Evaluation of Biological Activity of Natural Compounds: Current Trends and Methods
Abstract
1. Introduction
2. Cytotoxicity Activity in Cultured Mammalian Cells
3. Antihyperglycemic Activity
| Method Name | Assay Type | Description | Detection (Output) | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|---|
| α-amylase inhibition | Isolated enzymes | Measurement of the ability of novel molecules to inhibit the activity of α-amylase | Colorimetric and fluorometric assays |
|
| [89] |
| α-glucosidase inhibition | Isolated enzymes | Measurement of the ability of novel molecules to inhibit the activity of α-glucosidase | Colorimetric, fluorometric and bioluminescent assays |
|
| [89,90] |
| Dipeptidyl peptidase IV inhibition | Isolated enzymes | Measurement of the ability of molecules to inhibit the activity of dipeptidyl peptidase IV | Colorimetric, fluorometric, immunoassay |
|
| [89,90] |
| Tyrosine phosphatase 1B inhibition | Isolated enzymes | Measurement of the ability of molecules to inhibit the activity of tyrosine phosphatase 1B | Colorimetric, fluorometric |
|
| [74,77] |
| Glucose uptake | Cell-based | Measurement of the ability of molecules to modify glucose uptake into cells | Colorimetric, fluorometric, bioluminescent assays |
|
| [91,92] |
| Insulin secretion | Cell-based | Measurement of the ability of molecules to modulate insulin secretion | Bioluminescent assay, immuno-/radioimmunoassay |
|
| [87,88] |
4. Anti-Inflammatory Activity
5. Analgesic Activity
| Method Name | Assay Type | Description | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| Displacement of radioligands at receptors | 3H-N binding assay | Radiolabeled ligand: 3H-N Type of receptor: opiate Tissue culture: rat brain |
|
| [118,119] |
| 3H-DHM binding assay | Radiolabeled ligand: 3H-DHM Type of receptor: µ-opiate Tissue culture: rat brain | [124] | |||
| 3H-B binding assay | Radiolabeled ligand: 3H-B Type of receptor: κ-opiate Tissue culture: guinea pig cerebellum | [119,131] | |||
| 3H-NCNH2 binding assay | Radiolabeled ligand: 3H-NCNH2 assay Type of receptor: nociceptin Tissue culture: rat brain | [131] | |||
| 35S-GT binding assay | Radiolabeled ligand: 35S-GT Type of receptor: cannabinoid Tissue culture: human brain | [131,132] | |||
| 3H-R binding assay | Radiolabeled ligand: 3H-R Type of receptor: vanilloid Tissue culture: rat brain | [133,134] |
6. Anticoagulant Activity
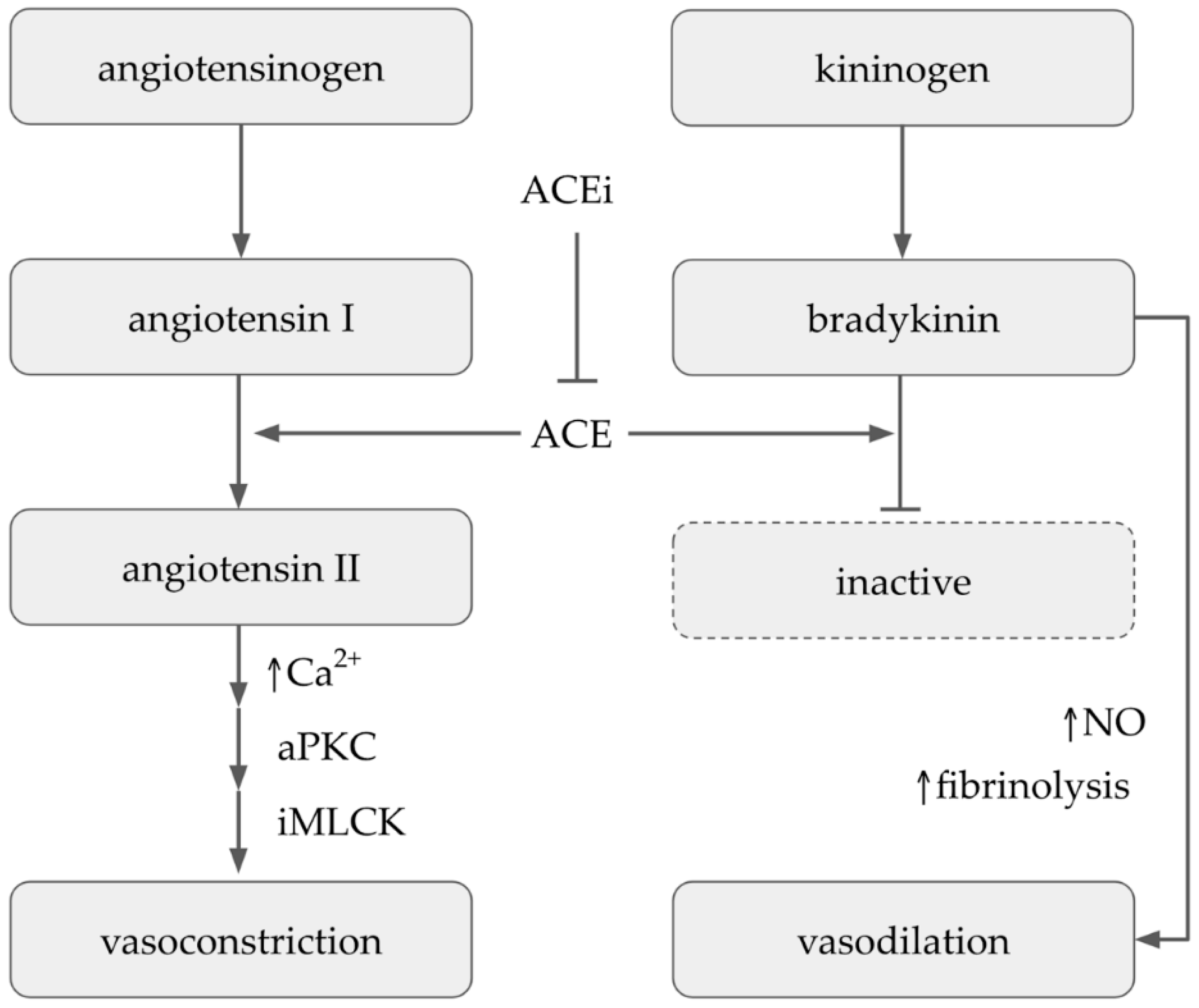
7. Antihypertensive Activity
8. Antioxidant Activity
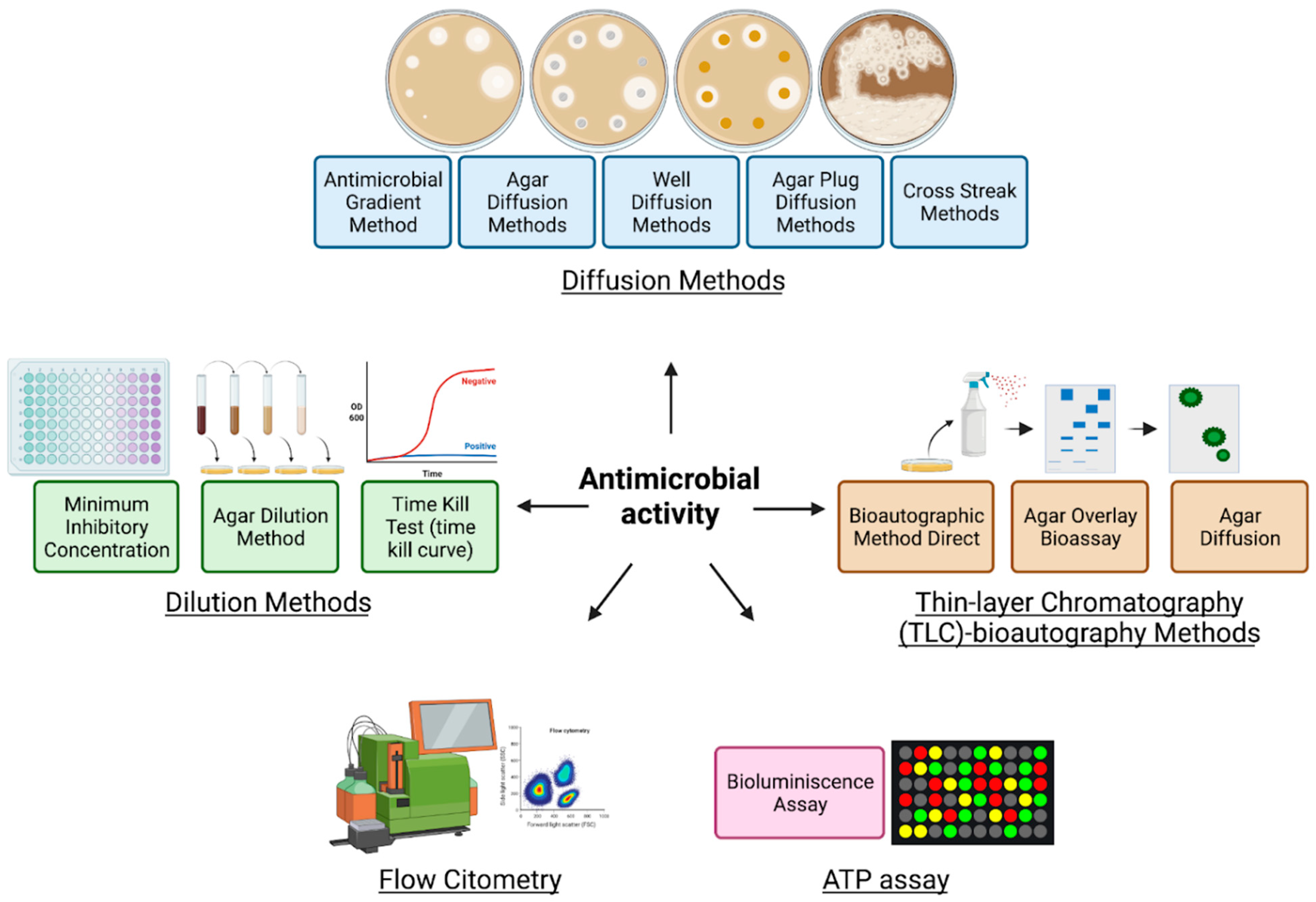
9. Antimicrobial Activity
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malaquias, G.; Santos Cerqueira, G.; Pinheiro Ferreira, P.M.; Landim Pacheco, A.C.; de Castro e Souza, J.M.; do Socorro Meireles de Deus, M.; Peron, A.P. Utilização na medicina popular, potencial terapêutico e toxicidade em nível celular das plantas Rosmarinus officinalis L., Salvia officinalis L. e Mentha piperita L. (Família Lamiaceae). Rev. Intertox Toxicol. Risco Ambient. Soc. 2015, 7, 50–68. [Google Scholar] [CrossRef][Green Version]
- Kingston, D.G.I. Modern natural products drug discovery and its relevance to biodiversity conservation. J. Nat. Prod. 2011, 74, 496–511. [Google Scholar] [CrossRef] [PubMed]
- Fabricant, D.S.; Farnsworth, N.R. The value of plants used in traditional medicine for drug discovery. Environ. Health Perspect. 2001, 109 (Suppl. S1), 69–75. [Google Scholar] [CrossRef] [PubMed]
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef] [PubMed]
- Martin, Y.C.; Kofron, J.L.; Traphagen, L.M. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002, 45, 4350–4358. [Google Scholar] [CrossRef]
- Rasoanaivo, P.; Wright, C.W.; Willcox, M.L.; Gilbert, B. Whole plant extracts versus single compounds for the treatment of malaria: Synergy and positive interactions. Malar. J. 2011, 10 (Suppl. S1), S4. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, B.; Alves, L. Synergy in plant medicines. Curr. Med. Chem. 2003, 10, 13–20. [Google Scholar] [CrossRef]
- Vinken, M.; Blaauboer, B.J. In vitro testing of basal cytotoxicity: Establishment of an adverse outcome pathway from chemical insult to cell death. Toxicol. Vitr. 2017, 39, 104–110. [Google Scholar] [CrossRef]
- Di Nunzio, M.; Valli, V.; Tomás-Cobos, L.; Tomás-Chisbert, T.; Murgui-Bosch, L.; Danesi, F.; Bordoni, A. Is cytotoxicity a determinant of the different in vitro and in vivo effects of bioactives? BMC Complement. Altern. Med. 2017, 17, 453. [Google Scholar] [CrossRef]
- Ling, T.; Lang, W.H.; Maier, J.; Quintana Centurion, M.; Rivas, F. Cytostatic and Cytotoxic Natural Products against Cancer Cell Models. Molecules 2019, 24, 2012. [Google Scholar] [CrossRef]
- Sachana, M.; Hargreaves, A.J. Chapter 9—Toxicological testing: In vivo and in vitro models. In Veterinary Toxicology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 145–161. [Google Scholar]
- Jablonská, E.; Kubásek, J.; Vojtěch, D.; Ruml, T.; Lipov, J. Test conditions can significantly affect the results of in vitro cytotoxicity testing of degradable metallic biomaterials. Sci. Rep. 2021, 11, 6628. [Google Scholar] [CrossRef]
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for cell viability assays. Food Front. 2020, 1, 332–349. [Google Scholar] [CrossRef]
- Riss, T.; Niles, A.; Moravec, R.; Karassina, N.; Vidugiriene, J. Cytotoxicity assays: In vitro methods to measure dead cells. In Assay Guidance Manual; Sittampalam, G.S., Coussens, N.P., Brimacombe, K., Grossman, A., Arkin, M., Auld, D., Austin, C., Baell, J., Bejcek, B., Caaveiro, J.M.M., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Gordon, J.L.; Brown, M.A.; Reynolds, M.M. Cell-Based Methods for Determination of Efficacy for Candidate Therapeutics in the Clinical Management of Cancer. Diseases 2018, 6, 85. [Google Scholar] [CrossRef]
- King, T.C. Cell injury, cellular responses to injury, and cell death. In Elsevier’s Integrated Pathology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–20. ISBN 9780323043281. [Google Scholar]
- McStay, G.P.; Green, D.R. Measuring apoptosis: Caspase inhibitors and activity assays. Cold Spring Harb. Protoc. 2014, 2014, 799–806. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Heller-Uszynska, K.; Kilian, A. Microarray TRAP—A high-throughput assay to quantitate telomerase activity. Biochem. Biophys. Res. Commun. 2004, 323, 465–472. [Google Scholar] [CrossRef]
- Menyhárt, O.; Harami-Papp, H.; Sukumar, S.; Schäfer, R.; Magnani, L.; de Barrios, O.; Győrffy, B. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochim. Biophys. Acta 2016, 1866, 300–319. [Google Scholar] [CrossRef]
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Karuppusamy Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The soft agar colony formation assay. J. Vis. Exp. 2014, 92, e51998. [Google Scholar] [CrossRef]
- Hulkower, K.I.; Herber, R.L. Cell migration and invasion assays as tools for drug discovery. Pharmaceutics 2011, 3, 107–124. [Google Scholar] [CrossRef]
- Grada, A.; Otero-Vinas, M.; Prieto-Castrillo, F.; Obagi, Z.; Falanga, V. Research techniques made simple: Analysis of collective cell migration using the wound healing assay. J. Investig. Dermatol. 2017, 137, e11–e16. [Google Scholar] [CrossRef]
- van Tonder, A.; Joubert, A.M.; Cromarty, A.D. Limitations of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay when compared to three commonly used cell enumeration assays. BMC Res. Notes 2015, 8, 47. [Google Scholar] [CrossRef]
- Akter, S.; Addepalli, R.; Netzel, M.E.; Tinggi, U.; Fletcher, M.T.; Sultanbawa, Y.; Osborne, S.A. Antioxidant-Rich Extracts of Terminalia ferdinandiana Interfere with Estimation of Cell Viability. Antioxidants 2019, 8, 191. [Google Scholar] [CrossRef]
