Recent Advances in Mass Spectrometry-Based Structural Elucidation Techniques
Abstract
1. Introduction to Mass Spectrometry and Overview
2. Technical Advances in MS-Based Structural Elucidation Techniques
2.1. Separation Techniques for MS-Based Mixture Analysis
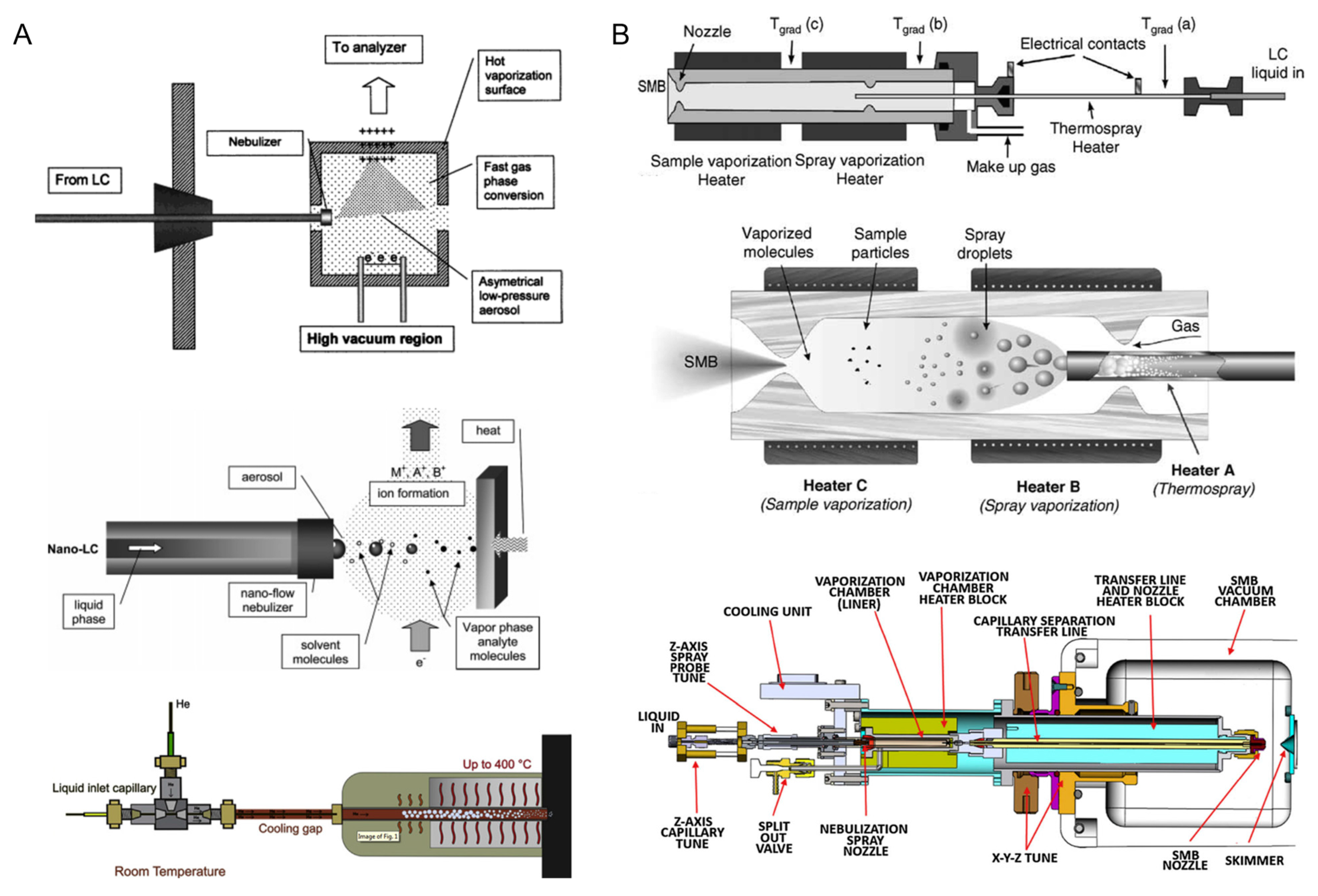
2.2. EI Mass Spectrometry
2.3. Soft Ionization and Tandem MS Based on Collisional Activation
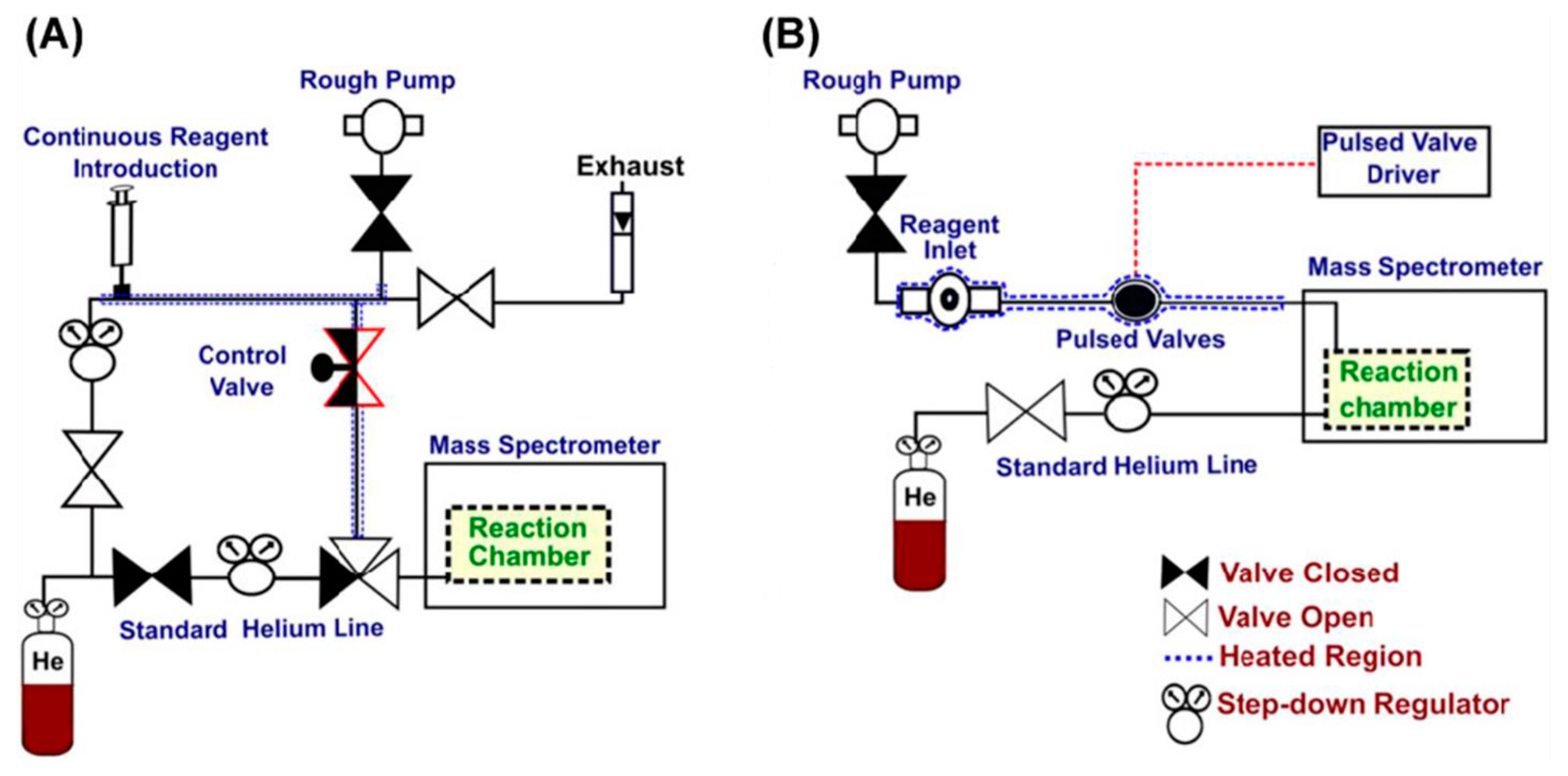
2.4. Ion–Molecule Reactions
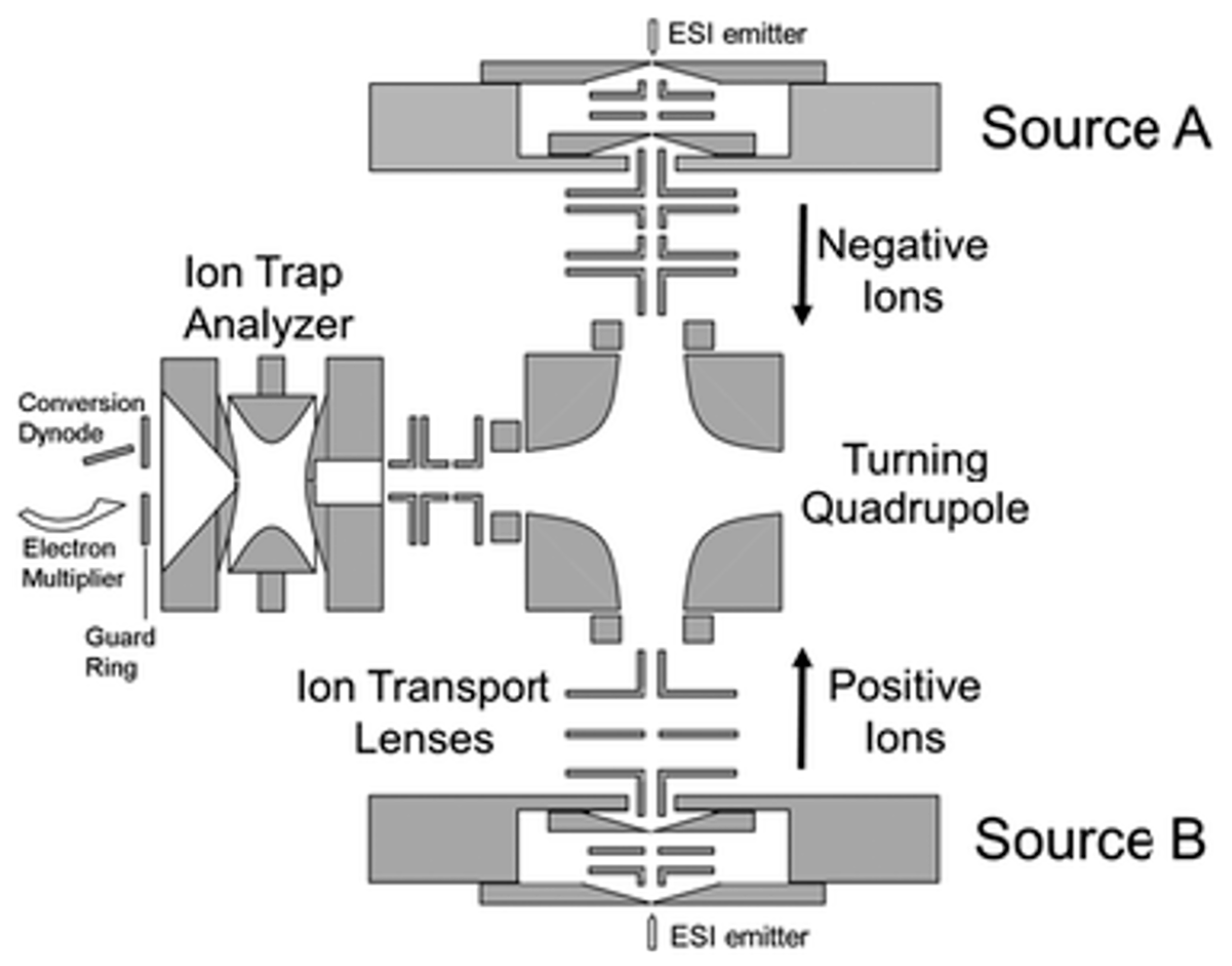
2.5. Ion–Ion Reactions
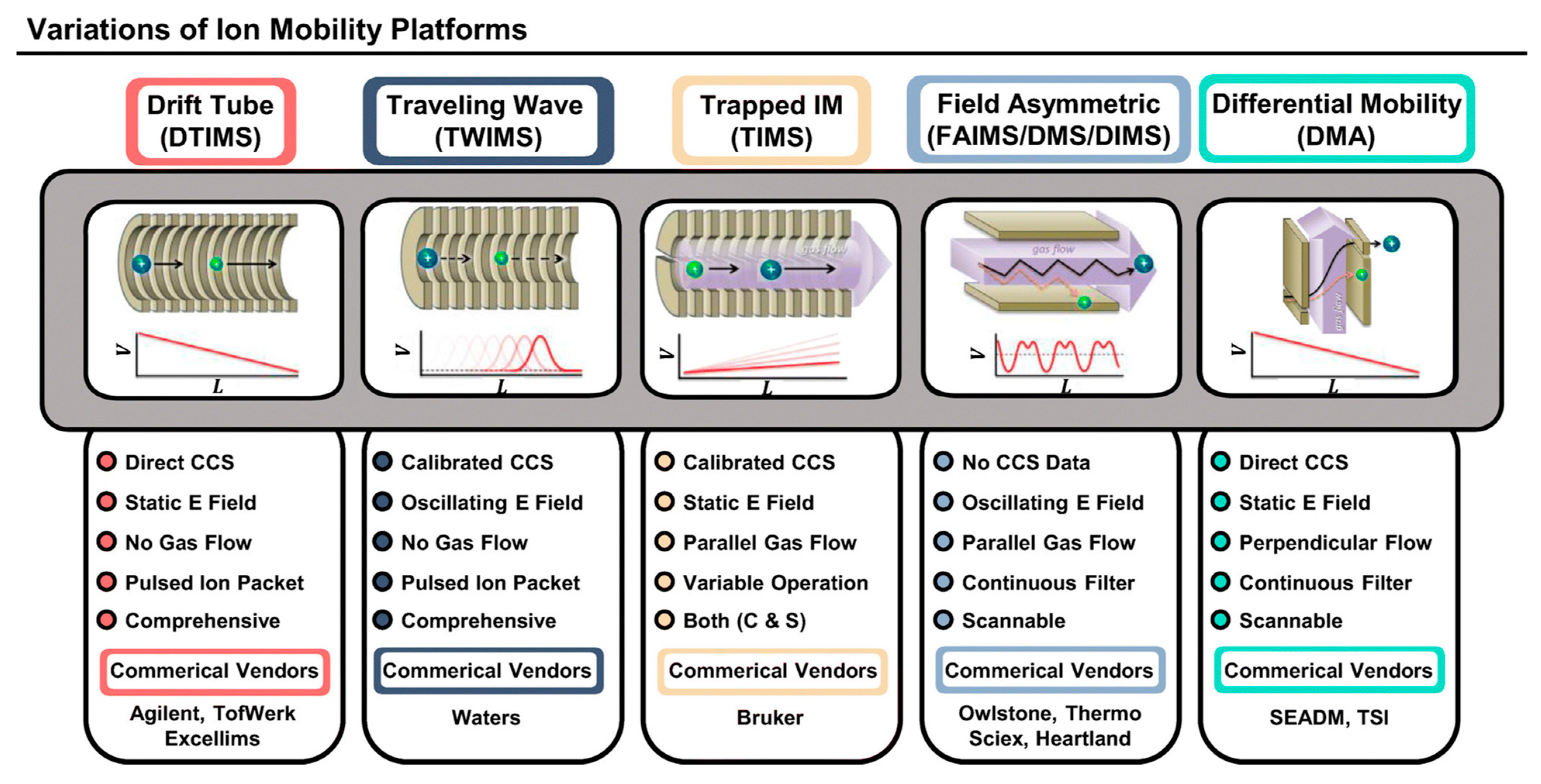
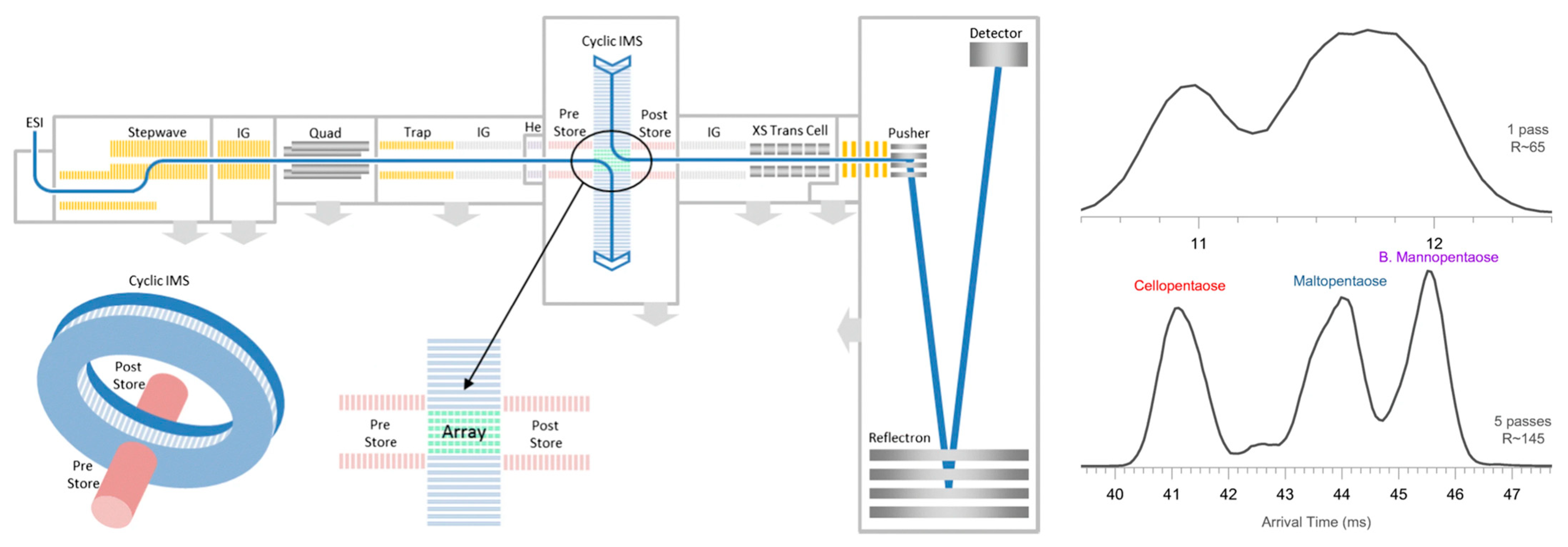
2.6. Ion Mobility Spectrometry-MS
2.7. Coupling MS with Other Analytical Tools
3. Applications of Mass Spectrometry-Based Techniques for Structural Analysis
3.1. Applications in Proteomics
3.1.1. Studies of PTMs
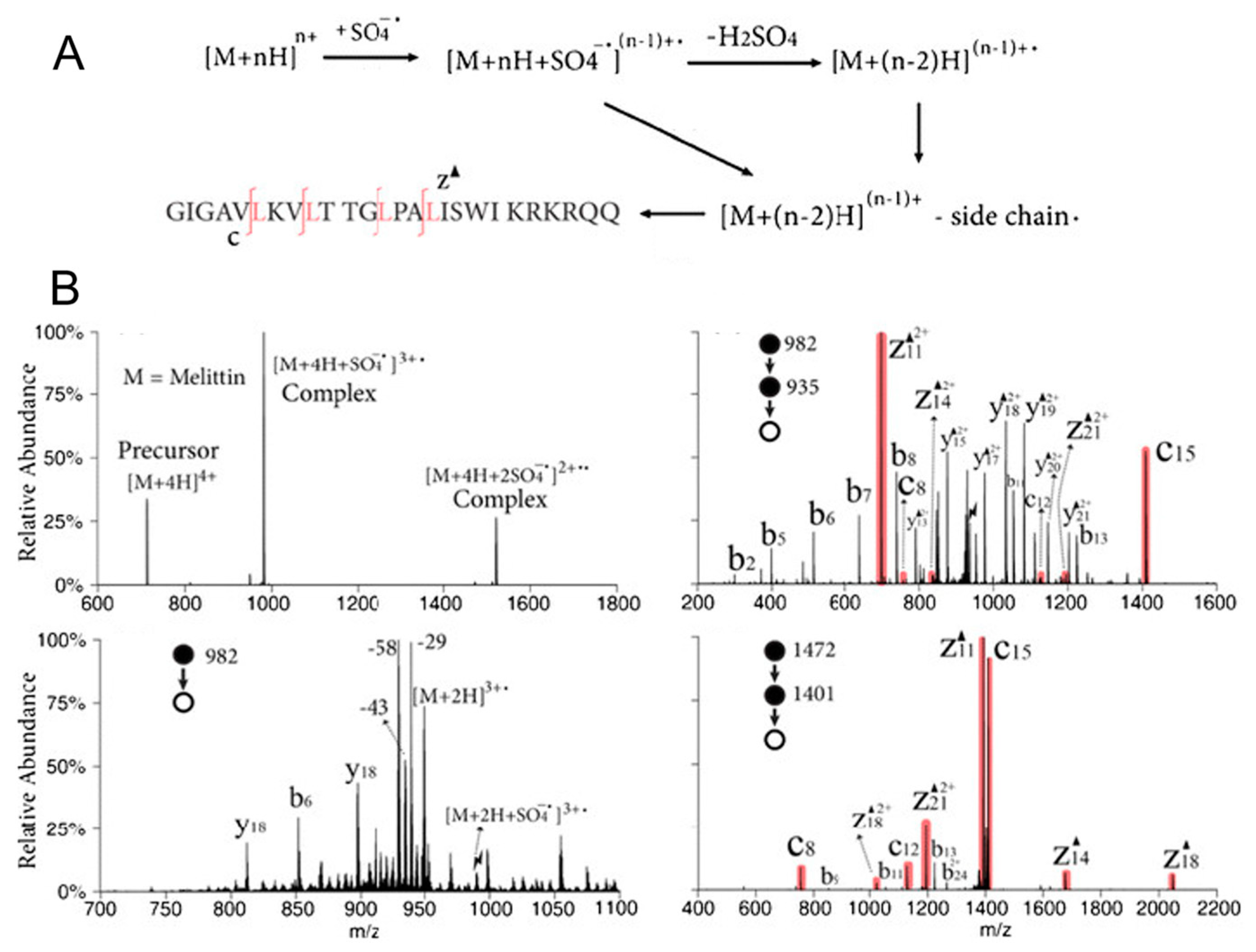
3.1.2. Gas-Phase Ion Chemistry for Protein Structural Elucidation
3.1.3. Ion Mobility in Protein Structural Elucidation
3.2. Applications in Lipidomics and Metabolomics
3.2.1. Structural Elucidation of Lipids
3.2.2. Structural Elucidation of Metabolites
3.3. Applications in Glycomics
3.4. Applications in Petroleomics
3.5. Applications in Natural Products Structural Analysis
4. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feider, C.L.; Krieger, A.; DeHoog, R.J.; Eberlin, L.S. Ambient Ionization Mass Spectrometry: Recent Developments and Applications. Anal. Chem. 2019, 91, 4266–4290. [Google Scholar] [CrossRef] [PubMed]
- de Hoffmann, E.; Stroobant, V. 2 Mass Analyzers. In Mass Spectrometry: Principles and Applications, 3rd ed.; Wiley: Hoboken, NJ, USA, 2013; pp. 85–173. [Google Scholar]
- Glish, G.L.; Burinsky, D.J. Hybrid Mass Spectrometers for Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2008, 19, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.K.-Y.; Niyonsaba, E.; Alzarieni, K.Z.; Boulos, V.M.; Yerabolu, R.; Kenttämaa, H.I. Determination of the Compound Class and Functional Groups in Protonated Analytes via Diagnostic Gas-Phase Ion-Molecule Reactions. Mass Spectrom. Rev. 2021, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Warneke, J.; Mayer, M.; Rohdenburg, M.; Ma, X.; Liu, J.K.Y.; Grellmann, M.; Debnath, S.; Azov, V.A.; Apra, E.; Young, R.P.; et al. Direct Functionalization of C−H Bonds by Electrophilic Anions. Proc. Natl. Acad. Sci. USA 2020, 117, 23374–23379. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Rohdenburg, M.; Knorke, H.; Kawa, S.; Liu, J.K.-Y.; Aprà, E.; Asmis, K.R.; Azov, V.A.; Laskin, J.; Jenne, C.; et al. Binding of Saturated and Unsaturated C6-Hydrocarbons to the Electrophilic Anion [B12Br11]−: A Systematic Mechanistic Study. Phys. Chem. Chem. Phys. 2022, 24, 21759–21772. [Google Scholar] [CrossRef]
- Ma, X.; Jin, C.; Wang, D.; Nash, J.J.; Kenttämaa, H.I. Relative Reactivities of Three Isomeric Aromatic Biradicals with a 1,4-Biradical Topology Are Controlled by Polar Effects. Chem. Eur. J. 2019, 25, 6355–6361. [Google Scholar] [CrossRef]
- Ma, X.; Feng, E.; Jiang, H.; Boulos, V.M.; Gao, J.; Nash, J.J.; Kenttämaa, H.I. Protonated Ground-State Singlet meta-Pyridynes React from an Excited Triplet State. J. Org. Chem. 2021, 86, 3249–3260. [Google Scholar] [CrossRef]
- Feng, E.; Yu, Z.J.; Jiang, H.; Ma, X.; Nash, J.J.; Kenttämaa, H.I. Gas-Phase Reactivity of Phenylcarbyne Anions. J. Am. Chem. Soc. 2022, 144, 8576–8590. [Google Scholar] [CrossRef]
- Jin, C.; Feng, E.; Ma, X.; Tang, W.; Sheng, H.; Wittrig, A.; Gao, J.; Kenttämaa, H.I. Reactivity of para-Benzynes in Solution and in the Gas Phase. Tetrahedron Lett. 2021, 74, 153161. [Google Scholar] [CrossRef]
- Max, J.P.; Ma, X.; Kotha, R.R.; Ding, D.; Milton, J.; Nash, J.J.; Kenttämaa, H.I. Reactivity of Organic σ,σ,σ,σ,σ-Pentaradicals. Int. J. Mass Spectrom. 2019, 435, 280–290. [Google Scholar] [CrossRef]
- Lubes, G.; Goodarzi, M. GC–MS Based Metabolomics Used for the Identification of Cancer Volatile Organic Compounds as Biomarkers. J. Pharm. Biomed. 2018, 147, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Asteggiano, A.; Occhipinti, A.; Capuzzo, A.; Mecarelli, E.; Aigotti, R.; Medana, C. Quali–Quantitative Characterization of Volatile and Non-Volatile Compounds in Protium heptaphyllum (Aubl.) Marchand Resin by GC–MS Validated Method, GC–FID and HPLC–HRMS2. Molecules 2021, 26, 1447. [Google Scholar] [CrossRef] [PubMed]
- Khakimov, B.; Bakhytkyzy, I.; Fauhl-Hassek, C.; Engelsen, S.B. Non-volatile Molecular Composition and Discrimination of Single Grape White Wines of Chardonnay, Riesling, Sauvignon Blanc and Silvaner Using Untargeted GC-MS Analysis. Food Chem. 2022, 369, 130878. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Zhao, N.; Lu, W.; Zhao, F.; Zheng, S.; Wang, W.; Chen, W. Effect of Different Lactobacillus Species on Volatile and Nonvolatile Flavor Compounds in Juices Fermentation. Food Sci. Nutr. 2019, 7, 2214–2223. [Google Scholar] [CrossRef]
- Dai, M.; Ma, T.; Niu, Y.; Zhang, M.; Zhu, Z.; Wang, S.; Liu, H. Analysis of Low-molecular-weight Metabolites in Stomach Cancer Cells by a Simplified and Inexpensive GC/MS Metabolomics Method. Anal. Bioanal. Chem. 2020, 412, 2981–2991. [Google Scholar] [CrossRef]
- Chen, L.; Dean, B.; Liang, X. A Technical Overview of Supercritical Fluid Chromatography-Mass Spectrometry (SFC-MS) and Its Recent Applications in Pharmaceutical Research and Development. Drug Discov. Today Technol. 2021, 40, 69–75. [Google Scholar] [CrossRef]
