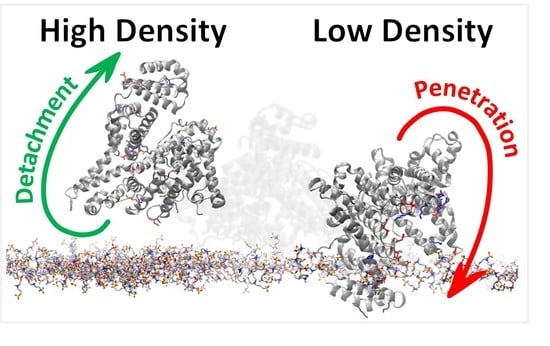
Distinct Antifouling Mechanisms on Different Chain Densities of Zwitterionic Polymers
Abstract
1. Introduction
2. Computational Methods
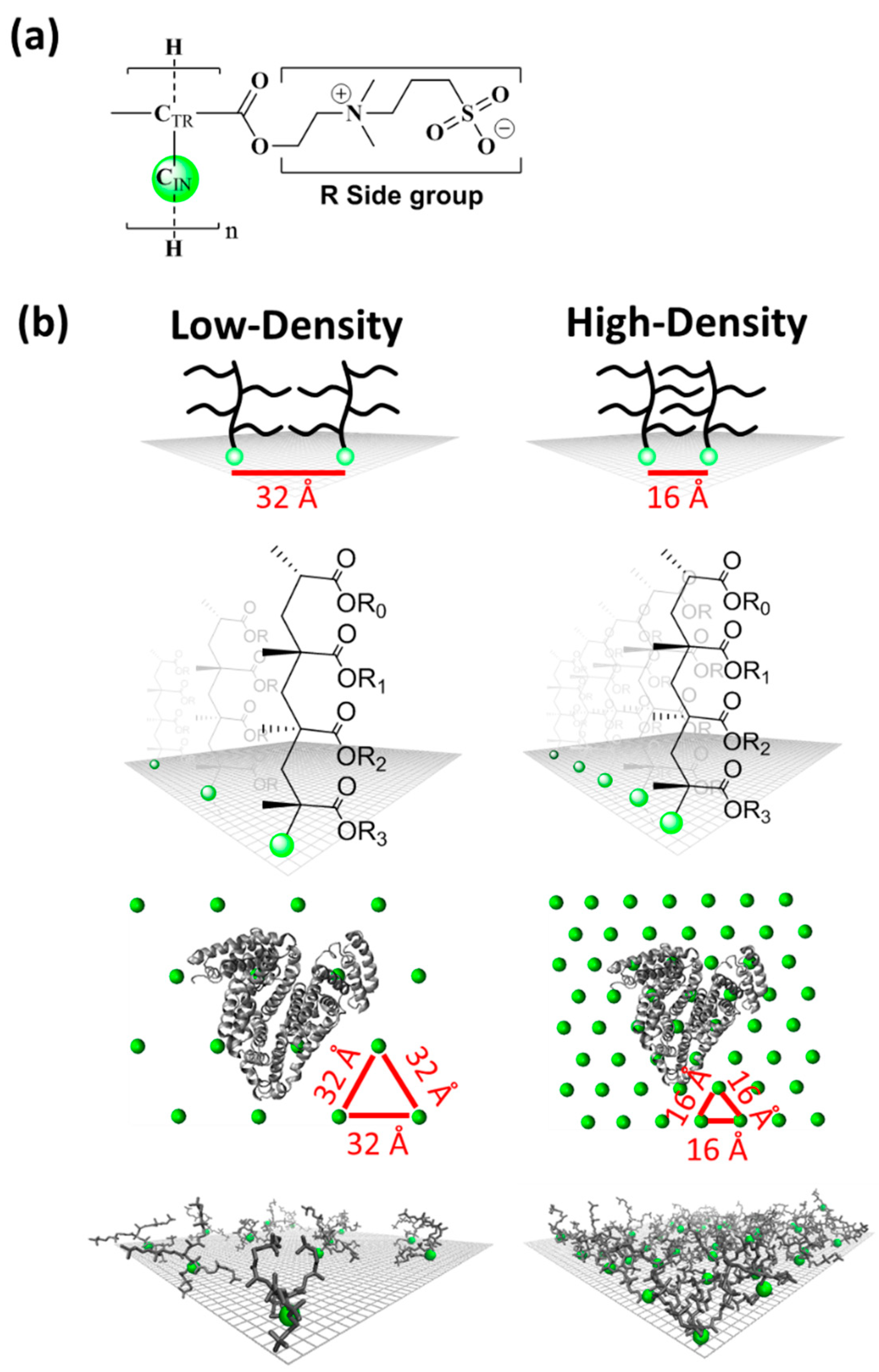
2.1. Constructions of Low-Density and High-Density Polymer Chain Surfaces
2.2. Constructions of BSA Protein on Poly(SPE) Low-Density and High-Density Polymer Chain Surfaces
2.3. Integration of the Poly(SPE) Chains within CHARMM Force-Field
2.4. All-Atom Explicit Molecular Dynamic (MD) Simulations Protocol
2.5. Root-Mean Square Deviation (RMSD) Analyses
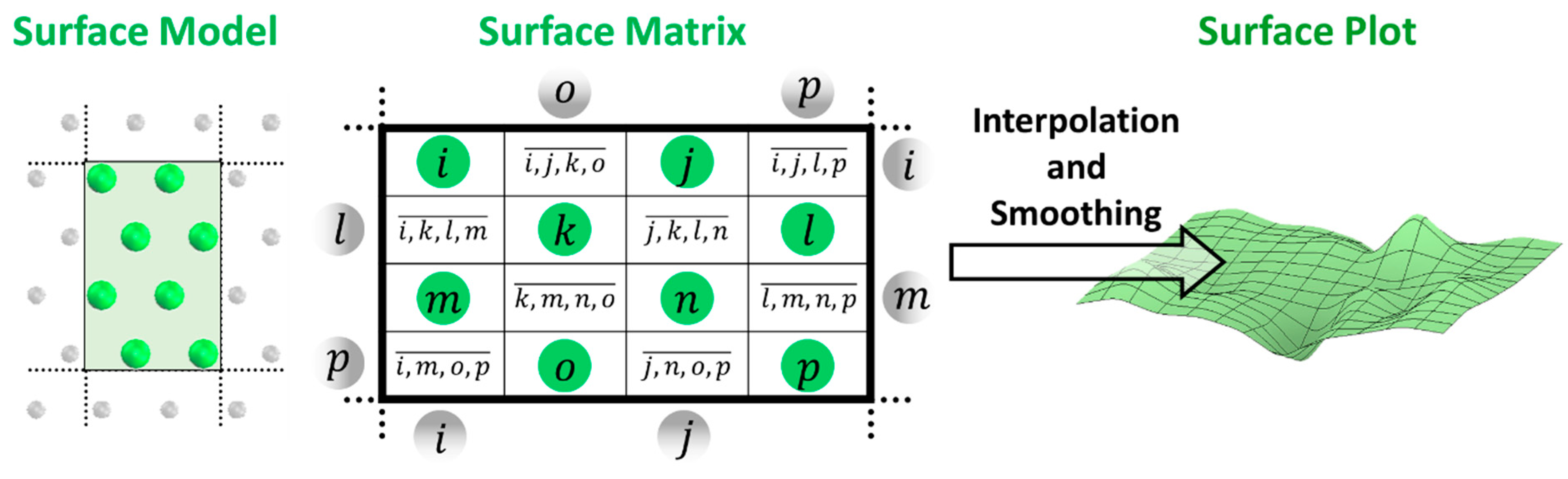
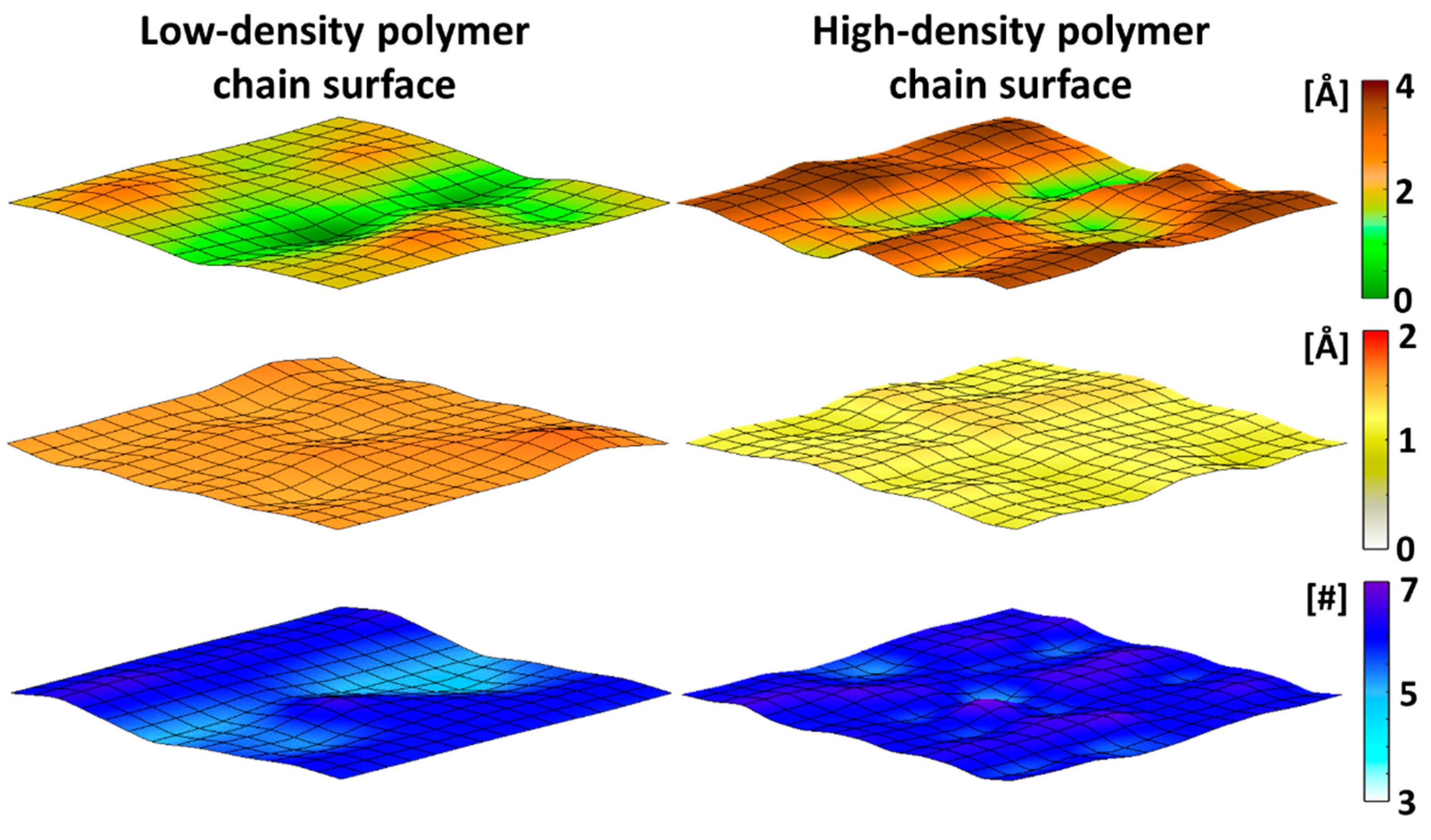
2.6. Characterization of Topology Heights of the Polymer Chains on the Surfaces
2.7. Characterization of the Root Mean Square Fluctuation (RMSF) of the Polymer Chains on the Surfaces
2.8. Characterization of the Water Hydration of the Polymer Chains on the Surfaces
2.9. Production of Surface Plots Images for Characterizations of Heights, RMSF and Water Hydration of the Polymer Chains on the Surfaces
2.10. Estimation of Detachment and/or Adsorption of BSA on the Polymer Chain Surfaces
2.11. Electrostatic Interactions Analyses
2.12. Hydrogen Bond Interactions Analyses
3. Results and Discussion
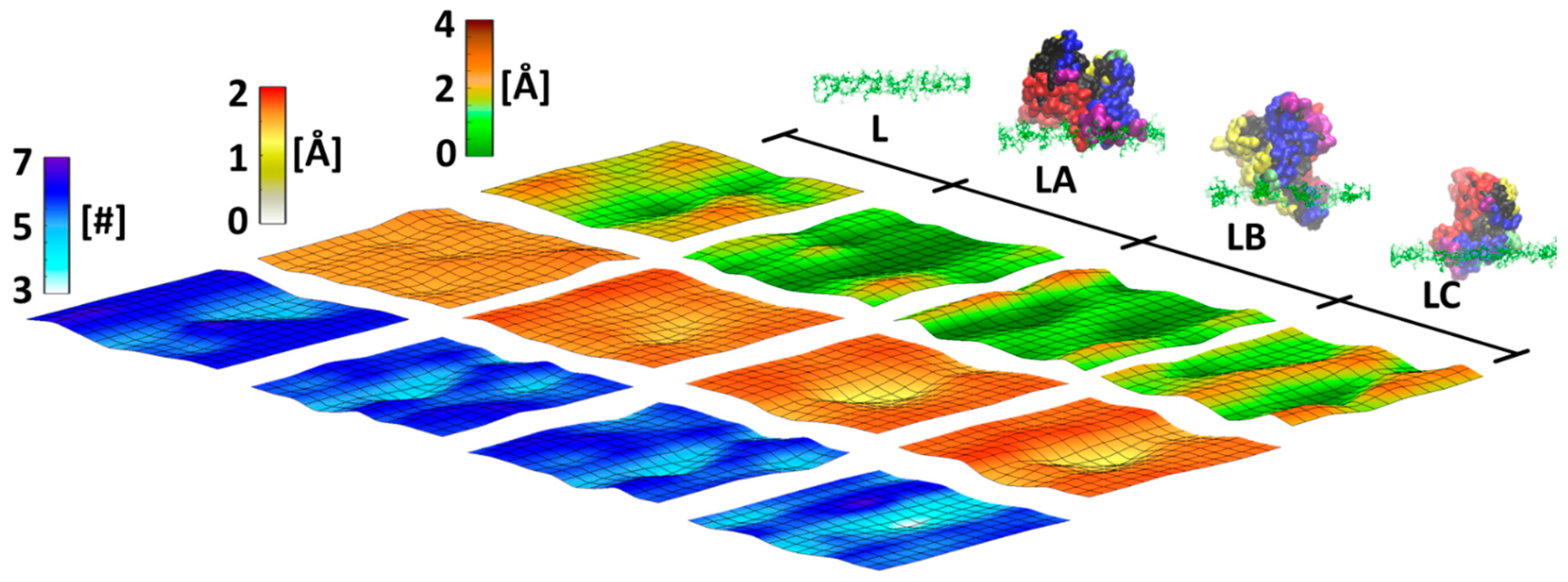
3.1. Characterization of Surfaces with High- and Low-Density Polymer Chain Coating
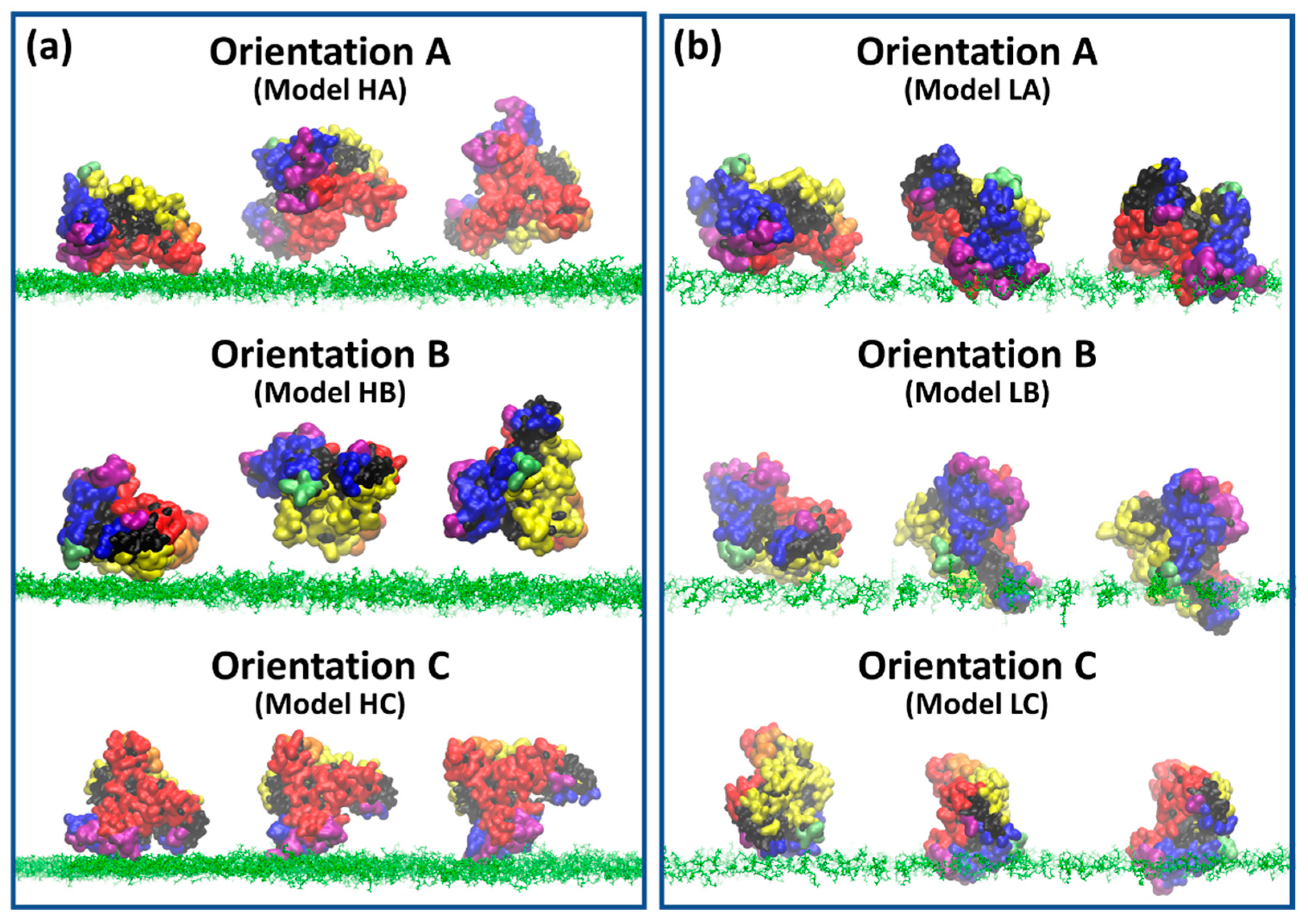
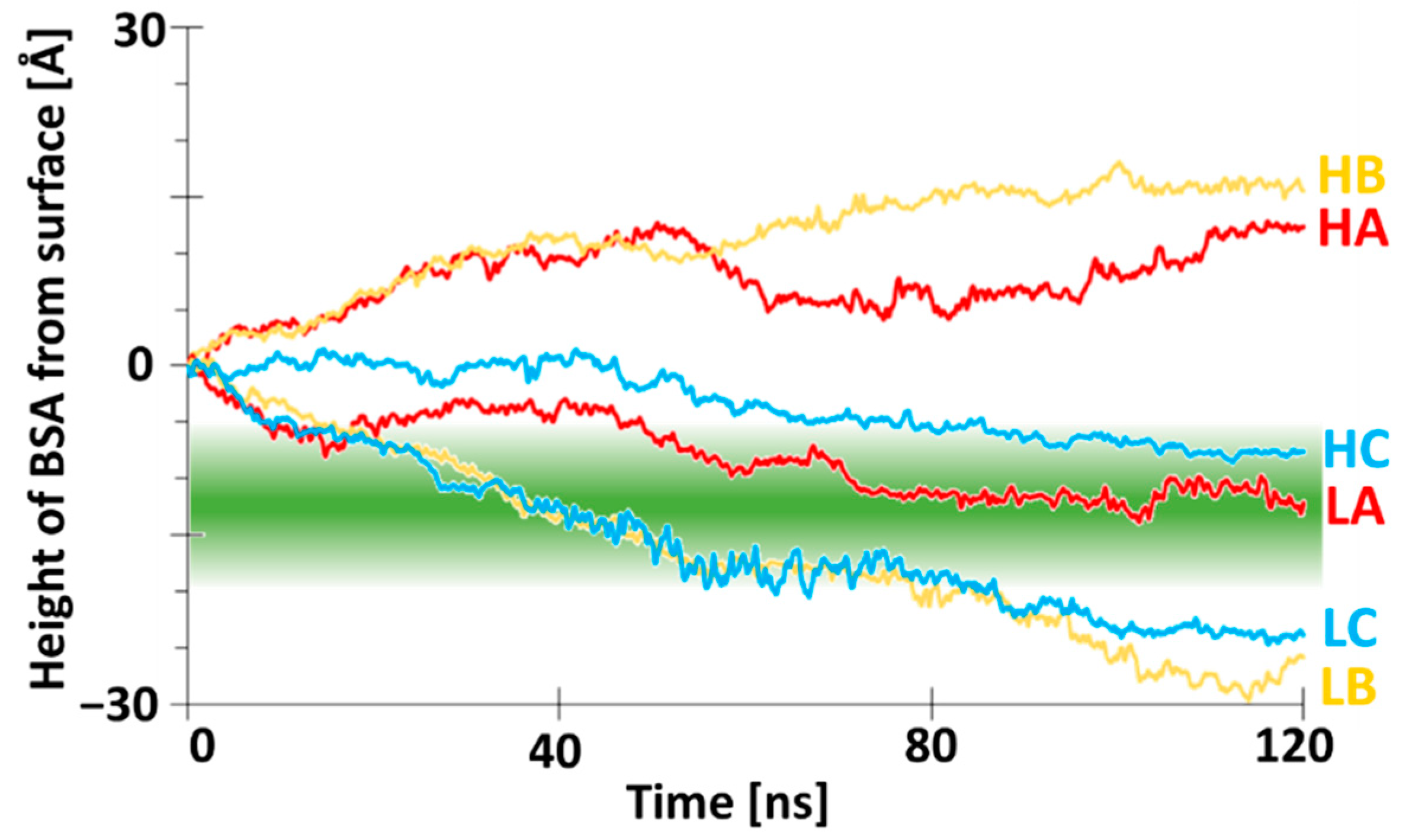
3.2. BSA Does Not Penetrate at High-Density Polymer Chains but Penetrates at Low-Density Polymer Chains
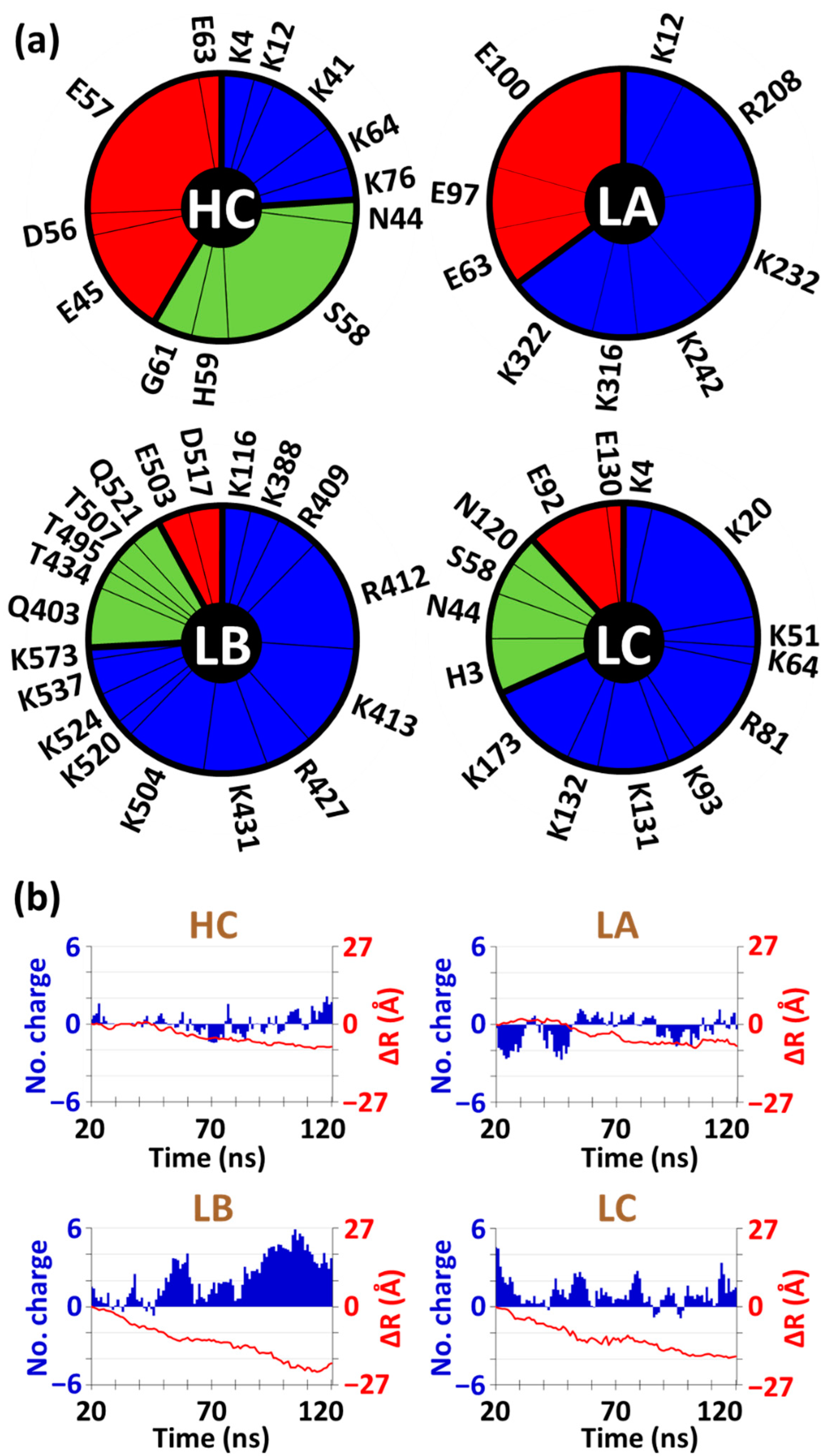
3.3. Two Distinct Mechanisms of BSA at High-Density Polymer Chain Coating Surface
3.4. Distinct Adhesion Mechanisms of BSA at Low-Density Polymer Chain Coating Surface
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hatakeyama, E.S.; Ju, H.; Gabriel, C.J.; Lohr, J.L.; Bara, J.E.; Noble, R.D.; Freeman, B.D.; Gin, D.L. New protein-resistant coatings for water filtration membranes based on quaternary ammonium and phosphonium polymers. J. Membr. Sci. 2009, 330, 104–116. [Google Scholar] [CrossRef]
