Molecular Insight into the Binding of Astilbin with Human Serum Albumin and Its Effect on Antioxidant Characteristics of Astilbin
Abstract
:1. Introduction
2. Results and Discussion
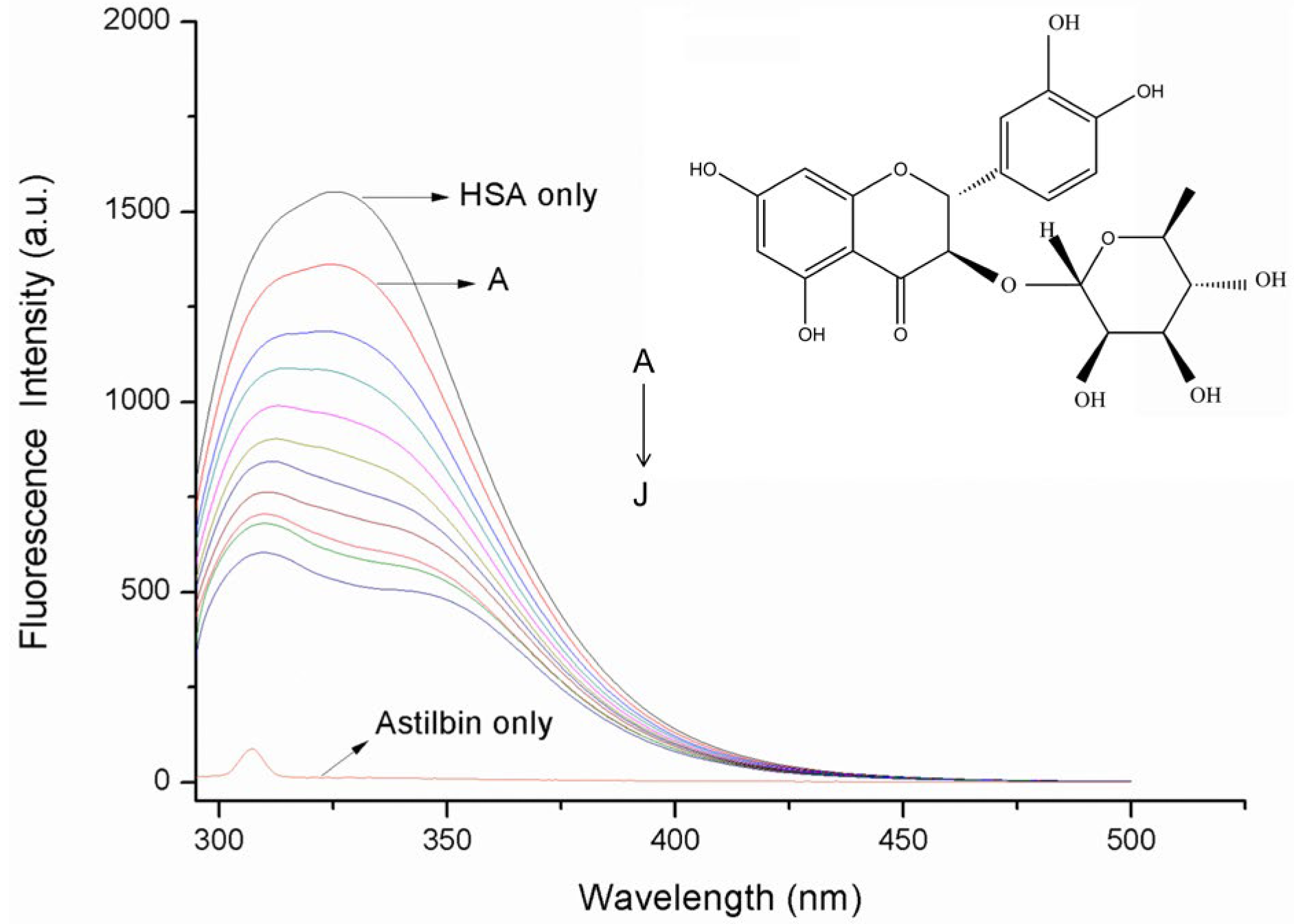
2.1. Fluorescence Quenching Studies
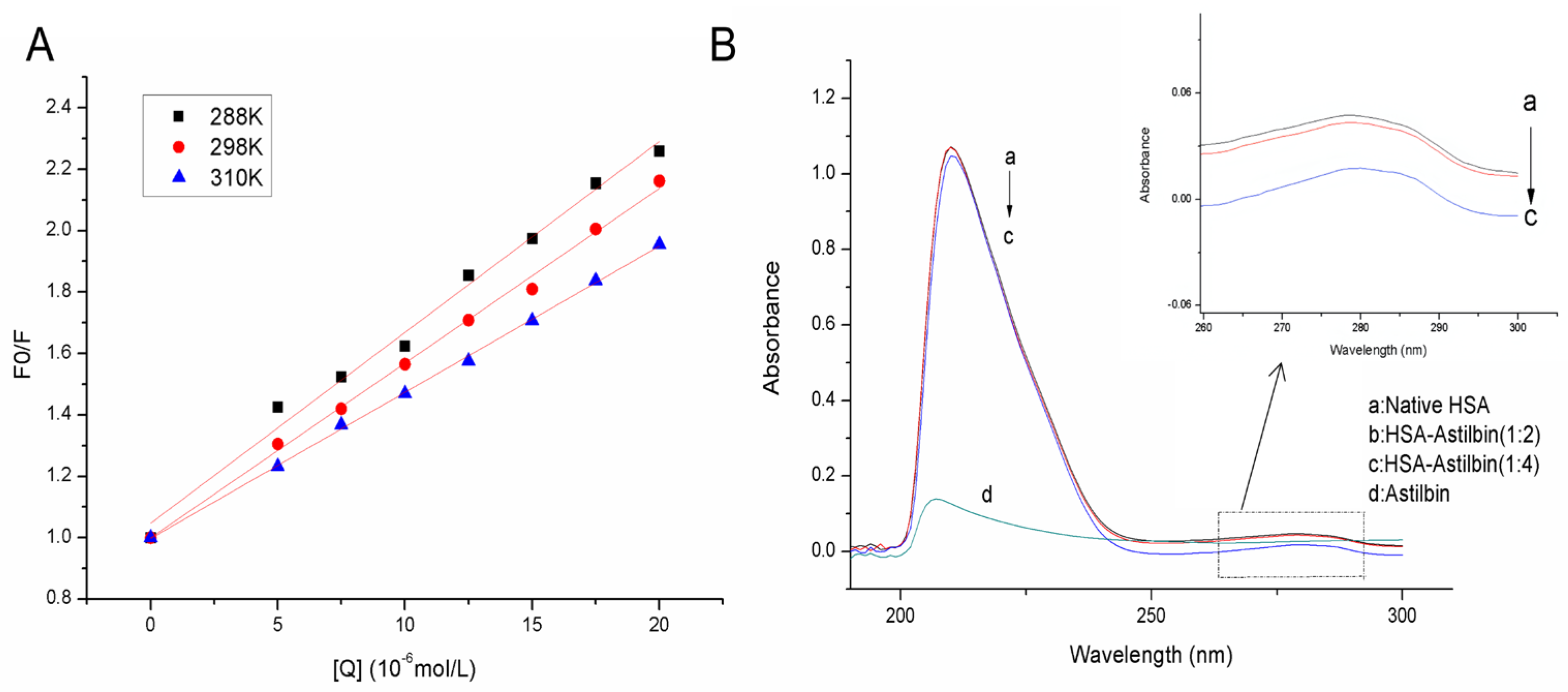
2.2. UV-Vis Absorption Spectrum
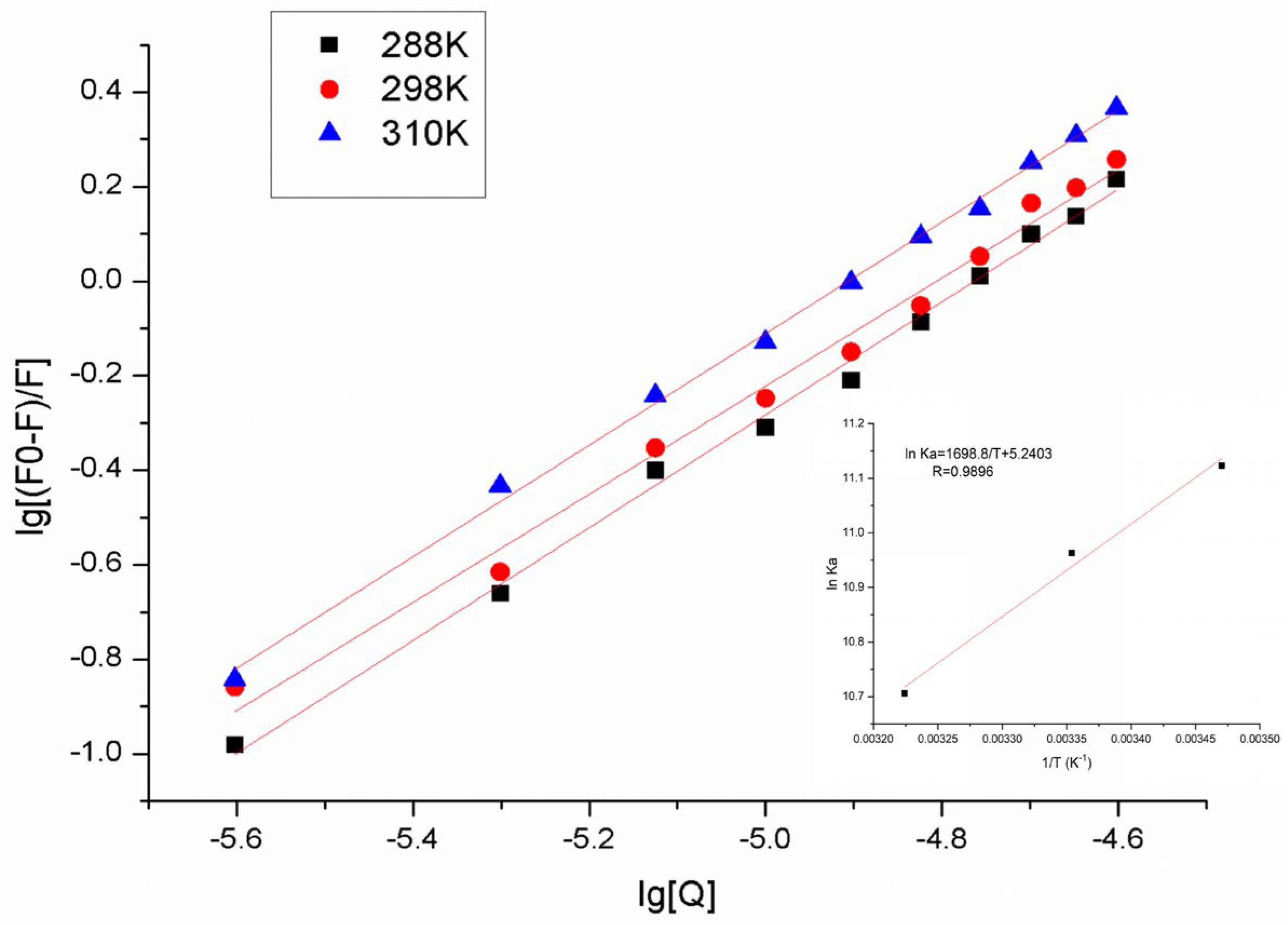
2.3. Binding Constant and the Number of Binding Sites
2.4. Thermodynamic Interaction
2.5. Determination of Energy Transfer
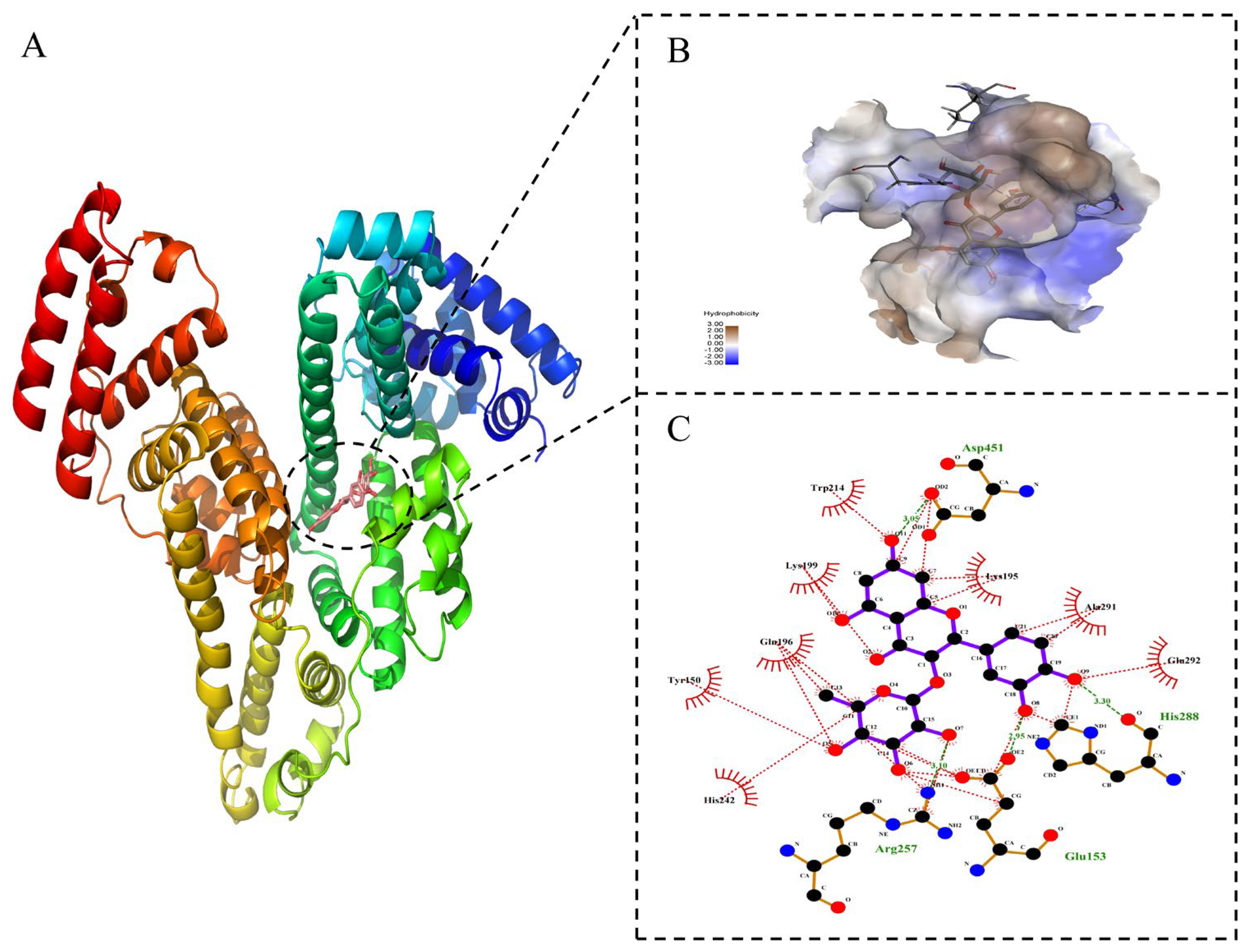
2.6. Identification of Binding Location
2.7. Effect of Astilbin on the Conformation of HSA
2.7.1. Synchronous Fluorescence Spectroscopy Studies
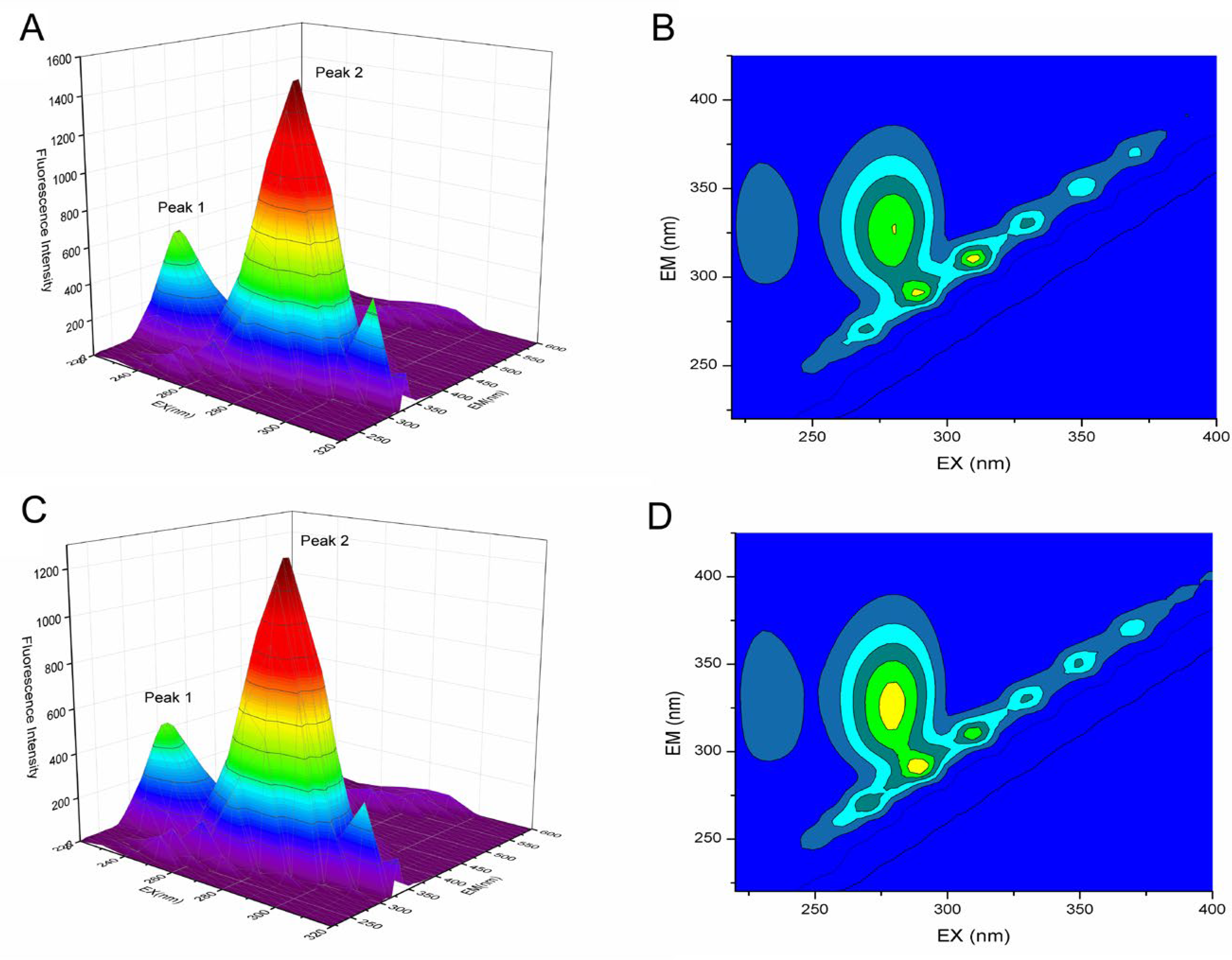
2.7.2. Three-Dimensional Fluorescence Spectroscopy
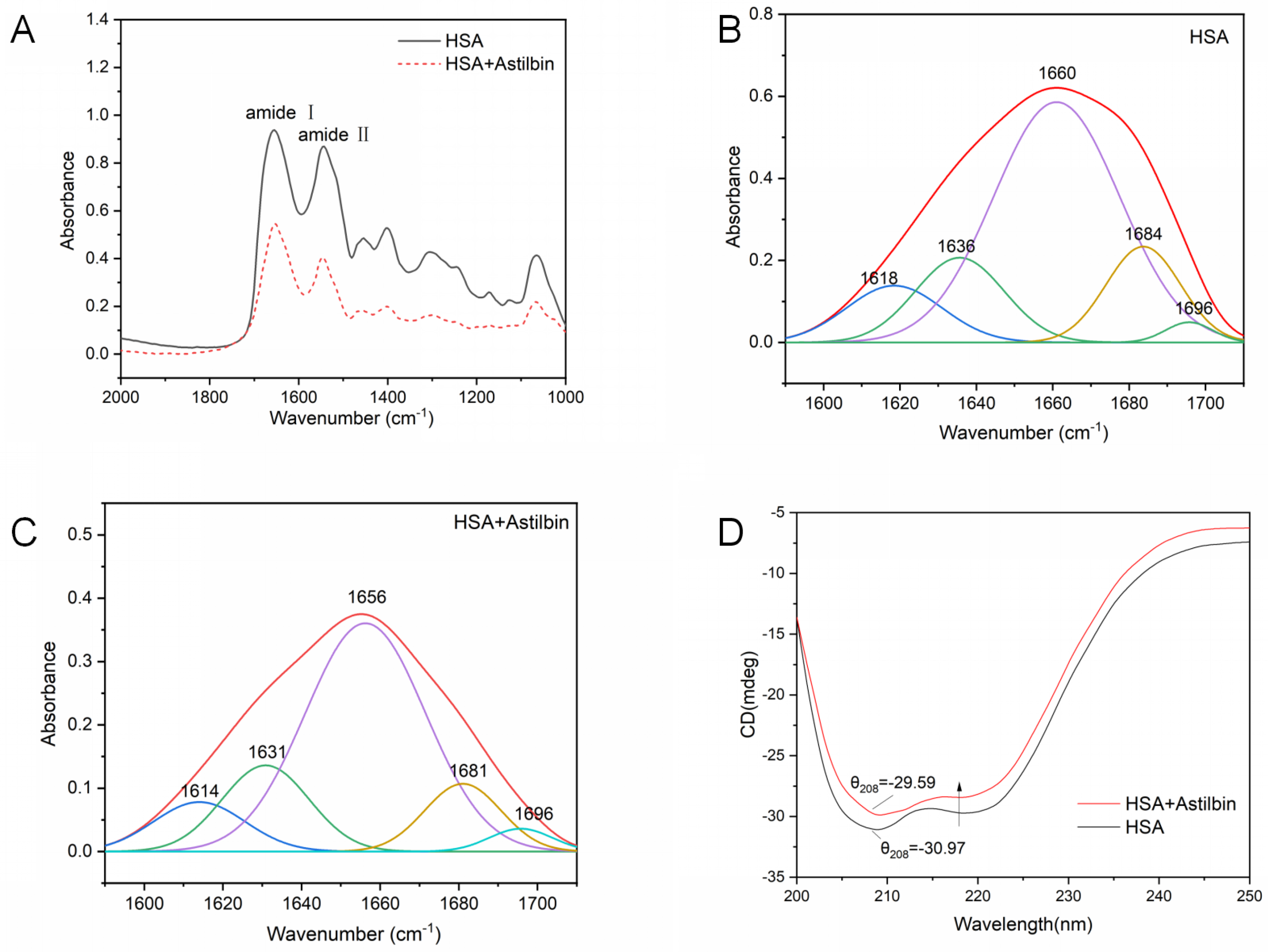
2.7.3. Micro-FTIR Spectrum
2.7.4. Circular Dichroism Spectra
2.8. Molecular Docking Technology
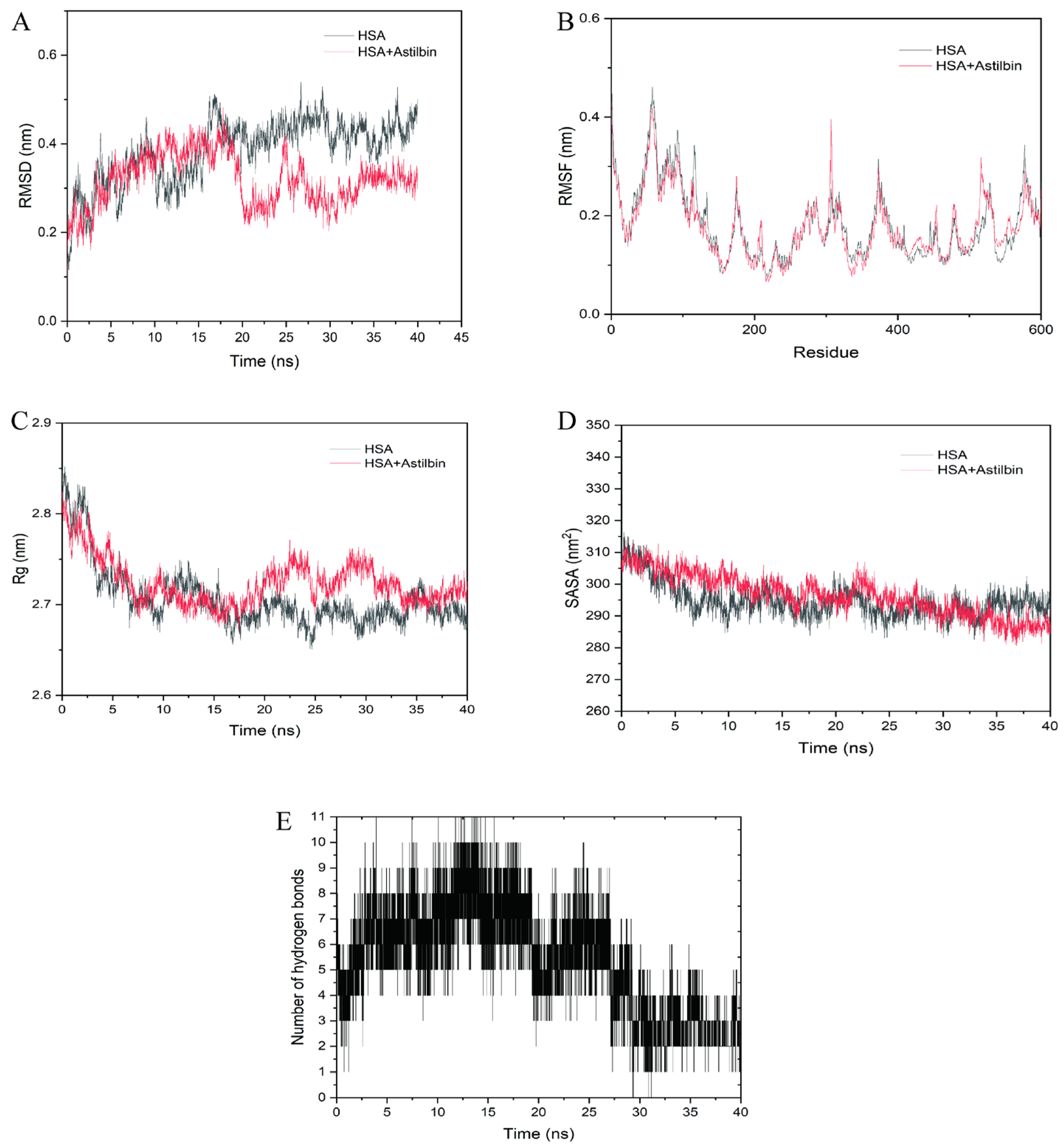
2.9. Molecular Dynamics Simulations
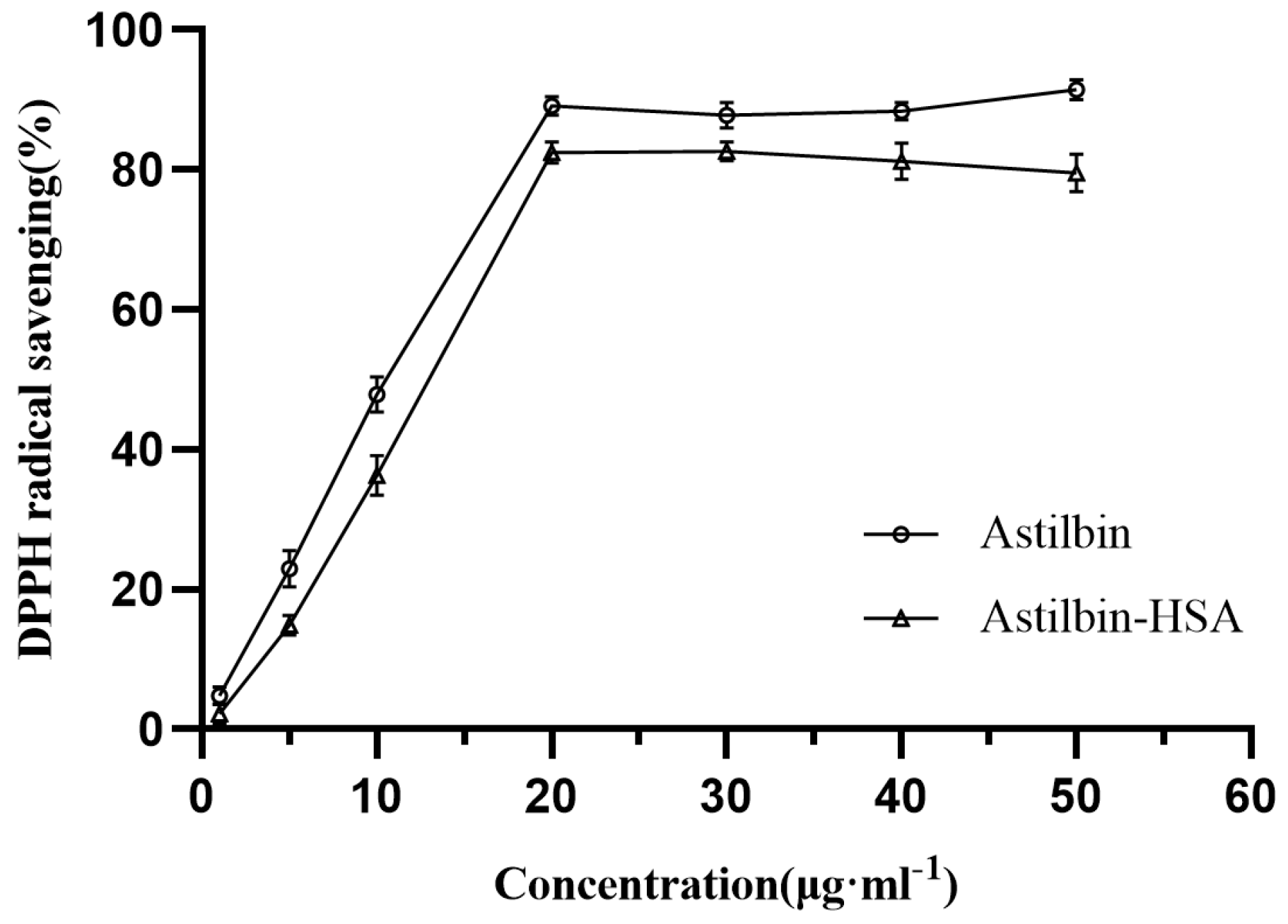
2.10. Effect of HSA on the Antioxidant Capability of Astilbin
3. Materials and Methods
3.1. Materials and Sample Preparation
3.2. Fluorescence Measurements
3.2.1. Fluorescence Spectroscopy Experiments
3.2.2. Synchronous Fluorescence Spectroscopy
3.2.3. Three-Dimensional Fluorescence Spectroscopy
3.2.4. Site Marker Competitive Binding Experiments
3.3. Ultraviolet-Visible Spectroscopy
3.4. Circular Dichroism Spectroscopy
3.5. Micro-FTIR Measurement
3.6. Molecular Docking Simulation
3.7. Molecular Dynamics Simulations
3.8. Effect of HSA on the Antioxidant Capacity of Astilbin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hostetler, G.L.; Ralston, R.A.; Schwartz, S.J. Flavones: Food Sources, Bioavailability, Metabolism, and Bioactivity. Adv. Nutr. 2017, 8, 423–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, S.; Wang, W. Spectroscopic methodologies and computational simulation studies on the characterization of the interaction between human serum albumin and astragalin. J. Biomol. Struct. Dyn. 2020, 39, 2959–2970. [Google Scholar] [CrossRef]
- Bozzo, G.G.; Unterlander, N. In through the out door: Biochemical mechanisms affecting flavonoid glycoside catabolism in plants. Plant Sci. 2021, 308, 110904. [Google Scholar] [CrossRef] [PubMed]
