Mass Spectrometry-Based Identification of Bioactive Bee Pollen Proteins: Evaluation of Allergy Risk after Bee Pollen Supplementation
Abstract
1. Introduction
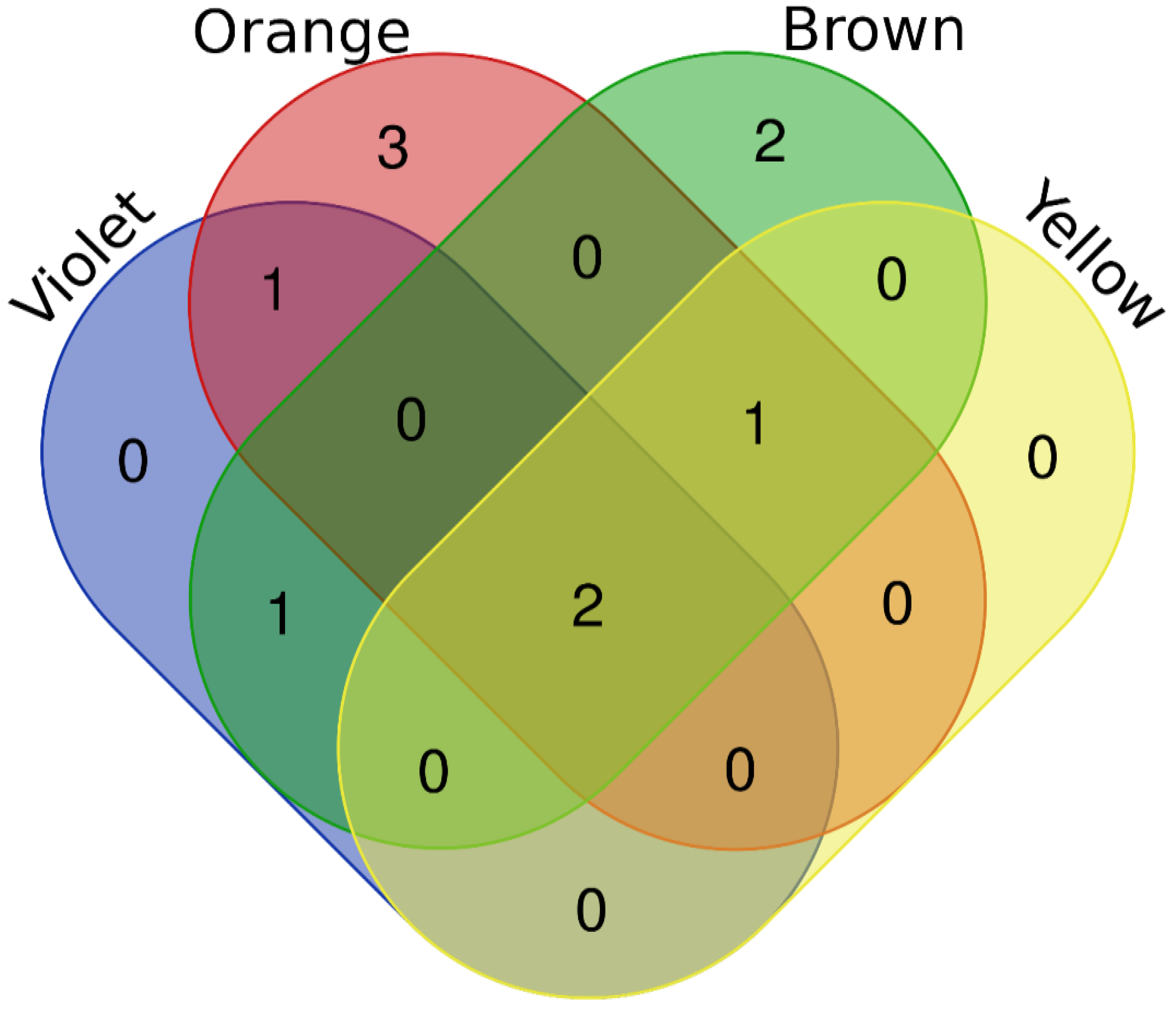
2. Results
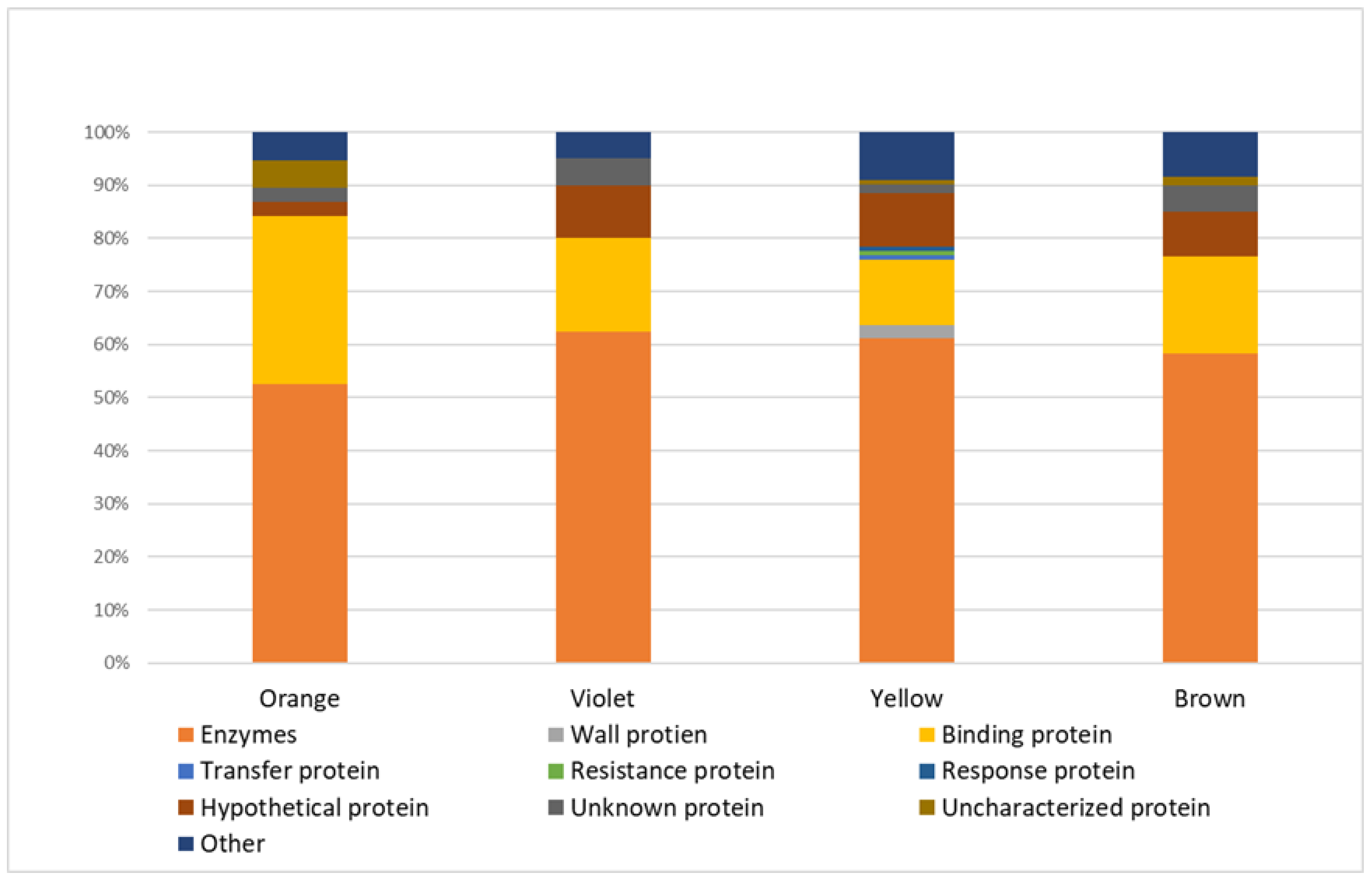
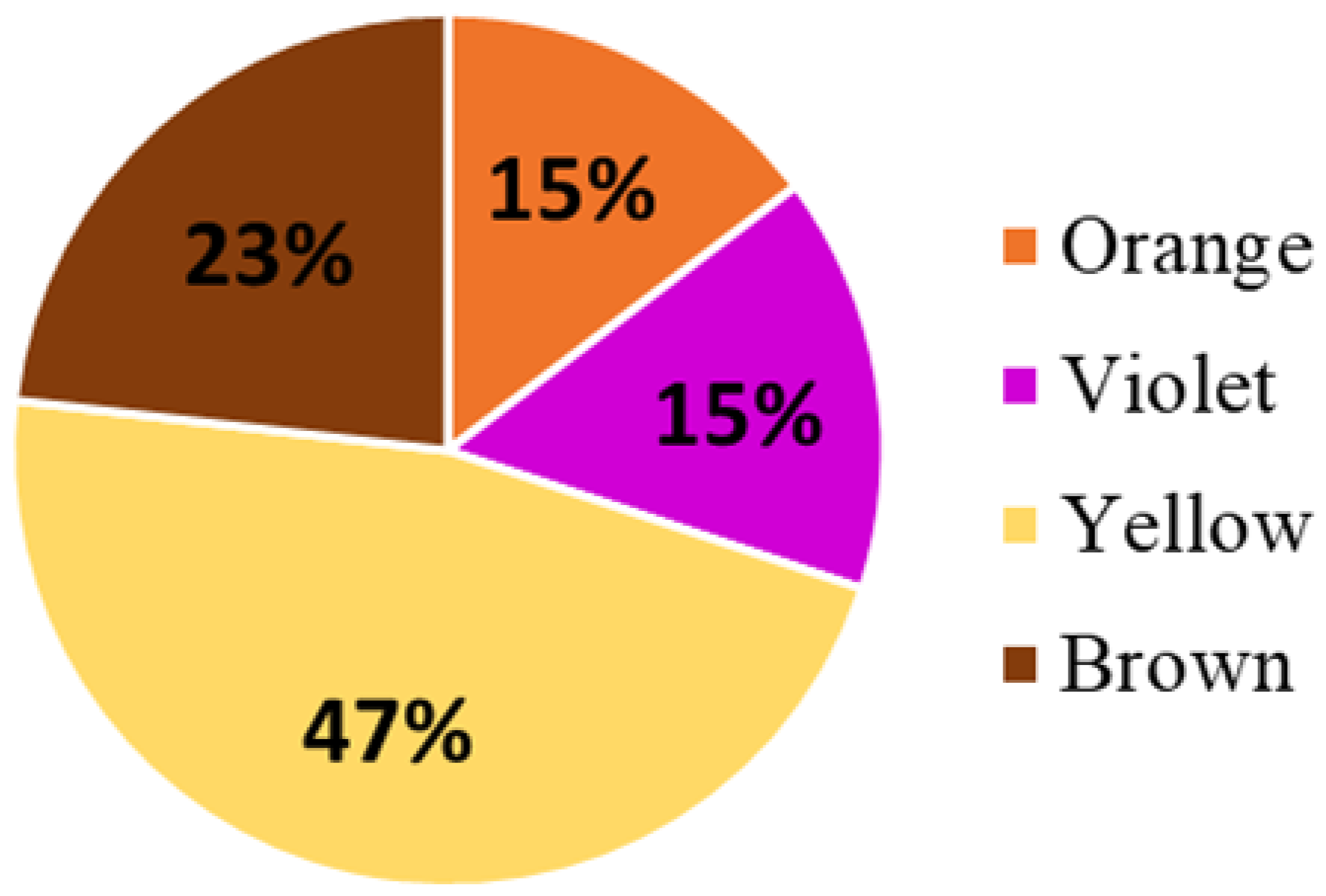
2.1. Proteins Taxonomically Classified to Viridiplantae Clade
- n = 74 in yellow bee pollen (≈61% of all proteins identified in yellow pollen);
- n = 35 in brown bee pollen (≈58% of proteins identified in brown samples);
- n = 25 in violet bee pollen (≈63% of proteins identified in violet pollen);
- n = 20 in orange bee pollen (≈53% of proteins identified in orange samples).
- n = 15 in yellow bee pollen (≈12% of all proteins identified in yellow pollen);
- n = 12 in orange bee pollen (≈32% of proteins identified in orange pollen);
- n = 11 in brown bee pollen (≈18% of proteins identified in brown pollen);
- n = 7 in violet bee pollen (≈18% of proteins identified in violet pollen).