- Niles, A.L.; Moravec, R.A.; Riss, T.L. Update on in vitro cytotoxicity assays for drug development. Expert Opin. Drug Discov. 2008, 3, 655–669. [Google Scholar] [CrossRef]
- Aslantürk, Ö.S. In vitro cytotoxicity and cell viability assays: Principles, advantages, and disadvantages. In Genotoxicity—A Predictable Risk to Our Actual World; Larramendy, M.L., Soloneski, S., Eds.; InTech: London, UK, 2018; ISBN 978-1-78923-418-3. [Google Scholar]
- Jain, A.K.; Singh, D.; Dubey, K.; Maurya, R.; Mittal, S.; Pandey, A.K. Models and methods for in vitro toxicity. In In Vitro Toxicology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 45–65. ISBN 9780128046678. [Google Scholar]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1-3. [Google Scholar] [CrossRef]
- Lebeau, P.F.; Chen, J.; Byun, J.H.; Platko, K.; Austin, R.C. The trypan blue cellular debris assay: A novel low-cost method for the rapid quantification of cell death. MethodsX 2019, 6, 1174–1180. [Google Scholar] [CrossRef]
- Śliwka, L.; Wiktorska, K.; Suchocki, P.; Milczarek, M.; Mielczarek, S.; Lubelska, K.; Cierpiał, T.; Łyżwa, P.; Kiełbasiński, P.; Jaromin, A.; et al. The comparison of MTT and CVS assays for the assessment of anticancer agent interactions. PLoS ONE 2016, 11, e0155772. [Google Scholar] [CrossRef]
- Bopp, S.K.; Lettieri, T. Comparison of four different colorimetric and fluorometric cytotoxicity assays in a zebrafish liver cell line. BMC Pharmacol. 2008, 8, 8. [Google Scholar] [CrossRef]
- Barile, F.A. Continuous cell lines as a model for drug toxicity assessment. In Vitro Methods in Pharmaceutical Research; Elsevier: Amsterdam, The Netherlands, 1997; pp. 33–54. ISBN 9780121633905. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Vega-Avila, E.; Pugsley, M.K. An overview of colorimetric assay methods used to assess survival or proliferation of mammalian cells. Proc. West. Pharmacol. Soc. 2011, 54, 10–14. [Google Scholar]
- Walzl, A.; Unger, C.; Kramer, N.; Unterleuthner, D.; Scherzer, M.; Hengstschläger, M.; Schwanzer-Pfeiffer, D.; Dolznig, H. The Resazurin Reduction Assay Can Distinguish Cytotoxic from Cytostatic Compounds in Spheroid Screening Assays. J. Biomol. Screen. 2014, 19, 1047–1059. [Google Scholar] [CrossRef]
- Präbst, K.; Engelhardt, H.; Ringgeler, S.; Hübner, H. Basic colorimetric proliferation assays: MTT, WST, and resazurin. Methods Mol. Biol. 2017, 1601, 1–17. [Google Scholar] [CrossRef]
- Jakštys, B.; Ruzgys, P.; Tamošiūnas, M.; Šatkauskas, S. Different cell viability assays reveal inconsistent results after bleomycin electrotransfer in vitro. J. Membr. Biol. 2015, 248, 857–863. [Google Scholar] [CrossRef]
- Gutiérrez, L.; Stepien, G.; Gutiérrez, L.; Pérez-Hernández, M.; Pardo, J.; Pardo, J.; Grazú, V.; de la Fuente, J.M. Nanotechnology in drug discovery and development. In Comprehensive Medicinal Chemistry III; Elsevier: Amsterdam, The Netherlands, 2017; pp. 264–295. ISBN 9780128032015. [Google Scholar]
- Weisenthal, L.M.; Lippman, M.E. Clonogenic and nonclonogenic in vitro chemosensitivity assays. Cancer Treat. Rep. 1985, 69, 615–632. [Google Scholar]
- Weisenthal, L.M.; Dill, P.L.; Kurnick, N.B.; Lippman, M.E. Comparison of dye exclusion assays with a clonogenic assay in the determination of drug-induced cytotoxicity. Cancer Res. 1983, 43, 258–264. [Google Scholar]
- Juan-García, A.; Taroncher, M.; Font, G.; Ruiz, M.-J. Micronucleus induction and cell cycle alterations produced by deoxynivalenol and its acetylated derivatives in individual and combined exposure on HepG2 cells. Food Chem. Toxicol. 2018, 118, 719–725. [Google Scholar] [CrossRef]
- Wu, H.; Chen, L.; Zhu, F.; Han, X.; Sun, L.; Chen, K. The cytotoxicity effect of resveratrol: Cell cycle arrest and induced apoptosis of breast cancer 4T1 cells. Toxins 2019, 11, 731. [Google Scholar] [CrossRef]
- Wlodkowic, D.; Telford, W.; Skommer, J.; Darzynkiewicz, Z. Apoptosis and beyond: Cytometry in studies of programmed cell death. Methods Cell Biol. 2011, 103, 55–98. [Google Scholar] [CrossRef]
- Rieger, A.M.; Nelson, K.L.; Konowalchuk, J.D.; Barreda, D.R. Modified annexin V/propidium iodide apoptosis assay for accurate assessment of cell death. J. Vis. Exp. 2011, 50, 2597. [Google Scholar] [CrossRef]
- Khazaei, S.; Esa, N.M.; Ramachandran, V.; Hamid, R.A.; Pandurangan, A.K.; Etemad, A.; Ismail, P. In vitro Antiproliferative and Apoptosis Inducing Effect of Allium atroviolaceum Bulb Extract on Breast, Cervical, and Liver Cancer Cells. Front. Pharmacol. 2017, 8, 5. [Google Scholar] [CrossRef]
- Ballav, S.; Jaywant Deshmukh, A.; Siddiqui, S.; Aich, J.; Basu, S. Two-Dimensional and Three-Dimensional Cell Culture and Their Applications. In Cell Culture—Advanced Technology and Applications in Medical and Life Sciences; Biochemistry; IntechOpen: London, UK, 2021. [Google Scholar]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- Berrouet, C.; Dorilas, N.; Rejniak, K.A.; Tuncer, N. Comparison of Drug Inhibitory Effects (IC50) in Monolayer and Spheroid Cultures. Bull. Math. Biol. 2020, 82, 68. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.Z.; Wagner, D.C.; Hörner, N.; Horst, D.; Lang, H.; Tagscherer, K.E.; Roth, W. Ex vivo tissue slice culture system to measure drug-response rates of hepatic metastatic colorectal cancer. BMC Cancer 2019, 19, 1030. [Google Scholar] [CrossRef]
- Roelants, C.; Pillet, C.; Franquet, Q.; Sarrazin, C.; Peilleron, N.; Giacosa, S.; Guyon, L.; Fontanell, A.; Fiard, G.; Long, J.-A.; et al. Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate Targeted Therapies for Patients with Renal Carcinoma. Cancers 2020, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Koerfer, J.; Kallendrusch, S.; Merz, F.; Wittekind, C.; Kubick, C.; Kassahun, W.T.; Schumacher, G.; Moebius, C.; Gaßler, N.; Schopow, N.; et al. Organotypic slice cultures of human gastric and esophagogastric junction cancer. Cancer Med. 2016, 5, 1444–1453. [Google Scholar] [CrossRef]
- Temblador, A.; Topalis, D.; van den Oord, J.; Andrei, G.; Snoeck, R. Organotypic Epithelial Raft Cultures as a Three-Dimensional In Vitro Model of Merkel Cell Carcinoma. Cancers 2022, 14, 1091. [Google Scholar] [CrossRef]
- Lin, R.-Z.; Lin, R.-Z.; Chang, H.-Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef]
- Cui, X.; Hartanto, Y.; Zhang, H. Advances in multicellular spheroids formation. J. R. Soc. Interface 2017, 14, 20160877. [Google Scholar] [CrossRef]
- Shankaran, A.; Prasad, K.; Chaudhari, S.; Brand, A.; Satyamoorthy, K. Advances in development and application of human organoids. 3 Biotech 2021, 11, 257. [Google Scholar] [CrossRef]
- Caipa Garcia, A.L.; Arlt, V.M.; Phillips, D.H. Organoids for toxicology and genetic toxicology: Applications with drugs and prospects for environmental carcinogenesis. Mutagenesis 2022, 37, 143–154. [Google Scholar] [CrossRef]
- Fey, S.J.; Wrzesinski, K. Determination of drug toxicity using 3D spheroids constructed from an immortal human hepatocyte cell line. Toxicol. Sci. 2012, 127, 403–411. [Google Scholar] [CrossRef]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef]
- Kitaeva, K.V.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Cell Culture Based in vitro Test Systems for Anticancer Drug Screening. Front. Bioeng. Biotechnol. 2020, 8, 322. [Google Scholar] [CrossRef]
- Garcia-Hernando, M.; Benito-Lopez, F.; Basabe-Desmonts, L. Advances in microtechnology for improved cytotoxicity assessment. Front. Mater. 2020, 7, 582030. [Google Scholar] [CrossRef]