- Tatsis, E.C.; Boeren, S.; Exarchou, V.; Troganis, A.N.; Vervoort, J.; Gerothanassis, I.P. Identification of the major constituents of Hypericum perforatum by LC/SPE/NMR and/or LC/MS. Phytochemistry 2006, 68, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Zhu, J.; Du, H.; Nong, H.; He, X.; Chen, X. Astilbin from Smilax glabra Roxb. Attenuates Inflammatory Responses in Complete Freund’s Adjuvant-Induced Arthritis Rats. Evid.-Based Complement. Altern. Med. 2017, 2017, 8246420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kedrina-Okutan, O.; Novello, V.; Hoffmann, T.; Hadersdorfer, J.; Occhipinti, A.; Schwab, W.; Ferrandino, A. Constitutive polyphenols in blades and veins of grapevine (Vitis vinifera L.) healthy leaves. J. Agric. Food Chem. 2018, 66, 10977–10990. [Google Scholar] [CrossRef]
- Syntia, F.; Marie, L.S.; Pierre, W.-T.; Axel, M. Understanding sweetness of dry wines: First evidence of astilbin isomers in red wines and quantitation in a one-century range of vintages. Food Chem. 2021, 352, 129293. [Google Scholar]
- Fayad, S.; Cretin, B.N.; Marchal, A. Development and Validation of an LC-FTMS Method for Quantifying Natural Sweeteners in Wine. Food Chem. 2020, 311, 125881. [Google Scholar] [CrossRef]
- Shuaibo, S.; Zijian, Y.; Xiaolong, S.; Weihui, Q.; Yanlin, C.; Xinxian, X.; Yuezheng, H.; Weijun, G.; Ping, S. Astilbin prevents osteoarthritis development through the TLR4/MD-2 pathway. J. Cell. Mol. Med. 2020, 24, 13104–13114. [Google Scholar]
- Shi, Y.; Xie, J.; Chen, R.; Liu, G.; Tao, Y.; Fan, Y.; Wang, X.; Li, L.; Xu, J. Inhibitory effects of astilbin, neoastilbin and isoastilbin on human cytochrome CYP3A4 and 2D6 activities. Biomed. Chromatogr. BMC 2021, 35, e5039. [Google Scholar] [CrossRef]
- Chi, X.; Shi, G.; Zhang, Q.; Liu, Q.; Yin, H.; Zhang, Y.; Li, S. Astilbin protects chicken blood lymphocytes from cadmium-induced necroptosis via oxidative stress and the PI3K/Akt pathway. Ecotoxicol. Environ. Saf. 2019, 190, 110064. [Google Scholar]
- Chen, F.; Sun, Z.; Zhu, X.; Ma, Y. Astilbin inhibits high glucose-induced autophagy and apoptosis through the PI3K/Akt pathway in human proximal tubular epithelial cells. Biomed. Pharmacother. 2018, 106, 1175–1181. [Google Scholar] [CrossRef]