- Dodds, J.N.; Baker, E.S. Ion Mobility Spectrometry: Fundamental Concepts, Instrumentation, Applications, and the Road Ahead. J. Am. Soc. Mass Spectrom. 2019, 30, 2185–2195. [Google Scholar] [CrossRef]
- Lindinger, W.; Howorka, F.; Shull, J.M.; Phelps, A.V.; Zipf, E.C.; Kim, Y.K.; Futrell, J.H. Applications. In Electron Impact Ionization; Märk, T.D., Dunn, G.H., Eds.; Springer: Vienna, Austria, 1985; pp. 320–375. [Google Scholar]
- de Hoffmann, E.; Stroobant, V. Electron Ionization. In Mass Spectrometry: Principles and Applications, 3rd ed.; Wiley: Hoboken, NJ, USA, 2013; pp. 15–17. [Google Scholar]
- Stein, S. Mass Spectral Reference Libraries: An Ever-Expanding Resource for Chemical Identification. Anal. Chem. 2012, 84, 7274–7282. [Google Scholar] [CrossRef]
- Niyonsaba, E.; Manheim, J.M.; Yerabolu, R.; Kenttämaa, H.I. Recent Advances in Petroleum Analysis by Mass Spectrometry. Anal. Chem. 2019, 91, 156–177. [Google Scholar] [CrossRef]
- Sutton, P.A.; Rowland, S.J. High Temperature Gas Chromatography–Time-of-Flight-Mass Spectrometry (HTGC–ToF-MS) for High-Boiling Compounds. J. Chromatogr. A 2012, 1243, 69–80. [Google Scholar] [CrossRef]
- Reynolds, K.A.; Luhavaya, H.; Li, J.; Dahesh, S.; Nizet, V.; Yamanaka, K.; Moore, B.S. Isolation and Structure Elucidation of Lipopeptide Antibiotic Taromycin B from the Activated Taromycin Biosynthetic Gene Cluster. J. Antibiot. 2018, 71, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Berman, P.; Futoran, K.; Lewitus, G.M.; Mukha, D.; Benami, M.; Shlomi, T.; Meiri, D. A New ESI-LC/MS Approach for Comprehensive Metabolic Profiling of Phytocannabinoids in Cannabis. Sci. Rep. 2018, 8, 14280. [Google Scholar] [CrossRef] [PubMed]
- Tsoupras, A.; Lordan, R.; Keefe, E.; Shiels, K.; Saha, S.K.; Zabetakis, I. Structural Elucidation of Irish Ale Bioactive Polar Lipids with Antithrombotic Properties. Biomolecules 2020, 10, 1075. [Google Scholar] [CrossRef] [PubMed]
- Rigano, F.; Tranchida, P.Q.; Dugo, P.; Mondello, L. High-Performance Liquid Chromatography Combined with Electron Ionization Mass Spectrometry: A Review. Trends Analyt. Chem. 2019, 118, 112–122. [Google Scholar] [CrossRef]
- Termopoli, V.; Famiglini, G.; Palma, P.; Piergiovanni, M.; Rocio-Bautista, P.; Ottaviani, M.F.; Cappiello, A.; Saeed, M.; Perry, S. Evaluation of a Liquid Electron Ionization Liquid Chromatography–Mass Spectrometry Interface. J. Chromatogr. A 2019, 1591, 120–130. [Google Scholar] [CrossRef]
- Seemann, B.; Alon, T.; Tsizin, S.; Fialkov, A.B.; Amirav, A. Electron Ionization LC-MS with Supersonic Molecular Beams—The New Concept, Benefits and Applications. J. Mass Spectrom. 2015, 50, 1252–1263. [Google Scholar] [CrossRef]
- Rigano, F.; Albergamo, A.; Sciarrone, D.; Beccaria, M.; Purcaro, G.; Mondello, L. Nano Liquid Chromatography Directly Coupled to Electron Ionization Mass Spectrometry for Free Fatty Acid Elucidation in Mussel. Anal. Chem. 2016, 88, 4021–4028. [Google Scholar] [CrossRef]
- Alon, T.; Amirav, A. How Enhanced Molecular Ions in Cold EI Improve Compound Identification by the NIST Library. Rapid Commun. Mass Spectrom. 2015, 29, 2287–2292. [Google Scholar] [CrossRef]
- Famiglini, G.; Palma, P.; Pierini, E.; Trufelli, H.; Cappiello, A. Organochlorine Pesticides by LC−MS. Anal. Chem. 2008, 80, 3445–3449. [Google Scholar] [CrossRef]
- Famiglini, G.; Palma, P.; Termopoli, V.; Trufelli, H.; Cappiello, A. Single-Step LC/MS Method for the Simultaneous Determination of GC-Amenable Organochlorine and LC-Amenable Phenoxy Acidic Pesticides. Anal. Chem. 2009, 81, 7373–7378. [Google Scholar] [CrossRef]
- Albergamo, A.; Rigano, F.; Purcaro, G.; Mauceri, A.; Fasulo, S.; Mondello, L. Free Fatty Acid Profiling of Marine Sentinels by nanoLC-EI-MS for the Assessment of Environmental Pollution Effects. Sci. Total Environ. 2016, 571, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Mazumdar, S. Electrospray Ionization Mass Spectrometry: A Technique to Access the Information beyond the Molecular Weight of the Analyte. Int. J. Anal. Chem. 2012, 2012, 282574. [Google Scholar] [CrossRef] [PubMed]
- Kaddis, C.S.; Loo, J.A. Native Protein MS and Ion Mobility: Large Flying Proteins with ESI. Anal. Chem. 2007, 79, 1778–1784. [Google Scholar] [CrossRef]
- Sleno, L.; Volmer, D.A. Ion Activation Methods for Tandem Mass Spectrometry. J. Mass Spectrom. 2004, 39, 1091–1112. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.T.; Hoffman, K.L.; Janiczek-Dolphin, N.; Bockbrader, H.; Parker, T.D. Tandem-in-Time Mass Spectrometry as a Quantitative Bioanalytical Tool. Anal. Chem. 1997, 69, 4519–4523. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.V.; Yost, R.A.; Kelley, P.E.; Bradford, D.C. Tandem-in-Space and Tandem-in-Time Mass Spectrometry: Triple Quadrupoles and Quadrupole Ion Traps. Anal. Chem. 1990, 62, 2162–2172. [Google Scholar] [CrossRef]
- Stingl, C.; Ihling, C.; Ammerer, G.; Sinz, A.; Mechtler, K. Application of Different Fragmentation Techniques for the Analysis of Phosphopeptides Using a Hybrid Linear Ion Trap-FTICR Mass Spectrometer. Biochim. Biophys. Acta Proteins Proteom. 2006, 1764, 1842–1852. [Google Scholar] [CrossRef]
- Makarov, A.; Denisov, E.; Kholomeev, A.; Balschun, W.; Lange, O.; Strupat, K.; Horning, S. Performance Evaluation of a Hybrid Linear Ion Trap/Orbitrap Mass Spectrometer. Anal. Chem. 2006, 78, 2113–2120. [Google Scholar] [CrossRef]
- Stiving, A.Q.; VanAernum, Z.L.; Busch, F.; Harvey, S.R.; Sarni, S.H.; Wysocki, V.H. Surface-Induced Dissociation: An Effective Method for Characterization of Protein Quaternary Structure. Anal. Chem. 2019, 91, 190–209. [Google Scholar] [CrossRef]