- Leckband, D.; Sheth, S.; Halperin, A. Grafted poly(ethylene oxide) brushes as nonfouling surface coatings. J. Biomater. Sci. Polym. Ed. 1999, 10, 1125–1147. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Sakiyama, T.; Imamura, K. On the adsorption of proteins on solid surfaces, a common but very complicated phenomenon. J. Biosci. Bioeng. 2001, 91, 233–244. [Google Scholar] [CrossRef]
- Charnley, M. Designed polymer structures with antifouling–antimicrobial properties Reactive and Functional Polymers. React. Funct. Polym. 2011, 71, 329–334. [Google Scholar] [CrossRef]
- Bhoj, Y.; Tharmavaram, M.; Rawtani, D. A comprehensive approach to antifouling strategies in desalination, marine environment, and wastewater treatment. Chem. Phys. Impact 2021, 2, 100008. [Google Scholar] [CrossRef]
- Rodríguez-Hernández, J. Polymers against Microorganisms; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Fu, J.; Zhang, H.; Guo, Z.; Feng, D.Q.; Thiyagarajan, V.; Yao, H. Combat biofouling with microscopic ridge-like surface morphology: A bioinspired study. J. R. Soc. Interface 2018, 15, 20170823. [Google Scholar] [CrossRef]
- Yang, W.J.; Neoh, K.G.; Kang, E.T.; Teo, S.L.M.; Rittschof, D. Polymer brush coatings for combating marine biofouling. Prog. Polym. Sci. 2014, 39, 1017–1042. [Google Scholar] [CrossRef]
- Krishnan, S.; Weinman, C.J.; Ober, C.K. Advances in polymers for anti-biofouling surfaces. J. Mater. Chem. 2008, 18, 3405–3413. [Google Scholar] [CrossRef]
- Ostuni, E.; Chapman, R.G.; Holmlin, R.E.; Takayama, S.; Whitesides, G.M. A Survey of Structure-Property Relationships of Surfaces that Resist the Adsorption of Proteins. Langmuir 2001, 17, 5605–5620. [Google Scholar] [CrossRef]
- He, M.; Gao, K.; Zhou, L.; Jiao, Z.; Wu, M.; Cao, J.; You, X.; Cai, Z.; Su, Y.; Jiang, Z. Zwitterionic materials for antifouling membrane surface construction. Acta Biomater. 2016, 40, 142–152. [Google Scholar] [CrossRef]
- Chen, S.; Li, L.; Zhao, C.; Zheng, J. Surface hydration: Principles and applications toward low-fouling/nonfouling biomaterials. Polymer 2010, 51, 5283–5293. [Google Scholar] [CrossRef]
- Ye, L.; Zhang, Y.; Wang, Q.; Zhou, X.; Yang, B.; Ji, F.; Dong, D.; Gao, L.; Cui, Y.; Yao, F. Physical Cross-Linking Starch-Based Zwitterionic Hydrogel Exhibiting Excellent Biocompatibility, Protein Resistance, and Biodegradability. ACS Appl. Mater. Interfaces 2016, 8, 15710–15723. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.; Aiba, Y.; Morimoto, N.; Nakabayashi, N.; Ishihara, K. Semi-interpenetrating polymer networks composed of biocompatible phospholipid polymer and segmented polyurethane. J. Biomed. Mater. Res. 2000, 51, 701–708. [Google Scholar] [CrossRef]
