schena, F.P.; Fanizzi, F.P. Multivariate analysis of 1H-NMR spectra of genetically characterized extra virgin olive oils and growth soil correlations. JAOCS J. Am. Oil Chem. Soc. 2011, 88, 1463–1475. [Google Scholar] [CrossRef]
- Brescia, M.A.; Košir, I.J.; Caldarola, V.; Kidrič, J.; Sacco, A. Chemometric classification of Apulian and Slovenian wines using 1H NMR and ICP-OES together with HPICE data. J. Agric. Food Chem. 2003, 51, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Gougeon, L.; da Costa, G.; Guyon, F.; Richard, T. 1H NMR metabolomics applied to Bordeaux red wines. Food Chem. 2019, 301, 125257. [Google Scholar] [CrossRef]
- Papotti, G.; Bertelli, D.; Graziosi, R.; Silvestri, M.; Bertacchini, L.; Durante, C.; Plessi, M. Application of one- and two-dimensional NMR spectroscopy for the characterization of protected designation of Origin Lambrusco wines of modena. J. Agric. Food Chem. 2013, 61, 1741–1746. [Google Scholar] [CrossRef]
- Mannina, L.; Marini, F.; Gobbino, M.; Sobolev, A.P.; Capitani, D. NMR and chemometrics in tracing European olive oils: The case study of Ligurian samples. Talanta 2010, 80, 2141–2148. [Google Scholar] [CrossRef]
- Mannina, L.; Patumi, M.; Proietti, N.; Bassi, D.; Segre, A.L. Geographical characterization of Italian extra virgin olive oils using high-field 1H NMR spectroscopy. J. Agric. Food Chem. 2001, 49, 2687–2696. [Google Scholar] [CrossRef]
- Sacchi, R.; Mannina, L.; Fiordiponti, P.; Barone, P.; Paolillo, L.; Patumi, M.; Segre, A. Characterization of Italian Extra Virgin Olive Oils Using 1H-NMR Spectroscopy. J. Agric. Food Chem. 1998, 46, 3947–3951. [Google Scholar] [CrossRef]
- Mannina, L.; Sobolev, A.P.; Di Lorenzo, A.; Vista, S.; Tenore, G.C.; Daglia, M. Chemical Composition of Different Botanical Origin Honeys Produced by Sicilian Black Honeybees (Apis mellifera ssp. sicula). J. Agric. Food Chem. 2015, 63, 5864–5874. [Google Scholar] [CrossRef]
- Ingallina, C.; Maccelli, A.; Spano, M.; Di Matteo, G.; Di Sotto, A.; Giusti, A.M.; Vinci, G.; Di Giacomo, S.; Rapa, M.; Ciano, S.; et al. Chemico-biological characterization of torpedino di fondi® tomato fruits: A comparison with san marzano cultivar at two ripeness stages. Antioxidants 2020, 9, 1027. [Google Scholar] [CrossRef]
- Sorrequieta, A.; Abriata, L.; Boggio, S.; Valle, E. Off-the-Vine Ripening of Tomato Fruit Causes Alteration in the Primary Metabolite Composition. Metabolites 2013, 3, 967. [Google Scholar] [CrossRef] [PubMed]
- Sciubba, F.; Avanzato, D.; Vaccaro, A.; Capuani, G.; Spagnoli, M.; Di Cocco, M.E.; Tzareva, I.N.; Delfini, M. Monitoring of pistachio (Pistacia Vera) ripening by high field nuclear magnetic resonance spectroscopy. Nat. Prod. Res. 2017, 31, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhao, Y.; Yang, J.; Jiang, Y.; Lu, F.; Jia, Y.; Yang, B. Metabolomic analyses of banana during postharvest senescence by 1H-high resolution-NMR. Food Chem. 2017, 218, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Palmioli, A.; Alberici, D.; Ciaramelli, C.; Airoldi, C. Metabolomic profiling of beers: Combining 1H NMR spectroscopy and chemometric approaches to discriminate craft and industrial products. Food Chem. 2020, 327, 127025. [Google Scholar] [CrossRef] [PubMed]
- Gougeon, L.; da Costa, G.; Richard, T.; Guyon, F. Wine authenticity by quantitative1H NMR versus multitechnique analysis: A case study. Food Anal. Methods 2019, 12, 956–965. [Google Scholar] [CrossRef]
- Gougeon, L.; Da Costa, G.; Le Mao, I.; Ma, W.; Teissedre, P.L.; Guyon, F.; Richard, T. Wine Analysis and Authenticity Using 1H-NMR Metabolomics Data: Application to Chinese Wines. Food Anal. Methods 2018, 11, 3425–3434. [Google Scholar] [CrossRef]
- Ehlers, M.; Horn, B.; Raeke, J.; Fauhl-Hassek, C.; Hermann, A.; Brockmeyer, J.; Riedl, J. Towards harmonization of non-targeted 1H NMR spectroscopy-based wine authentication: Instrument comparison. Food Control 2022, 132, 108508. [Google Scholar] [CrossRef]