- Zubarev, R.A. Electron-Capture Dissociation Tandem Mass Spectrometry. Curr. Opin. Biotechnol. 2004, 15, 12–16. [Google Scholar] [CrossRef]
- Kim, M.-S.; Pandey, A. Electron Transfer Dissociation Mass Spectrometry in Proteomics. Proteomics 2012, 12, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Zhurov, K.O.; Fornelli, L.; Wodrich, M.D.; Laskay, Ü.A.; Tsybin, Y.O. Principles of Electron Capture and Transfer Dissociation Mass Spectrometry Applied to Peptide and Protein Structure Analysis. Chem. Soc. Rev. 2013, 42, 5014–5030. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Liang, X.; McLuckey, S.A. Ion Trap versus Low-Energy Beam-Type Collision-Induced Dissociation of Protonated Ubiquitin Ions. Anal. Chem. 2006, 78, 1218–1227. [Google Scholar] [CrossRef]
- McLuckey, S.A.; Goeringer, D.E. Special Feature: Tutorial Slow Heating Methods in Tandem Mass Spectrometry. J. Mass Spectrom. 1997, 32, 461–474. [Google Scholar] [CrossRef]
- Bayat, P.; Lesage, D.; Cole, R.B. Low-Energy Collision-Induced Dissociation (Low-Energy CID), Collision-Induced Dissociation (CID), and Higher Energy Collision Dissociation (HCD) Mass Spectrometry for Structural Elucidation of Saccharides and Clarification of Their Dissolution Mechanism in DMAc/LiCl. J. Mass Spectrom. 2018, 53, 705–716. [Google Scholar]
- Wang, J.; Zhao, J.; Nie, S.; Xie, M.; Li, S. Mass Spectrometry for Structural Elucidation and Sequencing of Carbohydrates. Trends Analyt. Chem. 2021, 144, 116436. [Google Scholar] [CrossRef]
- Li, M.; Zhu, W.; Zheng, H.; Zhang, J. Efficient HCD-pd-EThcD Approach for N-Glycan Mapping of Therapeutic Antibodies at Intact Glycopeptide Level. Anal. Chim. Acta 2022, 1189, 339232. [Google Scholar] [CrossRef] [PubMed]
- Neves, B.; Duarte, S.; Domingues, P.; Pérez-Sala, D.; Oliveira, M.M.; Domingues, M.D.; Melo, T. Advancing Target Identification of Nitrated Phospholipids in Biological Systems by HCD Specific Fragmentation Fingerprinting in Orbitrap Platforms. Molecules 2020, 25, 2120. [Google Scholar] [CrossRef]
- Jora, M.; Burns, A.P.; Ross, R.L.; Lobue, P.A.; Zhao, R.; Palumbo, C.M.; Beal, P.A.; Addepalli, B.; Limbach, P.A. Differentiating Positional Isomers of Nucleoside Modifications by Higher-Energy Collisional Dissociation Mass Spectrometry (HCD MS). J. Am. Soc. Mass Spectrom. 2018, 29, 1745–1756. [Google Scholar] [CrossRef]
- Brodbelt, J.S.; Morrison, L.J.; Santos, I. Ultraviolet Photodissociation Mass Spectrometry for Analysis of Biological Molecules. Chem. Rev. 2020, 120, 3328–3380. [Google Scholar] [CrossRef]
- Cleland, T.P.; DeHart, C.J.; Fellers, R.T.; VanNispen, A.J.; Greer, J.B.; LeDuc, R.D.; Parker, W.R.; Thomas, P.M.; Kelleher, N.L.; Brodbelt, J.S. High-Throughput Analysis of Intact Human Proteins Using UVPD and HCD on an Orbitrap Mass Spectrometer. J. Proteome Res. 2017, 16, 2072–2079. [Google Scholar] [CrossRef] [PubMed]
- Quick, M.M.; Crittenden, C.M.; Rosenberg, J.A.; Brodbelt, J.S. Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry. Anal. Chem. 2018, 90, 8523–8530. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-F.; Zhao, J.; Cao, W.; Zhang, W.; Xia, Y.; Ouyang, Z. Site-Specific Photochemical Reaction for Improved C=C Location Analysis of Unsaturated Lipids by Ultraviolet Photodissociation. Research 2022, 2022, 9783602. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Rustam, Y.; Palmieri, M.; Sieber, O.M.; Reid, G.E. Evaluation of Ultraviolet Photodissociation Tandem Mass Spectrometry for the Structural Assignment of Unsaturated Fatty Acid Double Bond Positional Isomers. Anal. Bioanal. Chem. 2020, 412, 2339–2351. [Google Scholar] [CrossRef]
- Klein, D.R.; Leach, F.E.; Amster, I.J.; Brodbelt, J.S. Structural Characterization of Glycosaminoglycan Carbohydrates Using Ultraviolet Photodissociation. Anal. Chem. 2019, 91, 6019–6026. [Google Scholar] [CrossRef]
- Kong, J.Y.; Yu, Z.; Easton, M.W.; Niyonsaba, E.; Ma, X.; Yerabolu, R.; Sheng, H.; Jarrell, T.M.; Zhang, Z.; Ghosh, A.K.; et al. Differentiating Isomeric Deprotonated Glucuronide Drug Metabolites via Ion/Molecule Reactions in Tandem Mass Spectrometry. Anal. Chem. 2018, 90, 9426–9433. [Google Scholar] [CrossRef]
- Ma, X.; Chong, L.; Tian, R.; Shi, R.; Hu, T.Y.; Ouyang, Z.; Xia, Y. Identification and Quantitation of Lipid C=C Location Isomers: A Shotgun Lipidomics Approach Enabled by Photochemical Reaction. Proc. Natl. Acad. Sci. USA 2016, 113, 2573–2578. [Google Scholar] [CrossRef]
- Feng, E.; Ma, X.; Jiang, H.; Sheng, H.; Rowell, C.E.; Kenttämaa, H.I. Differentiation of Protonated Sulfonate Esters from Isomeric Sulfite Esters and Sulfones by Gas-Phase Ion–Molecule Reactions Followed by Diagnostic Collision-Activated Dissociation in Tandem Mass Spectrometry Experiments. Anal. Chem. 2022, 94, 7928–7935. [Google Scholar] [CrossRef]
- Feng, E.; Ma, X.; Kenttämaa, H.I. Characterization of Protonated Substituted Ureas by Using Diagnostic Gas-Phase Ion-Molecule Reactions Followed by Collision-Activated Dissociation in Tandem Mass Spectrometry Experiments. Anal. Chem. 2021, 93, 7851–7859. [Google Scholar] [CrossRef]
- Jarrell, T.; Riedeman, J.; Carlsen, M.; Replogle, R.; Selby, T.; Kenttämaa, H.I. Multiported Pulsed Valve Interface for a Linear Quadrupole Ion Trap Mass Spectrometer to Enable Rapid Screening of Multiple Functional-Group Selective Ion–Molecule Reactions. Anal. Chem. 2014, 86, 6533–6539. [Google Scholar] [CrossRef]