2.2. Proteins Taxonomically Classified to Apis Spp.
2.3. The Total Quantitative Protein Content of Bee Pollen, Based on Bradford’s Analysis
3. Discussion
4. Materials and Methods
4.1. Bee Pollen
4.2. Bee Pollen Samples Pre-Treatment
4.3. Analysis of a Total Protein Concentration in Bee Pollen Aqueous Extracts
4.4. Nanolc-MALDI-TOF/TOF MS Qualitative Proteomic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Denisow, B.; Denisow-Pietrzyk, M. Biological and therapeutic properties of bee pollen: A review. J. Sci. Food Agric. 2016, 96, 4303–4309. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.A.; Ripari, N.; Conte, F.L.; da Silva Honorio, M.; Sartori, A.A.; Matucci, R.H.; Sforcin, J.M. An overview about apitherapy and its clinical applications. Phytomed. Plus 2022, 2, 100239. [Google Scholar] [CrossRef]
- Albaridi, N.A. Antibacterial potency of honey. Int. J. Microbiol. 2019, 2019, 2464507. [Google Scholar] [CrossRef] [PubMed]
- Salatino, A. Perspectives for uses of propolis in therapy against infectious diseases. Molecules 2022, 27, 4594. [Google Scholar] [CrossRef]
- Ratajczak, M.; Kaminska, D.; Matuszewska, E.; Hołderna-Kedzia, E.; Rogacki, J.; Matysiak, J. Promising antimicrobial properties of bioactive compounds from different honeybee products. Molecules 2021, 26, 4007. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Nanda, V. Composition and functionality of bee pollen: A review. Trends Food Sci. Technol. 2020, 98, 82–106. [Google Scholar] [CrossRef]
- Ares, A.M.; Valverde, S.; Bernal, J.L.; Nozal, M.J.; Bernal, J. Extraction and determination of bioactive compounds from bee pollen. J. Pharm. Biomed. Anal. 2018, 147, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, S.A.M.; Elashal, M.H.; Yosri, N.; Du, M.; Musharraf, S.G.; Nahar, L.; Sarker, S.D.; Guo, Z.; Cao, W.; Zou, X.; et al. Bee pollen: Current status and therapeutic potential. Nutrients 2021, 13, 1876. [Google Scholar] [CrossRef]
- Brochard, M.; Correia, P.; Barroca, M.J.; Guiné, R.P.F. Development of a new pasta product by the incorporation of chestnut flour and bee pollen. Appl. Sci. 2021, 11, 6617. [Google Scholar] [CrossRef]
- Estevinho, L.M.; Rodrigues, S.; Pereira, A.P.; Feás, X. Portuguese bee pollen: Palynological study, nutritional and microbiological evaluation. Int. J. Food Sci. Technol. 2012, 47, 429–435. [Google Scholar] [CrossRef]
- Giampieri, F.; Quiles, J.L.; Cianciosi, D.; Forbes-Hernández, T.Y.; Orantes-Bermejo, F.J.; Alvarez-Suarez, J.M.; Battino, M. Bee products: An emblematic example of underutilized sources of bioactive compounds. J. Agric. Food Chem. 2022, 70, 6833–6848. [Google Scholar] [CrossRef] [PubMed]
- Komosinska-Vassev, K.; Olczyk, P.; Kaźmierczak, J.; Mencner, L.; Olczyk, K. Bee pollen: Chemical composition and therapeutic application. Evid. Based. Complement. Alternat. Med. 2015, 2015, 297425. [Google Scholar] [CrossRef] [PubMed]
- Karasiewicz, M.; Bogacz, A.; Krzysztoszek, J.; Pędziwiatr, D.; Czerny, B. Products of plant origin—benefits and the potential risk for the consumer. J. Med. Sci. 2017, 86, 47–57. [Google Scholar] [CrossRef][Green Version]