- Spiteri, M.; Jamin, E.; Thomas, F.; Rebours, A.; Lees, M.; Rogers, K.M.; Rutledge, D.N. Fast and global authenticity screening of honey using 1H-NMR profiling. Food Chem. 2015, 189, 60–66. [Google Scholar] [CrossRef]
- Toci, A.T.; de Moura Ribeiro, M.V.; de Toledo, P.R.A.B.; Boralle, N.; Pezza, H.R.; Pezza, L. Fingerprint and authenticity roasted coffees by 1H-NMR: The Brazilian coffee case. Food Sci. Biotechnol. 2018, 27, 19–26. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Ridge, C.D.; Harnly, J.; Mazzola, E.P.; Luthria, D.L. Spectroscopic analysis of wheat fractions and reconstituted whole wheat mixtures by 1H-NMR and NIR. Cereal Chem. 2017, 94, 471–479. [Google Scholar] [CrossRef]
- Girelli, C.R.; Del Coco, L.; Papadia, P.; De Pascali, S.A.; Fanizzi, F.P. Harvest year effects on apulian EVOOs evaluated by 1H NMR based metabolomics. PeerJ 2016, 4, e2740. [Google Scholar] [CrossRef] [PubMed]
- del Coco, L.; de Pascali, S.A.; Fanizzi, F. 1H NMR Metabolic Profiling of Apulian EVOOs: Fine Pedoclimatic Influences in Salento Cultivars; RSC: London, UK, 2015; pp. 154–160. [Google Scholar]
- Godelmann, R.; Fang, F.; Humpfer, E.; Schütz, B.; Bansbach, M.; Schäfer, H.; Spraul, M. Targeted and nontargeted wine analysis by 1H NMR spectroscopy combined with multivariate statistical analysis. differentiation of important parameters: Grape variety, geographical origin, year of vintage. J. Agric. Food Chem. 2013, 61, 5610–5619. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhong, Q.; Fauhl-Hassek, C.; Pfister, M.K.H.; Horn, B.; Huang, Z. Classification of Chinese wine varieties using 1H NMR spectroscopy combined with multivariate statistical analysis. Food Control 2018, 88, 113–122. [Google Scholar] [CrossRef]
- Girelli, C.R.; Del Coco, L.; Zelasco, S.; Salimonti, A.; Conforti, F.L.; Biagianti, A.; Barbini, D.; Fanizzi, F.P. Traceability of “Tuscan PGI” extra virgin olive oils by 1H NMR metabolic profiles collection and analysis. Metabolites 2018, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Longobardi, F.; Ventrella, A.; Napoli, C.; Humpfer, E.; Schütz, B.; Schäfer, H.; Kontominas, M.G.; Sacco, A. Classification of olive oils according to geographical origin by using 1H NMR fingerprinting combined with multivariate analysis. Food Chem. 2012, 130, 177–183. [Google Scholar] [CrossRef]
- Girelli, C.R.; Calò, F.; Angilè, F.; Mazzi, L.; Barbini, D.; Fanizzi, F.P. 1h nmr spectroscopy to characterize italian extra virgin olive oil blends, using statistical models and databases based on monocultivar reference oils. Foods 2020, 9, 1797. [Google Scholar] [CrossRef]
- Caligiani, A.; Palla, L.; Acquotti, D.; Marseglia, A.; Palla, G. Application of 1H NMR for the characterisation of cocoa beans of different geographical origins and fermentation levels. Food Chem. 2014, 157, 94–99. [Google Scholar] [CrossRef]
- Ordoudi, S.A.; Cagliani, L.R.; Lalou, S.; Naziri, E.; Tsimidou, M.Z.; Consonni, R. 1H NMR-based metabolomics of saffron reveals markers for its quality deterioration. Food Res. Int. 2015, 70, 1–6. [Google Scholar] [CrossRef]
- Klare, J.; Rurik, M.; Rottmann, E.; Bollen, A.; Kohlbacher, O.; Fischer, M.; Hackl, T. Determination of the Geographical Origin of Asparagus officinalis L. By 1H NMR Spectroscopy. J. Agric. Food Chem. 2020, 68, 14353–14363. [Google Scholar] [CrossRef]
- Schmitt, C.; Schneider, T.; Rumask, L.; Fischer, M.; Hackl, T. Food Profiling: Determination of the Geographical Origin of Walnuts by 1H NMR Spectroscopy Using the Polar Extract. J. Agric. Food Chem. 2020, 68, 15526–15534. [Google Scholar] [CrossRef]
- Bachmann, R.; Klockmann, S.; Haerdter, J.; Fischer, M.; Hackl, T. 1H NMR Spectroscopy for Determination of the Geographical Origin of Hazelnuts. J. Agric. Food Chem. 2018, 66, 11873–11879. [Google Scholar] [CrossRef] [PubMed]
- Luong, D.V.; Tam, N.Q.; Xuan, D.T.T.; Tai, N.T. NMR based metabolomic approach for evaluation of Vietnamese honey. Vietnam J. Chem. 2019, 57, 712–716. [Google Scholar] [CrossRef]
