Weak Noncovalent Interactions in Three Closely Related Adamantane-Linked 1,2,4-Triazole N-Mannich Bases: Insights from Energy Frameworks, Hirshfeld Surface Analysis, In Silico 11β-HSD1 Molecular Docking and ADMET Prediction
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis and Crystallization
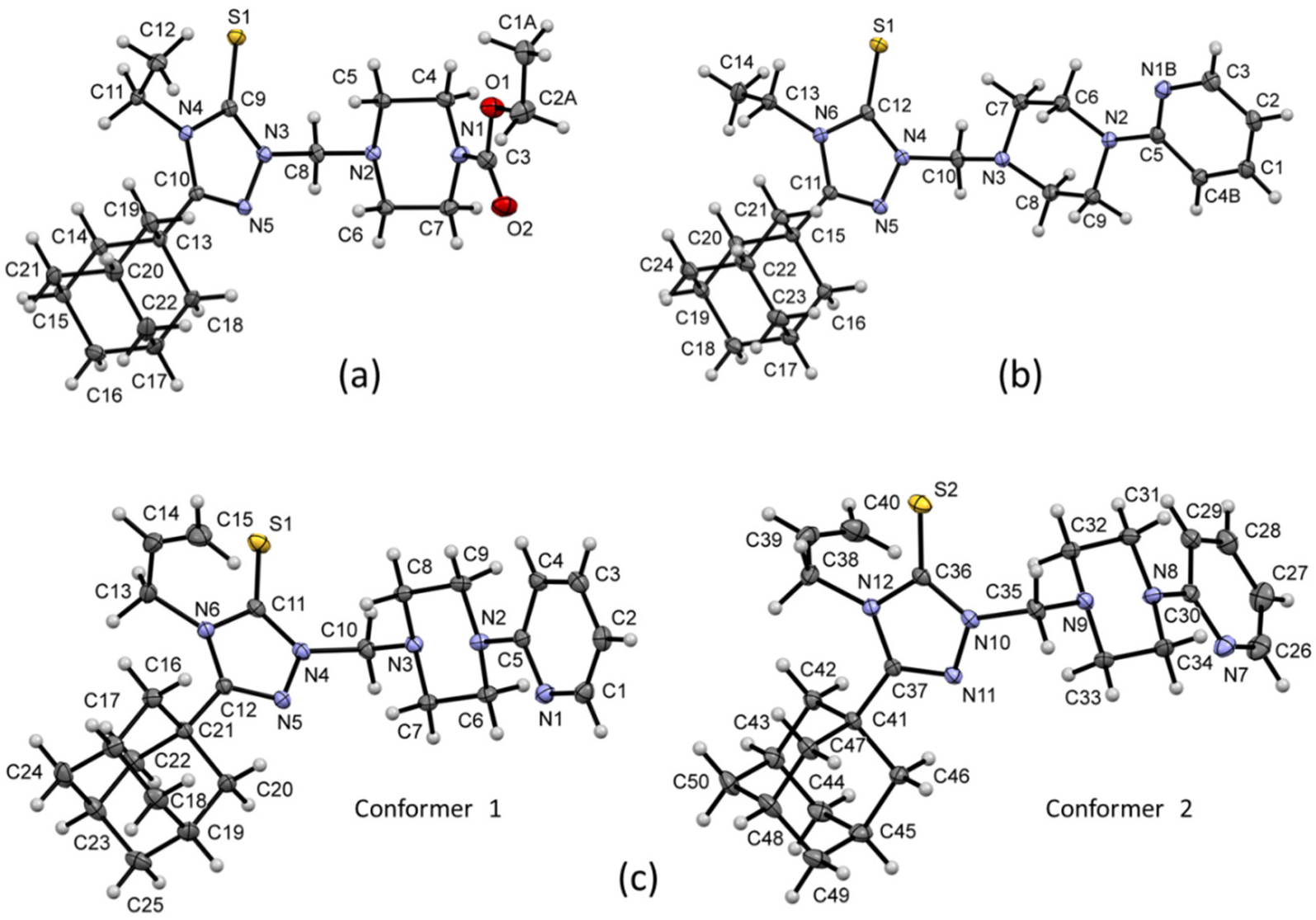
2.2. Description of Molecular and Crystal Structures
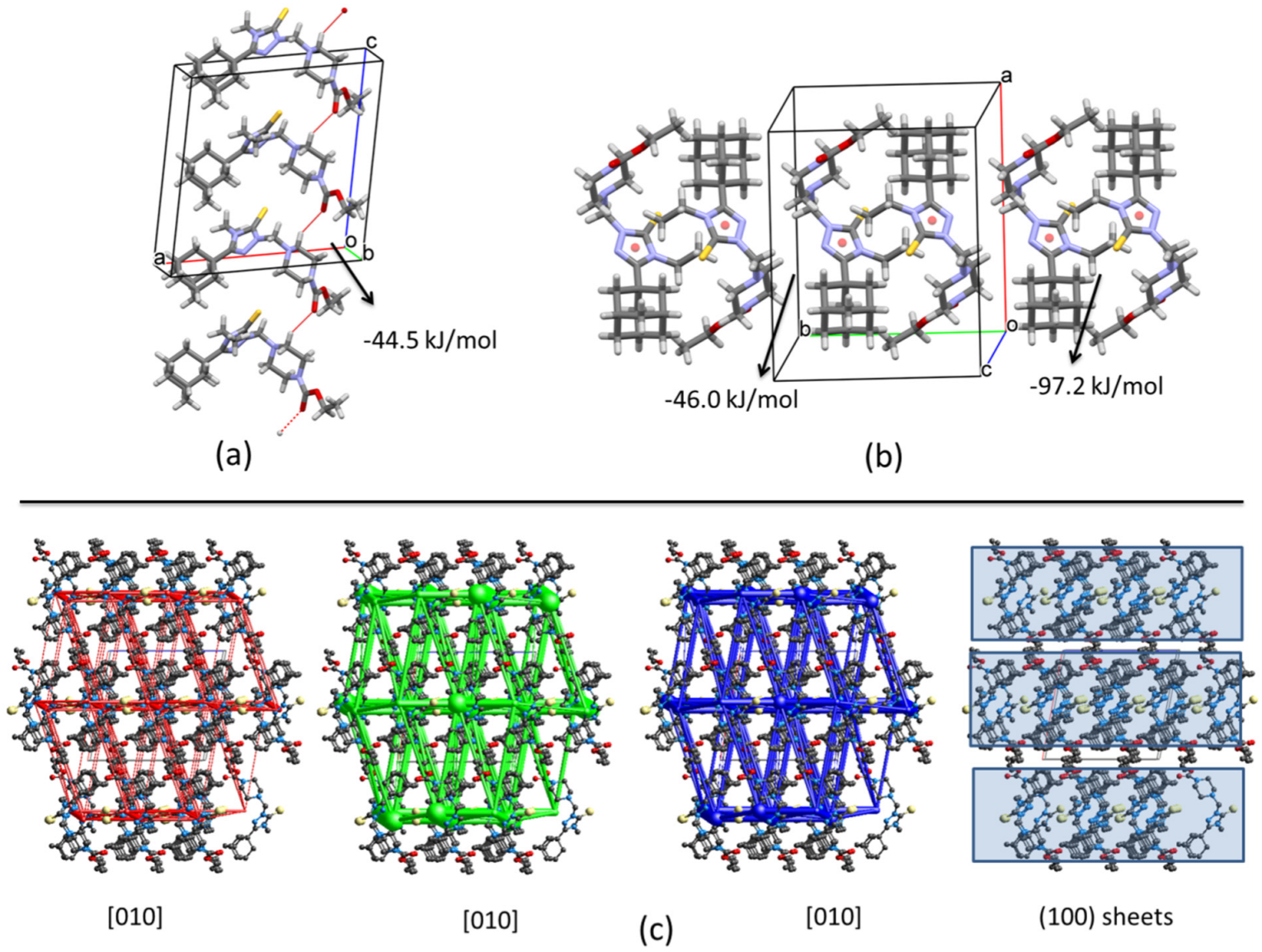
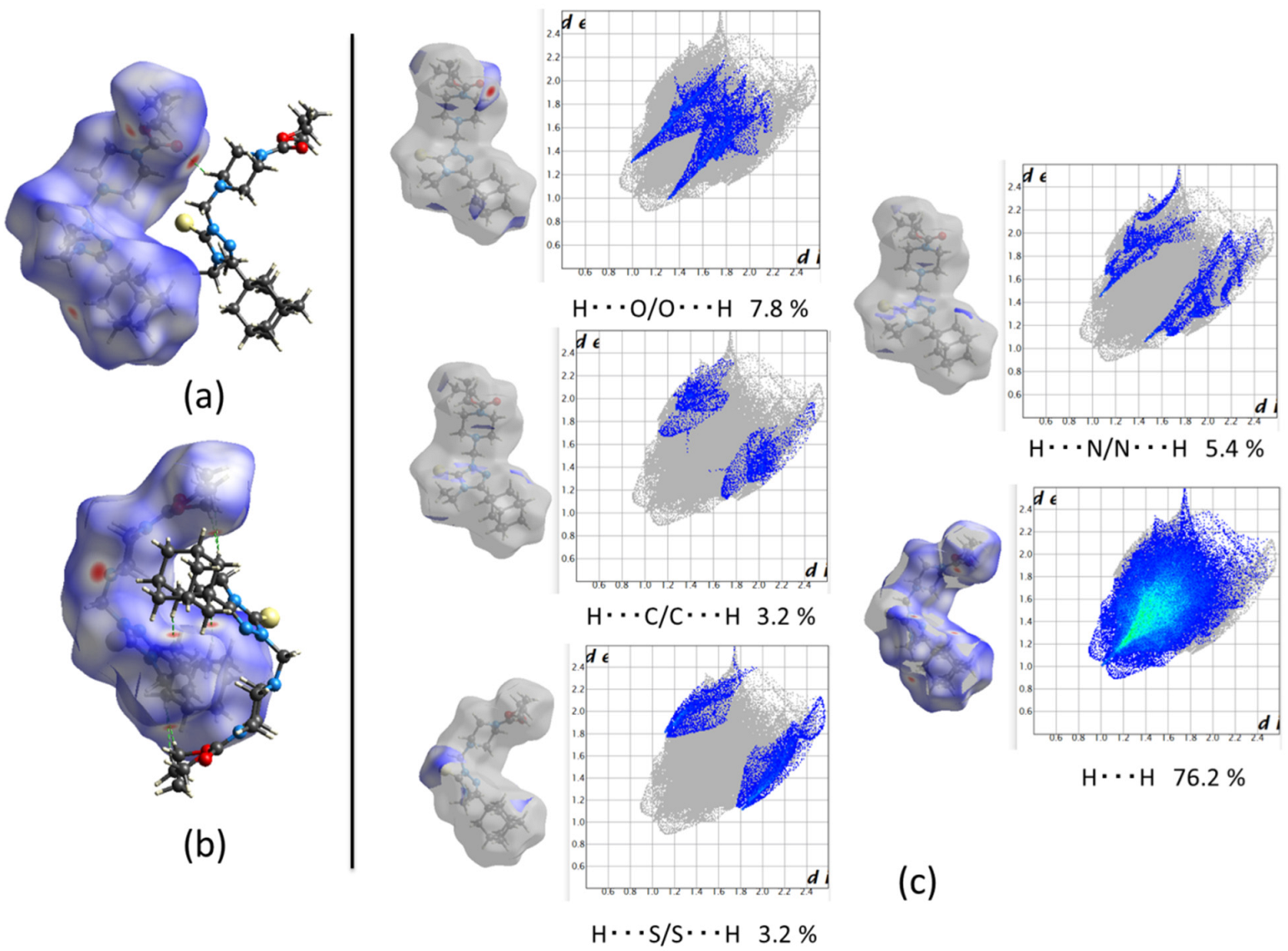
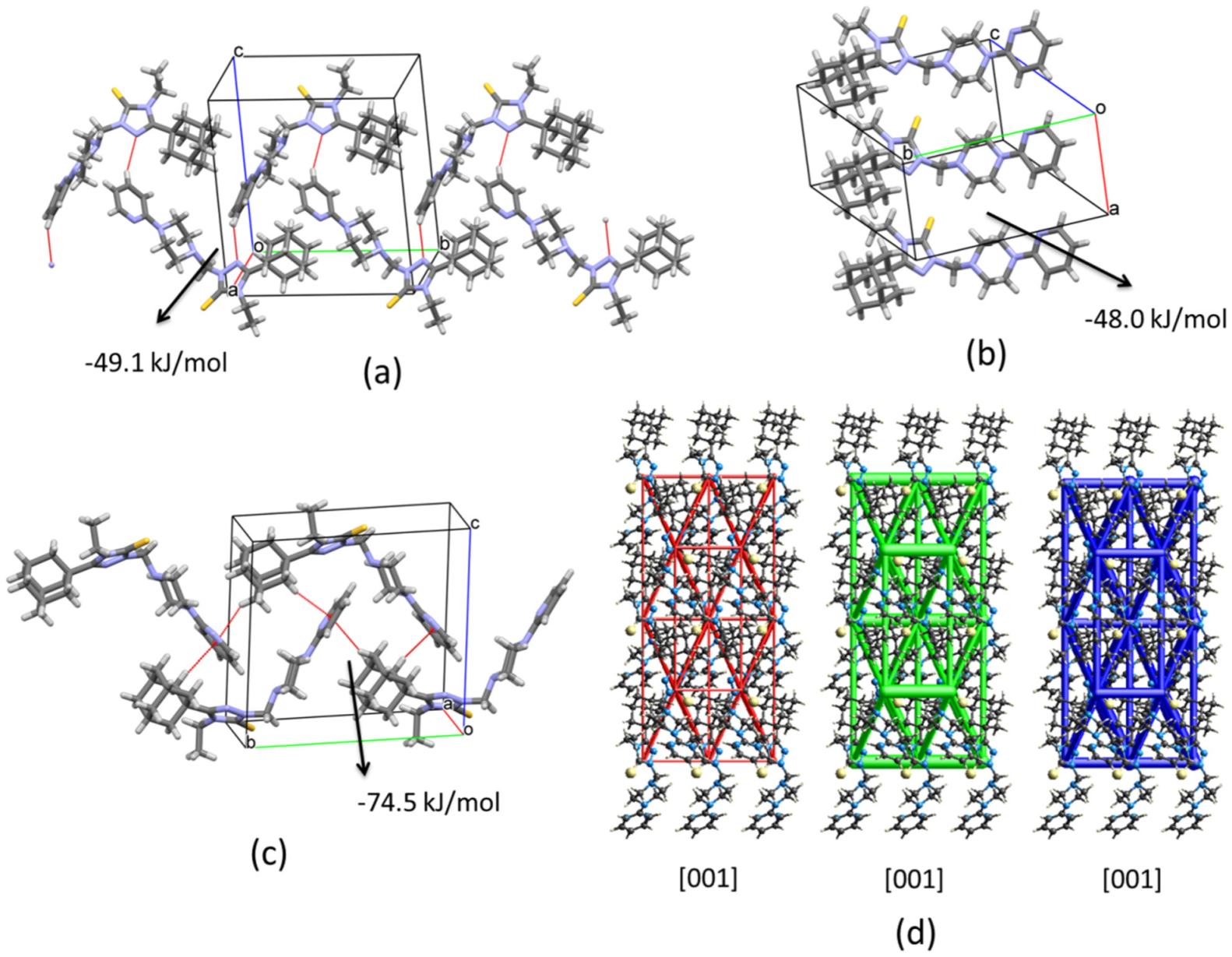
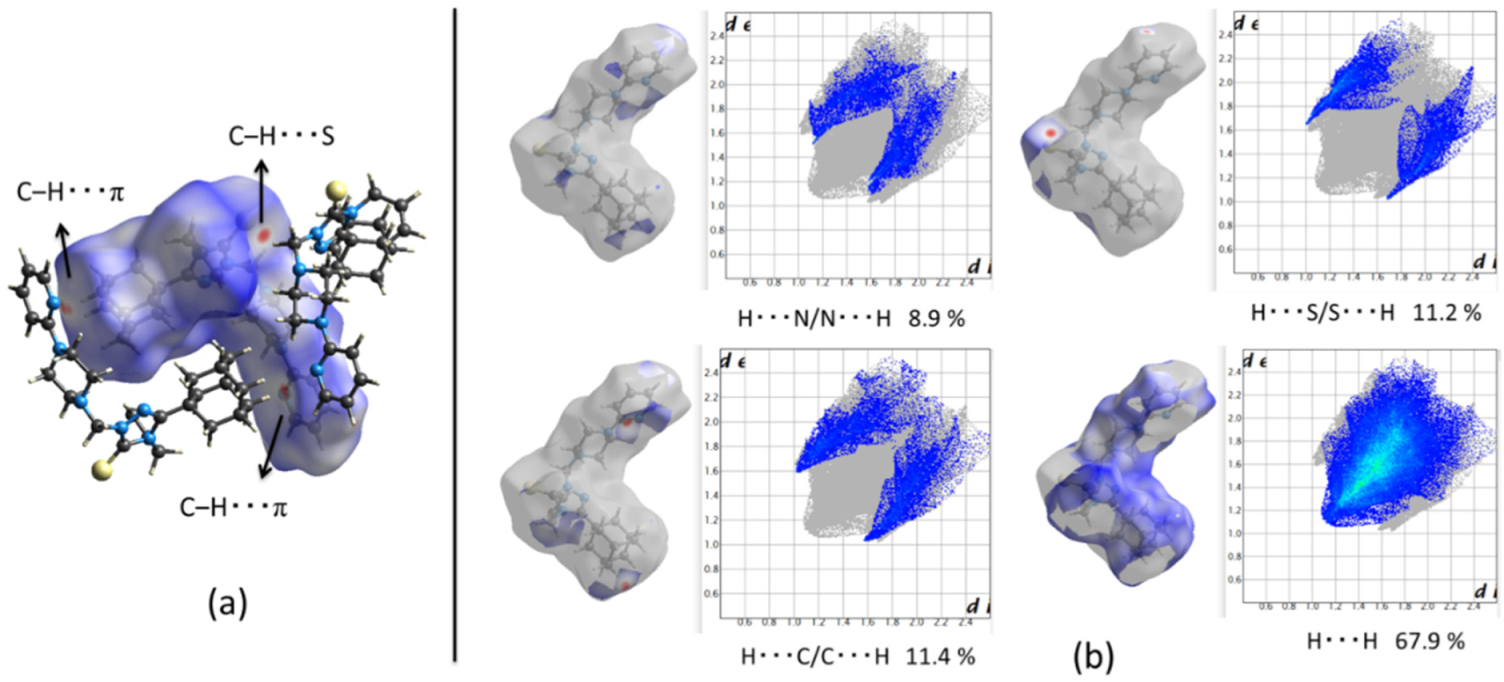
2.2.1. Crystal Packing of Compound 1 and Interactions
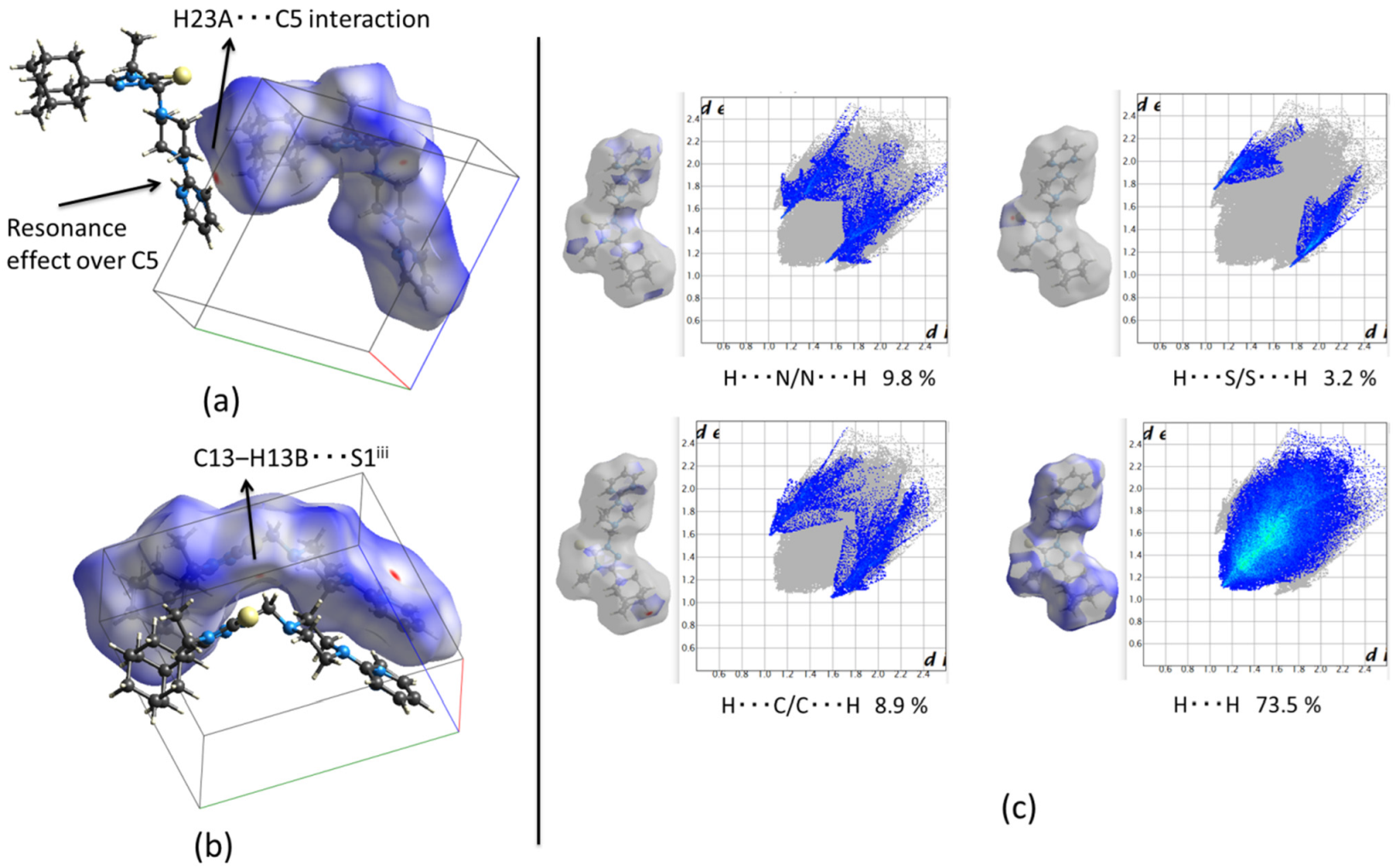
2.2.2. Crystal Packing of Compound 2 and Interactions
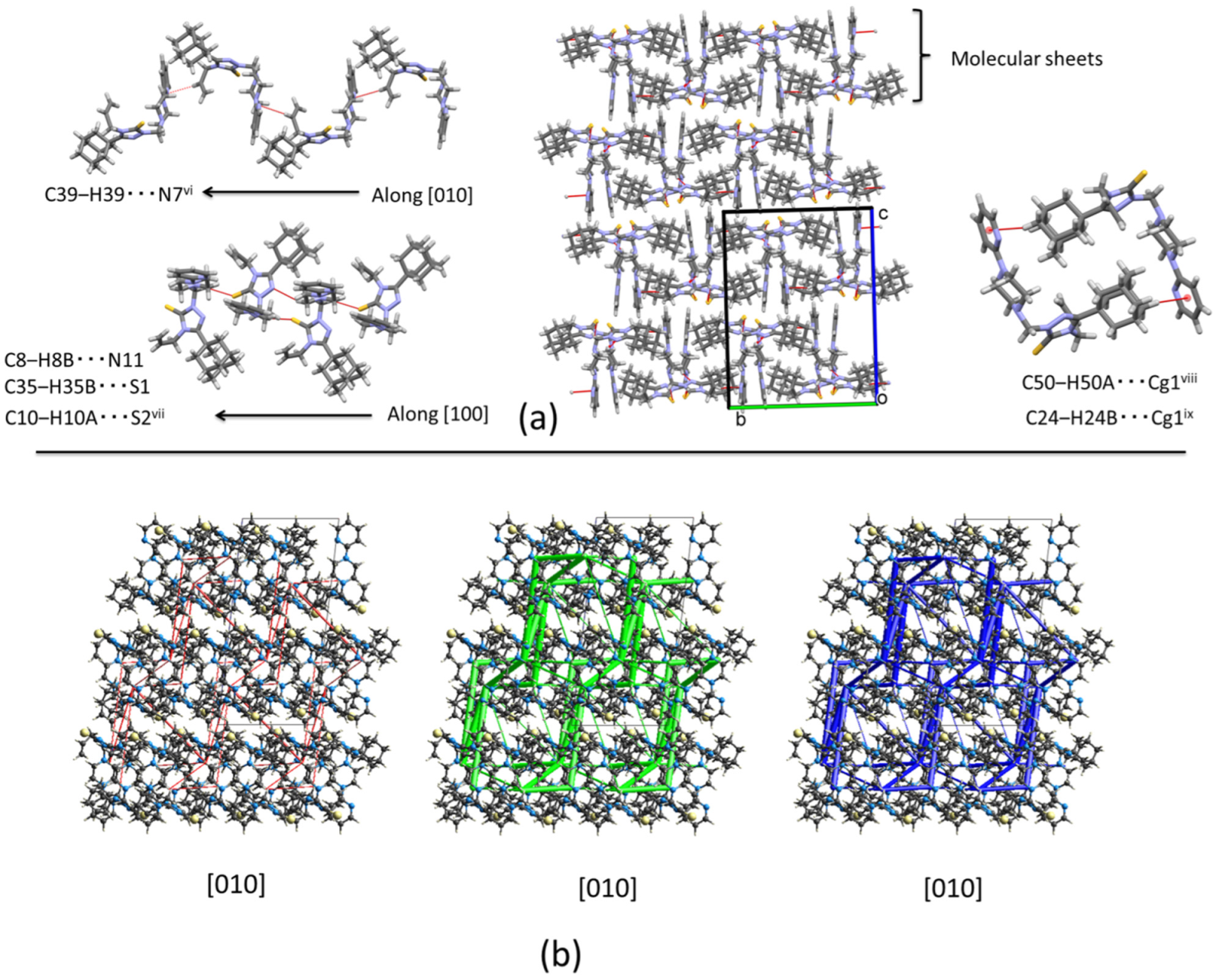
2.2.3. Crystal Packing of Compound 3 and Interactions
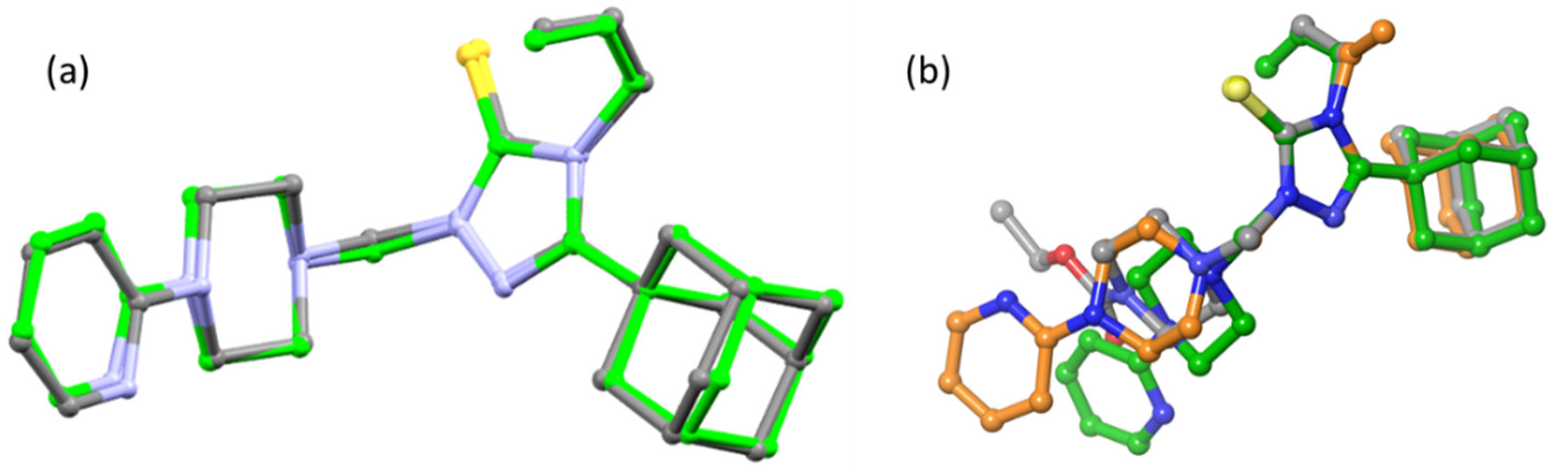
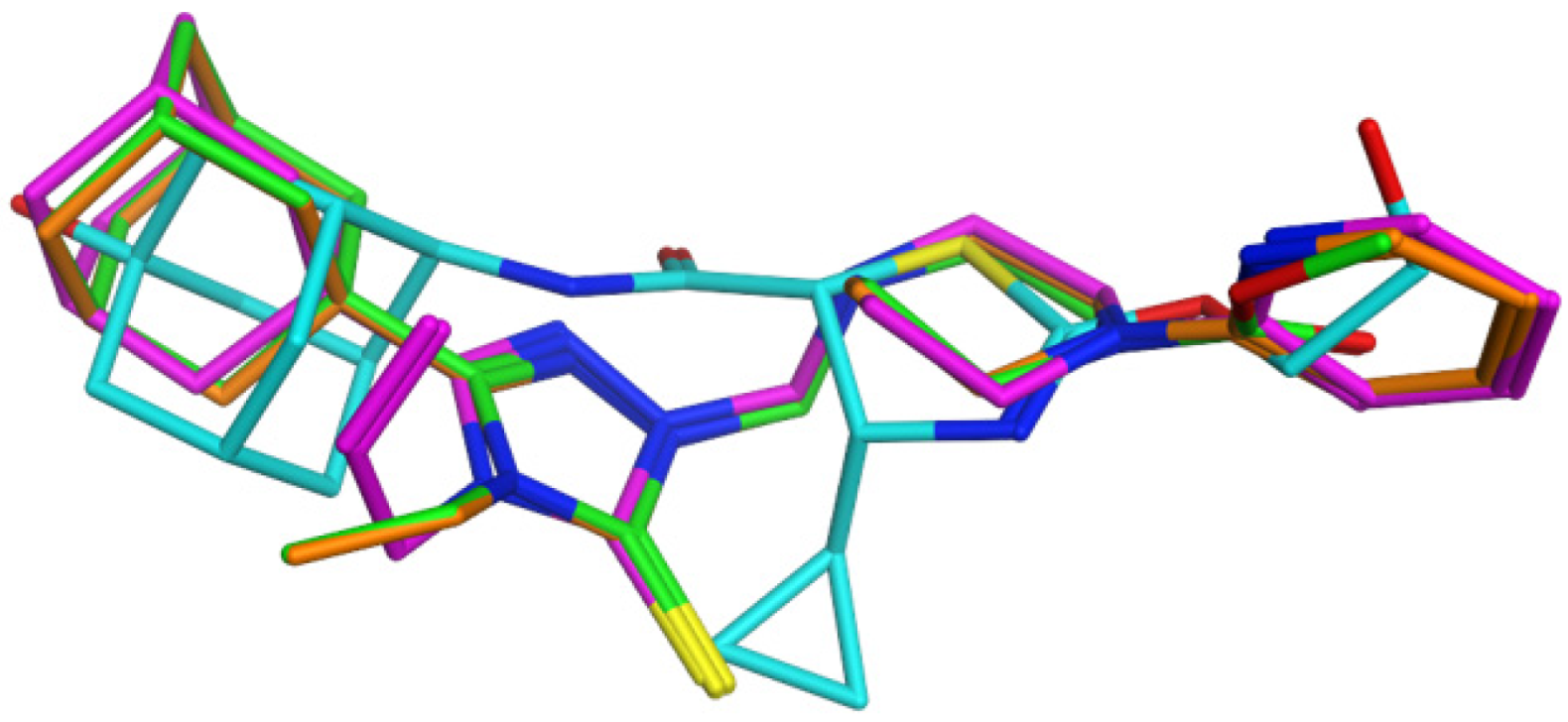
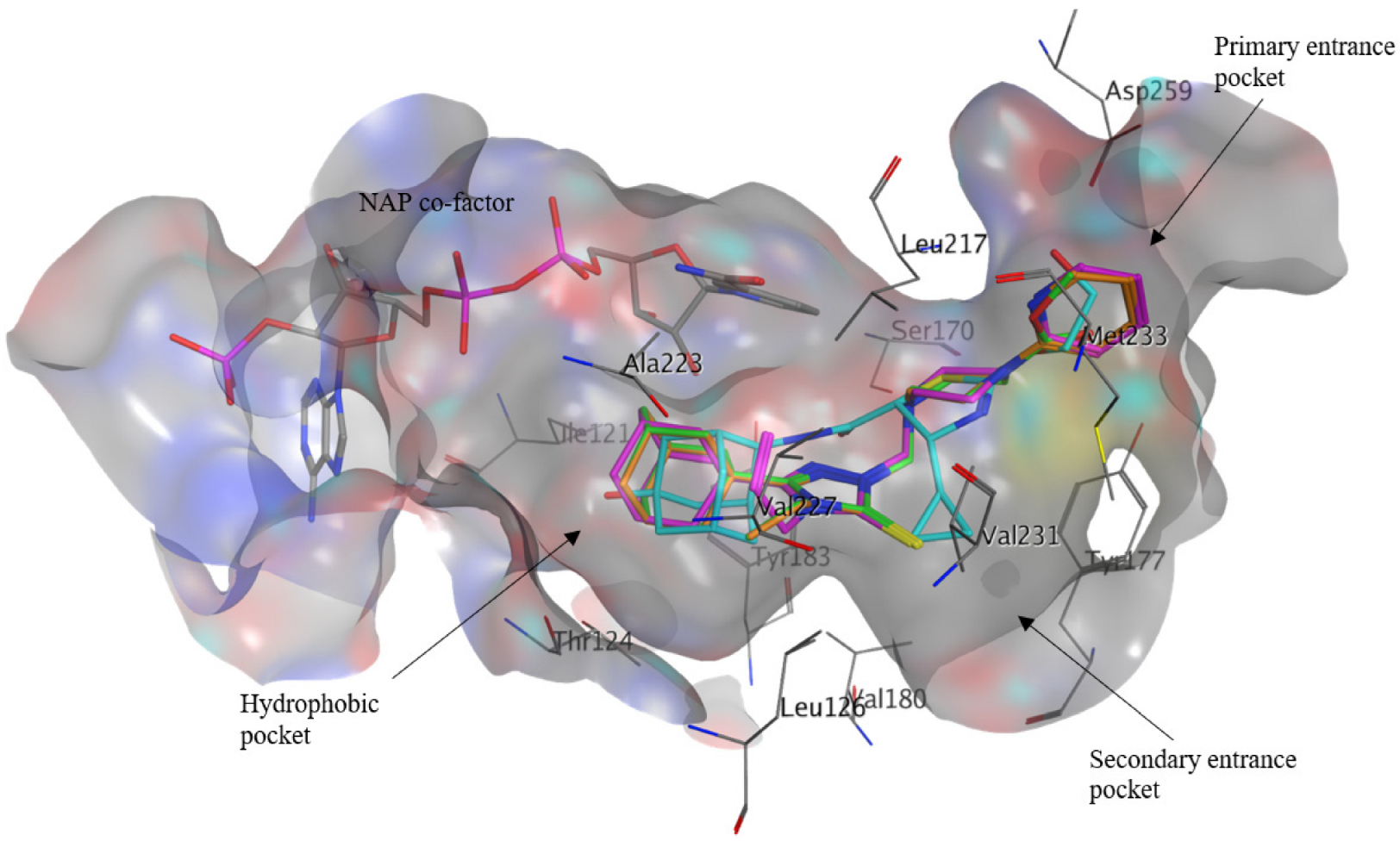
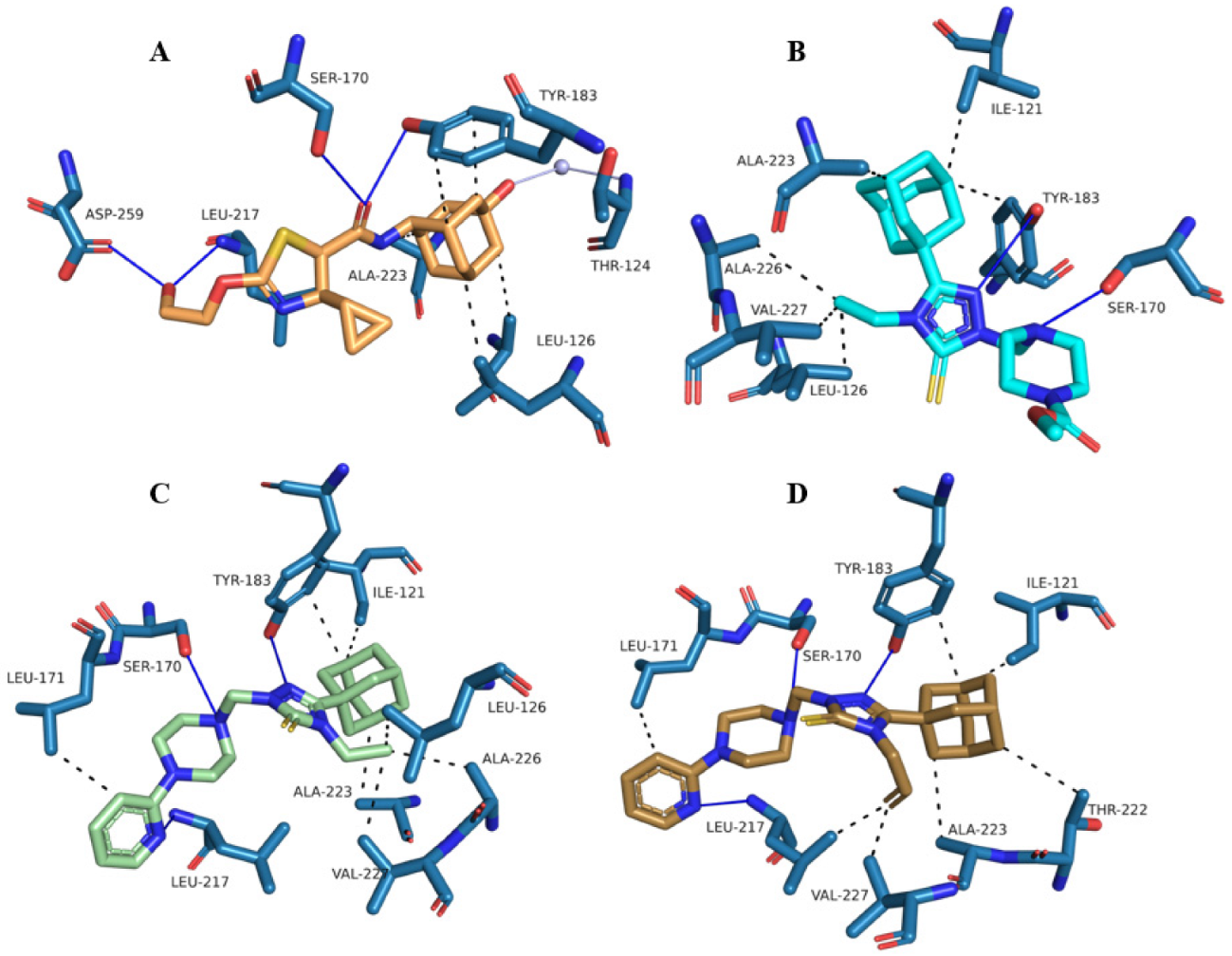
2.3. Molecular Docking Analysis
2.4. ADMET Analysis
3. Materials and Methods
3.1. Single-Crystal X-ray Diffraction
3.2. Hirshfeld Surface and Energy Frameworks Analysis
3.3. Molecular Docking Studies
3.3.1. Preparation of Protein and Ligands
3.3.2. Molecular Docking of Compounds 1–3
3.4. In Silico Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Spilovska, K.; Zemek, F.; Korabecny, J.; Nepovimova, E.; Soukup, O.; Windisch, M.; Kuca, K. Adamantane—A lead structure for drugs in clinical practice. Curr. Med. Chem. 2016, 23, 3245–3266. [Google Scholar] [CrossRef] [PubMed]
- Wanka, L.; Iqbal, K.; Schreiner, P.R. The lipophilic bullet hits the targets: Medicinal chemistry of adamantane derivatives. Chem. Rev. 2013, 113, 3516–3604. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Obando, D.; Liao, V.; Lifa, T.; Codd, R. The many faces of the adamantyl group in drug design. Eur. J. Med. Chem. 2011, 46, 1949–1963. [Google Scholar] [CrossRef] [PubMed]
- Lamoureux, G.; Artavia, G. Use of the adamantane structure in medicinal chemistry. Curr. Med. Chem. 2010, 17, 2967–2978. [Google Scholar] [CrossRef] [PubMed]
- Davies, W.L.; Grunnert, R.R.; Haff, R.F.; McGahen, J.W.; Neumeyer, E.M.; Paulshock, M.; Watts, J.C.; Wood, T.R.; Hermann, E.C.; Hoffmann, C.E. Antiviral activity of 1-adamantamine (amantadine). Science 1964, 144, 862–863. [Google Scholar] [CrossRef]
- Wendel, H.A.; Snyder, M.T.; Pell, S. Trial of amantadine in epidemic influenza. Clin. Pharmacol. Ther. 1966, 7, 38–43. [Google Scholar] [CrossRef]
- Rosenthal, K.S.; Sokol, M.S.; Ingram, R.L.; Subramanian, R.; Fort, R.C. Tromantadine: Inhibitor of early and late events in herpes simplex virus replication. Antimicrob. Agents Chemother. 1982, 22, 1031–1036. [Google Scholar] [CrossRef]