- Kong, J.Y.; Hilger, R.T.; Jin, C.; Yerabolu, R.; Zimmerman, J.R.; Replogle, R.W.; Jarrell, T.M.; Easterling, L.; Kumar, R.; Kenttämaa, H.I. Integration of a Multichannel Pulsed-Valve Inlet System to a Linear Quadrupole Ion Trap Mass Spectrometer for the Rapid Consecutive Introduction of Nine Reagents for Diagnostic Ion/Molecule Reactions. Anal. Chem. 2019, 91, 15652–15660. [Google Scholar] [CrossRef] [PubMed]
- Habicht, S.C.; Vinueza, N.R.; Archibold, E.F.; Duan, P.; Kenttämaa, H.I. Identification of the Carboxylic Acid Functionality by Using Electrospray Ionization and Ion−Molecule Reactions in a Modified Linear Quadrupole Ion Trap Mass Spectrometer. Anal. Chem. 2008, 80, 3416–3421. [Google Scholar] [CrossRef] [PubMed]
- Feng, E.; Fu, Y.; Ma, X.; Kotha, R.R.; Ding, D.; Kenttämaa, H.I. A Portable Reagent Inlet System Designed to Diminish the Impact of Air and Water to Ion–Molecule Reactions Studied in a Linear Quadrupole Ion Trap. J. Amer. Soc. Mass Spectrom. 2022, 33, 1794–1798. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Y.; Lei, H.-R.; Kenttämaa, H.I. Laser-Induced Acoustic Desorption. MRS Bull. 2019, 44, 372–381. [Google Scholar] [CrossRef]
- McLuckey, S.A.; Huang, T.-Y. Ion/Ion Reactions: New Chemistry for Analytical MS. Anal. Chem. 2009, 81, 8669–8676. [Google Scholar] [CrossRef]
- Prentice, B.M.; McLuckey, S.A. Gas-Phase Ion/Ion Reactions of Peptides and Proteins: Acid/Base, Redox, and Covalent Chemistries. Chem. Commun. 2013, 49, 947–965. [Google Scholar] [CrossRef]
- Richardson, K.; Langridge, D.; Dixit, S.M.; Ruotolo, B.T. An Improved Calibration Approach for Traveling Wave Ion Mobility Spectrometry: Robust, High-Precision Collision Cross Sections. Anal. Chem. 2021, 93, 3542–3550. [Google Scholar] [CrossRef]
- Ridgeway, M.; Woods, L.; Park, M. CHAPTER 5 Trapped Ion Mobility Spectrometry—Basics and Calibration. In Ion Mobility-Mass Spectrometry: Fundamentals and Applications; The Royal Society of Chemistry: London, UK, 2022; pp. 105–131. [Google Scholar]
- Dodds, J.N.; May, J.C.; McLean, J.A. Correlating Resolving Power, Resolution, and Collision Cross Section: Unifying Cross-Platform Assessment of Separation Efficiency in Ion Mobility Spectrometry. Anal. Chem. 2017, 89, 12176–12184. [Google Scholar] [CrossRef]
- Lietz, C.B.; Yu, Q.; Li, L. Large-Scale Collision Cross-Section Profiling on a Traveling Wave Ion Mobility Mass Spectrometer. J. Am. Soc. Mass Spectrom. 2014, 25, 2009–2019. [Google Scholar] [CrossRef]
- Schroeder, M.; Meyer, S.W.; Heyman, H.M.; Barsch, A.; Sumner, L.W. Generation of a Collision Cross Section Library for Multi-Dimensional Plant Metabolomics Using UHPLC-Trapped Ion Mobility-MS/MS. Metabolites 2020, 10, 13. [Google Scholar] [CrossRef]
- Zhou, Z.; Luo, M.; Chen, X.; Yin, Y.; Xiong, X.; Wang, R.; Zhu, Z.-J. Ion Mobility Collision Cross-Section Atlas for Known and Unknown Metabolite Annotation in Untargeted Metabolomics. Nat. Commun. 2020, 11, 4334. [Google Scholar] [CrossRef] [PubMed]
- Plante, P.-L.; Francovic-Fontaine, É.; May, J.C.; McLean, J.A.; Baker, E.S.; Laviolette, F.; Marchand, M.; Corbeil, J. Predicting Ion Mobility Collision Cross-Sections Using a Deep Neural Network: DeepCCS. Anal. Chem. 2019, 91, 5191–5199. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Köhler, N.D.; Brunner, A.-D.; Wanka, J.-M.H.; Voytik, E.; Strauss, M.T.; Theis, F.J.; Mann, M. Deep Learning the Collisional Cross Sections of the Peptide Universe from a Million Experimental Values. Nat. Commun. 2021, 12, 1185. [Google Scholar] [CrossRef]
- Rainey, M.A.; Watson, C.A.; Asef, C.K.; Foster, M.R.; Baker, E.S.; Fernández, F.M. CCS Predictor 2.0: An Open-Source Jupyter Notebook Tool for Filtering Out False Positives in Metabolomics. bioRxiv 2022. [Google Scholar] [CrossRef]
- Gabelica, V.; Shvartsburg, A.A.; Afonso, C.; Barran, P.; Benesch, J.L.P.; Bleiholder, C.; Bowers, M.T.; Bilbao, A.; Bush, M.F.; Campbell, J.L.; et al. Recommendations for Reporting Ion Mobility Mass Spectrometry Measurements. Mass Spectrom. Rev. 2019, 38, 291–320. [Google Scholar] [CrossRef] [PubMed]
- Elyashberg, M. Identification and Structure Elucidation by NMR Spectroscopy. Trends Analyt. Chem. 2015, 69, 88–97. [Google Scholar] [CrossRef]
- Maveyraud, L.; Mourey, L. Protein X-ray Crystallography and Drug Discovery. Molecules 2020, 25, 1030. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.; Vonrüti, N.; Novák, P.; Aschauer, U.; Berg, E.J. Elucidation of LixNi0.8Co0.15Al0.05O2 Redox Chemistry by Operando Raman Spectroscopy. Chem. Mater. 2018, 30, 4694–4703. [Google Scholar] [CrossRef]
- Makarem, M.; Lee, C.M.; Kafle, K.; Huang, S.; Chae, I.; Yang, H.; Kubicki, J.D.; Kim, S.H. Probing Cellulose Structures with Vibrational Spectroscopy. Cellulose 2019, 26, 35–79. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Huck, C.W. Near-Infrared Spectroscopy in Bio-Applications. Molecules 2020, 25, 2948. [Google Scholar] [CrossRef]
- Gross, L.; Schuler, B.; Pavliček, N.; Fatayer, S.; Majzik, Z.; Moll, N.; Peña, D.; Meyer, G. Atomic Force Microscopy for Molecular Structure Elucidation. Angew. Chem. Int. Ed. 2018, 57, 3888–3908. [Google Scholar] [CrossRef] [PubMed]
- Commodo, M.; Kaiser, K.; De Falco, G.; Minutolo, P.; Schulz, F.; D’Anna, A.; Gross, L. On the Early Stages of Soot Formation: Molecular Structure Elucidation by High-Resolution Atomic Force Microscopy. Combust. Flame 2019, 205, 154–164. [Google Scholar] [CrossRef]
- Xue, M.; Kim, C.S.; Healy, A.R.; Wernke, K.M.; Wang, Z.; Frischling, M.C.; Shine, E.E.; Wang, W.; Herzon, S.B.; Crawford, J.M. Structure Elucidation of Colibactin and Its DNA Cross-Links. Science 2019, 365, eaax2685. [Google Scholar] [CrossRef]