- Mi, S.; Du, Z.; Xu, Y.; Wu, Z.; Qian, X.; Zhang, M.; Sun, W. Microfluidic co-culture system for cancer migratory analysis and anti-metastatic drugs screening. Sci. Rep. 2016, 6, 35544. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Bhise, N.S.; Ribas, J.; Manoharan, V.; Zhang, Y.S.; Polini, A.; Massa, S.; Dokmeci, M.R.; Khademhosseini, A. Organ-on-a-chip platforms for studying drug delivery systems. J. Control. Release 2014, 190, 82–93. [Google Scholar] [CrossRef]
- Lubamba, B.; Jensen, T.; McClelland, R. Rapid Detection of Direct Compound Toxicity and Trailing Detection of Indirect Cell Metabolite Toxicity in a 96-Well Fluidic Culture Device for Cell-Based Screening Environments: Tactics in Six Sigma Quality Control Charts. Appl. Sci. 2022, 12, 2786. [Google Scholar] [CrossRef]
- Roglic, G.; World Health Organization. Global Report on Diabetes; World Health Organization: Geneva, Switzerland, 2016; ISBN 9789241565257. [Google Scholar]
- Reyes, B.A.S.; Dufourt, E.C.; Ross, J.; Warner, M.J.; Tanquilut, N.C.; Leung, A.B. Selected phyto and marine bioactive compounds: Alternatives for the treatment of type 2 diabetes. In Studies in Natural Products Chemistry; Elsevier: Amsterdam, The Netherlands, 2017; Volume 55, pp. 111–143. ISBN 9780444640680. [Google Scholar]
- Thete, M.; Dilip, A. Recent advances and methods for in-vitro evaluation of antidiabetic activity: A review. Int. J. Eng. Appl. Sci. Technol. 2020, 4, 194–198. [Google Scholar]
- Gromova, L.V.; Fetissov, S.O.; Gruzdkov, A.A. Mechanisms of glucose absorption in the small intestine in health and metabolic diseases and their role in appetite regulation. Nutrients 2021, 13, 2474. [Google Scholar] [CrossRef]
- Xue, B.; Kim, Y.-B.; Lee, A.; Toschi, E.; Bonner-Weir, S.; Kahn, C.R.; Neel, B.G.; Kahn, B.B. Protein-tyrosine phosphatase 1B deficiency reduces insulin resistance and the diabetic phenotype in mice with polygenic insulin resistance. J. Biol. Chem. 2007, 282, 23829–23840. [Google Scholar] [CrossRef] [PubMed]
- Deacon, C.F. Physiology and Pharmacology of DPP-4 in Glucose Homeostasis and the Treatment of Type 2 Diabetes. Front. Endocrinol. 2019, 10, 80. [Google Scholar] [CrossRef]
- Prabhakar, P.K.; Sivakumar, P.M. Protein Tyrosine Phosphatase 1B Inhibitors: A Novel Therapeutic Strategy for the Management of type 2 Diabetes Mellitus. Curr. Pharm. Des. 2019, 25, 2526–2539. [Google Scholar] [CrossRef] [PubMed]
- Mulvihill, E.E.; Drucker, D.J. Pharmacology, physiology, and mechanisms of action of dipeptidyl peptidase-4 inhibitors. Endocr. Rev. 2014, 35, 992–1019. [Google Scholar] [CrossRef]
- Omar, B.; Ahrén, B. Pleiotropic mechanisms for the glucose-lowering action of DPP-4 inhibitors. Diabetes 2014, 63, 2196–2202. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.-C.; Saha, S.; Chernoff, J. PTP1B: A double agent in metabolism and oncogenesis. Trends Biochem. Sci. 2010, 35, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Elchebly, M.; Payette, P.; Michaliszyn, E.; Cromlish, W.; Collins, S.; Loy, A.L.; Normandin, D.; Cheng, A.; Himms-Hagen, J.; Chan, C.C.; et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 1999, 283, 1544–1548. [Google Scholar] [CrossRef]
- Csepregi, R.; Temesfői, V.; Sali, N.; Poór, M.; Needs, P.W.; Kroon, P.A.; Kőszegi, T. A One-Step Extraction and Luminescence Assay for Quantifying Glucose and ATP Levels in Cultured HepG2 Cells. Int. J. Mol. Sci. 2018, 19, 2670. [Google Scholar] [CrossRef] [PubMed]
- Blodgett, A.B.; Kothinti, R.K.; Kamyshko, I.; Petering, D.H.; Kumar, S.; Tabatabai, N.M. A fluorescence method for measurement of glucose transport in kidney cells. Diabetes Technol. Ther. 2011, 13, 743–751. [Google Scholar] [CrossRef]
- Dong, S.; Alahari, S. FACS-based Glucose Uptake Assay of Mouse Embryonic Fibroblasts and Breast Cancer Cells Using 2-NBDG Probe. Bio Protoc. 2018, 8, e2816. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, V.P. Mechanism of glucose transport across the yeast cell membrane. J. Bacteriol. 1962, 84, 485–491. [Google Scholar] [CrossRef]
- Schmidl, S.; Iancu, C.V.; Reifenrath, M.; Choe, J.-Y.; Oreb, M. A label-free real-time method for measuring glucose uptake kinetics in yeast. FEMS Yeast Res. 2021, 21, foaa069. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Noh, S.; Lim, S.; Kim, B. Plant extracts for type 2 diabetes: From traditional medicine to modern drug discovery. Antioxidants 2021, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Jakab, M.; Jav, S.; Streif, D.; Pitschmann, A.; Zehl, M.; Purevsuren, S.; Glasl, S.; Ritter, M. Extracts from Leonurus sibiricus L. increase insulin secretion and proliferation of rat INS-1E insulinoma cells. J. Ethnopharmacol. 2013, 150, 85–94. [Google Scholar] [CrossRef]
- Ansari, P.; Flatt, P.R.; Harriott, P.; Abdel-Wahab, Y.H.A. Insulin secretory and antidiabetic actions of Heritiera fomes bark together with isolation of active phytomolecules. PLoS ONE 2022, 17, e0264632. [Google Scholar] [CrossRef] [PubMed]
- Hager, R.; Pitsch, J.; Kerbl-Knapp, J.; Neuhauser, C.; Ollinger, N.; Iken, M.; Ranner, J.; Mittermeier-Kleßinger, V.; Dawid, C.; Lanzerstorfer, P.; et al. A High-Content Screen for the Identification of Plant Extracts with Insulin Secretion-Modulating Activity. Pharmaceuticals 2021, 14, 809. [Google Scholar] [CrossRef]
- Kalwat, M.A.; Wichaidit, C.; Nava Garcia, A.Y.; McCoy, M.K.; McGlynn, K.; Hwang, I.H.; MacMillan, J.B.; Posner, B.A.; Cobb, M.H. Insulin promoter-driven Gaussia luciferase-based insulin secretion biosensor assay for discovery of β-cell glucose-sensing pathways. ACS Sens. 2016, 1, 1208–1212. [Google Scholar] [CrossRef]
- Bhatia, A.; Singh, B.; Arora, R.; Arora, S. In vitro evaluation of the α-glucosidase inhibitory potential of methanolic extracts of traditionally used antidiabetic plants. BMC Complement. Altern. Med. 2019, 19, 74. [Google Scholar] [CrossRef]
- Mechchate, H.; Es-Safi, I.; Louba, A.; Alqahtani, A.S.; Nasr, F.A.; Noman, O.M.; Farooq, M.; Alharbi, M.S.; Alqahtani, A.; Bari, A.; et al. In Vitro Alpha-Amylase and Alpha-Glucosidase Inhibitory Activity and In Vivo Antidiabetic Activity of Withania frutescens L. Foliar Extract. Molecules 2021, 26, 293. [Google Scholar] [CrossRef]
- Yamamoto, N.; Ueda-Wakagi, M.; Sato, T.; Kawasaki, K.; Sawada, K.; Kawabata, K.; Akagawa, M.; Ashida, H. Measurement of glucose uptake in cultured cells. Curr. Protoc. Pharmacol. 2015, 71, 12.14.1–12.14.26. [Google Scholar] [CrossRef] [PubMed]
- Maric, T.; Mikhaylov, G.; Khodakivskyi, P.; Bazhin, A.; Sinisi, R.; Bonhoure, N.; Yevtodiyenko, A.; Jones, A.; Muhunthan, V.; Abdelhady, G.; et al. Bioluminescent-based imaging and quantification of glucose uptake in vivo. Nat. Methods 2019, 16, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef]
- Shehata, I.A.; El-harshany, E.; Abdallah, H.M.; Esmat, A.; Abdel-sattar, E.A. Anti-inflammatory activity of Kleinia odora. Eur. J. Integr. Med. 2018, 23, 64–69. [Google Scholar] [CrossRef]
- Ondua, M.; Njoya, E.M.; Abdalla, M.A.; McGaw, L.J. Anti-inflammatory and antioxidant properties of leaf extracts of eleven South African medicinal plants used traditionally to treat inflammation. J. Ethnopharmacol. 2019, 234, 27–35. [Google Scholar] [CrossRef]
- Chan, P.-M.; Tan, Y.-S.; Chua, K.-H.; Sabaratnam, V.; Kuppusamy, U.R. Attenuation of Inflammatory Mediators (TNF-α and Nitric Oxide) and Up-Regulation of IL-10 by Wild and Domesticated Basidiocarps of Amauroderma rugosum (Blume & T. Nees) Torrend in LPS-Stimulated RAW264.7 Cells. PLoS ONE 2015, 10, e0139593. [Google Scholar] [CrossRef]
- Abdulkhaleq, L.A.; Assi, M.A.; Abdullah, R.; Zamri-Saad, M.; Taufiq-Yap, Y.H.; Hezmee, M.N.M. The crucial roles of inflammatory mediators in inflammation: A review. Vet. World 2018, 11, 627–635. [Google Scholar] [CrossRef]
- Oguntibeju, O.O. Medicinal plants with anti-inflammatory activities from selected countries and regions of Africa. J. Inflamm. Res. 2018, 11, 307–317. [Google Scholar] [CrossRef]