- Jia, L.; Tomaszewski, J.E.; Hanrahan, C.; Coward, L.; Noker, P.; Gorman, G.; Nikonenko, B.; Protopopova, M. Parmacodynamics and pharmacokinetics of SQ109, a new diamine-based antitubercular drug. Br. J. Pharmacol. 2005, 144, 80–87. [Google Scholar] [CrossRef]
- Protopopova, M.; Hanrahan, C.; Nikonenko, B.; Samala, R.; Chen, P.; Gearhart, J.; Einck, L.; Nacy, C.A. Identification of a new antitubercular drug candidate, SQ109, from a combinatorial library of 1,2-ethylenediamines. J. Antimicrob. Chemother. 2005, 56, 968–974. [Google Scholar] [CrossRef]
- Britten, C.D.; Garrett-Mayer, E.; Chin, S.H.; Shirai, K.; Ogretmen, B.; Bentz, T.A.; Brisendine, A.; Anderton, K.; Cusack, S.L.; Maines, L.W.; et al. A phase I study of ABC294640, a first-in-class sphingosine kinase-2 inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 2017, 23, 4642–4650. [Google Scholar] [CrossRef]
- Han, T.; Goralski, M.; Capota, E.; Padrick, S.B.; Kim, J.; Xie, Y.; Nijhawan, D. The antitumor toxin CD437 is a direct inhibitor of DNA polymerase α. Nat. Chem. Biol. 2016, 12, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Nasr, R.R.; Hmadi, R.A.; El-Eit, R.M.; Iskandarani, A.N.; Jabbour, M.N.; Zaatari, G.S.; Mahon, F.X.; Pisano, C.C.; Darwiche, N.D. ST1926, an orally active synthetic retinoid, induces apoptosis in chronic myeloid leukemia cells and prolongs survival in a murine model. Int. J. Cancer 2015, 137, 698–709. [Google Scholar] [CrossRef] [PubMed]
- Augeri, D.J.; Robl, J.A.; Betebenner, D.A.; Magnin, D.R.; Khanna, A.; Robertson, J.G.; Wang, A.; Simpkins, L.M.; Taunk, P.; Huang, Q.; et al. Discovery and preclinical profile of saxagliptin (BMS-477118): A highly potent, long-acting, orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. J. Med. Chem. 2005, 48, 5025–5037. [Google Scholar] [CrossRef] [PubMed]
- Villhauer, E.B.; Brinkman, J.A.; Naderi, G.B.; Burkey, B.F.; Dunning, B.E.; Prasad, K.; Mangold, B.L.; Russell, M.E.; Hughes, T.E. 1-(3-Hydroxy-1-adamantyl)aminoacetyl-2-cyano-(S)-pyrrolidine: A potent, selective, and orally bioavailable dipeptidyl peptidase IV inhibitor with antihyperglycemic properties. J. Med. Chem. 2003, 46, 2774–2789. [Google Scholar] [CrossRef] [PubMed]
- Balkovec, J.M.; Thieringer, R.; Mundt, S.S.; Hermanowski-Vosatka, A.; Graham, D.W.; Donald, W.; Patel, G.F.; Susan, S.D.; Waddell, S.T.; Sherman, T.; et al. Preparation of 1,2,4-triazole derivatives as 11β-hydroxysteroid dehydrogenase 1 inhibitors useful for treatment of diabetes, obesity and dyslipidemia. WO 20030814 (2003). Chem. Abstr. 2003, 139, 180065. [Google Scholar]
- Olson, S.; Aster, S.D.; Brown, K.; Carbin, L.; Graham, D.W.; Hermanowski-Vosatka, A.; LeGrand, C.B.; Mundt, S.S.; Robbins, M.A.; Schaeffer, J.M.; et al. Adamantyl triazoles as selective inhibitors of 11β-hydroxysteroid dehydrogenase type 1. Bioorganic Med. Chem. Lett. 2005, 15, 4359–4362. [Google Scholar] [CrossRef]