- Mándi, A.; Kurtán, T. Applications of OR/ECD/VCD to the Structure Elucidation of Natural Products. Nat. Prod. Rep. 2019, 36, 889–918. [Google Scholar] [CrossRef]
- Boiteau, R.M.; Hoyt, D.W.; Nicora, C.D.; Kinmonth-Schultz, H.A.; Ward, J.K.; Bingol, K. Structure Elucidation of Unknown Metabolites in Metabolomics by Combined NMR and MS/MS Prediction. Metabolites 2018, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Bingol, K. Recent Advances in Targeted and Untargeted Metabolomics by NMR and MS/NMR Methods. High-Throughput 2018, 7, 9. [Google Scholar] [CrossRef]
- Elyashberg, M.; Argyropoulos, D. Computer Assisted Structure Elucidation (CASE): Current and Future Perspectives. Magn. Reson. Chem. 2021, 59, 669–690. [Google Scholar] [CrossRef]
- Marcarino, M.O.; Zanardi, M.a.M.; Cicetti, S.; Sarotti, A.M. NMR Calculations with Quantum Methods: Development of New Tools for Structural Elucidation and Beyond. Acc. Chem. Res. 2020, 53, 1922–1932. [Google Scholar] [CrossRef]
- Bhinderwala, F.; Wase, N.; DiRusso, C.; Powers, R. Combining Mass Spectrometry and NMR Improves Metabolite Detection and Annotation. J. Proteome Res. 2018, 17, 4017–4022. [Google Scholar] [CrossRef]
- Garcia-Perez, I.; Posma, J.M.; Serrano-Contreras, J.I.; Boulangé, C.L.; Chan, Q.; Frost, G.; Stamler, J.; Elliott, P.; Lindon, J.C.; Holmes, E.; et al. Identifying Unknown Metabolites Using NMR-Based Metabolic Profiling Techniques. Nat. Protoc. 2020, 15, 2538–2567. [Google Scholar] [CrossRef]
- Zhang, P.; Zhu, S.; Zhao, M.; Dai, Y.; Zhang, L.; Ding, S.; Zhao, P.; Li, J. Integration of 1H NMR- and UPLC-Q-TOF/MS-Based Plasma Metabonomics Study to Identify Diffuse Axonal Injury Biomarkers in Rat. Brain Res. Bull. 2018, 140, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Kosmopoulou, M.; Giannopoulou, A.F.; Iliou, A.; Benaki, D.; Panagiotakis, A.; Velentzas, A.D.; Konstantakou, E.G.; Papassideri, I.S.; Mikros, E.; Stravopodis, D.J.; et al. Human Melanoma-Cell Metabolic Profiling: Identification of Novel Biomarkers Indicating Metastasis. Int. J. Mol. Sci. 2020, 21, 2436. [Google Scholar] [CrossRef] [PubMed]
- Přichystal, J.; Schug, K.A.; Lemr, K.; Novák, J.; Havlíček, V. Structural Analysis of Natural Products. Anal. Chem. 2016, 88, 10338–10346. [Google Scholar] [CrossRef] [PubMed]
- Marth, J.D. A Unified Vision of the Building Blocks of Life. Nat. Cell Biol. 2008, 10, 1015. [Google Scholar] [CrossRef] [PubMed]
- Catherman, A.D.; Skinner, O.S.; Kelleher, N.L. Top down Proteomics: Facts and Perspectives. Biochem. Biophys. Res. Commun. 2014, 445, 683–693. [Google Scholar] [CrossRef]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.-C.; Yates, J.R. Protein Analysis by Shotgun/Bottom-up Proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef]
- Huo, B.; Zhang, W.; Zhao, X.; Dong, H.; Yu, Y.; Wang, J.; Qian, X.; Qin, W. A Triarylphosphine–Trimethylpiperidine Reagent for the One-Step Derivatization and Enrichment of Protein Post-Translational Modifications and Identification by Mass Spectrometry. Chem. Commun. 2018, 54, 13790–13793. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.; Sun, H.; Maroto, R.; Brasier, A.R. Selective Affinity Enrichment of Nitrotyrosine-Containing Peptides for Quantitative Analysis in Complex Samples. J. Proteome Res. 2017, 16, 2983–2992. [Google Scholar] [CrossRef]
- Brandi, J.; Noberini, R.; Bonaldi, T.; Cecconi, D. Advances in Enrichment Methods for Mass Spectrometry-Based Proteomics Analysis of Post-Translational Modifications. J. Chromatogr. A 2022, 1678, 463352. [Google Scholar] [CrossRef]
- Lehner, M.; Rieth, S.; Höllmüller, E.; Spliesgar, D.; Mertes, B.; Stengel, F.; Marx, A. Profiling of the ADP-Ribosylome in Living Cells. Angew. Chem. Int. Ed. 2022, 61, e202200977. [Google Scholar] [CrossRef]
- Yu, Q.; Wang, B.; Chen, Z.; Urabe, G.; Glover, M.S.; Shi, X.; Guo, L.-W.; Kent, K.C.; Li, L. Electron-Transfer/Higher-Energy Collision Dissociation (EThcD)-Enabled Intact Glycopeptide/Glycoproteome Characterization. J. Am. Soc. Mass Spectrom. 2017, 28, 1751–1764. [Google Scholar] [CrossRef] [PubMed]
- Pegg, C.L.; Zacchi, L.F.; Recinos, D.R.; Howard, C.B.; Schulz, B.L. Identification of Novel Glycosylation Events on Human Serum-Derived Factor IX. Glycoconj. J. 2020, 37, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Brodbelt, J.S. Deciphering Combinatorial Post-Translational Modifications by Top-Down Mass Spectrometry. Curr. Opin. Chem. Biol. 2022, 70, 102180. [Google Scholar] [CrossRef] [PubMed]
- Greer, S.M.; Brodbelt, J.S. Top-Down Characterization of Heavily Modified Histones Using 193 nm Ultraviolet Photodissociation Mass Spectrometry. J. Proteome Res. 2018, 17, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.; McLuckey, S.A. Ion/Ion Charge Inversion/Attachment in Conjunction with Dipolar DC Collisional Activation as a Selective Screen for Sulfo- and Phosphopeptides. Int. J. Mass Spectrom. 2019, 444, 116181. [Google Scholar] [CrossRef]
- Pitts-McCoy, A.M.; Harrilal, C.P.; McLuckey, S.A. Gas-Phase Ion/Ion Chemistry as a Probe for the Presence of Carboxylate Groups in Polypeptide Cations. J. Am. Soc. Mass Spectrom. 2019, 30, 329–338. [Google Scholar] [CrossRef]
- Peng, Z.; Bu, J.; McLuckey, S.A. The Generation of Dehydroalanine Residues in Protonated Polypeptides: Ion/Ion Reactions for Introducing Selective Cleavages. J. Am. Soc. Mass Spectrom. 2017, 28, 1765–1774. [Google Scholar] [CrossRef]