- Alamgir, A.N.M. Phytoconstituents—Active and inert constituents, metabolic pathways, chemistry and application of phytoconstituents, primary metabolic products, and bioactive compounds of primary metabolic origin. In Therapeutic Use of Medicinal Plants and Their Extracts: Volume 2; Progress in Drug Research; Springer: Cham, Switzerland, 2018; Volume 74, pp. 25–164. ISBN 978-3-319-92386-4. [Google Scholar]
- Ożarowski, M.; Karpiński, T.M. Extracts and Flavonoids of Passiflora Species as Promising Anti-inflammatory and Antioxidant Substances. Curr. Pharm. Des. 2021, 27, 2582–2604. [Google Scholar] [CrossRef]
- Xu, Y.-B.; Chen, G.-L.; Guo, M.-Q. Antioxidant and Anti-Inflammatory Activities of the Crude Extracts of Moringa oleifera from Kenya and Their Correlations with Flavonoids. Antioxidants 2019, 8, 296. [Google Scholar] [CrossRef]
- Blando, F.; Calabriso, N.; Berland, H.; Maiorano, G.; Gerardi, C.; Carluccio, M.A.; Andersen, Ø.M. Radical Scavenging and Anti-Inflammatory Activities of Representative Anthocyanin Groupings from Pigment-Rich Fruits and Vegetables. Int. J. Mol. Sci. 2018, 19, 169. [Google Scholar] [CrossRef] [PubMed]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A review of its effects on human health. Foods 2017, 6, 92. [Google Scholar] [CrossRef]
- Studzińska-Sroka, E.; Dubino, A. Lichens as a source of chemical compounds with anti-inflammatory activity. Herba Pol. 2018, 64, 56–64. [Google Scholar] [CrossRef]
- Wu, G.-J.; Shiu, S.-M.; Hsieh, M.-C.; Tsai, G.-J. Anti-inflammatory activity of a sulfated polysaccharide from the brown alga Sargassum cristaefolium. Food Hydrocoll. 2016, 53, 16–23. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Zeng, K.-W. Natural products as a crucial source of anti-inflammatory drugs: Recent trends and advancements—ProQuest. TMR 2019, 4, 257–268. [Google Scholar] [CrossRef]
- Chan, P.-M.; Kanagasabapathy, G.; Tan, Y.-S.; Sabaratnam, V.; Kuppusamy, U.R. Amauroderma rugosum (Blume & T. Nees) Torrend: Nutritional Composition and Antioxidant and Potential Anti-Inflammatory Properties. Evid. Based Complement. Alternat. Med. 2013, 2013, 304713. [Google Scholar] [CrossRef] [PubMed]
- Benevides Bahiense, J.; Marques, F.M.; Figueira, M.M.; Vargas, T.S.; Kondratyuk, T.P.; Endringer, D.C.; Scherer, R.; Fronza, M. Potential anti-inflammatory, antioxidant and antimicrobial activities of Sambucus australis. Pharm. Biol. 2017, 55, 991–997. [Google Scholar] [CrossRef]
- Rao, U.M. In vitro nitric oxide scavenging and anti inflammatory activities of different solvent extracts of various parts of Musa paradisiaca. Malays. J. Anal. Sci. 2016, 20, 1191–1202. [Google Scholar] [CrossRef]
- Hunter, R.A.; Storm, W.L.; Coneski, P.N.; Schoenfisch, M.H. Inaccuracies of nitric oxide measurement methods in biological media. Anal. Chem. 2013, 85, 1957–1963. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, X.; Broderick, M.; Fein, H. Measurement of nitric oxide production in biological systems by using griess reaction assay. Sensors 2003, 3, 276–284. [Google Scholar] [CrossRef]
- Bryan, N.S.; Grisham, M.B. Methods to detect nitric oxide and its metabolites in biological samples. Free Radic. Biol. Med. 2007, 43, 645–657. [Google Scholar] [CrossRef]
- Crowther, J.R. The ELISA Guidebook; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 516, ISBN 978-1-60327-253-7. [Google Scholar]
- Deepak, S.; Kottapalli, K.; Rakwal, R.; Oros, G.; Rangappa, K.; Iwahashi, H.; Masuo, Y.; Agrawal, G. Real-Time PCR: Revolutionizing Detection and Expression Analysis of Genes. Curr. Genom. 2007, 8, 234–251. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, Q.-L.; Hou, S.-B.; Chen, G. Chemical constituents from the rhizomes of Polygonatum sibiricum Red. and anti-inflammatory activity in RAW264.7 macrophage cells. Nat. Prod. Res. 2019, 33, 2359–2362. [Google Scholar] [CrossRef]
- del Carmen Pinto, M.; Tejeda, A.; Duque, A.L.; Macías, P. Determination of lipoxygenase activity in plant extracts using a modified ferrous oxidation-xylenol orange assay. J. Agric. Food Chem. 2007, 55, 5956–5959. [Google Scholar] [CrossRef]
- Gunathilake, K.D.P.P.; Ranaweera, K.K.D.S.; Rupasinghe, H.P.V. Influence of Boiling, Steaming and Frying of Selected Leafy Vegetables on the In Vitro Anti-inflammation Associated Biological Activities. Plants 2018, 7, 22. [Google Scholar] [CrossRef]
- Marzouk, B.; Marzouk, Z.; Haloui, E.; Fenina, N.; Bouraoui, A.; Aouni, M. Screening of analgesic and anti-inflammatory activities of Citrullus colocynthis from southern Tunisia. J. Ethnopharmacol. 2010, 128, 15–19. [Google Scholar] [CrossRef]
- Auld, D.S.; Farmen, M.W.; Kahl, S.D.; Kriauciunas, A.; McKnight, K.L.; Montrose, C.; Weidner, J.R. Receptor Binding Assays for HTS and Drug Discovery. In Assay Guidance Manual; Sittampalam, G.S., Coussens, N.P., Nelson, H., Arkin, M., Auld, D., Austin, C., Bejcek, B., Glicksman, M., Inglese, J., Iversen, P.W., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Koyyalagunta, D. Opioid Analgesics. In Pain Management; Elsevier: Amsterdam, The Netherlands, 2007; pp. 939–964. ISBN 9780721603346. [Google Scholar]
- Vardanyan, R.S.; Hruby, V.J. Analgesics. In Synthesis of Essential Drugs; Elsevier: Amsterdam, The Netherlands, 2006; pp. 19–55. ISBN 9780444521668. [Google Scholar]
- Sharma, A. Various biological activities of DHA derivatives. In Dehydroacetic Acid and Its Derivatives; Elsevier: Amsterdam, The Netherlands, 2017; pp. 81–118. ISBN 9780081019269. [Google Scholar]
- Sullivan, K.R.; Cammarano, W.B.; Wiener-Kronish, J.P. Analgesics, tranquilizers, and sedatives. In Cardiac Intensive Care; Elsevier: Amsterdam, The Netherlands, 2010; pp. 504–515. ISBN 9781416037736. [Google Scholar]
- Eguchi, K.; Makimura, M.; Murakoshi, Y. Properties of opiate receptor binding in an opiate tolerant state. In Advances in Endogenous and Exogenous Opioids; Elsevier: Amsterdam, The Netherlands, 1981; pp. 428–430. ISBN 9780444804020. [Google Scholar]
- Yunes, R.A.; Filho, V.C.; Ferreira, J.; Calixto, J.B. The use of natural products as sources of new analgesic drugs. In Bioactive Natural Products (Part K); Studies in natural products chemistry; Elsevier: Amsterdam, The Netherlands, 2005; Volume 30, pp. 191–212. ISBN 9780444518545. [Google Scholar]
- Pert, C.B.; Snyder, S.H. Properties of opiate-receptor binding in rat brain. Proc. Natl. Acad. Sci. USA 1973, 70, 2243–2247. [Google Scholar] [CrossRef]
- Vogel, H.G.; Vogel, W.H.; Schölkens, B.A.; Sandow, J.; Müller, G.; Vogel, W.F. Analgesic, anti-inflammatory, and anti-pyretic activity1. In Drug Discovery and Evaluation; Vogel, H.G., Vogel, W.H., Schölkens, B.A., Sandow, J., Müller, G., Vogel, W.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 670–773. ISBN 978-3-540-29837-3. [Google Scholar]
- Wang, T.-X.; Wu, G.-J.; Jiang, J.-G. Natural Products with Analgesic Effect from Herbs and Nutraceuticals Used in Traditional Chinese Medicines. Curr. Mol. Med. 2020, 20, 461–483. [Google Scholar] [CrossRef]
- Sajid-Ur-Rehman, M.; Ishtiaq, S.; Khan, M.A.; Alshamrani, M.; Younus, M.; Shaheen, G.; Abdullah, M.; Sarwar, G.; Khan, M.S.; Javed, F. Phytochemical profiling, in vitro and in vivo anti-inflammatory, analgesic and antipyretic potential of Sesuvium sesuvioides (Fenzl) Verdc. (Aizoaceae). Inflammopharmacology 2021, 29, 789–800. [Google Scholar] [CrossRef]
- Alvarez-Perez, B.; Poras, H.; Maldonado, R. The inhibition of enkephalin catabolism by dual enkephalinase inhibitor: A novel possible therapeutic approach for opioid use disorders. Br. J. Pharmacol. 2021, 1, 1–15. [Google Scholar] [CrossRef]
- Vogel, H.G. (Ed.) Drug Discovery and Evaluation: Pharmacological Assays; Springer: Berlin/Heidelberg, Germany, 2008; ISBN 978-3-540-70995-4. [Google Scholar]