- Hirota, K.; Murakami, K.; Nemoto, K.; Miyake, Y. Coating of a surface with 2-methacryloyloxyethyl phosphorylcholine (MPC) co-polymer significantly reduces retention of human pathogenic microorganisms. FEMS Microbiol. Lett. 2005, 248, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Doncom, K.E.B.; Warren, N.J.; Armes, S.P. Polysulfobetaine-based diblock copolymer nano-objects via polymerization-induced self-assembly. Polmer Chem. 2015, 6, 7264–7273. [Google Scholar] [CrossRef]
- Liu, Q.S.; Li, W.C.; Wang, H.; Liu, L.Y. A Facile Method of Using Sulfobetaine-Containing Copolymers for Biofouling Resistance. J. Appl. Polym. Sci. 2014, 131, 40789. [Google Scholar] [CrossRef]
- Men, Y.; Peng, S.; Yang, P.; Jiang, Q.; Zhang, Y.; Shen, B.; Dong, P.; Pang, Z.; Yang, W. Biodegradable Zwitterionic Nanogels with Long Circulation for Antitumor Drug Delivery. ACS Appl. Mater. Interfaces 2018, 10, 23509–23521. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Yu, B.; Wang, S.; Shen, Y.Q.; Cong, H.L. Review of the research on anti-protein fouling coatings materials. Prog. Org. Coat. 2020, 147, 105860. [Google Scholar] [CrossRef]
- Haro-Mares, N.B.; Meza-Contreras, J.C.; Lopez-Dellamary Toral, F.A.; Gonzalez-Cruz, R.; Silva-Guzman, J.A.; Manriquez-Gonzalez, R. A Simplified Method of Synthesis to Obtain Zwitterionic Cellulose under Mild Conditions with Active Ionic Moieties. Molecules 2020, 25, 3065. [Google Scholar] [CrossRef]
- Liu, L.; Li, W.; Liu, Q. Recent development of antifouling polymers: Structure, evaluation, and biomedical applications in nano/micro-structures. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014, 6, 599–614. [Google Scholar] [CrossRef]
- Ziemann, E.; Coves, T.; Levin, O.; Bernstein, R. Zwitterion Polymer Brushes on Porous Membranes: Characterization, Tribology, Performance, and the Effect of Electrolyte Anions. ACS Appl. Polym. Mater. 2020, 2, 4613–4625. [Google Scholar] [CrossRef]