- Campos, M.G.R.; Frigerio, C.; Lopes, J.; Bogdanov, S. What is the future of Bee-Pollen? J. ApiProduct ApiMedical Sci. 2010, 2, 131–144. [Google Scholar] [CrossRef]
- Végh, R.; Csóka, M.; Sörös, C.; Sipos, L. Food safety hazards of bee pollen—A review. Trends Food Sci. Technol. 2021, 114, 490–509. [Google Scholar] [CrossRef]
- Leang, Z.X.; Thalayasingam, M.; O’Sullivan, M. A paediatric case of exercise-augmented anaphylaxis following bee pollen ingestion in western australia. Asia Pac. Allergy 2022, 12, e23. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, M.; Saburi, W.; Mori, H.; Kimura, A. α-Glucosidases and α-1,4-glucan lyases: Structures, functions, and physiological actions. Cell. Mol. Life Sci. 2016, 73, 2727–2751. [Google Scholar] [CrossRef]
- Zimmer, B.M.; Barycki, J.J.; Simpson, M.A. Integration of sugar metabolism and proteoglycan synthesis by udp-glucose dehydrogenase. J. Histochem. Cytochem. 2021, 69, 13–23. [Google Scholar] [CrossRef]
- Buttstedt, A.; Moritz, R.F.A.; Erler, S. Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family. Biol. Rev. 2014, 89, 255–269. [Google Scholar] [CrossRef]
- Bauer, J.A.; Zámocká, M.; Majtán, J.; Bauerová-Hlinková, V. Glucose oxidase, an enzyme “Ferrari”: Its structure, function, production and properties in the light of various industrial and biotechnological applications. Biomolecules 2022, 12, 472. [Google Scholar] [CrossRef]
- de Oliveira, R.C.; do Nascimento Queiroz, S.C.; da Luz, C.F.P.; Porto, R.S.; Rath, S. Bee pollen as a bioindicator of environmental pesticide contamination. Chemosphere 2016, 163, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Kasiotis, K.M.; Anagnostopoulos, C.; Anastasiadou, P.; Machera, K. Pesticide residues in honeybees, honey and bee pollen by LC–MS/MS screening: Reported death incidents in honeybees. Sci. Total Environ. 2014, 485–486, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Matuszewska, E.; Klupczynska, A.; Maciołek, K.; Kokot, Z.J.; Matysiak, J. Multielemental analysis of bee pollen, propolis, and royal jelly collected in west-central poland. Molecules 2021, 26, 2415. [Google Scholar] [CrossRef]
- De-Melo, A.A.M.; Estevinho, M.L.M.F.; Almeida-Muradian, L.B. A diagnosis of the microbiological quality of dehydrated bee-pollen produced in Brazil. Lett. Appl. Microbiol. 2015, 61, 477–483. [Google Scholar] [CrossRef]
- de Arruda, V.A.S.; Vieria dos Santos, A.; Figueiredo Sampaio, D.; da Silva Araújo, E.; de Castro Peixoto, A.L.; Estevinho, M.L.F.; Bicudo de Almeida-Muradian, L. Microbiological quality and physicochemical characterization of brazilian bee pollen. J. Apic. Res. 2017, 56, 231–238. [Google Scholar] [CrossRef]