- Schievano, E.; Peggion, E.; Mammi, S. 1H nuclear magnetic resonance spectra of chloroform extracts of honey for chemometric determination of its botanical origin. J. Agric. Food Chem. 2010, 58, 57–65. [Google Scholar] [CrossRef]
- Santucci, C.; Tenori, L.; Luchinat, C. NMR fingerprinting as a tool to evaluate post-harvest time-related changes of peaches, tomatoes and plums. Food Res. Int. 2015, 75, 106–114. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.A.; Flumignan, D.L.; Pezza, H.R.; Pezza, L. 1H NMR spectroscopy combined with multivariate data analysis for differentiation of Brazilian lager beer according to brewery. Eur. Food Res. Technol. 2019, 245, 2365–2372. [Google Scholar] [CrossRef]
- Brigante, F.I.; García, M.E.; López Radcenco, A.; Moyna, G.; Wunderlin, D.A.; Baroni, M.V. Identification of chia, flax and sesame seeds authenticity markers by NMR-based untargeted metabolomics and their validation in bakery products containing them. Food Chem. 2022, 387, 132925. [Google Scholar] [CrossRef]
- Guyader, S.; Thomas, F.; Portaluri, V.; Jamin, E.; Akoka, S.; Silvestre, V.; Remaud, G. Authentication of edible fats and oils by non-targeted 13C INEPT NMR spectroscopy. Food Control 2018, 91, 216–224. [Google Scholar] [CrossRef]
- da Silva, L.A.; Flumignan, D.L.; Tininis, A.G.; Pezza, H.R.; Pezza, L. Discrimination of Brazilian lager beer by 1H NMR spectroscopy combined with chemometrics. Food Chem. 2019, 272, 488–493. [Google Scholar] [CrossRef]
- Yong, C.H.; Muhammad, S.A.; Aziz, F.A.; Nasir, F.I.; Mustafa, M.Z.; Ibrahim, B.; Kelly, S.D.; Cannavan, A.; Seow, E.K. Detecting adulteration of stingless bee honey using untargeted 1H NMR metabolomics with chemometrics. Food Chem. 2022, 368, 130808. [Google Scholar] [CrossRef]
- Tang, F.; Green, H.S.; Wang, S.C.; Hatzakis, E. Analysis and authentication of avocado oil using high resolution nmr spectroscopy. Molecules 2021, 26, 310. [Google Scholar] [CrossRef]
- Andre, C.M.; Soukoulis, C. Food quality assessed by chemometrics. Foods 2020, 9, 897. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Carvelo, A.M. Data mining/machine learning methods in foodomics. Curr. Opin. Food Sci. 2021, 37, 76–82. [Google Scholar] [CrossRef]
- Corsaro, C.; Vasi, S.; Neri, F.; Mezzasalma, A.M.; Neri, G.; Fazio, E. NMR in Metabolomics: From Conventional Statistics to Machine Learning and Neural Network Approaches. Appl. Sci. 2022, 12, 2824. [Google Scholar] [CrossRef]
- Picone, G.; Mengucci, C.; Capozzi, F. The NMR added value to the green foodomics perspective: Advances by machine learning to the holistic view on food and nutrition. Magn. Reson. Chem. 2022, 60, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Giberson, J.; Scicluna, J.; Legge, N.; Longstaffe, J. Developments in Benchtop NMR Spectroscopy 2015–2020, 1st ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2021; Volume 102, ISBN 9780128246092. [Google Scholar]
- Blümich, B. Low-field and benchtop NMR. J. Magn. Reson. 2019, 306, 27–35. [Google Scholar] [CrossRef]
- Grootveld, M.; Percival, B.; Gibson, M.; Osman, Y.; Edgar, M.; Molinari, M.; Mather, M.L.; Casanova, F.; Wilson, P.B. Progress in low-field benchtop NMR spectroscopy in chemical and biochemical analysis. Anal. Chim. Acta 2019, 1067, 11–30. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobolev, A.P.; Ingallina, C.; Spano, M.; Di Matteo, G.; Mannina, L. NMR-Based Approaches in the Study of Foods. Molecules 2022, 27, 7906. https://doi.org/10.3390/molecules27227906
Sobolev AP, Ingallina C, Spano M, Di Matteo G, Mannina L. NMR-Based Approaches in the Study of Foods. Molecules. 2022; 27(22):7906. https://doi.org/10.3390/molecules27227906
Chicago/Turabian StyleSobolev, Anatoly P., Cinzia Ingallina, Mattia Spano, Giacomo Di Matteo, and Luisa Mannina. 2022. "NMR-Based Approaches in the Study of Foods" Molecules 27, no. 22: 7906. https://doi.org/10.3390/molecules27227906
APA StyleSobolev, A. P., Ingallina, C., Spano, M., Di Matteo, G., & Mannina, L. (2022). NMR-Based Approaches in the Study of Foods. Molecules, 27(22), 7906. https://doi.org/10.3390/molecules27227906