- Lorusso, E.; Ali, W.; Leniart, M.; Gebert, B.; Oberthur, M.; Gutmann, J.S. Tuning the Density of Zwitterionic Polymer Brushes on PET Fabrics by Aminolysis: Effect on Antifouling Performances. Polymers 2019, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.T.; Leckband, D.E. Protein Adsorption on Grafted Zwitterionic Polymers Depends on Chain Density and Molecular Weight. Adv. Funct. Mater. 2020, 30, 2000757. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, D.; Ren, B.; Gong, X.; Xu, L.; Feng, Z.Q.; Chang, Y.; He, Y.; Zheng, J. Molecular simulations and understanding of antifouling zwitterionic polymer brushes. J. Mater. Chem. B 2020, 8, 3814–3828. [Google Scholar] [CrossRef]
- Xiang, Y.; Xu, R.G.; Leng, Y. Molecular Simulations of the Hydration Behavior of a Zwitterion Brush Array and Its Antifouling Property in an Aqueous Environment. Langmuir 2018, 34, 2245–2257. [Google Scholar] [CrossRef]
- Belinskaia, D.A.; Voronina, P.A.; Shmurak, V.I.; Vovk, M.A.; Batalova, A.A.; Jenkins, R.O.; Goncharov, N.V. The Universal Soldier: Enzymatic and Non-Enzymatic Antioxidant Functions of Serum Albumin. Antioxidants 2020, 9, 966. [Google Scholar] [CrossRef]
- Oshiba, Y.; Harada, Y.; Yamaguchi, T. Precise surface modification of porous membranes with well-defined zwitterionic polymer for antifouling applications. J. Membr. Sci. 2021, 619, 118772. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, Y.; Li, Y. Control of Protein (BSA) Fouling in RO System by Antiscalants. J. Memb. Sci. 2010, 364, 372–379. [Google Scholar] [CrossRef]
- Hildebrand, V.; Laschewsky, A.; Pach, M.; Muller-Buschbaum, P.; Papadakis, C.M. Effect of the zwitterion structure on the thermo-responsive behaviour of poly(sulfobetaine methacrylates). Polym. Chem. 2017, 8, 310–322. [Google Scholar] [CrossRef]
- Isobe, Y.; Yamada, K.; Nakano, T.; Okamoto, Y. Stereospecific free-radical polymerization of methacrylates using fluoroalcohols as solvents. Macromolecules 1999, 32, 5979–5981. [Google Scholar] [CrossRef]
- Rao, Z.Z.; Takayanagi, M.; Nagaoka, M. Ab Initio Quantitative Prediction of Tacticity in Radical Polymerization of Poly(methyl methacrylate) by a Molecular Simulation Technique with the Conformation Indexing for Multiple Transition States. J. Phys. Chem. C 2020, 124, 16895–16901. [Google Scholar] [CrossRef]