- Foreman, D.J.; Bhanot, J.S.; Lee, K.W.; McLuckey, S.A. Valet Parking for Protein Ion Charge State Concentration: Ion/Molecule Reactions in Linear Ion Traps. Anal. Chem. 2020, 92, 5419–5425. [Google Scholar] [CrossRef]
- Dixit, S.M.; Polasky, D.A.; Ruotolo, B.T. Collision Induced Unfolding of Isolated Proteins in the Gas Phase: Past, Present, and Future. Curr. Opin. Chem. Biol. 2018, 42, 93–100. [Google Scholar] [CrossRef]
- Fantin, S.M.; Parson, K.F.; Niu, S.; Liu, J.; Polasky, D.A.; Dixit, S.M.; Ferguson-Miller, S.M.; Ruotolo, B.T. Collision Induced Unfolding Classifies Ligands Bound to the Integral Membrane Translocator Protein. Anal. Chem. 2019, 91, 15469–15476. [Google Scholar] [CrossRef]
- Hernandez-Alba, O.; Wagner-Rousset, E.; Beck, A.; Cianférani, S. Native Mass Spectrometry, Ion Mobility, and Collision-Induced Unfolding for Conformational Characterization of IgG4 Monoclonal Antibodies. Anal. Chem. 2018, 90, 8865–8872. [Google Scholar] [CrossRef] [PubMed]
- Turzo, S.M.B.A.; Seffernick, J.T.; Rolland, A.D.; Donor, M.T.; Heinze, S.; Prell, J.S.; Wysocki, V.H.; Lindert, S. Protein Shape Sampled by Ion Mobility Mass Spectrometry Consistently Improves Protein Structure Prediction. Nat. Commun. 2022, 13, 4377. [Google Scholar] [CrossRef] [PubMed]
- El-Baba, T.J.; Woodall, D.W.; Raab, S.A.; Fuller, D.R.; Laganowsky, A.; Russell, D.H.; Clemmer, D.E. Melting Proteins: Evidence for Multiple Stable Structures upon Thermal Denaturation of Native Ubiquitin from Ion Mobility Spectrometry-Mass Spectrometry Measurements. J. Am. Chem. Soc. 2017, 139, 6306–6309. [Google Scholar] [CrossRef] [PubMed]
- Welte, M.A.; Gould, A.P. Lipid Droplet Functions Beyond Energy Storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1260–1272. [Google Scholar] [CrossRef]
- Coones, R.T.; Green, R.J.; Frazier, R.A. Investigating Lipid Headgroup Composition within Epithelial Membranes: A Systematic Review. Soft Matter 2021, 17, 6773–6786. [Google Scholar] [CrossRef] [PubMed]
- Monson, E.A.; Crosse, K.M.; Das, M.; Helbig, K.J. Lipid Droplet Density Alters the Early Innate Immune Response to Viral Infection. PLoS ONE 2018, 13, e0190597. [Google Scholar] [CrossRef]
- Soto-Avellaneda, A.; Morrison, B.E. Signaling and Other Functions of Lipids in Autophagy: A Review. Lipids Health Dis. 2020, 19, 214. [Google Scholar] [CrossRef]
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and Cancer: Emerging Roles in Pathogenesis, Diagnosis and Therapeutic Intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293. [Google Scholar] [CrossRef]
- Zhang, W.; Jian, R.; Zhao, J.; Liu, Y.; Xia, Y. Deep-Lipidotyping by Mass Spectrometry: Recent Technical Advances and Applications. J. Lipid Res. 2022, 63, 100219. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, B.; Yu, Q.; Li, L. Identification of Double Bond Position Isomers in Unsaturated Lipids by m-CPBA Epoxidation and Mass Spectrometry Fragmentation. Anal. Chem. 2019, 91, 1791–1795. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, M.; Shi, X.; Liu, Y.; Li, Z.; Jagodinsky, J.C.; Ma, M.; Welham, N.V.; Morris, Z.S.; Li, L. Quantification and Molecular Imaging of Fatty Acid Isomers from Complex Biological Samples by Mass Spectrometry. Chem. Sci. 2021, 12, 8115–8122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, D.; Chen, Q.; Wu, J.; Ouyang, Z.; Xia, Y. Online Photochemical Derivatization Enables Comprehensive Mass Spectrometric Analysis of Unsaturated Phospholipid Isomers. Nat. Commun. 2019, 10, 79. [Google Scholar] [CrossRef]
- Randolph, C.E.; Shenault, D.S.M.; Blanksby, S.J.; McLuckey, S.A. Localization of Carbon–Carbon Double Bond and Cyclopropane Sites in Cardiolipins via Gas-Phase Charge Inversion Reactions. J. Am. Soc. Mass Spectrom. 2021, 32, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Randolph, C.E.; Shenault, D.S.M.; Blanksby, S.J.; McLuckey, S.A. Structural Elucidation of Ether Glycerophospholipids Using Gas-Phase Ion/Ion Charge Inversion Chemistry. J. Am. Soc. Mass Spectrom. 2020, 31, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Specker, J.T.; Van Orden, S.L.; Ridgeway, M.E.; Prentice, B.M. Identification of Phosphatidylcholine Isomers in Imaging Mass Spectrometry Using Gas-Phase Charge Inversion Ion/Ion Reactions. Anal. Chem. 2020, 92, 13192–13201. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.R.; Brodbelt, J.S. Structural Characterization of Phosphatidylcholines Using 193 nm Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem. 2017, 89, 1516–1522. [Google Scholar] [CrossRef]
- Macias, L.A.; Feider, C.L.; Eberlin, L.S.; Brodbelt, J.S. Hybrid 193 nm Ultraviolet Photodissociation Mass Spectrometry Localizes Cardiolipin Unsaturations. Anal. Chem. 2019, 91, 12509–12516. [Google Scholar] [CrossRef]
- Vu, N.; Brown, J.; Giles, K.; Zhang, Q. Ozone-Induced Dissociation on a Traveling Wave High-Resolution Mass Spectrometer for Determination of Double-Bond Position in Lipids. Rapid Commun. Mass Spectrom. 2017, 31, 1415–1423. [Google Scholar] [CrossRef]
- Berthias, F.; Poad, B.L.J.; Thurman, H.A.; Bowman, A.P.; Blanksby, S.J.; Shvartsburg, A.A. Disentangling Lipid Isomers by High-Resolution Differential Ion Mobility Spectrometry/Ozone-Induced Dissociation of Metalated Species. J. Am. Soc. Mass Spectrom. 2021, 32, 2827–2836. [Google Scholar] [CrossRef]
- Harris, R.A.; May, J.C.; Stinson, C.A.; Xia, Y.; McLean, J.A. Determining Double Bond Position in Lipids Using Online Ozonolysis Coupled to Liquid Chromatography and Ion Mobility-Mass Spectrometry. Anal. Chem. 2018, 90, 1915–1924. [Google Scholar] [CrossRef]