- Diao, H.; Kang, Z.; Han, F.; Jiang, W. Astilbin protects diabetic rat heart against ischemia–reperfusion injury via blockade of HMGB1-dependent NF-κB signaling pathway. Food Chem. Toxicol. 2014, 63, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, G.; Ahn, S.N. Structure, enzymatic activities, glycation and therapeutic potential of human serum albumin: A natural cargo. Int. J. Biol. Macromol. 2019, 123, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Hashempour, S.; Shahabadi, N.; Adewoye, A.; Murphy, B.; Rouse, C.; Salvatore, B.A.; Stratton, C.; Mahdavian, E. Binding Studies of AICAR and Human Serum Albumin by Spectroscopic, Theoretical, and Computational Methodologies. Molecules 2020, 25, 5410. [Google Scholar] [CrossRef]
- Chugh, H.; Kumar, P.; Tomar, V.; Kaur, N.; Sood, D.; Chandra, R. Interaction of noscapine with human serum albumin (HSA): A spectroscopic and molecular modelling approach. J. Photochem. Photobiol. A Chem. 2019, 372, 168–176. [Google Scholar] [CrossRef]
- Xiaodan, Y.; Dongjie, Y.; Dandan, P. Interactions of Bromocarbazoles with Human Serum Albumin Using Spectroscopic Methods. Molecules 2018, 23, 3120. [Google Scholar]
- Shamsi, A.; Shahwan, M.; Khan, M.S.; Alhumaydhi, F.A.; Alsagaby, S.A.; Al Abdulmonem, W.; Abdullaev, B.; Yadav, D.K. Mechanistic Insight into Binding of Huperzine A with Human Serum Albumin: Computational and Spectroscopic Approaches. Molecules 2022, 27, 797. [Google Scholar] [CrossRef]
- Yuai, Z.; Yiqing, C.; Yan, L.; Xuemei, Z. Interactions between Human Serum Albumin and Sulfadimethoxine Determined Using Spectroscopy and Molecular Docking. Molecules 2022, 27, 1526. [Google Scholar]
- Dan, Z.; Qingfeng, Z. Study on the interaction betweem astilbin and bovine serum albumin. Food Ferment. Ind. 2018, 44, 83–87. [Google Scholar] [CrossRef]
- Liu, J.; He, Y.; Liu, D.; He, Y.; Tang, Z.; Lou, H.; Huo, Y.; Cao, X. Characterizing the binding interaction of astilbin with bovine serum albumin: A spectroscopic study in combination with molecular docking technology. RSC Adv. 2018, 8, 7280–7286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, S.; Peng, X.; Yu, Q.; Bian, H.; Huang, F.; Liang, H. Multi-spectroscopic studies on the interaction of human serum albumin with astilbin: Binding characteristics and structural analysis. J. Lumin. 2013, 136, 422–429. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, C.; Cai, Y.; Chen, X.; Zhao, Y.; Liu, X.; Zhao, J.; Sun, S.; Liu, W. Astilbin ameliorates oxidative stress and apoptosis in d-galactose-induced senescence by regulating the PI3K/Akt/m-TOR signaling pathway in the brains of mice. Int. Immunopharmacol. 