- Tanaka, Y.; Noguchi, K.; Shibata, T.; Okada, A. Effect of High-Pressure on Radical Polymerizability of Acrylamides and Methacrylamides Capable of Forming Stable Radicals. Macromolecules 1978, 11, 1017–1021. [Google Scholar] [CrossRef]
- Zheng, J.; He, Y.; Chen, S.; Li, L.; Bernards, M.T.; Jiang, S. Molecular simulation studies of the structure of phosphorylcholine self-assembled monolayers. J. Chem. Phys. 2006, 125, 174714. [Google Scholar] [CrossRef] [PubMed]
- Majorek, K.A.; Porebski, P.J.; Dayal, A.; Zimmerman, M.D.; Jablonska, K.; Stewart, A.J.; Chruszcz, M.; Minor, W. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012, 52, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Kalé, L.; Skeel, R.; Bhandarkar, M.; Brunner, R.; Gursoy, A.; Krawetz, N.; Phillips, J.; Shinozaki, A.; Varadarajan, K.; Schulten, K. NAMD2: Greater Scalability for Parallel Molecular Dynamics. J. Comput. Phys. 1999, 151, 283–312. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Mahoney, M.W.; Jorgensen, W.L. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000, 112, 8910–8922. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.H.; Pastor, R.W.; Brooks, B.R. Constant-Pressure Molecular-Dynamics Simulation—The Langevin Piston Method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Tu, K.; Tobias, D.J.; Klein, M.L. Constant pressure and temperature molecular dynamics simulation of a fully hydrated liquid crystal phase dipalmitoylphosphatidylcholine bilayer. Biophys. J. 1995, 69, 2558–2562. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Torrens-Fontanals, M.; Stepniewski, T.M.; Aranda-Garcia, D.; Morales-Pastor, A.; Medel-Lacruz, B.; Selent, J. How Do Molecular Dynamics Data Complement Static Structural Data of GPCRs. Int. J. Mol. Sci. 2020, 21, 5933. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.mathworks.com/help/matlab/ref/interp2.html (accessed on 12 September 2022).
- Available online: https://www.mathworks.com/help/curvefit/smooth.html (accessed on 12 September 2022).
- Bao, X.; She, Q.; Long, W.; Wu, Q. Ammonium ultra-selective membranes for wastewater treatment and nutrient enrichment: Interplay of surface charge and hydrophilicity on fouling propensity and ammonium rejection. Water Res. 2021, 190, 116678. [Google Scholar] [CrossRef]