- Harrison, C.; Traynor, J.R. The [35S]GTPγS binding assay: Approaches and applications in pharmacology. Life Sci. 2003, 74, 489–508. [Google Scholar] [CrossRef] [PubMed]
- Finn, D. TRPV1 (VR-1) Vanilloid Receptor. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–8. ISBN 9780080552323. [Google Scholar]
- Messeguer, A.; Planells-Cases, R.; Ferrer-Montiel, A. Physiology and pharmacology of the vanilloid receptor. Curr. Neuropharmacol. 2006, 4, 1–15. [Google Scholar] [CrossRef]
- Thanawala, V.; Kadam, V.J.; Ghosh, R. Enkephalinase inhibitors: Potential agents for the management of pain. Curr. Drug Targets 2008, 9, 887–894. [Google Scholar] [CrossRef]
- Benyamin, R.; Trescot, A.M.; Datta, S.; Buenaventura, R.; Adlaka, R.; Sehgal, N.; Glaser, S.E.; Vallejo, R. Opioid complications and side effects. Pain Physician 2008, 11, S105–S120. [Google Scholar] [CrossRef]
- González-Rodríguez, S.; Poras, H.; Menéndez, L.; Lastra, A.; Ouimet, T.; Fournié-Zaluski, M.-C.; Roques, B.P.; Baamonde, A. Synergistic combinations of the dual enkephalinase inhibitor PL265 given orally with various analgesic compounds acting on different targets, in a murine model of cancer-induced bone pain. Scand. J. Pain 2017, 14, 25–38. [Google Scholar] [CrossRef]
- Wang, K.-H.; Li, S.-F.; Zhao, Y.; Li, H.-X.; Zhang, L.-W. In vitro anticoagulant activity and active components of safflower injection. Molecules 2018, 23, 170. [Google Scholar] [CrossRef]
- de Sousa, D.P. Analgesic-like activity of essential oils constituents. Molecules 2011, 16, 2233–2252. [Google Scholar] [CrossRef]
- Vetter, I.; Deuis, J.R.; Mueller, A.; Israel, M.R.; Starobova, H.; Zhang, A.; Rash, L.D.; Mobli, M. NaV1.7 as a pain target—From gene to pharmacology. Pharmacol. Ther. 2017, 172, 73–100. [Google Scholar] [CrossRef]
- Mueller, A.; Starobova, H.; Morgan, M.; Dekan, Z.; Cheneval, O.; Schroeder, C.I.; Alewood, P.F.; Deuis, J.R.; Vetter, I. Antiallodynic effects of the selective NaV1.7 inhibitor Pn3a in a mouse model of acute postsurgical pain: Evidence for analgesic synergy with opioids and baclofen. Pain 2019, 160, 1766–1780. [Google Scholar] [CrossRef] [PubMed]
- Jin, N.Z.; Gopinath, S.C.B. Potential blood clotting factors and anticoagulants. Biomed. Pharmacother. 2016, 84, 356–365. [Google Scholar] [CrossRef]
- Brinkman, H.J.M. Global assays and the management of oral anticoagulation. Thromb. J. 2015, 13, 9. [Google Scholar] [CrossRef] [PubMed]
- Leite, P.M.; Miranda, A.P.N.; Amorim, J.M.; Duarte, R.C.F.; Bertolucci, S.K.V.; Carvalho, M.; das Gracas Carvalho, M.; Castilho, R.O. In Vitro Anticoagulant Activity of Mikania laevigata: Deepening the Study of the Possible Interaction between Guaco and Anticoagulants. J. Cardiovasc. Pharmacol. 2019, 74, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.A.; Mehta, A. Target specific anticoagulant peptides: A review. Int. J. Pept. Res. Ther. 2018, 24, 1–12. [Google Scholar] [CrossRef]
- Hirsh, J.; Anand, S.S.; Halperin, J.L.; Fuster, V. Mechanism of action and pharmacology of unfractionated heparin. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Z.; Linhardt, R.J. Lessons learned from the contamination of heparin. Nat. Prod. Rep. 2009, 26, 313–321. [Google Scholar] [CrossRef]
- Onishi, A.; St Ange, K.; Dordick, J.S.; Linhardt, R.J. Heparin and anticoagulation. Front. Biosci. 2016, 21, 1372–1392. [Google Scholar] [CrossRef]
- Gogoi, D.; Ramani, S.; Bhartari, S.; Chattopadhyay, P.; Mukherjee, A.K. Characterization of active anticoagulant fraction and a fibrin(ogen)olytic serine protease from leaves of Clerodendrum colebrookianum, a traditional ethno-medicinal plant used to reduce hypertension. J. Ethnopharmacol. 2019, 243, 112099. [Google Scholar] [CrossRef]
- Cai, W.; Xu, H.; Xie, L.; Sun, J.; Sun, T.; Wu, X.; Fu, Q. Purification, characterization and in vitro anticoagulant activity of polysaccharides from Gentiana scabra Bunge roots. Carbohydr. Polym. 2016, 140, 308–313. [Google Scholar] [CrossRef]
- Ghlissi, Z.; Krichen, F.; Kallel, R.; Amor, I.B.; Boudawara, T.; Gargouri, J.; Zeghal, K.; Hakim, A.; Bougatef, A.; Sahnoun, Z. Sulfated polysaccharide isolated from Globularia alypum L.: Structural characterization, in vivo and in vitro anticoagulant activity, and toxicological profile. Int. J. Biol. Macromol. 2019, 123, 335–342. [Google Scholar] [CrossRef]
- Ryu, R.; Jung, U.J.; Kim, H.-J.; Lee, W.; Bae, J.-S.; Park, Y.B.; Choi, M.-S. Anticoagulant and Antiplatelet Activities of Artemisia princeps Pampanini and Its Bioactive Components. Prev. Nutr. Food Sci. 2013, 18, 181–187. [Google Scholar] [CrossRef]
- Skalski, B.; Pawelec, S.; Jedrejek, D.; Rolnik, A.; Pietukhov, R.; Piwowarczyk, R.; Stochmal, A.; Olas, B. Antioxidant and anticoagulant effects of phenylpropanoid glycosides isolated from broomrapes (Orobanche caryophyllacea, Phelipanche arenaria, and P. ramosa). Biomed. Pharmacother. 2021, 139, 111618. [Google Scholar] [CrossRef]
- Brinks, H.L.; Eckhart, A.D. Regulation of GPCR signaling in hypertension. Biochim. Biophys. Acta 2010, 1802, 1268–1275. [Google Scholar] [CrossRef]
- NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: A pooled analysis of 1201 population-representative studies with 104 million participants. Lancet 2021, 398, 957–980. [Google Scholar] [CrossRef]
- Herman, L.L.; Padala, S.A.; Ahmed, I.; Bashir, K. Angiotensin converting enzyme inhibitors (ACEI). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Masuyer, G.; Douglas, R.G.; Sturrock, E.D.; Acharya, K.R. Structural basis of Ac-SDKP hydrolysis by Angiotensin-I converting enzyme. Sci. Rep. 2015, 5, 13742. [Google Scholar] [CrossRef]
- Murray, B.A.; Walsh, D.J.; FitzGerald, R.J. Modification of the furanacryloyl-L-phenylalanylglycylglycine assay for determination of angiotensin-I-converting enzyme inhibitory activity. J. Biochem. Biophys. Methods 2004, 59, 127–137. [Google Scholar] [CrossRef]
- Lam, L.H.; Shimamura, T.; Sakaguchi, K.; Noguchi, K.; Ishiyama, M.; Fujimura, Y.; Ukeda, H. Assay of angiotensin I-converting enzyme-inhibiting activity based on the detection of 3-hydroxybutyric acid. Anal. Biochem. 2007, 364, 104–111. [Google Scholar] [CrossRef]
- Li, J.; Liu, Z.; Zhao, Y.; Zhu, X.; Yu, R.; Dong, S.; Wu, H. Novel Natural Angiotensin Converting Enzyme (ACE)-Inhibitory Peptides Derived from Sea Cucumber-Modified Hydrolysates by Adding Exogenous Proline and a Study of Their Structure−Activity Relationship. Mar. Drugs 2018, 16, 271. [Google Scholar] [CrossRef]
- Elbl, G.; Wagner, H. A new method for the in vitro screening of inhibitors of angiotensin-converting enzyme (ACE), using the chromophore- and fluorophore-labelled substrate, dansyltriglycine. Planta Med. 1991, 57, 137–141. [Google Scholar] [CrossRef]
- Baudin, B.; Bénéteau-Burnat, B.; Baumann, F.C.; Giboudeau, J. A reliable radiometric assay for the determination of angiotensin I-converting enzyme activity in urine. J. Clin. Chem. Clin. Biochem. 1990, 28, 857–861. [Google Scholar] [CrossRef]
- Jimsheena, V.K.; Gowda, L.R. Colorimetric, high-throughput assay for screening Angiotensin I-converting enzyme inhibitors. Anal. Chem. 2009, 81, 9388–9394. [Google Scholar] [CrossRef]
- Schwager, S.L.; Carmona, A.K.; Sturrock, E.D. A high-throughput fluorimetric assay for angiotensin I-converting enzyme. Nat. Protoc. 2006, 1, 1961–1964. [Google Scholar] [CrossRef] [PubMed]
- Francenia Santos-Sánchez, N.; Salas-Coronado, R.; Villanueva-Cañongo, C.; Hernández-Carlos, B. Antioxidant compounds and their antioxidant mechanism. In Antioxidants; Shalaby, E., Ed.; IntechOpen: London, UK, 2019; ISBN 978-1-78923-919-5. [Google Scholar]
- Munteanu, I.G.; Apetrei, C. Analytical methods used in determining antioxidant activity: A review. Int. J. Mol. Sci. 2021, 22, 3380. [Google Scholar] [CrossRef] [PubMed]