- Joharapurkar, A.; Dhanesha, N.; Shah, G.; Kharul, R.; Jain, M. 11β-Hydroxysteroid dehydrogenase type 1: Potential therapeutic target for metabolic syndrome. Pharmacol. Rep. 2012, 64, 1055–1065. [Google Scholar] [CrossRef]
- Stewart, P.M.; Tomlinson, J.W. Selective inhibitors of 11β-hydroxysteroid dehydrogenase type 1 for patients with metabolic syndrome. Diabetes 2009, 58, 14–15. [Google Scholar] [CrossRef][Green Version]
- Gathercole, L.L.; Stewart, P.M. Targeting the pre-receptor metabolism of cortisol as a novel therapy in obesity and diabetes. J. Steroid Biochem. Mol. Biol. 2010, 122, 21–27. [Google Scholar] [CrossRef]
- Bujalska, I.J.; Gathercole, L.L.; Tomlinson, J.W.; Darimont, C.; Ermolieff, J.; Fanjul, A.N.; Rejto, P.A.; Stewart, P.M. A novel selective 11β-hydroxysteroid dehydrogenase type 1 inhibitor prevents human adipogenesis. J. Endocrinol. 2008, 197, 297–307. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, Y.; Olson, S.H.; Graham, D.; Patel, G.; Hermanowski-Vosatka, A.; Mundt, S.; Shah, K.; Springer, M.; Thieringer, R.; Wright, S.; et al. Phenylcyclobutyl triazoles as selective inhibitors of 11β-hydroxysteroid dehydrogenase type 1. Bioorganic Med. Chem. Lett. 2008, 18, 3412–3416. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.; Powers, J.P.; Liu, J.; Ursu, S.; Sudom, A.; Yan, X.; Xu, H.; Meininger, D.; DeGraffenreid, M.; He, X.; et al. Distinctive molecular inhibition mechanisms for selective inhibitors of human 11β-hydroxysteroid dehydrogenase type 1. Bioorg. Med. Chem. 2008, 16, 8922–8931. [Google Scholar] [CrossRef] [PubMed]
- Cott, J.S.; Choormun, J. 11β-Hydroxysteroid dehydrogenase Type 1 (11β-HSD-1) inhibitors in development. In New Therapeutic Strategies for Type-2 Diabetes, Small Molecule Approaches; Jones, R.N., Ed.; RSC: Cambridge, UK, 2012; Chapter 5; pp. 109–133. [Google Scholar]
- Al-Wahaibi, L.H.; Joubert, J.; Blacque, O.; Al-Shaalan, N.H.; El-Emam, A.A. Crystal structure, Hirshfeld surface analysis and DFT studies of 5-(adamantan-1-yl)-3-[(4-chlorobenzyl)sulfanyl]-4-methyl-4H-1,2,4-triazole, a potential 11β-HSD1 inhibitor. Sci. Rep. 2019, 9, 19745. [Google Scholar] [CrossRef] [PubMed]
- Osman, D.A.; Macías, M.A.; Al-Wahaibi, L.H.; Al-Shaalan, N.H.; Zondagh, L.S.; Joubert, J.; Garcia-Granda, S.; El-Emam, A.A. Structural insights and docking analysis of adamantane-linked 1,2,4-triazole derivatives as potential 11β-HSD1 inhibitors. Molecules 2021, 26, 5335. [Google Scholar] [CrossRef]
- Sakkiah, S.; Meganathan, C.; Sohn, Y.S.; Namadevan, S.; Lee, K.W. Identification of important chemical features of 11β-hydroxysteroid dehydrogenase type1 inhibitors: Application of ligand based virtual screening and density functional theory. Int. J. Mol. Sci. 2012, 13, 5138–5162. [Google Scholar] [CrossRef]