- Poad, B.L.J.; Green, M.R.; Kirk, J.M.; Tomczyk, N.; Mitchell, T.W.; Blanksby, S.J. High-Pressure Ozone-Induced Dissociation for Lipid Structure Elucidation on Fast Chromatographic Timescales. Anal. Chem. 2017, 89, 4223–4229. [Google Scholar] [CrossRef] [PubMed]
- Paine, M.R.L.; Poad, B.L.J.; Eijkel, G.B.; Marshall, D.L.; Blanksby, S.J.; Heeren, R.M.A.; Ellis, S.R. Mass Spectrometry Imaging with Isomeric Resolution Enabled by Ozone-Induced Dissociation. Angew. Chem. Int. Ed. 2018, 57, 10530–10534. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kora, G.; Bowen, B.P.; Pan, C. MIDAS: A Database-Searching Algorithm for Metabolite Identification in Metabolomics. Anal. Chem. 2014, 86, 9496–9503. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An International Repository for Metabolomics Data and Metadata, Metabolite Standards, Protocols, Tutorials and Training, and Analysis Tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef]
- Ji, H.; Xu, Y.; Lu, H.; Zhang, Z. Deep MS/MS-Aided Structural-Similarity Scoring for Unknown Metabolite Identification. Anal. Chem. 2019, 91, 5629–5637. [Google Scholar] [CrossRef]
- Zhu, H.; Ma, X.; Kong, J.Y.; Zhang, M.; Kenttämaa, H.I. Identification of Carboxylate, Phosphate, and Phenoxide Functionalities in Deprotonated Molecules Related to Drug Metabolites via Ion–Molecule Reactions with Water and Diethylhydroxyborane. J. Am. Soc. Mass Spectrom. 2017, 28, 2189–2200. [Google Scholar] [CrossRef]
- Alzarieni, K.Z.; Max, J.P.; Easton, M.; Milton, J.R.; Ma, X.; Dong, X.; Gu, C.; Kenttämaa, H.I. Identification of the Carboxylic Acid Functionality in Protonated Drug Metabolite Model Compounds by Using Tandem Mass Spectrometry Based on Ion-Molecule Reactions Coupled with High Performance Liquid Chromatography. Int. J. Mass Spectrom. 2021, 463, 116551. [Google Scholar] [CrossRef]
- Fine, J.; Kuan-Yu Liu, J.; Beck, A.; Alzarieni, K.Z.; Ma, X.; Boulos, V.M.; Kenttämaa, H.I.; Chopra, G. Graph-Based Machine Learning Interprets and Predicts Diagnostic Isomer-Selective Ion–Molecule Reactions in Tandem Mass Spectrometry. Chem. Sci. 2020, 11, 11849–11858. [Google Scholar] [CrossRef]
- Nardy, A.F.F.R.; Freire-de-Lima, L.; Freire-de-Lima, C.G.; Morrot, A. The Sweet Side of Immune Evasion: Role of Glycans in the Mechanisms of Cancer Progression. Front. Oncol. 2016, 6, 54. [Google Scholar] [CrossRef]
- Wolfert, M.A.; Boons, G.-J. Adaptive Immune Activation: Glycosylation Does Matter. Nat. Chem. Biol. 2013, 9, 776–784. [Google Scholar] [CrossRef]
- Aizpurua-Olaizola, O.; Sastre Toraño, J.; Falcon-Perez, J.M.; Williams, C.; Reichardt, N.; Boons, G.J. Mass Spectrometry for Glycan Biomarker Discovery. Trends Analyt. Chem. 2018, 100, 7–14. [Google Scholar] [CrossRef]
- Tsai, S.-T.; Liew, C.Y.; Hsu, C.; Huang, S.-P.; Weng, W.-C.; Kuo, Y.-H.; Ni, C.-K. Automatic Full Glycan Structural Determination through Logically Derived Sequence Tandem Mass Spectrometry. ChemBioChem 2019, 20, 2351–2359. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.Y.; Yen, C.-C.; Chen, J.-L.; Tsai, S.-T.; Pawar, S.; Wu, C.-Y.; Ni, C.-K. Structural Identification of N-Glycan Isomers Using Logically Derived Sequence Tandem Mass Spectrometry. Commun. Chem. 2021, 4, 92. [Google Scholar] [CrossRef]
- Sun, S.; Huang, C.; Wang, Y.; Liu, Y.; Zhang, J.; Zhou, J.; Gao, F.; Yang, F.; Chen, R.; Mulloy, B.; et al. Toward Automated Identification of Glycan Branching Patterns Using Multistage Mass Spectrometry with Intelligent Precursor Selection. Anal. Chem. 2018, 90, 14412–14422. [Google Scholar] [CrossRef]
- She, Y.-M.; Tam, R.Y.; Li, X.; Rosu-Myles, M.; Sauvé, S. Resolving Isomeric Structures of Native Glycans by Nanoflow Porous Graphitized Carbon Chromatography–Mass Spectrometry. Anal. Chem. 2020, 92, 14038–14046. [Google Scholar] [CrossRef]
- Snyder, C.M.; Zhou, X.; Karty, J.A.; Fonslow, B.R.; Novotny, M.V.; Jacobson, S.C. Capillary Electrophoresis–Mass Spectrometry for Direct Structural Identification of Serum N-Glycans. J. Chromatogr. A 2017, 1523, 127–139. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, X.; Schocker, N.S.; Renslow, R.S.; Orton, D.J.; Khamsi, J.; Ashmus, R.A.; Almeida, I.C.; Tang, K.; Costello, C.E.; et al. Enhancing Glycan Isomer Separations with Metal Ions and Positive and Negative Polarity Ion Mobility Spectrometry-Mass Spectrometry Analyses. Anal. Bioanal. Chem. 2017, 409, 467–476. [Google Scholar] [CrossRef]
- Wei, J.; Tang, Y.; Ridgeway, M.E.; Park, M.A.; Costello, C.E.; Lin, C. Accurate Identification of Isomeric Glycans by Trapped Ion Mobility Spectrometry-Electronic Excitation Dissociation Tandem Mass Spectrometry. Anal. Chem. 2020, 92, 13211–13220. [Google Scholar] [CrossRef]
- Ujma, J.; Ropartz, D.; Giles, K.; Richardson, K.; Langridge, D.; Wildgoose, J.; Green, M.; Pringle, S. Cyclic Ion Mobility Mass Spectrometry Distinguishes Anomers and Open-Ring Forms of Pentasaccharides. J. Am. Soc. Mass Spectrom. 2019, 30, 1028–1037. [Google Scholar] [CrossRef]
- Giri, A.; Coutriade, M.; Racaud, A.; Stefanuto, P.-H.; Okuda, K.; Dane, J.; Cody, R.B.; Focant, J.-F. Compositional Elucidation of Heavy Petroleum Base Oil by GC × GC-EI/PI/CI/FI-TOFMS. J. Mass Spectrom. 2019, 54, 148–157. [Google Scholar] [CrossRef]