- Martín-Muñoz, M.F.; Bartolome, B.; Caminoa, M.; Bobolea, I.; Garcia Ara, M.C.; Quirce, S. Bee pollen: A dangerous food for allergic children. identification of responsible allergens. Allergol. Immunopathol. 2010, 38, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Rhim, T.; Choi, Y.S.; Nam, B.Y.; Uh, S.T.; Park, J.S.; Kim, Y.H.; Paik, Y.K.; Park, C.S. Plasma protein profiles in early asthmatic responses to inhalation allergen challenge. Allergy Eur. J. Allergy Clin. Immunol. 2009, 64, 47–54. [Google Scholar] [CrossRef]
- Pablos, I.; Wildner, S.; Asam, C.; Wallner, M.; Gadermaier, G. Pollen allergens for molecular diagnosis. Curr. Allergy Asthma Rep. 2016, 16, 31. [Google Scholar] [CrossRef]
- Mayda, N.; Özkök, A.; Ecem Bayram, N.; Gerçek, Y.C.; Sorkun, K. Bee bread and bee pollen of different plant sources: Determination of phenolic content, antioxidant activity, fatty acid and element profiles. J. Food Meas. Charact. 2020, 14, 1795–1809. [Google Scholar] [CrossRef]
- Cai, G.; Serafini-Fracassini, D.; Del Duca, S. Regulation of pollen tube growth by transglutaminase. Plants 2013, 2, 87–106. [Google Scholar] [CrossRef]
- Voisin, M.R.; Borici-Mazi, R. Anaphylaxis to supplemental oral lactase enzyme. Allergy Asthma Clin. Immunol. 2016, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Weryszko-Chmielewska, E. Pollen development, biology and function. Adv. Dermatol. Allergol. Dermatol. I Alergol. 2003, 20, 212–217. [Google Scholar]
- Virdi, A.S.; Singh, S.; Singh, P. Abiotic stress responses in plants: Roles of calmodulin-regulated proteins. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Perochon, A.; Aldon, D.; Galaud, J.-P.; Ranty, B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 2011, 93, 2048–2053. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Esquivel, M.L.; Guidos-Fogelbach, G.A.; Rojo-Gutiérrez, M.I.; Mellado-Abrego, J.; Bermejo-Guevara, M.A.; Castillo-Narváez, G.; Velázquez-Sámano, G.; Velasco-Medina, A.A.; Moya-Almonte, M.G.; Vallejos-Pereira, C.M.; et al. Identification of an allergenic calmodulin from Amaranthus palmeri pollen. Mol. Immunol. 2021, 132, 150–156. [Google Scholar] [CrossRef]
- Poncet, P.; Senechal, H.; Clement, G.; Purohit, A.; Sutra, J.-P.; Desvaux, F.-X.; Wal, J.-M.; Pauli, G.; Peltre, G.; Gougeon, M.-L. Evaluation of ash pollen sensitization pattern using proteomic approach with individual sera from allergic patients. Allergy 2010, 65, 571–580. [Google Scholar] [CrossRef]
- Sun, T.; Li, S.; Ren, H. Profilin as a regulator of the membrane-actin cytoskeleton interface in plant cells. Front. Plant Sci. 2013, 4, 1–7. [Google Scholar] [CrossRef]
- del Río, P.R.; Díaz-Perales, A.; Sánchez-García, S.; Escudero, C.; Ibáñez, M.D.; Méndez-Brea, P.; Barber, D. Profilin, a change in the paradigm. J. Investig. Allergol. Clin. Immunol. 2018, 28, 1–12. [Google Scholar] [CrossRef]
- Yang, Y.-S.; Xu, Z.-Q.; Zhu, W.; Zhu, D.-X.; Jiao, Y.-X.; Zhang, L.-S.; Hou, Y.-B.; Wei, J.-F.; Sun, J.-L. Molecular and immunochemical characterization of profilin as major allergen from Platanus acerifolia pollen. Int. Immunopharmacol. 2022, 106, 108601. [Google Scholar] [CrossRef]
- WHO/IUIS Allergen Nomenclature Home Page [Internet]. Available online: http://www.allergen.org/ (accessed on 2 September 2022).