- Okazaki, S.; Takahashi, T.; Iwamura, T.; Nakaki, J.; Sekiya, Y.; Yagi, M.; Kumagai, H.; Sato, M.; Sakami, S.; Nitta, A.; et al. HIS-388, a novel orally active and long-acting 11β-hydroxysteroid dehydrogenase type 1 inhibitor, ameliorates insulin sensitivity and glucose intolerance in diet-induced obesity and nongenetic type 2 diabetic murine models. J. Pharmacol. Exp. Ther. 2014, 351, 181–189. [Google Scholar] [CrossRef]
- Goldberg, F.W.; Dossetter, A.G.; Scott, J.S.; Robb, G.R.; Boyd, S.; Groombridge, S.D.; Kemmitt, P.D.; Sjögren, T.; Gutierrez, P.M.; deSchoolmeester, J.; et al. Optimization of brain penetrant 11β-hydroxysteroid dehydrogenase type I inhibitors and in vivo testing in diet-induced obese mice. J. Med. Chem. 2014, 57, 970–986. [Google Scholar] [CrossRef]
- Studzińska, R.; Kupczyk, D.; Płaziński, W.; Baumgart, S.; Bilski, R.; Paprocka, R.; Kołodziejska, R. Novel 2-(adamantan-1-ylamino)thiazol-4(5H)-one derivatives and their inhibitory activity towards 11β-HSD1-synthesis, molecular docking and in vitro studies. Int. J. Mol. Sci. 2021, 22, 8609. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Grandhi, D.S.; Tawfik, S.S.; Al-Shaalan, N.H.; Elmorsy, M.A.; El-Emam, A.A.; Percino, M.J.; Thamotharan, S. Probing the effect of halogen substituents (Br, Cl, and F) on the non-covalent interactions in 1-(adamantan-1-yl)-3-arylthiourea derivatives: A theoretical study. ACS Omega 2021, 6, 4816–4830. [Google Scholar] [CrossRef]
- El-Emam, A.A.; Kumar, E.S.; Janani, K.; Al-Wahaibi, L.H.; Blacque, O.; El-Awady, M.I.; Al-Shaalan, N.H.; Percino, M.J.; Thamotharan, S. Quantitative assessment of the nature of noncovalent interactions in N-substituted-5-(adamantan-1-yl)- 1,3,4-thiadiazole-2-amines: Insights from crystallographic and QTAIM analysis. RSC Adv. 2020, 10, 9840–9853. [Google Scholar] [CrossRef]
- Al-Mutairi, A.A.; Alagappan, K.; Blacque, O.; Al-Alshaikh, M.A.; El-Emam, A.A.; Percino, M.J.; Thamotharan, S. Crystallographic and theoretical exploration of weak hydrogen bonds in arylmethyl N’-(adamantan-1-yl)piperidine-1-carbothioimidates and molecular docking analysis. ACS Omega 2021, 6, 27026–27037. [Google Scholar] [CrossRef] [PubMed]
- Al-Wahaibi, L.H.; Alsfouk, A.; El-Emam, A.A.; Blacque, O. Crystal structures and Hirshfeld surface analysis of 2-(adamantan-1-yl)-5-(4-fluorophenyl)-1,3,4-oxadiazole and 2-(adamantan-1-yl)-5-(4-chlorophenyl)-1,3,4-oxadiazole. Acta Crystallogr. Sect. E Crystallogr. Commun. 2019, 75, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Al-Wahaibi, L.H.; Sujay, S.; Muthu, G.G.; El-Emam, A.A.; Venkataramanan, N.S.; Al-Omary, F.A.M.; Ghabbour, H.A.; Percino, J.; Thamotharan, S. Theoretical investigations of two adamantane derivatives: A combined X-ray, DFT, QTAIM analysis and molecular docking. J. Mol. Struct. 2018, 1159, 233–245. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Alvarez, N.; Blacque, O.; Veiga, N.; Al-Mutairi, A.A.; El-Emam, A.A. Synthesis and structure insights of two novel broad-spectrum antibacterial candidates based on (E)-N’-[(heteroaryl)methylene]adamantane-1-carbohydrazides. Molecules 2020, 25, 1934. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.M.; Al-Wahaibi, L.H.; Shehatou, G.S.; El-Emam, A.A. Adamantane-linked isothiourea derivatives suppress the growth of experimental hepatocellular carcinoma via inhibition of TLR4-MyD88-NF-κB signaling. Am. J. Cancer Res. 2021, 11, 350–369. [Google Scholar]
- Al-Abdullah, E.S.; Asiri, H.H.; Lahsasni, S.; Habib, E.E.; Ibrahim, T.M.; El-Emam, A.A. Synthesis, antimicrobial, and anti-inflammatory activity, of novel S-substituted and N-substituted 5-(1-adamantyl)-1,2,4-triazole-3-thiols. Drug Des. Dev. Ther. 2014, 8, 505–518. [Google Scholar] [CrossRef]
- El-Emam, A.A.; Al-Tamimi, A.M.S.; Al-Omar, M.A.; Alrashood, K.A.; Habib, E.E. Synthesis and antimicrobial activity of novel 5-(1-adamantyl)-2-aminomethyl-4-substituted-1,2,4-triazoline-3-thiones. Eur. J. Med. Chem. 2013, 68, 96–102. [Google Scholar] [CrossRef]
- Al-Abdullah, E.S.; Al-Tuwaijri, H.M.; Hassan, H.M.; Haiba, M.E.; Habib, E.E.; El-Emam, A.A. Antimicrobial and hypoglycemic activities of novel N-Mannich bases derived from 5-(1-adamantyl)-4-substituted-1,2,4-triazoline-3-thiones. Int. J. Mol. Sci. 2014, 15, 22995–23010. [Google Scholar] [CrossRef]
- El-Emam, A.A.; El-Brollosy, N.R.; Ghabbour, H.A.; Quah, C.K.; Fun, H.K. 3-(Adamantan-1-yl)-4-ethyl-1H-1,2,4-triazole-5(4H)-thione. Acta Crystallogr. Sect. E Struct. Rep. Online 2012, 68, o1347. [Google Scholar] [CrossRef]
- Almutairi, M.S.; Al-Shehri, M.M.; El-Emam, A.A.; Ng, S.W.; Tiekink, E.R.T. 3-(Adamantan-1-yl)-4-(prop-2-en-1-yl)-1H-1,2,4-triazole-5(4H)-thione. Acta Crystallogr. Sect. E Struct. Rep. Online 2012, 68, o656. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Cremer, D.; Pople, J.A. General definition of ring puckering coordinates. J. Am. Chem. Soc. 1975, 97, 1354–1358. [Google Scholar] [CrossRef]
- Wzgarda-Raj, K.; Palusiak, M.; Wojtulewski, S.; Rybarczyk-Pirek, A.J. The role of sulfur interactions in crystal architecture: Experimental and quantum theoretical studies on hydrogen, halogen, and chalcogen bonds in trithiocyanuric acid-pyridine N-oxide co-crystals. CrystEngComm 2021, 23, 324–334. [Google Scholar] [CrossRef]
- Peng, K.; Pan, Y.; Li, J.; Khan, Z.; Fan, M.; Yin, H.; Tong, C.; Zhao, Y.; Liang, G.; Zheng, C. 11β-Hydroxysteroid dehydrogenase type 1(11β-HSD1) mediates insulin resistance through JNK activation in adipocytes. Sci. Rep. 2016, 6, 37160. [Google Scholar] [CrossRef] [PubMed]