- Florin Danet, A. Recent advances in antioxidant capacity assays. In Antioxidants—Benefits, Sources, Mechanisms of Action; Waisundara, V., Ed.; IntechOpen: London, UK, 2021; ISBN 978-1-83968-864-5. [Google Scholar]
- Niu, L.; Han, D. Chemical Analysis of Antioxidant Capacity: Mechanisms and Techniques; De Gruyter: Berlin, Germany, 2020; ISBN 9783110573763. [Google Scholar]
- Ivanova, A.; Gerasimova, E.; Gazizullina, E. Study of Antioxidant Properties of Agents from the Perspective of Their Action Mechanisms. Molecules 2020, 25, 4251. [Google Scholar] [CrossRef] [PubMed]
- Basyal, D.; Neupane, A.; Pandey, D.P.; Pandeya, S. Phytochemical Screening and In Vitro Antioxidant and Anti-inflammatory Activities of Aerial Parts of Euphorbia hirta L. J. Nepal Chem. Soc. 2021, 42, 115–124. [Google Scholar] [CrossRef]
- Lesjak, M.; Beara, I.; Simin, N.; Pintać, D.; Majkić, T.; Bekvalac, K.; Orčić, D.; Mimica-Dukić, N. Antioxidant and anti-inflammatory activities of quercetin and its derivatives. J. Funct. Foods 2018, 40, 68–75. [Google Scholar] [CrossRef]
- Ma, Y.; Feng, Y.; Diao, T.; Zeng, W.; Zuo, Y. Experimental and theoretical study on antioxidant activity of the four anthocyanins. J. Mol. Struct. 2020, 1204, 127509. [Google Scholar] [CrossRef]
- Su, Y.; Li, L. Structural characterization and antioxidant activity of polysaccharide from four auriculariales. Carbohydr. Polym. 2020, 229, 115407. [Google Scholar] [CrossRef]
- Pohl, F.; Kong Thoo Lin, P. The Potential Use of Plant Natural Products and Plant Extracts with Antioxidant Properties for the Prevention/Treatment of Neurodegenerative Diseases: In Vitro, In Vivo and Clinical Trials. Molecules 2018, 23, 3283. [Google Scholar] [CrossRef]
- Hirayama, O.; Takagi, M.; Hukumoto, K.; Katoh, S. Evaluation of antioxidant activity by chemiluminescence. Anal. Biochem. 1997, 247, 237–241. [Google Scholar] [CrossRef]
- Arslan Burnaz, N.; Küçük, M.; Akar, Z. An on-line HPLC system for detection of antioxidant compounds in some plant extracts by comparing three different methods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1052, 66–72. [Google Scholar] [CrossRef]
- Yan, R.; Cao, Y.; Yang, B. HPLC-DPPH screening method for evaluation of antioxidant compounds extracted from Semen Oroxyli. Molecules 2014, 19, 4409–4417. [Google Scholar] [CrossRef] [PubMed]
- Amatatongchai, M.; Laosing, S.; Chailapakul, O.; Nacapricha, D. Simple flow injection for screening of total antioxidant capacity by amperometric detection of DPPH radical on carbon nanotube modified-glassy carbon electrode. Talanta 2012, 97, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Alasalvar, C.; Liyana-Pathirana, C.M. Antioxidant phytochemicals in hazelnut kernel (Corylus avellana L.) and hazelnut byproducts. J. Agric. Food Chem. 2007, 55, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Becker, E.M.; Nissen, L.R.; Skibsted, L.H. Antioxidant evaluation protocols: Food quality or health effects. Eur. Food Res. Technol. 2004, 219, 561–571. [Google Scholar] [CrossRef]
- Munteanu, I.G.; Apetrei, C. A review on electrochemical sensors and biosensors used in assessing antioxidant activity. Antioxidants 2022, 11, 584. [Google Scholar] [CrossRef]
- Dong, J.-W.; Cai, L.; Xing, Y.; Yu, J.; Ding, Z.-T. Re-evaluation of ABTS*+ Assay for Total Antioxidant Capacity of Natural Products. Nat. Prod. Commun. 2015, 10, 2169–2172. [Google Scholar] [CrossRef] [PubMed]
- Prior, R.L.; Wu, X.; Schaich, K. Standardized methods for the determination of antioxidant capacity and phenolics in foods and dietary supplements. J. Agric. Food Chem. 2005, 53, 4290–4302. [Google Scholar] [CrossRef]
- Huang, D.; Ou, B.; Prior, R.L. The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 2005, 53, 1841–1856. [Google Scholar] [CrossRef]
- Srivastava, S.; Adholeya, A.; Conlan, X.A.; Cahill, D.M. Acidic Potassium Permanganate Chemiluminescence for the Determination of Antioxidant Potential in Three Cultivars of Ocimum basilicum. Plant Foods Hum. Nutr. 2016, 71, 72–80. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Measurement of antioxidant activity. J. Funct. Foods 2015, 18, 757–781. [Google Scholar] [CrossRef]
- Ashida, S.; Okazaki, S.; Tsuzuki, W.; Suzuki, T. Chemiluminescent method for the evaluation of antioxidant activity using lipid hydroperoxide-luminol. Anal. Sci. 1991, 7, 93–96. [Google Scholar] [CrossRef]
- Bunaciu, A.A.; Danet, A.F.; Fleschin, Ş.; Aboul-Enein, H.Y. Recent applications for in vitro antioxidant activity assay. Crit. Rev. Anal. Chem. 2016, 46, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Antolovich, M.; Prenzler, P.D.; Patsalides, E.; McDonald, S.; Robards, K. Methods for testing antioxidant activity. Analyst 2002, 127, 183–198. [Google Scholar] [CrossRef]
- Pascualreguera, M.; Ortegacarmona, I.; Molinadiaz, A. Spectrophotometric determination of iron with ferrozine by flow-injection analysis. Talanta 1997, 44, 1793–1801. [Google Scholar] [CrossRef]
- Baba, S.A.; Malik, S.A. Determination of total phenolic and flavonoid content, antimicrobial and antioxidant activity of a root extract of Arisaema jacquemontii Blume. J. Taibah Univ. Sci. 2015, 9, 449–454. [Google Scholar] [CrossRef]
- Apak, R.; Güçlü, K.; Ozyürek, M.; Karademir, S.E. Novel total antioxidant capacity index for dietary polyphenols and vitamins C and E, using their cupric ion reducing capability in the presence of neocuproine: CUPRAC method. J. Agric. Food Chem. 2004, 52, 7970–7981. [Google Scholar] [CrossRef]
- Ribeiro, J.P.N.; Magalhães, L.M.; Reis, S.; Lima, J.L.F.C.; Segundo, M.A. High-throughput total cupric ion reducing antioxidant capacity of biological samples determined using flow injection analysis and microplate-based methods. Anal. Sci. 2011, 27, 483. [Google Scholar] [CrossRef]
- Özyürek, M.; Güçlü, K.; Tütem, E.; Başkan, K.S.; Erçağ, E.; Esin Çelik, S.; Baki, S.; Yıldız, L.; Karaman, Ş.; Apak, R. A comprehensive review of CUPRAC methodology. Anal. Methods 2011, 3, 2439. [Google Scholar] [CrossRef]
- Mamedov, N. Medicinal plants studies: History, challenges and prospective. Med. Aromat. Plants 2012, 1, 1000e133. [Google Scholar] [CrossRef]
- Arastehfar, A.; Gabaldón, T.; Garcia-Rubio, R.; Jenks, J.D.; Hoenigl, M.; Salzer, H.J.F.; Ilkit, M.; Lass-Flörl, C.; Perlin, D.S. Drug-Resistant Fungi: An Emerging Challenge Threatening Our Limited Antifungal Armamentarium. Antibiotics 2020, 9, 877. [Google Scholar] [CrossRef]
- Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). EUCAST definitive document EDef 7.1: Method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 2008, 14, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Caron, F. Antimicrobial susceptibility testing: A four facets tool for the clinician. J. Des Anti-Infect. 2012, 14, 168–174. [Google Scholar] [CrossRef]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms 2021, 9, 41. [Google Scholar] [CrossRef]
- Aldholmi, M.; Marchand, P.; Ourliac-Garnier, I.; Le Pape, P.; Ganesan, A. A Decade of Antifungal Leads from Natural Products: 2010–2019. Pharmaceuticals 2019, 12, 182. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, D.; Lee, C.C.; Sheh-Hong, L. Antimicrobial Activity of Endophytic Fungi Isolated from Swietenia macrophylla Leaves. Nat. Prod. Commun. 2014, 9, 1934578X1400900. [Google Scholar] [CrossRef]
- Ruban, P.; Gajalakshmi, K. In vitro antibacterial activity of Hibiscus rosa-sinensis flower extract against human pathogens. Asian Pac. J. Trop. Biomed. 2012, 2, 399–403. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [CrossRef]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef]
- Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Dannaoui, E.; Espinel-Ingroff, A. Antifungal susceptibly testing by concentration gradient strip etest method for fungal isolates: A review. J. Fungi 2019, 5, 108. [Google Scholar] [CrossRef]
- Songtham, S. A Comparison of Two Methods Used for Measuring the Antagonistic Activity of Bacillus Species. Walailak J. Sci. Technol. (WJST) 2011, 5, 161–171. [Google Scholar]
- Lorelli, J.P.; Held, A.A. Screening of Blastocladialean Fungi for Antibiotic Production by a Modified “Cross-Streak” Test. Mycologia 1983, 75, 909. [Google Scholar] [CrossRef]
- Homans, A.L.; Fuchs, A. Direct bioautography on thin-layer chromatograms as a method for detecting fungitoxic substances. J. Chromatogr. A 1970, 51, 327–329. [Google Scholar] [CrossRef]
- Suleimana, M.M.; McGaw, L.J.; Naidoo, V.; Eloff, J.N. Detection of antimicrobial compounds by bioautography of different extracts of leaves of selected South African tree species. Afr. J. Tradit. Complement. Altern. Med. 2009, 7, 64–78. [Google Scholar] [CrossRef]
- Dewanjee, S.; Gangopadhyay, M.; Bhattacharya, N.; Khanra, R.; Dua, T.K. Bioautography and its scope in the field of natural product chemistry. J. Pharm. Anal. 2015, 5, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Marston, A. Thin-layer chromatography with biological detection in phytochemistry. J. Chromatogr. A 2011, 1218, 2676–2683. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.; Santos, A.; Reis, F.; Tavares, R.M.; Baptista, P.; Lino-Neto, T.; Almeida-Aguiar, C. A new effective assay to detect antimicrobial activity of filamentous fungi. Microbiol. Res. 2013, 168, 1–5. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Y.; Wang, R.; Wang, Z.; Yang, B.; Kuang, H. An Evolving Technology That Integrates Classical Methods with Continuous Technological Developments: Thin-Layer Chromatography Bioautography. Molecules 2021, 26, 4647. [Google Scholar] [CrossRef]
- Ericsson, H.M.; Sherris, J.C. Antibiotic sensitivity testing. Report of an international collaborative study. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 1971, 217, 90. [Google Scholar]
- Alexander, B.D. Reference Method For Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; p. 50. ISBN 1-56238-831-2. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Marroki, A.; Bousmaha-Marroki, L. Antibiotic resistance diagnostic methods for pathogenic bacteria. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2021; ISBN 9780128012383. [Google Scholar]
- Therese, K.L.; Bagyalakshmi, R.; Madhavan, H.N.; Deepa, P. In-vitro susceptibility testing by agar dilution method to determine the minimum inhibitory concentrations of amphotericin b, fluconazole and ketoconazole against ocular fungal isolates. Indian J. Med. Microbiol. 2006, 24, 273–279. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Sheehan, D.J.; Rex, J.H. Determination of fungicidal activities against yeasts and molds: Lessons learned from bactericidal testing and the need for standardization. Clin. Microbiol. Rev. 2004, 17, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Klepser, M.E.; Ernst, E.J.; Lewis, R.E.; Ernst, M.E.; Pfaller, M.A. Influence of test conditions on antifungal time-kill curve results: Proposal for standardized methods. Antimicrob. Agents Chemother. 1998, 42, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Vojtek, L.; Dobes, P.; Buyukguzel, E.; Atosuo, J.; Hyrsl, P. Bioluminescent assay for evaluating antimicrobial activity in insect haemolymph. Eur. J. Entomol. 2014, 111, 335–340. [Google Scholar] [CrossRef]
- Finger, S.; Wiegand, C.; Buschmann, H.-J.; Hipler, U.-C. Antibacterial properties of cyclodextrin-antiseptics-complexes determined by microplate laser nephelometry and ATP bioluminescence assay. Int. J. Pharm. 2013, 452, 188–193. [Google Scholar] [CrossRef]
- Ke, H.-M.; Tsai, I.J. Understanding and using fungal bioluminescence—Recent progress and future perspectives. Curr. Opin. Green Sustain. Chem. 2021, 33, 100570. [Google Scholar] [CrossRef]
- Green, L.; Petersen, B.; Steimel, L.; Haeber, P.; Current, W. Rapid determination of antifungal activity by flow cytometry. J. Clin. Microbiol. 1994, 32, 1088–1091. [Google Scholar] [CrossRef]
- Ramani, R.; Chaturvedi, V. Flow cytometry antifungal susceptibility testing of pathogenic yeasts other than Candida albicans and comparison with the NCCLS broth microdilution test. Antimicrob. Agents Chemother. 2000, 44, 2752–2758. [Google Scholar] [CrossRef]
- Mani-López, E.; Cortés-Zavaleta, O.; López-Malo, A. A review of the methods used to determine the target site or the mechanism of action of essential oils and their components against fungi. SN Appl. Sci. 2021, 3, 44. [Google Scholar] [CrossRef]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef]
- Famuyide, I.M.; Aro, A.O.; Fasina, F.O.; Eloff, J.N.; McGaw, L.J. Antibacterial and antibiofilm activity of acetone leaf extracts of nine under-investigated south African Eugenia and Syzygium (Myrtaceae) species and their selectivity indices. BMC Complement. Altern. Med. 2019, 19, 141. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Mishra, A.; Jha, B. Anti-quorum Sensing and Anti-biofilm Activity of Delftia tsuruhatensis Extract by Attenuating the Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2017, 7, 337. [Google Scholar] [CrossRef] [PubMed]
- Choo, J.H.; Rukayadi, Y.; Hwang, J.K. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 2006, 42, 637–641. [Google Scholar] [CrossRef]
- Hayashi, M.A.; Bizerra, F.C.; Da Silva, P.I. Antimicrobial compounds from natural sources. Front. Microbiol. 2013, 4, 195. [Google Scholar] [CrossRef] [PubMed]
- Masota, N.E.; Vogg, G.; Ohlsen, K.; Holzgrabe, U. Reproducibility challenges in the search for antibacterial compounds from nature. PLoS ONE 2021, 16, e0255437. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T.; International Natural Product Sciences Taskforce. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Wilson, B.A.P.; Thornburg, C.C.; Henrich, C.J.; Grkovic, T.; O’Keefe, B.R. Creating and screening natural product libraries. Nat. Prod. Rep. 2020, 37, 893–918. [Google Scholar] [CrossRef]
- Santana, K.; do Nascimento, L.D.; Lima E Lima, A.; Damasceno, V.; Nahum, C.; Braga, R.C.; Lameira, J. Applications of Virtual Screening in Bioprospecting: Facts, Shifts, and Perspectives to Explore the Chemo-Structural Diversity of Natural Products. Front. Chem. 2021, 9, 662688. [Google Scholar] [CrossRef]
- Forry, S.P.; Madonna, M.C.; López-Pérez, D.; Lin, N.J.; Pasco, M.D. Automation of antimicrobial activity screening. AMB Express 2016, 6, 20. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Yong, J.N. The chemistry and biological activities of natural products from Northern African plant families: From Aloaceae to Cupressaceae. RSC Adv. 2014, 4, 61975–61991. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, P.; Cheng, G.; Zhang, Y. A Brief Review of Phenolic Compounds Identified from Plants: Their Extraction, Analysis, and Biological Activity. Nat. Prod. Commun. 2022, 17, 1934578X2110697. [Google Scholar] [CrossRef]
- Schneider, G. Automating drug discovery. Nat. Rev. Drug Discov. 2018, 17, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Nyamundanda, G.; Brennan, L.; Gormley, I.C. Probabilistic principal component analysis for metabolomic data. BMC Bioinform. 2010, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Hinzman, A.A.; Kang, E.L.; Szczesniak, R.D.; Lu, L.J. Computational and statistical analysis of metabolomics data. Metabolomics 2015, 11, 1492–1513. [Google Scholar] [CrossRef]
- Bartel, J.; Krumsiek, J.; Theis, F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013, 4, e201301009. [Google Scholar] [CrossRef]