2021, 99, 108035. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Liu, Z.; Cao, Z.; Shi, Y.; Yang, N.; Cao, G.; Zhang, C.; Sun, R.; Zhang, C. Topical astilbin ameliorates imiquimod-induced psoriasis-like skin lesions in SKH-1 mice via suppression dendritic cell-Th17 inflammation axis. J. Cell. Mol. Med. 2022, 26, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, X.; Bao, Y.; Zhang, C.; Liu, H.; Li, Z.; Chen, M.; Wang, C.; Guo, Q.; Peng, X. Molecular insight on the binding of monascin to bovine serum albumin (BSA) and its effect on antioxidant characteristics of monascin. Food Chem. 2020, 315, 126228. [Google Scholar] [CrossRef]
- Munteanu, I.G.; Apetrei, C. Analytical Methods Used in Determining Antioxidant Activity: A Review. Int. J. Mol. Sci. 2021, 22, 3380. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, T.; Sun, Y.; Diao, M.; Zhang, J.; Ren, L. Multi-spectroscopic and molecular modeling studies on the interactions of serum albumin with 20(S,R)-protopanaxadiol and 20(S,R)-protopanaxatriol that inhibit HCT-116 cells proliferation. Food Biosci. 2021, 40, 100913. [Google Scholar] [CrossRef]
- Gu, J.; Zheng, S.; Zhao, H.; Sun, T. Investigation on the interaction between triclosan and bovine serum albumin by spectroscopic methods. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2020, 55, 52–59. [Google Scholar] [CrossRef]
- Razzak, M.A.; Lee, J.E.; Choi, S.S. Structural insights into the binding behavior of isoflavonoid glabridin with human serum albumin. Food Hydrocoll. 2019, 91, 290–300. [Google Scholar] [CrossRef]
- Ma, R.; Guo, D.-X.; Li, H.-F.; Liu, H.-X.; Zhang, Y.-R.; Ji, J.-B.; Xing, J.; Wang, S.-Q. Spectroscopic methodologies and molecular docking studies on the interaction of antimalarial drug piperaquine and its metabolites with human serum albumin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 222, 117158. [Google Scholar] [CrossRef]
- Suryawanshi, V.D.; Walekar, L.S.; Gore, A.H.; Anbhule, P.V.; Kolekar, G.B. Spectroscopic analysis on the binding interaction of biologically active pyrimidine derivative with bovine serum albumin. J. Pharm. Anal. 2016, 6, 56–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Zheng, S.; Huang, X.; He, Q.; Sun, T. Exploring the mode of binding between butylated hydroxyanisole with bovine serum albumin: Multispectroscopic and molecular docking study. Food Chem. 2021, 357, 129771. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, N.; Shareghi, B.; Farhadian, S.; Hosseini-Koupaei, M. A comparative study of the interaction of naringenin with lysozyme by multi-spectroscopic methods, activity comparisons, and molecular modeling procedures. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 271, 120931. [Google Scholar] [CrossRef]
- Shahabadi, N.; Zendehcheshm, S. Evaluation of ct-DNA and HSA binding propensity of antibacterial drug chloroxine: Multi-spectroscopic analysis, atomic force microscopy and docking simulation. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 230, 118042. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Yang, Q.; Mao, X.; Chai, W.-M.; Peng, Y. Study on the interaction between 4-(1H-indol-3-yl)-2-(p-tolyl)quinazoline-3-oxide and human serum albumin. Bioorganic Med. Chem. 2020, 28, 115720. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, L.; Wang, Y.; Zhou, J.; Ding, J.; Li, W.; Xin, Y.; Fan, S.; Wang, Z.; Wang, Y. Biointeractions of Herbicide Atrazine with Human Serum Albumin: UV-Vis, Fluorescence and Circular Dichroism Approaches. Int. J. Environ. Res. Public Health 2018, 15, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szkudlarek, A.; Pożycka, J.; Kulig, K.; Owczarzy, A.; Rogóż, W.; Maciążek-Jurczyk, M. Changes in Glycated Human Serum Albumin Binding Affinity for Losartan in the Presence of Fatty Acids In Vitro Spectroscopic Analysis. Molecules 2022, 27, 401. [Google Scholar] [CrossRef]
- Qi, X.; Xu, D.; Zhu, J.; Wang, S.; Peng, J.; Gao, W.; Cao, Y. Studying the interaction mechanism between bovine serum albumin and lutein dipalmitate: Multi-spectroscopic and molecular docking techniques. Food Hydrocoll. 2021, 113, 106513. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, L.; Sun, Q.; Gan, N.; Zhang, Q.; Yang, J.; Yi, B.; Liao, X.; Zhu, D.; Li, H. Study on the interaction between 2,6-dihydroxybenzoic acid nicotine salt and human serum albumin by multi-spectroscopy and molecular dynamics simulation. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 270, 120868. [Google Scholar] [CrossRef]
- Manjushree, M.; Revanasiddappa, H.D. Interpretation of the binding interaction between bupropion hydrochloride with human serum albumin: A collective spectroscopic and computational approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 209, 264–273. [Google Scholar]