- Dzyakanchuk, A.A.; Balázs, Z.; Nashev, L.G.; Amrein, K.E.; Odermatt, A. 11β-Hydroxysteroid dehydrogenase 1 reductase activity is dependent on a high ratio of NADPH/NADP(+) and is stimulated by extracellular glucose. Mol. Cell Endocrinol. 2009, 301, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.; Holmes, M.; Seckl, J. 11β-hydroxysteroid dehydrogenases: Intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 2013, 93, 1139–1206. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Tice, C.M.; Zhao, W.; Cacatian, S.; Ye, Y.J.; Singh, S.B.; Lindblom, P.; McKeever, B.M.; Krosky, P.M.; Kruk, B.A.; et al. Structure-based design and synthesis of 1,3-oxazinan-2-one inhibitors of 11β-hydroxysteroid dehydrogenase type 1. J. Med. Chem. 2011, 54, 6050–6062. [Google Scholar] [CrossRef]
- Patel, J.R.; Shuai, Q.; Dinges, J.; Winn, M.; Pliushchev, M.; Fung, S.; Monzon, K.; Chiou, W.; Wang, J.; Pan, L.; et al. Discovery of adamantane ethers as inhibitors of 11β-HSD-1: Synthesis and biological evaluation. Bioorg. Med. Chem. Lett. 2007, 17, 750–755. [Google Scholar] [CrossRef]
- Roberts, B.C.; Mancera, R.L. Ligand-protein docking with water molecules. J. Chem. Inf. Model. 2008, 48, 397–408. [Google Scholar] [CrossRef]
- Borba, J.V.V.B.; Alves, V.; Braga, R.; Korn, D.; Overdahl, K.; Silva, A.C.; Hall, S.; Overdahl, E.; Strickland, J.; Allen, D.; et al. STopTox: An in-silico alternative to animal testing for acute systemic and TOPical TOXicity. Environ. Health Perspect. 2022, 130, 27012. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Deodhar, M.; Al Rihani, S.B.; Arwood, M.J.; Darakjian, L.; Dow, P.; Turgeon, J.; Michaud, V. Mechanisms of CYP450 inhibition: Understanding drug-drug interactions due to mechanism-based inhibition in clinical practice. Pharmaceutics 2020, 12, 846. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Joubert, J.; Geldenhuys, W.J.; Van der Schyf, C.J.; Oliver, D.W.; Kruger, H.G.; Govender, T.; Malan, S.F. Polycyclic cage structures as lipophilic scaffolds for neuroactive drugs. ChemMedChem 2012, 7, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Webster, S.P.; McBride, A.; Binnie, M.; Sooy, K.; Seckl, J.R.; Andrew, R.; Pallin, T.D.; Hunt, H.J.; Perrior, T.R.; Ruffles, V.S.; et al. Selection and early clinical evaluation of the brain-penetrant 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) inhibitor UE2343 (Xanamem™). Br. J. Pharmacol. 2017, 174, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Sooy, K.; Noble, J.; McBride, A.; Binnie, M.; Yau, J.L.; Seckl, J.R.; Walker, B.R.; Webster, S.P. Cognitive and disease-modifying effects of 11β-hydroxysteroid dehydrogenase type 1 inhibition in male Tg2576 mice, a model of Alzheimer’s disease. Endocrinology 2015, 156, 4592–4603. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.C.; Reid, J.S. The analytical calculation of absorption in multifaceted crystals. Acta Crystallogr. Sect. A Found. Crystallogr. 1995, 51, 887–897. [Google Scholar] [CrossRef]
- CrysAlisPro, Version 1.171.40.68a; Rigaku Oxford Diffraction Ltd.: Yarnton, UK, 2019.