- Hatzler, L.; Panetta, V.; Lau, S.; Wagner, P.; Bergmann, R.L.; Illi, S.; Bergmann, K.E.; Keil, T.; Hofmaier, S.; Rohrbach, A.; et al. Molecular spreading and predictive value of preclinical IgE response to Phleum pratense in children with hay fever. J. Allergy Clin. Immunol. 2012, 130, 894–901. [Google Scholar] [CrossRef]
- Asero, R.; Tripodi, S.; Dondi, A.; Di Rienzo Businco, A.; Sfika, I.; Bianchi, A.; Candelotti, P.; Caffarelli, C.; Povesi Dascola, C.; Ricci, G.; et al. Prevalence and clinical relevance of IgE sensitization to profilin in childhood: A multicenter study. Int. Arch. Allergy Immunol. 2015, 168, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.; Díaz-Perales, A.; Villalba, M.; Chivato, T. Challenges for allergy diagnosis in regions with complex pollen exposures. Curr. Allergy Asthma Rep. 2015, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.; De La Torre, F.; Lombardero, M.; Antépara, I.; Colas, C.; Dávila, I.; Tabar, A.I.; Vidal, C.; Villalba, M.; Salcedo, G.; et al. Component-resolved diagnosis of pollen allergy based on skin testing with profilin, polcalcin and lipid transfer protein pan-allergens. Clin. Exp. Allergy 2009, 39, 1764–1773. [Google Scholar] [CrossRef] [PubMed]
- Nucera, E.; Aruanno, A.; Rizzi, A.; Pecora, V.; Patriarca, G.; Buonomo, A.; Mezzacappa, S.; Schiavino, D. Profilin desensitization: A case series. Int. J. Immunopathol. Pharmacol. 2016, 29, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Md, R.I.; Polash, A.H.; Sakib, M.S.; Saha, C.; Rahman, A. Computational identification of Brassica napus pollen specific protein Bnm1 as an allergen. Int. J. Bioinforma. Biosci. 2013, 3, 45–57. [Google Scholar] [CrossRef]
- Assarehzadegan, M.A.; Sankian, M.; Jabbari, F.; Tehrani, M.; Falak, R.; Varasteh, A. Identification of methionine synthase (Sal k 3), as a novel allergen of Salsola kali pollen. Mol. Biol. Rep. 2011, 38, 65–73. [Google Scholar] [CrossRef]
- Gadermaier, G.; Hauser, M.; Ferreira, F. Allergens of weed pollen: An overview on recombinant and natural molecules. Methods 2014, 66, 55–66. [Google Scholar] [CrossRef]
- Aksüt, Y.; Pekmez, M.; Uzuner, S.K.; Gelincik, A.; Büyüköztürk, S.; Arda, N. Molecular cloning, expression, purification and in silico epitope prediction of cobalamin-independent methionine synthase (Mor a 2), as a novel allergen from Morus alba pollen. Arch. Med. Sci. 2021. [Google Scholar] [CrossRef]
- Chardin, H.; Mayer, C.; Sénéchal, H.; Tepfer, M.; Desvaux, F.X.; Peltre, G. Characterization of high-molecular-mass allergens in oilseed rape pollen. Int. Arch. Allergy Immunol. 2001, 125, 128–134. [Google Scholar] [CrossRef]
- Cornara, L.; Biagi, M.; Xiao, J.; Burlando, B. Therapeutic properties of bioactive compounds from different honeybee products. Front. Pharmacol. 2017, 8, 412. [Google Scholar] [CrossRef]
- Matuszewska, E.; Matysiak, J.; Rosiński, G.; Kędzia, E.; Ząbek, W.; Zawadziński, J.; Matysiak, J. Mining the royal jelly proteins: Combinatorial hexapeptide ligand library significantly improves the ms-based proteomic identification in complex biological samples. Molecules 2021, 26, 2762. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Kim, B.Y.; Park, H.G.; Deng, Y.; Yoon, H.J.; Choi, Y.S.; Lee, K.S.; Jin, B.R. Major royal jelly protein 2 acts as an antimicrobial agent and antioxidant in royal jelly. J. Asia Pac. Entomol. 2019, 22, 684–689. [Google Scholar] [CrossRef]
- Park, H.G.; Kim, B.Y.; Park, M.J.; Deng, Y.; Choi, Y.S.; Lee, K.S.; Jin, B.R. Antibacterial activity of major royal jelly proteins of the honeybee (Apis mellifera) royal jelly. J. Asia Pac. Entomol. 2019, 22, 737–741. [Google Scholar] [CrossRef]
- Lin, Y.; Shao, Q.; Zhang, M.; Lu, C.; Fleming, J.; Su, S. Royal jelly-derived proteins enhance proliferation and migration of human epidermal keratinocytes in an in vitro scratch wound model. BMC Complement. Altern. Med. 2019, 19, 175. [Google Scholar] [CrossRef]