- Hadichegeni, S.; Goliaei, B.; Taghizadeh, M.; Davoodmanesh, S.; Taghavi, F.; Hashemi, M. Characterization of the interaction between human serum albumin and diazinon via spectroscopic and molecular docking methods. Hum. Exp. Toxicol. 2018, 37, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Kang, S.; Tian, J.; Li, M.; Liang, X.; Yang, M.; Zheng, Y.; Pi, Y.; Cao, X.; Liu, Y.; et al. Interaction of xylitol with whey proteins: Multi-spectroscopic techniques and docking studies. Food Chem. 2020, 326, 126804. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Gao, G.; Zhang, L.; Cai, Q.; Lu, N.; Cheng, L.; Li, S.; Hou, X. Drug-protein binding mechanism of juglone for early pharmacokinetic profiling: Insights from ultrafiltration, multi-spectroscopic and molecular docking methods. J. Pharm. Biomed. Anal. 2017, 141, 262–269. [Google Scholar] [CrossRef]
- Tayyab, S.; Sam, S.E.; Kabir, M.Z.; Ridzwan, N.F.W.; Mohamad, S.B. Molecular interaction study of an anticancer drug, ponatinib with human serum albumin using spectroscopic and molecular docking methods. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 214, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Ganorkar, K.; Mukherjee, S.; Singh, P.; Ghosh, S.K. Stabilization of a potential anticancer thiosemicarbazone derivative in Sudlow site I of human serum albumin: In vitro spectroscopy coupled with molecular dynamics simulation. Biophys. Chem. 2021, 269, 106509. [Google Scholar] [CrossRef]
- Vanaei, S.; Parizi, M.S.; Abdolhosseini, S.; Katouzian, I. Spectroscopic, molecular docking and molecular dynamic simulation studies on the complexes of β-lactoglobulin, safranal and oleuropein. Int. J. Biol. Macromol. 2020, 165, 2326–2337. [Google Scholar] [CrossRef]
- Xue, P.; Zhang, G.; Zhao, H.; Wang, W.; Zhang, J.; Ren, L. Serum albumin complexed with ellagic acid from pomegranate peel and its metabolite urolithin B. Food Biosci. 2022, 46, 101618. [Google Scholar] [CrossRef]
- Alsaif, N.A.; Wani, T.A.; Bakheit, A.H.; Zargar, S. Multi-spectroscopic investigation, molecular docking and molecular dynamic simulation of competitive interactions between flavonoids (quercetin and rutin) and sorafenib for binding to human serum albumin. Int. J. Biol. Macromol. 2020, 165, 2451–2461. [Google Scholar] [CrossRef]