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H.A. complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. Mercury CSD 2.0—New features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J. CrystalExplorer17; University of Western Australia: Perth, WA, Australia, 2017. [Google Scholar]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Turner, M.J.; Grabowsky, S.; Jayatilaka, D.; Spackman, M.A. Accurate and efficient model energies for exploring intermolecular interactions in molecular crystals. J. Phys. Chem. Lett. 2014, 5, 4249–4255. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, C.F.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer model energies and energy frameworks: Extension to metal coordination compounds, organic salts, solvates and open-shell systems. IUCrJ 2017, 4, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE), Version 2020.09; Chemical Computing Group: Montreal, QC, Canada. Available online: http://www.chemcomp.com (accessed on 28 January 2020).
- Bhattacharya, D.; Cheng, J. 3Drefine: Consistent protein structure refinement by optimizing hydrogen bonding network and atomic-level energy minimization. Proteins 2013, 81, 119–131. [Google Scholar] [CrossRef]
- ACD/ChemSketch, Version 2020.2.1; Advanced Chemistry Development, Inc.: Toronto, ON, Canada, 2021. Available online: http://www.acdlabs.com (accessed on 28 January 2020).
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef]
- Lukac, I.; Wyatt, P.G.; Gilbert, I.H.; Zuccotto, F. Ligand binding: Evaluating the contribution of the water molecules network using the Fragment Molecular Orbital method. J. Comput.-Aided Mol. Des. 2008, 35, 1025–1036. [Google Scholar] [CrossRef]
- Corbeil, C.R.; Williams, C.I.; Labute, P. Variability in docking success rates due to dataset preparation. J. Comput.-Aided Mol. Des. 2012, 26, 775–786. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]
- Fassio, A.V.; Santos, L.H.; Silveira, S.A.; Ferreira, R.S.; de Melo-Minardi, R.C. nAPOLI: A graph-based strategy to detect and visualize conserved protein-ligand interactions in large-scale. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1317–1328. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, Version 2.0; Schrodinger, LLC: New York, NY, USA, 2015.
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef]

| Compound 1 | Compound 2 | Compound 3 | |
|---|---|---|---|
| Empirical formula | C22H35N5O2S | C24H34N6S | C25H34N6S |
| Formula weight | 433.61 | 438.63 | 450.64 |
| Temperature (K) | 160 (1) | 160 (1) | 160 (1) |
| Crystal system | Monoclinic | Monoclinic | Monoclinic |
| Space group | P21/c | P21 | P21/n |
| a, b, c (Å) | 13.73179 (19), 1.46565 (15), 14.2961 (2) | 6.33861 (5), 13.40028 (10), 13.82724 (9) | 11.10030 (10), 7.8744 (2), 23.7572 (2) |
| α, β, γ (°) | 90, 100.5825 (15), 90 | 90, 102.1174 (7), 90 | 90, 96.8490 (10), 90 |
| Volume/Å3 | 2212.54 (6) | 1148.307 (14) | 4680.06 (8) |
| Z | 4 | 2 | 8 |
| Radiation type | Cu Kα (λ = 1.54184 Å) | Cu Kα (λ = 1.54184 Å) | Cu Kα (λ = 1.54184 Å) |
| Calculated density (g/cm3) | 1.302 | 1.269 | 1.279 |
| µ (mm−1) | 1.527 | 1.426 | 1.415 |
| Crystal size (mm3) | 0.05 × 0.04 × 0.02 | 0.2 × 0.12 × 0.06 | 0.2 × 0.11 × 0.08 |
| Diffractometer | Rigaku OD XtaLAB Synergy, Dualflex, Pilatus 200K | Rigaku OD SuperNova/Atlas area-detector | |
| Tmin, Tmax | 0.481, 0.731 | 0.831, 0.933 | 0.806, 0.926 |
| 2Θ range for data collection (°) | 6.548–149 | 6.538–149 | 6.204–149.006 |
| Index ranges | −17 ≤ h ≤ 17, −6 ≤ k ≤ 14, −17 ≤ l ≤ 17 | −7 ≤ h ≤ 7, −16 ≤ k ≤ 16, −16 ≤ l ≤ 17 | −13 ≤ h ≤ 13, −22 ≤ k ≤ 22, −27 ≤ l ≤ 29 |
| Reflections collected | 24,276 | 23,690 | 48,761 |
| Independent reflections | 4529 [Rint = 0.0333, Rsigma = 0.0257] | 4681 [Rint = 0.0152, Rsigma = 0.0096] | 9565 [Rint = 0.0308, Rsigma = 0.0199] |
| Data/restraints/parameters | 4529/38/293 | 4681/65/301 | 9565/0/577 |
| Goodness-of-fit on F2 | 1.050 | 1.049 | 1.034 |
| Final R indexes [I ≥ 2σ (I)] | R1 = 0.0344, wR2 = 0.0878 | R1 = 0.0255, wR2 = 0.0702 | R1 = 0.0336, wR2 = 0.0864 |
| Final R indexes [all data] | R1 = 0.0397, wR2 = 0.0912 | R1 = 0.0259, wR2 = 0.0704 | R1 = 0.0406, wR2 = 0.0908 |
| Δρmax/Δρmin (e Å−3) | 0.26/−0.25 | 0.15/−0.16 | 0.22/−0.22 |
| H-atom treatment | H-atom parameters constrained | ||
| CCDC number | 2,053,088 | 2,053,089 | 2,053,090 |
| D-H⋯A | D-H | H⋯A | D⋯A | D-H⋯A |
|---|---|---|---|---|
| Compound 1 | ||||
| C5-H5B‧‧‧O2i | 0.99 | 2.40 | 3.2952(17) | 150 |
| C4-H4B⋯O1 (intramolecular) | 0.99 | 2.31 | 2.7330(18) | 105 |
| C7-H7A⋯O2 (intramolecular) | 0.99 | 2.37 | 2.7781(19) | 104 |
| Compound 2 | ||||
| C1-H1A⋯N5ii | 0.95 | 2.75 | 3.633(3) | 155 |
| C13-H13B⋯S1iii | 0.99 | 3.19 | 3.845(3) | 125 |
| C21-H21A⋯* Cg1iv | 0.99 | 2.72 | 3.683(3) | 165 |
| C23-H23A⋯Cg1v | 0.99 | 2.74 | 3.610(3) | 147 |
| Compound 3 | ||||
| C39-H39⋯N7vi | 0.95 | 2.74 | 3.599(3) | 151 |
| C8-H8B⋯N11 between conformers | 0.99 | 2.75 | 3.712(3) | 163 |
| C35-H35B″ïS1 between conformers | 0.99 | 2.80 | 3.649(3) | 144 |
| C10-H10A⋯S2vii | 0.99 | 2.76 | 3.642(3) | 150 |
| C50-H50A⋯Cg1viii | 0.99 | 2.78 | 3.501(2) | 130 |
| C24-H24B⋯Cg1ix | 0.99 | 2.72 | 3.537(2) | 140 |
| N | Symop | R | Eele | Epol | Edis | Erep | Etot |
|---|---|---|---|---|---|---|---|
| Compound 1 | |||||||
| 2 | x, −y + 1/2, z + 1/2 C5-H5B⋯O2i | 7.25 | −16.3 | −8.4 | −53.6 | 41.4 | −44.5 |
| 1 | −x, −y, −z pair of inversion related molecules | 4.72 | −36.9 | −6.7 | −151.9 | 128.0 | −97.2 |
| 1 | −x, −y, −z interaction between pairs of inversion related molecules | 7.08 | −15.2 | −2.4 | −57.8 | 35.8 | −46.0 |
| 2 | −x, y + 1/2, −z + 1/2 | 14.19 | −6.3 | −1.6 | −27.0 | 23.2 | −17.0 |
| 2 | x, −y + 1/2, z + 1/2 | 14.32 | −9.5 | −1.7 | −24.1 | 20.8 | −19.5 |
| 2 | x, y, z | 13.73 | −5.2 | −0.8 | −32.3 | 22.0 | −20.6 |
| 2 | −x, y + 1/2, −z + 1/2 | 8.59 | −8.7 | −2.0 | −31.4 | 16.7 | −27.7 |
| 2 | −x, y + 1/2, −z + 1/2 | 9.90 | −8.9 | −2.8 | −18.6 | 14.8 | −18.5 |
| 1 | −x, −y, −z | 15.36 | 0.4 | −0.2 | −11.1 | 7.2 | −5.0 |
| Compound 2 | |||||||
| 2 | −x, y + 1/2, −z C1-H1A⋯N5ii along [010] | 9.97 | −21.9 | −5.6 | −44.6 | 27.5 | −49.1 |
| 2 | x, y, z along [100] | 6.34 | −14.1 | −9.9 | −73.7 | 62.2 | −48.0 |
| 2 | −x, y + 1/2, −z C21-H21A⋯Cg1iii along [010] | 9.09 | −27.4 | −6.4 | −69.1 | 31.3 | −74.5 |
| 2 | x, y, z | 13.40 | −11.1 | −1.9 | −37.4 | 14.6 | −36.7 |
| 2 | −x, y + 1/2, −z | 11.23 | −12.0 | −4.6 | −24.2 | 21.3 | −24.0 |
| 2 | −x, y + 1/2, −z | 10.30 | −6.5 | −3.5 | −33.3 | 22.0 | −24.9 |
| Compound 3 | |||||||
| 1 | Between conformers | 6.56 | −5.1 | −1.2 | −37.1 | 21.7 | −25.2 |