- Wang, S.Y.; Fang, L.F.; Zhu, B.K.; Matsuyama, H. Enhancing the antifouling property of polymeric membrane via surface charge regulation. J. Colloid Interface Sci. 2021, 593, 315–322. [Google Scholar] [CrossRef]
- Wang, X.L.; Wei, J.F.; Dai, Z.; Zhao, K.Y.; Zhang, H. Preparation and characterization of negatively charged hollow fiber nanofiltration membrane by plasma-induced graft polymerization. Desalination 2012, 286, 138–144. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, Y.; Yu, H.; Zhang, X.T.; Tian, X.X.; Fu, S.C.; Zhang, H.F. Improving the Biofouling Resistance of Polyamide Thin-Film Composite Membrane via Grafting Polyacrylamide Brush on the Surface by In-Situ Atomic Transfer Radical Polymerization. J. Membr. Sci. 2021, 629, 119283. [Google Scholar] [CrossRef]
- Wei, X.; Wang, Z.; Zhang, Z.; Wang, J.; Wang, S. Surface Modification of Commercial Aromatic Polyamide Reverse Osmosis Membranes by Graft Polymerization of 3-allyl-5,5-Dimethylhydantoin. J. Membr. Sci. 2010, 351, 222–233. [Google Scholar] [CrossRef]
- Castrillon, S.R.V.; Lu, X.L.; Shaffer, D.L.; Elimelech, M. Amine Enrichment and Poly(ethylene glycol) (PEG) Surface Modification of Thin-Film Composite Forward Osmosis Membranes for Organic Fouling Control. J. Membr. Sci. 2014, 450, 331–339. [Google Scholar] [CrossRef]
- Regev, C.; Belfer, S.; Holenberg, M.; Fainstein, R.; Parola, A.H.; Kasher, R. Fabrication of poly(ethylene glycol) particles with a micro-spherical morphology on polymeric fibers and its application in high flux water filtration. Sep. Purif. Technol. 2019, 210, 729–736. [Google Scholar] [CrossRef]
- Shan, X.Y.; Li, S.L.; Fu, W.M.; Hu, Y.L.; Gong, G.H.; Hu, Y.X. Preparation of high performance TFC RO membranes by surface grafting of small-molecule zwitterions. J. Membr. Sci. 2020, 608, 118209. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, X.Y.; Xie, M.; Wu, H.C.; Yoshioka, T.; Saeki, D.; Matsuyama, H. Antifouling thin-film composite membranes with multi-defense properties by controllably constructing amphiphilic diblock copolymer brush layer. J. Membr. Sci. 2020, 614, 118515. [Google Scholar] [CrossRef]