- Cakır, T.; Hendriks, M.M.W.B.; Westerhuis, J.A.; Smilde, A.K. Metabolic network discovery through reverse engineering of metabolome data. Metabolomics 2009, 5, 318–329. [Google Scholar] [CrossRef]
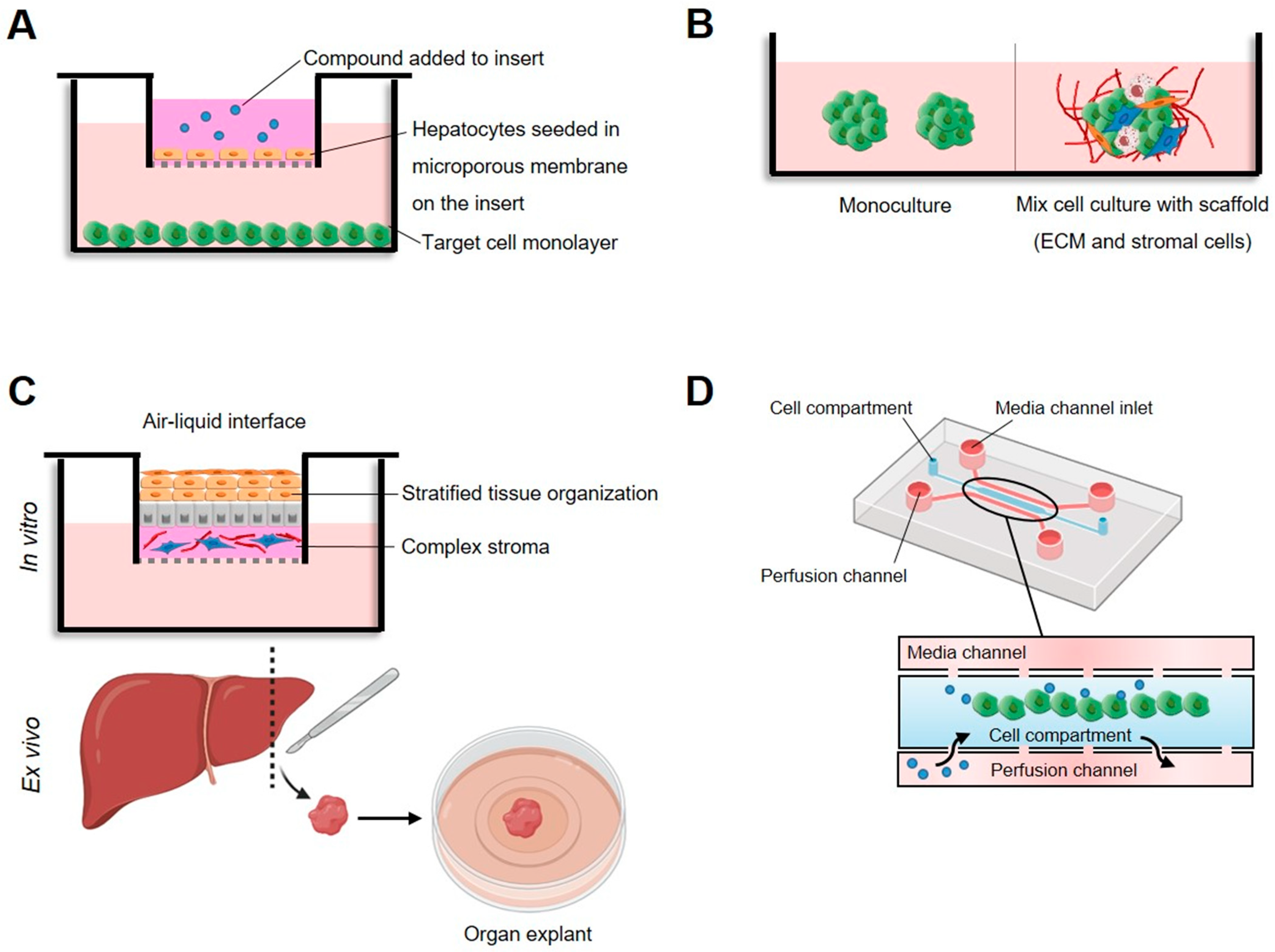
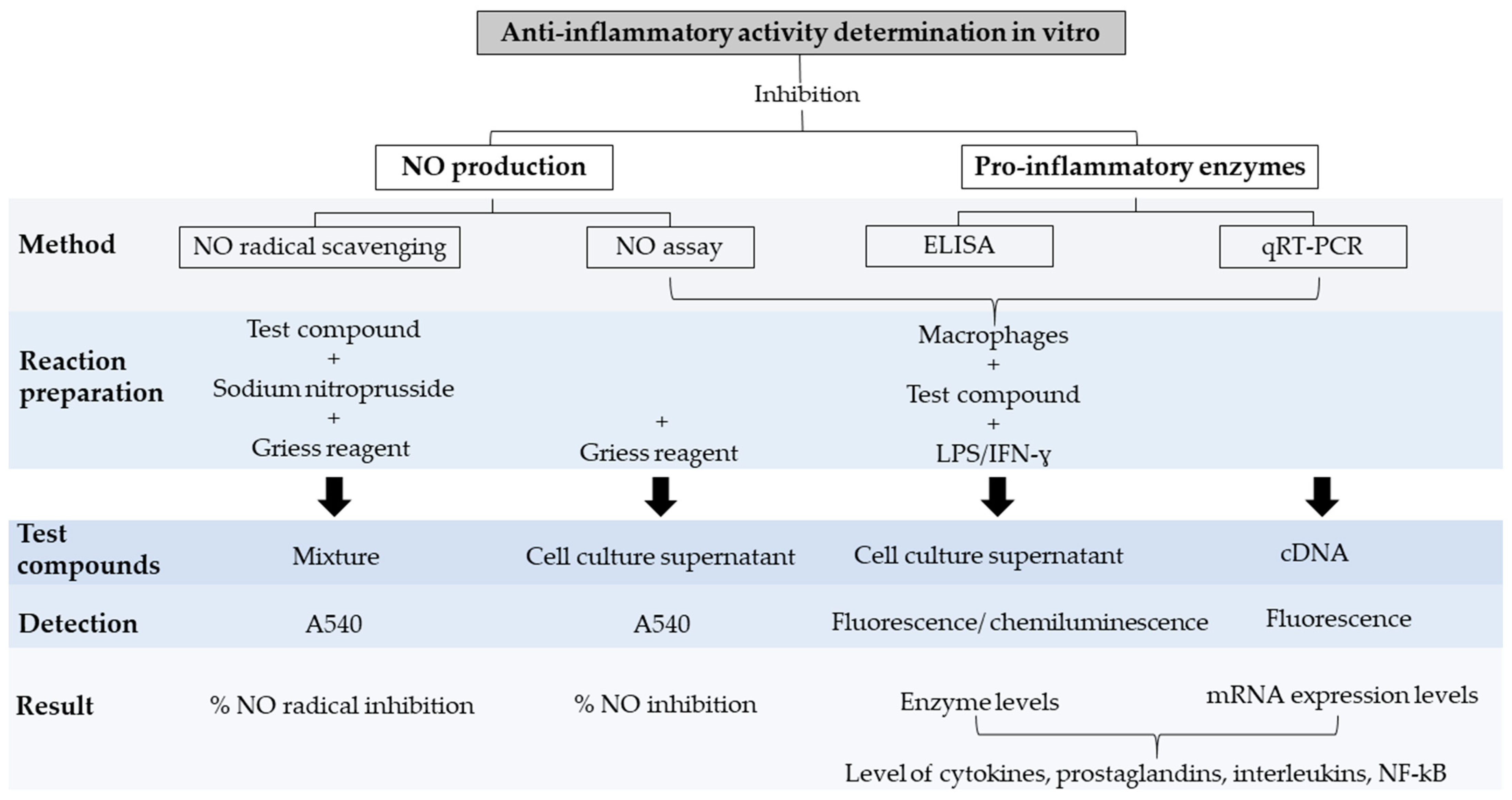
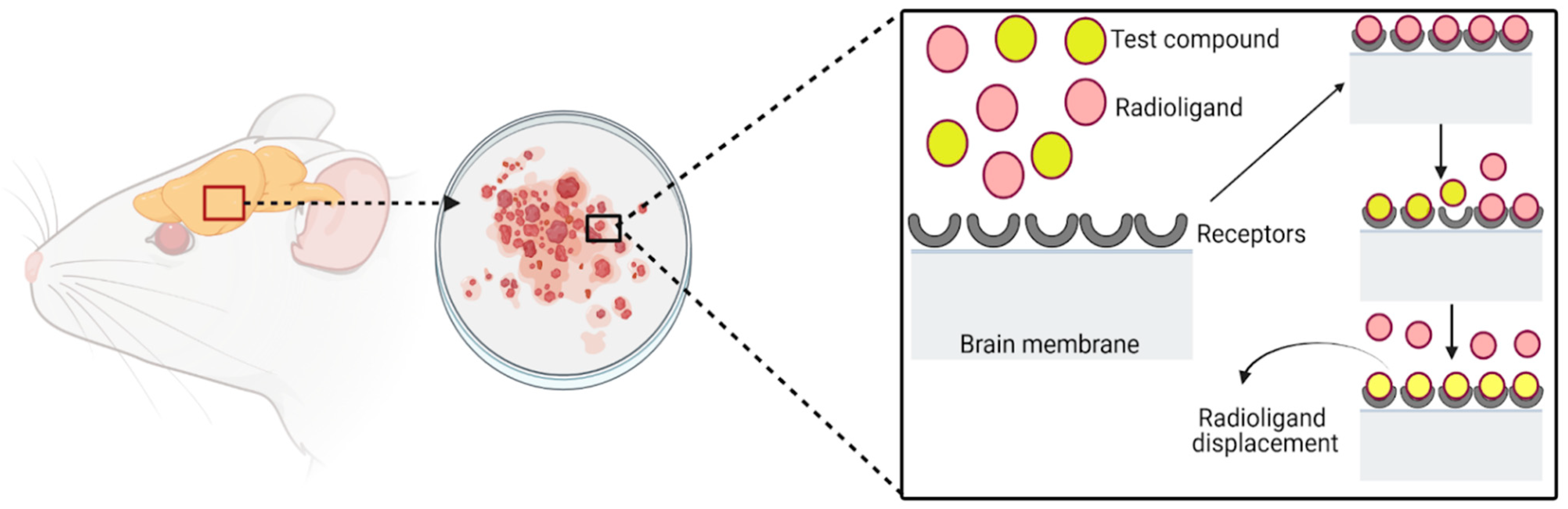
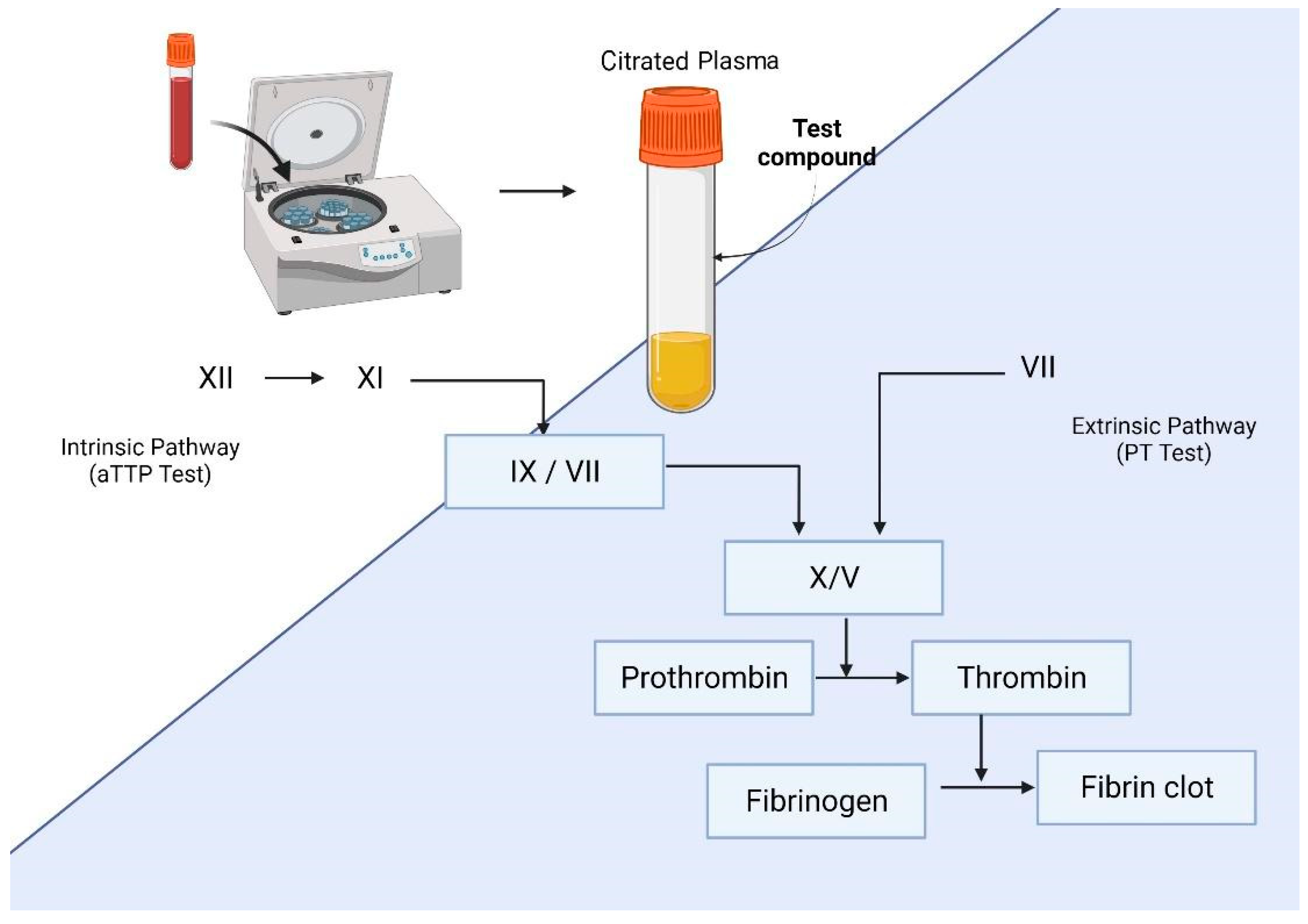

| Method | Principle | Detection | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| Cell Viability and Proliferation | |||||
| Dye exclusion | Detection of plasma membrane integrity | Trypan blue colorimetric/H + M |
|
| [27,28,29,30] |
| Crystal violet colorimetric/M, PR |
|
| [13,31] | ||
| Metabolic activity | Detection of mitochondrial dehydrogenase and oxidoreductase activity | Tetrazolium salts (MTT, XTT, MTS, WST) colorimetric/PR |
|
| [27,31,32,33,34] |
| Resazurin fluorescent/PR |
|
| [35,36,37] | ||
| Energy metabolism | Correlation between a bioluminescent reaction and the ability to synthesize ATP | Luciferase and luciferin luminescent/PR |
|
| [14,27] |
| Enzyme release-based | Determination of plasma membrane integrity (LDH release) | Lactate + tetrazolium salts/fluorescent probe colorimetric, fluorescent/PR |
|
| [27,32,38] |
| Colony formation | Determination of clonogenic growth | Low-adherence plates/M |
|
| [38,39,40,41] |
| Cell Cycle and Apoptosis | |||||
| Cell cycle arrest | Distribution of cell population in each cell cycle phase | PI, 7-AAD fluorescent/FC |
|
| [42,43] |
| Apoptosis/ necrosis | Detection of membrane integrity | PI/7-AAD, annexin V fluorescent/F |
|
| [44,45,46] |
| Method Type | Detection (Output) | Description | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| Enzyme inhibition | Fluorometric detection using fluorogenic peptide DANGPG |
|
|
| [131,135,136] |
| Method Name | Description | Detection Method | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| Radical/ROS-based scavenging assays | |||||
| 2,2′-azinobis (3-ethylbenzo-thiazoline 6- sulphonate) (ABTS)/Trolox equivalent antioxidant capacity (TEAC) test |
| Spectrophotometry (A734) |
|
| [167,177,178,179] |
| N, N-diphenyl-N’-picrylhydrazyl (DPPH) free radical |
|
|
|
| [174,177] |
| Oxygen radical absorbance capacity (ORAC) |
| Fluorometry |
|
| [175,180,181,182] |
| Chemiluminescence |
| Fluorometry |
|
| [178,179,180,183,184] |
| Non-radical redox-potential-based assays | |||||
| Ferric reducing antioxidant power (FRAP) |
|
|
|
| [173,178,179,185,186,187] |
| Cupric reducing antioxidant capacity (CUPRAC) |
| Spectrophotometry (A450) |
|
| [164,166,188,189,190] |
| Method Name | Description | Advantages | Disadvantages | Ref. | |
|---|---|---|---|---|---|
| Diffusion methods | Agar diffusion method | The antimicrobial agent diffuses from disks or strips into the solid culture medium that has been seeded with a pure culture |
|
| [198,199,204] |
| Well diffusion method | Diffusion of a liquid antimicrobial agent placed in a well punched into the solid culture medium that has been seeded with a pure culture | ||||
| Agar plug diffusion method | The first bacterium or fungus is grown on agar plates, here it will secrete molecules that diffuse in the agar which then is cut and placed on another agar plate inoculated with a different microorganism |
|
| [199,200,201] | |
| Antimicrobial gradient method (Etest) | Based on creating a concentration gradient of the antimicrobial agent tested in the agar medium where it is exposed to the selected microorganism |
|
| [205,206] | |
| Cross streak method | The first microbial strain is streaked in the center of the agar plate and incubated then in the same plate is seeded the second microorganism by a single streak perpendicular to the central streak |
|
| [207,208] | |
| Thin-layer chromatography (TLC)–bioautography methods | Bioautographic method direct | The antimicrobial activity is assessed directly onto the TLC plates where the extracts are separated by chromatography across a TLC plate, then the microorganisms are also applied by spray-identifying the localization of the fraction with antimicrobial potential |
|
| [209,210] |
| Agar overlay bioassay | The TLC plate is covered with agar seeded with the test microbe and the antimicrobial compounds are diffused onto the agar medium |
|
| [211,212,213] | |
| Agar diffusion | The antimicrobial agent is transferred from a TLC to an agar plate previously inoculated with the test microorganism |
|
| [212,214] | |
| Dilution methods | Broth dilution method Minimum inhibitory concentration (MIC) determination | Uses tubes or microdilution plates to measures the lowest concentration of antimicrobial agent that completely inhibits the growth of the bacteria/fungi |
|
| [215,216] |
| Agar dilution method | Similarly, to the procedure used in the disk diffusion method, a desired concentration of the antimicrobial agent is placed into an agar medium |
|
| [217,218,219] | |
| Time-kill test (time-kill curve) | Based on a time/concentration-dependent analysis of antimicrobial effects. Several tubes containing varying concentrations of the antimicrobial agent are seeded with the bacteria/fungi and the percentage of dead cells is determined along with the assay |
|
| [220,221] | |
| ATP bioluminescence assay | Bioluminescence ATP based | Based on the capacity to measure adenosine triphosphate (ATP) produced by bacteria or fungi |
|
| [222,223,224] |
| Flow cytofluorometric method | Flow cytofluorometric | Based on the capacity of damaged cells to emit a positive signal that is detected by flow cytometry analysis. The microorganism is exposed to antimicrobial agents and then stained with the intercalating agent propidium iodide. |
|
| [225,226,227] |
| Biofilm inhibition | Microtiter plate biofilm production assay | Based on the potential of the extracts to prevent initial cell attachment to microtiter plates. Biofilm formation/inhibition is observed through (OD) of the crystal violet present in the destaining solution measured at 595 nm |
|
| [228,229] |
| Quorum sensing inhibition | Violacein quantification | The inhibitory activity is measured by quantifying violacein production in a microplate reader using a spectrophotometer at 585 nm where the microorganism and extracts are cultured at different concentrations. |
|
| [230,231] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barba-Ostria, C.; Carrera-Pacheco, S.E.; Gonzalez-Pastor, R.; Heredia-Moya, J.; Mayorga-Ramos, A.; Rodríguez-Pólit, C.; Zúñiga-Miranda, J.; Arias-Almeida, B.; Guamán, L.P. Evaluation of Biological Activity of Natural Compounds: Current Trends and Methods. Molecules 2022, 27, 4490. https://doi.org/10.3390/molecules27144490
Barba-Ostria C, Carrera-Pacheco SE, Gonzalez-Pastor R, Heredia-Moya J, Mayorga-Ramos A, Rodríguez-Pólit C, Zúñiga-Miranda J, Arias-Almeida B, Guamán LP. Evaluation of Biological Activity of Natural Compounds: Current Trends and Methods. Molecules. 2022; 27(14):4490. https://doi.org/10.3390/molecules27144490
Chicago/Turabian StyleBarba-Ostria, Carlos, Saskya E. Carrera-Pacheco, Rebeca Gonzalez-Pastor, Jorge Heredia-Moya, Arianna Mayorga-Ramos, Cristina Rodríguez-Pólit, Johana Zúñiga-Miranda, Benjamin Arias-Almeida, and Linda P. Guamán. 2022. "Evaluation of Biological Activity of Natural Compounds: Current Trends and Methods" Molecules 27, no. 14: 4490. https://doi.org/10.3390/molecules27144490
APA StyleBarba-Ostria, C., Carrera-Pacheco, S. E., Gonzalez-Pastor, R., Heredia-Moya, J., Mayorga-Ramos, A., Rodríguez-Pólit, C., Zúñiga-Miranda, J., Arias-Almeida, B., & Guamán, L. P. (2022). Evaluation of Biological Activity of Natural Compounds: Current Trends and Methods. Molecules, 27(14), 4490. https://doi.org/10.3390/molecules27144490