- Maity, A.; Pal, U.; Chakraborty, B.; Sengupta, C.; Sau, A.; Chakraborty, S.; Basu, S. Preferential photochemical interaction of Ru (III) doped carbon nano dots with bovine serum albumin over human serum albumin. Int. J. Biol. Macromol. 2019, 137, 483–494. [Google Scholar] [CrossRef]
- Xinyu, Z.; Ruyi, C.; Yueyue, S.; Xiaoxi, Z.; Chongmei, T.; Daozong, X. Antioxidant and Anti-Inflammatory Activities of Six Flavonoids from Smilax glabra Roxb. Molecules 2020, 25, 5295. [Google Scholar]
- Petitpas, I.; Bhattacharya, A.A.; Twine, S.; East, M.; Curry, S. Crystal structure analysis of warfarin binding to human serum albumin: Anatomy of drug site I. J. Biol. Chem. 2001, 276, 22804–22809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, W.W.; Siu, S.W. PyMOL mControl: Manipulating molecular visualization with mobile devices. Biochem. Mol. Biol. Educ. 2017, 45, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, P.A.; Forli, S.; Goodsell, D.S.; Olson, A.J.; Sanner, M.F. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Comput. Biol. 2015, 11, e1004586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vina, A. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Zhang, G.; Li, N.; Zhang, Y.; Pan, J.; Gong, D. Binding mechanism of 4−octylphenol with human serum albumin: Spectroscopic investigations, molecular docking and dynamics simulation. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 255, 119662. [Google Scholar] [CrossRef]
- Anwer, R.; AlQumaizi, K.I.; Haque, S.; Somvanshi, P.; Ahmad, N.; AlOsaimi, S.M.; Fatma, T. Unravelling the interaction of glipizide with human serum albumin using various spectroscopic techniques and molecular dynamics studies. J. Biomol. Struct. Dyn. 2020, 39, 336–347. [Google Scholar] [CrossRef]
- Wu, S.; Sun, Y.; Chen, D.; Liu, H.; Li, Z.; Chen, M.; Wang, C.; Cheng, L.; Guo, Q.; Peng, X. The noncovalent conjugations of human serum albumin (HSA) with MS/AK and the effect on anti-oxidant capacity as well as anti-glycation activity of Monascus yellow pigments. Food Funct. 2021, 12, 3692–3704. [Google Scholar] [CrossRef]

| pH | T (K) | Ksv (×104 L·mol−1) | Kq (×1012 L·mol−1·s−1) | R | SD |
|---|---|---|---|---|---|
| 288 | 6.44 | 6.44 | 0.9897 | 0.14 | |
| 7.4 | 298 | 5.70 | 5.70 | 0.9980 | 0.12 |
| 310 | 4.70 | 4.70 | 0.9991 | 0.09 |
| pH | T (K) | Ka (×104 L·mol−1) | n | R | ΔH0 (kJ·mol−1) | ΔG0 (kJ·mol−1) | ΔS0 (J·mol−1·K−1) |