| 1 | Between conformers | 6.66 | −3.1 | −0.9 | −14.5 | 7.0 | −12.3 |
| 2 | −x + 1/2, y + 1/2, −z + 1/2 | 12.00 | −3.1 | −0.9 | −14.5 | 7.0 | −12.3 |
| 2 | −x + 1/2, y + 1/2, −z + 1/2 | 10.24 | −5.1 | −1.2 | −37.1 | 21.7 | −25.2 |
| 1 | Between conformers | 11.93 | −7.1 | −4.6 | −45.6 | 20.3 | −38.1 |
| 1 | −x, −y, −z | 11.86 | −12.6 | −3.1 | −10.3 | 6.6 | −20.6 |
| Compounds | Binding Affinity Scores (kcal/mol) | Hydrogen Bond Interactions | Aromatic Stacking Interactions | Water Bridge Interactions |
|---|---|---|---|---|
| 4YQ | −8.48 | Thr124 b Ser170 c Tyr183 c Leu217 c Asp259 c | None c | Thr122 b Thr124 a Leu217 b Gln234 b Ala236 b Ser260 b |
| Compound 1 | −8.46 | Ser170 a Tyr183 c Leu217 b | Tyr183 b | None c |
| Compound 2 | −8.92 | Ser170 a Tyr183 c Leu217 c | Tyr177 b Tyr183 b | None c |
| Compound 3 | −7.50 | Ser170 a Tyr183 c Leu217 c | Tyr177 b Tyr183 b | None c |
| Compound 1 | ||||||
|---|---|---|---|---|---|---|
| Toxicity and Target Classification | ProTox-II | Probability | STopTox | Confidence | admetSAR | Probability |
| Oral LD50 (mg/kg) a | 1000 | - | - | - | - | - |
| Oral Toxicity class | IV b | - | - | - | II c | 0.45 |
| Acute oral toxicity | - | - | Yes | 78% | - | - |
| Hepatotoxicity | No | 0.60 | - | - | No | 0.70 |
| Carcinogenicity | No | 0.55 | - | - | No | 0.90 |
| Immunotoxicity | No | 0.99 | - | - | - | - |
| Mutagenicity | No | 0.62 | - | - | No | 0.51 |
| Cytotoxicity | No | 0.73 | - | - | - | - |
| Acute inhalation toxicity | - | - | No | 54% | - | - |
| Acute dermal toxicity | - | - | No | 64% | - | - |
| Eye irritation and corrosion | - | - | Yes | 71% | No | >0.95 |
| Skin irritation and corrosion | - | - | No | 60% | - | - |
| Skin sensitization | - | - | No | 60% | - | - |
| Compound 2 | ||||||
| Oral LD50 (mg/kg) a | 162 | - | - | - | - | - |
| Oral Toxicity class | III b | - | - | - | III c | 0.45 |
| Acute oral toxicity | - | - | Yes | 78% | - | - |
| Hepatotoxicity | No | 0.73 | - | - | Yes | 0.53 |
| Carcinogenicity | Yes | 0.51 | - | - | No | 0.94 |
| Immunotoxicity | No | 0.95 | - | - | - | - |
| Mutagenicity | No | 0.59 | - | - | No | 0.52 |
| Cytotoxicity | No | 0.77 | - | - | - | - |
| Acute inhalation toxicity | - | - | Yes | 53% | - | - |
| Acute dermal toxicity | - | - | No | 65% | - | - |
| Eye irritation and corrosion | - | - | Yes | 54% | No | >0.97 |
| Skin irritation and corrosion | - | - | Yes | 50% | - | - |
| Skin sensitization | - | - | No | 60% | - | - |
| Compound 3 | ||||||
| Oral LD50 (mg/kg) a | 162 | - | - | - | - | - |
| Oral Toxicity class | III b | - | - | - | III c | 0.52 |
| Acute oral toxicity | - | - | Yes | 65% | - | - |
| Hepatotoxicity | No | 0.72 | - | - | Yes | 0.58 |
| Carcinogenicity | Yes | 0.50 | - | - | No | 0.94 |
| Immunotoxicity | No | 0.96 | - | - | - | - |
| Mutagenicity | No | 0.60 | - | - | No | 0.55 |
| Cytotoxicity | No | 0.77 | - | - | - | - |
| Acute inhalation toxicity | - | - | Yes | 55% | - | - |
| Acute dermal toxicity | - | - | No | 66% | - | - |
| Eye irritation and corrosion | - | - | Yes | 62% | No | >0.95 |
| Skin irritation and corrosion | - | - | Yes | 50% | - | - |
| Skin sensitization | - | - | Yes | 60% | - | - |
| Compound 1 | ||
|---|---|---|
| Pharmacokinetic Target Classification | SwissADME | admetSAR |
| Human oral bioavailability | 0.55 | Yes |
| GI a absorption | High | Yes |
| Caco-2 permeability b | - | No |
| BBB c | Yes | Yes |
| P-gp d substrate | No | No |
| P-gp d inhibitor | - | No |
| CYP1A2 e inhibitor | No | No |
| CYP2C9 e inhibitor | Yes | No |
| CYP2C19 e inhibitor | Yes | No |
| CYP2D6 e inhibitor | Yes | No |
| CYP3A4 e inhibitor | Yes | Yes |
| CYP2C9 e Substrate | - | No |
| CYP2D6 e Substrate | - | No |
| CYP3A4 e Substrate | - | Yes |
| Compound 2 | ||
| Human oral bioavailability | 0.55 | Yes |
| GI a absorption | High | Yes |
| Caco-2 permeability b | - | No |
| BBB c | Yes | Yes |
| P-gp d substrate | No | Yes |
| P-gp d inhibitor | - | No |
| CYP1A2 e inhibitor | No | No |
| CYP2C9 e inhibitor | Yes | No |
| CYP2C19 e inhibitor | Yes | Yes |
| CYP2D6 e inhibitor | Yes | No |
| CYP3A4 e inhibitor | Yes | Yes |
| CYP2C9 e substrate | - | No |
| CYP2D6 e substrate | - | No |
| CYP3A4 e substrate | - | Yes |
| Compound 3 | ||
| Human oral bioavailability | 0.55 | Yes |
| GI a absorption | High | Yes |
| Caco-2 permeability b | - | No |
| BBB c | Yes | Yes |
| P-gp d substrate | No | No |
| P-gp d inhibitor | - | Yes |
| CYP1A2 e inhibitor | No | No |
| CYP2C9 e inhibitor | Yes | Yes |
| CYP2C19 e inhibitor | Yes | Yes |
| CYP2D6 e inhibitor | Yes | No |
| CYP3A4 e inhibitor | Yes | Yes |
| CYP2C9 e substrate | - | No |
| CYP2D6 e substrate | - | No |
| CYP3A4 e substrate | - | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Wahaibi, L.H.; Macías, M.A.; Blacque, O.; Zondagh, L.S.; Joubert, J.; Thamotharan, S.; Percino, M.J.; Mohamed, A.A.B.; El-Emam, A.A. Weak Noncovalent Interactions in Three Closely Related Adamantane-Linked 1,2,4-Triazole N-Mannich Bases: Insights from Energy Frameworks, Hirshfeld Surface Analysis, In Silico 11β-HSD1 Molecular Docking and ADMET Prediction. Molecules 2022, 27, 7403. https://doi.org/10.3390/molecules27217403
Al-Wahaibi LH, Macías MA, Blacque O, Zondagh LS, Joubert J, Thamotharan S, Percino MJ, Mohamed AAB, El-Emam AA. Weak Noncovalent Interactions in Three Closely Related Adamantane-Linked 1,2,4-Triazole N-Mannich Bases: Insights from Energy Frameworks, Hirshfeld Surface Analysis, In Silico 11β-HSD1 Molecular Docking and ADMET Prediction. Molecules. 2022; 27(21):7403. https://doi.org/10.3390/molecules27217403
Chicago/Turabian StyleAl-Wahaibi, Lamya H., Mario A. Macías, Olivier Blacque, Luke S. Zondagh, Jacques Joubert, Subbiah Thamotharan, María Judith Percino, Ahmed A. B. Mohamed, and Ali A. El-Emam. 2022. "Weak Noncovalent Interactions in Three Closely Related Adamantane-Linked 1,2,4-Triazole N-Mannich Bases: Insights from Energy Frameworks, Hirshfeld Surface Analysis, In Silico 11β-HSD1 Molecular Docking and ADMET Prediction" Molecules 27, no. 21: 7403. https://doi.org/10.3390/molecules27217403
APA StyleAl-Wahaibi, L. H., Macías, M. A., Blacque, O., Zondagh, L. S., Joubert, J., Thamotharan, S., Percino, M. J., Mohamed, A. A. B., & El-Emam, A. A. (2022). Weak Noncovalent Interactions in Three Closely Related Adamantane-Linked 1,2,4-Triazole N-Mannich Bases: Insights from Energy Frameworks, Hirshfeld Surface Analysis, In Silico 11β-HSD1 Molecular Docking and ADMET Prediction. Molecules, 27(21), 7403. https://doi.org/10.3390/molecules27217403