- Matysiak, J.; Matuszewska, E.; Packi, K.; Klupczyńska-Gabryszak, A. Diagnosis of hymenoptera venom allergy: State of the art, challenges, and perspectives. Biomedicines 2022, 10, 2170. [Google Scholar] [CrossRef]
- Saisavoey, T.; Sangtanoo, P.; Chanchao, C.; Reamtong, O.; Karnchanatat, A. Identification of novel anti-inflammatory peptides from bee pollen (Apis mellifera) hydrolysate in lipopolysaccharide-stimulated RAW264.7 macrophages. J. Apic. Res. 2021, 60, 280–289. [Google Scholar] [CrossRef]
- Pełka, K.; Worobo, R.W.; Walkusz, J.; Szweda, P. Bee pollen and bee bread as a source of bacteria producing antimicrobials. Antibiotics 2021, 10, 713. [Google Scholar] [CrossRef]
- Hoover, S.E.; Ovinge, L.P. Pollen Collection, Honey production, and pollination services: Managing honey bees in an agricultural setting. J. Econ. Entomol. 2018, 111, 1509–1516. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
| Accession | Protein Name | Number of Peptides Identified as a Protein Fragment | Classes/Functions | |||
|---|---|---|---|---|---|---|
| Orange | Violet | Yellow | Brown | |||
| gi|126842411 | alpha-glucosidase [1] | 3 | Carbohydrate metabolism [17] | |||
| gi|283105164 | alpha-glucosidase III [2] | 3 | Carbohydrate metabolism [17] | |||
| gi|284812514 | MRJP5 [3] | 6 | Nutrition and development [19] | |||
| gi|33358394 | major royal jelly protein MRJP1 [4] | 7 | Nutrition and development [19] | |||
| gi|380025661 | PREDICTED: glucose dehydrogenase [acceptor]-like [5] | 3 | 3 | Carbohydrate metabolism [18] | ||
| gi|40557703 | major royal jelly protein MRJP1 precursor [6] | 11 | 8 | Nutrition and development [19] | ||
| gi|58585090 | glucose oxidase [3] | 5 | 15 | 6 | 6 | Carbohydrate metabolism [20] |
| gi|58585098 | major royal jelly protein 1 precursor [3] | 82 | 57 | 40 | 39 | Nutrition and development [19] |
| gi|58585108 | major royal jelly protein 2 precursor [3] | 4 | 4 | 5 | Nutrition and development [19] | |
| gi|62198227 | major royal jelly protein 7 precursor [3] | 11 | Nutrition and development [19] | |||
| Sample No | Harvest Date | Proteins Concentration [mg/mL] |
|---|---|---|
| 1 | 14 June 2018 | 0.91252 |
| 2 | 23 June 2018 | 0.97429 |
| 3 | 8 July 2018 | 0.73127 |
| 4 | 20 July 2018 | 0.68932 |
| Mean | 0.82685 | |
| SD | 0.137995096 | |
| RSD | 16.68925395 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matuszewska, E.; Plewa, S.; Pietkiewicz, D.; Kossakowski, K.; Matysiak, J.; Rosiński, G.; Matysiak, J. Mass Spectrometry-Based Identification of Bioactive Bee Pollen Proteins: Evaluation of Allergy Risk after Bee Pollen Supplementation. Molecules 2022, 27, 7733. https://doi.org/10.3390/molecules27227733
Matuszewska E, Plewa S, Pietkiewicz D, Kossakowski K, Matysiak J, Rosiński G, Matysiak J. Mass Spectrometry-Based Identification of Bioactive Bee Pollen Proteins: Evaluation of Allergy Risk after Bee Pollen Supplementation. Molecules. 2022; 27(22):7733. https://doi.org/10.3390/molecules27227733
Chicago/Turabian StyleMatuszewska, Eliza, Szymon Plewa, Dagmara Pietkiewicz, Kacper Kossakowski, Joanna Matysiak, Grzegorz Rosiński, and Jan Matysiak. 2022. "Mass Spectrometry-Based Identification of Bioactive Bee Pollen Proteins: Evaluation of Allergy Risk after Bee Pollen Supplementation" Molecules 27, no. 22: 7733. https://doi.org/10.3390/molecules27227733
APA StyleMatuszewska, E., Plewa, S., Pietkiewicz, D., Kossakowski, K., Matysiak, J., Rosiński, G., & Matysiak, J. (2022). Mass Spectrometry-Based Identification of Bioactive Bee Pollen Proteins: Evaluation of Allergy Risk after Bee Pollen Supplementation. Molecules, 27(22), 7733. https://doi.org/10.3390/molecules27227733