- Liu, P.S.; Chen, Q.; Liu, X.; Yuan, B.; Wu, S.S.; Shen, J.; Lin, S.C. Grafting of zwitterion from cellulose membranes via ATRP for improving blood compatibility. Biomacromolecules 2009, 10, 2809–2816. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Z.; Wang, J.X.; Wang, S.C. Improving the water flux and bio-fouling resistance of reverse osmosis (RO) membrane through surface modification by zwitterionic polymer. J. Membr. Sci. 2015, 493, 188–199. [Google Scholar] [CrossRef]
- Lange, S.C.; van Andel, E.; Smulders, M.M.; Zuilhof, H. Efficient and Tunable Three-Dimensional Functionalization of Fully Zwitterionic Antifouling Surface Coatings. Langmuir 2016, 32, 10199–10205. [Google Scholar] [CrossRef]
- van Andel, E.; de Bus, I.; Tijhaar, E.J.; Smulders, M.M.J.; Savelkoul, H.F.J.; Zuilhof, H. Highly Specific Binding on Antifouling Zwitterionic Polymer-Coated Microbeads as Measured by Flow Cytometry. ACS Appl. Mater. Interfaces 2017, 9, 38211–38221. [Google Scholar] [CrossRef]
- Jaramillo, H.; Boo, C.; Hashmi, S.M.; Elimelech, M. Zwitterionic coating on thin-film composite membranes to delay gypsum scaling in reverse osmosis. J. Membr. Sci. 2021, 618, 118568. [Google Scholar] [CrossRef]
- Kaner, P.; Rubakh, E.; Kim, D.H.; Asatekin, A. Zwitterion-containing polymer additives for fouling resistant ultrafiltration membranes. J. Membr. Sci. 2017, 533, 141–159. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Wee, K.H.; Bai, R. Highly hydrophilic and low-protein-fouling polypropylene membrane prepared by surface modification with sulfobetaine-based zwitterionic polymer through a combined surface polymerization method. J. Membr. Sci. 2010, 362, 326–333. [Google Scholar] [CrossRef]
- Li, L.Y.; Chen, S.F.; Jiang, S.Y. Protein interactions with oligo(ethylene glycol) (OEG) self-assembled monolayers: OEG stability, surface packing density and protein adsorption. J. Biomater. Sci. -Polym. Ed. 2007, 18, 1415–1427. [Google Scholar] [CrossRef] [PubMed]
- Vaisocherova, H.; Yang, W.; Zhang, Z.; Cao, Z.; Cheng, G.; Piliarik, M.; Homola, J.; Jiang, S. Ultralow fouling and functionalizable surface chemistry based on a zwitterionic polymer enabling sensitive and specific protein detection in undiluted blood plasma. Anal. Chem. 2008, 80, 7894–7901. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Xue, H.; Li, W.; Zhang, J.; Jiang, S. Pursuing „zero” protein adsorption of poly(carboxybetaine) from undiluted blood serum and plasma. Langmuir 2009, 25, 11911–11916. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Imbrogno, J.; Belfort, G.; Wang, X.L. Making polymeric membranes antifouling via „grafting from” polymerization of zwitterions. J. Appl. Polym. Sci. 2015, 132, 41781. [Google Scholar] [CrossRef]
- Tripathi, B.P.; Das, P.; Simon, F.; Stamm, M. Ultralow fouling membranes by surface modification with functional polydopamine. Eur. Polym. J. 2018, 99, 80–89. [Google Scholar] [CrossRef]
- Walker, E.J.; Pandiyarajan, C.K.; Efimenko, K.; Genzer, J. Generating Surface-Anchored Zwitterionic Networks and Studying Their Resistance to Bovine Serum Albumin Adsorption. ACS Appl. Polym. Mater. 2019, 1, 3323–3333. [Google Scholar] [CrossRef]
- Wang, P.; Meng, J.Q.; Xu, M.L.; Yuan, T.; Yang, N.; Sun, T.; Zhang, Y.F.; Feng, X.S.; Cheng, B.W. A simple but efficient zwitterionization method towards cellulose membrane with superior antifouling property and biocompatibility. J. Membr. Sci. 2015, 492, 547–558. [Google Scholar] [CrossRef]
- Brzozowska, A.M.; Hofs, B.; de Keizer, A.; Fokkink, R.; Stuart, M.A.C.; Norde, W. Reduction of protein adsorption on silica and polystyrene surfaces due to coating with Complex Coacervate Core Micelles. Colloids Surf. A-Physicochem. Eng. Asp. 2009, 347, 146–155. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Regev, C.; Jiang, Z.; Kasher, R.; Miller, Y. Distinct Antifouling Mechanisms on Different Chain Densities of Zwitterionic Polymers. Molecules 2022, 27, 7394. https://doi.org/10.3390/molecules27217394
Regev C, Jiang Z, Kasher R, Miller Y. Distinct Antifouling Mechanisms on Different Chain Densities of Zwitterionic Polymers. Molecules. 2022; 27(21):7394. https://doi.org/10.3390/molecules27217394
Chicago/Turabian StyleRegev, Clil, Zhongyi Jiang, Roni Kasher, and Yifat Miller. 2022. "Distinct Antifouling Mechanisms on Different Chain Densities of Zwitterionic Polymers" Molecules 27, no. 21: 7394. https://doi.org/10.3390/molecules27217394
APA StyleRegev, C., Jiang, Z., Kasher, R., & Miller, Y. (2022). Distinct Antifouling Mechanisms on Different Chain Densities of Zwitterionic Polymers. Molecules, 27(21), 7394. https://doi.org/10.3390/molecules27217394