- Olanrewaju, C.A.; Ramirez, C.E.; Fernandez-Lima, F. Comprehensive Screening of Polycyclic Aromatic Hydrocarbons and Similar Compounds Using GC–APLI–TIMS–TOFMS/GC–EI–MS. Anal. Chem. 2021, 93, 6080–6087. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Viidanoja, J.; Li, M.; Zhang, Y.; Ikonen, E.; Root, A.; Romanczyk, M.; Manheim, J.; Dziekonski, E.; Kenttämaa, H.I. Comparison of Atmospheric Pressure Chemical Ionization and Field Ionization Mass Spectrometry for the Analysis of Large Saturated Hydrocarbons. Anal. Chem. 2016, 88, 10592–10598. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhang, Y.; Milton, J.; Yerabolu, R.; Easterling, L.; Kenttämaa, H.I. Investigation of the Relative Abundances of Single-Core and Multicore Compounds in Asphaltenes by Using High-Resolution In-Source Collision-Activated Dissociation and Medium-Energy Collision-Activated Dissociation Mass Spectrometry with Statistical Considerations. Fuel 2019, 246, 126–132. [Google Scholar]
- Blaudeau, L.; Kenttämaa, H.I. Tandem Mass Spectrometric Characterization of the Molecular Radical Cations of Asphaltenes. Energy Fuels 2022, 36, 8684–8691. [Google Scholar] [CrossRef]
- Schmidt, E.M.; Pudenzi, M.A.; Santos, J.M.; Angolini, C.F.F.; Pereira, R.C.L.; Rocha, Y.S.; Denisov, E.; Damoc, E.; Makarov, A.; Eberlin, M.N. Petroleomics via Orbitrap Mass Spectrometry with Resolving Power above 1,000,000 at m/z 200. RSC Adv. 2018, 8, 6183–6191. [Google Scholar] [CrossRef]
- Folli, G.S.; Souza, L.M.; Araújo, B.Q.; Vaz, B.G.; Filgueiras, P.R.; Romão, W. Comparing the Intermediate Precision in Petroleomics by Ultrahigh-Resolution Mass Spectrometry. Energy Fuels 2021, 35, 16465–16481. [Google Scholar] [CrossRef]
- Pereira, I.; de Aguiar, D.V.A.; Vasconselos, G.; Vaz, B.G. Chapter 16—Fourier transform mass spectrometry applied to petroleomics. In Fundamentals and Applications of Fourier Transform Mass Spectrometry; Kanawati, B., Schmitt-Kopplin, P., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 509–528. [Google Scholar]
- Budzikiewicz, H. Mass Spectrometry in Natural Product Structure Elucidation. In Progress in the Chemistry of Organic Natural Products 100; Kinghorn, A.D., Falk, H., Kobayashi, J., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 77–221. [Google Scholar]
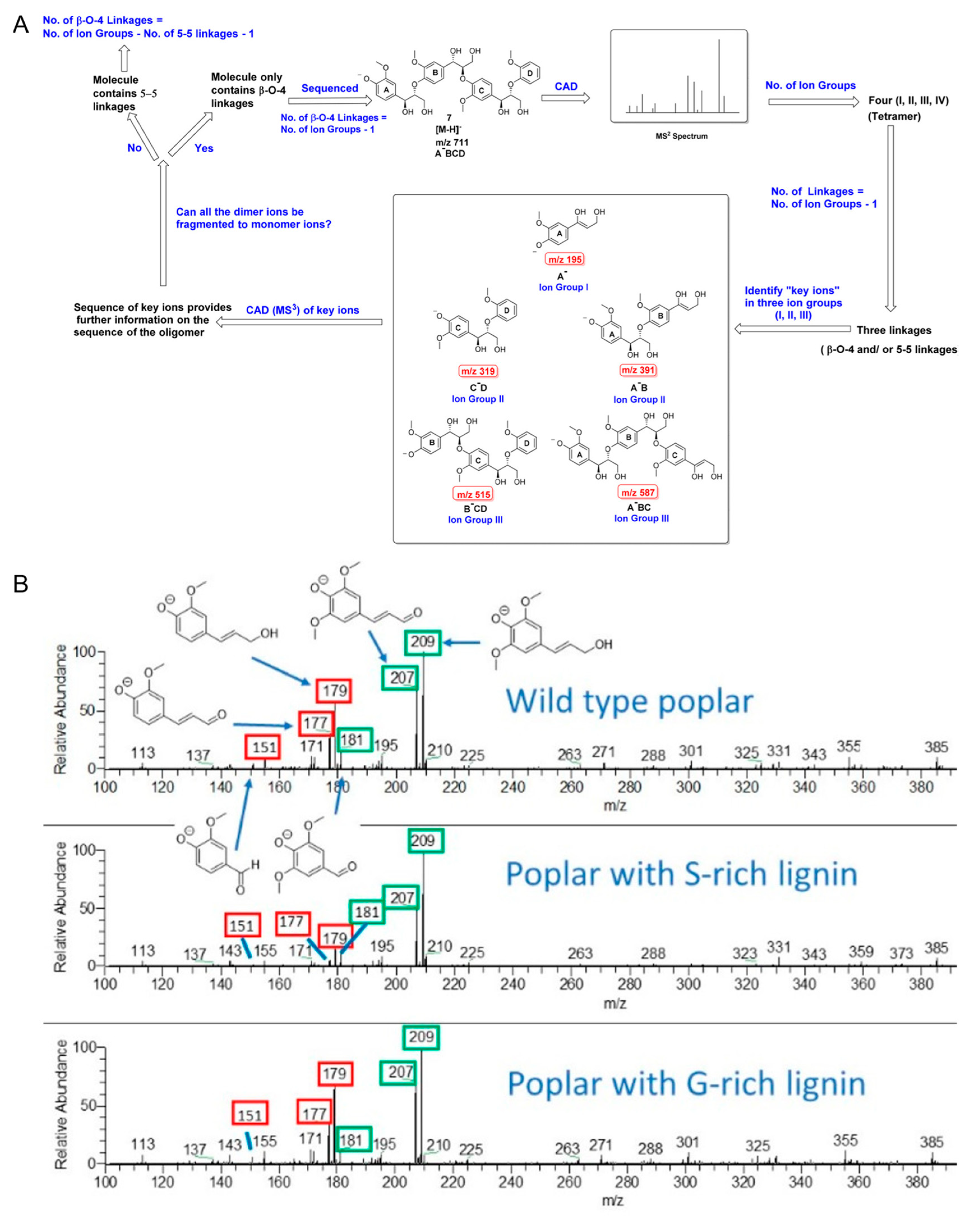
- Sheng, H.; Tang, W.; Gao, J.; Riedeman, J.S.; Li, G.; Jarrell, T.M.; Hurt, M.R.; Yang, L.; Murria, P.; Ma, X.; et al. (−)ESI/CAD MSn Procedure for Sequencing Lignin Oligomers Based on a Study of Synthetic Model Compounds with β-O-4 and 5-5 Linkages. Anal. Chem. 2017, 89, 13089–13096. [Google Scholar] [CrossRef]
- Letourneau, D.R.; Volmer, D.A. Mass Spectrometry-Based Methods for the Advanced Characterization and Structural Analysis of Lignin: A Review. Mass Spectrom. Rev. 2021, 1–45. [Google Scholar] [CrossRef]
- Xu, L.; Ma, X.; Murria, P.; Talpade, A.; Sheng, H.; Meilan, R.; Chapple, C.; Agrawal, R.; Delgass, W.N.; Ribeiro, F.H.; et al. Fast Determination of the Lignin Monomer Compositions of Genetic Variants of Poplar via Fast Pyrolysis/Atmospheric Pressure Chemical Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2021, 32, 2546–2551. [Google Scholar] [CrossRef]
- Cuendet, M.; Pezzuto, J.M. Antitumor Alkaloids in Clinical Use or in Clinical Trials. Mod. Alkaloids 2007, 25–52. [Google Scholar] [CrossRef]
- Ma, C.; Wang, J.; Chu, H.; Zhang, X.; Wang, Z.; Wang, H.; Li, G. Purification and Characterization of Aporphine Alkaloids from Leaves of Nelumbo nucifera Gaertn and Their Effects on Glucose Consumption in 3T3-L1 Adipocytes. Int. J. Mol. Sci. 2014, 15, 3481–3494. [Google Scholar] [CrossRef]
- Carnevale Neto, F.; Andréo, M.A.; Raftery, D.; Lopes, J.L.C.; Lopes, N.P.; Castro-Gamboa, I.; Lameiro de Noronha Sales Maia, B.H.; Costa, E.V.; Vessecchi, R. Characterization of Aporphine Alkaloids by Electrospray Ionization Tandem Mass Spectrometry and Density Functional Theory Calculations. Rapid Commun. Mass Spectrom. 2020, 34, e8533. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, L.; Chen, H.; Chen, S.; Liu, Y. Analysis of Flavonoid Metabolites in Citrus Peels (Citrus reticulata “Dahongpao”) Using UPLC-ESI-MS/MS. Molecules 2019, 24, 2680. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Hou, X.; Wei, Y. Fast Screening of Flavonoids from Switchgrass and Mikania micrantha by Liquid Chromatography Hybrid-Ion Trap Time-of-Flight Mass Spectrometry. Anal. Methods 2018, 10, 109–122. [Google Scholar] [CrossRef]
- Vuckovic, D. Current Trends and Challenges in Sample Preparation for Global Metabolomics Using Liquid Chromatography–Mass Spectrometry. Anal. Bioanal. Chem. 2012, 403, 1523–1548. [Google Scholar] [CrossRef]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from Sample Preparation to Data Analysis: A Primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef]
- Ma, X.; Fernández, F.M. Advances in Mass Spectrometry Imaging for Spatial Cancer Metabolomics. Mass Spectrom. Rev. 2022, e21804. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X. Recent Advances in Mass Spectrometry-Based Structural Elucidation Techniques. Molecules 2022, 27, 6466. https://doi.org/10.3390/molecules27196466
Ma X. Recent Advances in Mass Spectrometry-Based Structural Elucidation Techniques. Molecules. 2022; 27(19):6466. https://doi.org/10.3390/molecules27196466
Chicago/Turabian StyleMa, Xin. 2022. "Recent Advances in Mass Spectrometry-Based Structural Elucidation Techniques" Molecules 27, no. 19: 6466. https://doi.org/10.3390/molecules27196466
APA StyleMa, X. (2022). Recent Advances in Mass Spectrometry-Based Structural Elucidation Techniques. Molecules, 27(19), 6466. https://doi.org/10.3390/molecules27196466