|---|---|---|---|---|---|---|---|
| 288 | 6.77 ± 0.15 | 1.02 | 0.9856 | −26.67 ± 0.05 | |||
| 7.4 | 298 | 5.77 ± 0.02 | 1.00 | 0.9985 | −14.12 ± 0.54 | −27.11 ± 0.01 | 43.57 ± 1.75 |
| 310 | 4.46 ± 0.03 | 0.99 | 0.9959 | −27.63 ± 0.02 |
| pH | Site Marker | Ka (×104 L·mol−1) | R |
|---|---|---|---|
| Blank | 22.74 ± 0.09 | 0.9957 | |
| 7.4 | Ibuprofen | 19.93 ± 0.12 | 0.9932 |
| Warfarin sodium | 5.31 ± 0.15 | 0.9927 |
| System | Peaks | Peaks Position λex/λem (nm/nm) | Stokes Shift Δλ (nm) | Fluorescence Intensity (a.u.) |
|---|---|---|---|---|
| Native HSA | Fluorescence peak 1 | 280.0/330.0 | 50 | 1539 |
| Fluorescence peak 2 | 230.0/330.0 | 100 | 623.2 | |
| Astilbin−HSA (2:1) | Fluorescence peak 1 | 280.0/320.0 | 40 | 1291 |
| Fluorescence peak 2 | 230.0/330.0 | 100 | 478.6 |
| System | Structure Component (%) | |||||
|---|---|---|---|---|---|---|
| α-Helix | β-Sheet | β-Turn | β-Antiparallel | Random Coil | ||
| Method | FTIR | CD | FTIR | |||
| Native HSA | 59.06 ± 0.18 | 59.24 ± 0.16 | 10.60 ± 0.14 | 13.86 ± 0.09 | 1.85 ± 0.09 | 14.63 ± 0.13 |
| Astilbin–HSA | 56.31 ± 0.17 | 55.98 ± 0.15 | 7.36 ± 0.08 | 14.01 ± 0.05 | 2.33 ± 0.04 | 20.00 ± 0.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Sun, J.; Niu, T.; Mao, B.; Gao, S.; Zhao, P.; Sun, L. Molecular Insight into the Binding of Astilbin with Human Serum Albumin and Its Effect on Antioxidant Characteristics of Astilbin. Molecules 2022, 27, 4487. https://doi.org/10.3390/molecules27144487
Han X, Sun J, Niu T, Mao B, Gao S, Zhao P, Sun L. Molecular Insight into the Binding of Astilbin with Human Serum Albumin and Its Effect on Antioxidant Characteristics of Astilbin. Molecules. 2022; 27(14):4487. https://doi.org/10.3390/molecules27144487
Chicago/Turabian StyleHan, Xiangyu, Jing Sun, Tianmei Niu, Beibei Mao, Shijie Gao, Pan Zhao, and Linlin Sun. 2022. "Molecular Insight into the Binding of Astilbin with Human Serum Albumin and Its Effect on Antioxidant Characteristics of Astilbin" Molecules 27, no. 14: 4487. https://doi.org/10.3390/molecules27144487
APA StyleHan, X., Sun, J., Niu, T., Mao, B., Gao, S., Zhao, P., & Sun, L. (2022). Molecular Insight into the Binding of Astilbin with Human Serum Albumin and Its Effect on Antioxidant Characteristics of Astilbin. Molecules, 27(14), 4487. https://doi.org/10.3390/molecules27144487