Microbial Natural Products with Antiviral Activities, Including Anti-SARS-CoV-2: A Review
Abstract
1. Introduction
2. Anti-Human Immunodeficiency Virus
3. Anti-Hepatitis Virus
3.1. Anti-Hepatitis C Virus
3.2. Anti-Hepatitis B Virus
4. Anti-Herpes Simplex Virus
5. Anti-Influenza
6. Anti-Respiratory Syncytial Virus
7. Anti-SARS-CoV-2
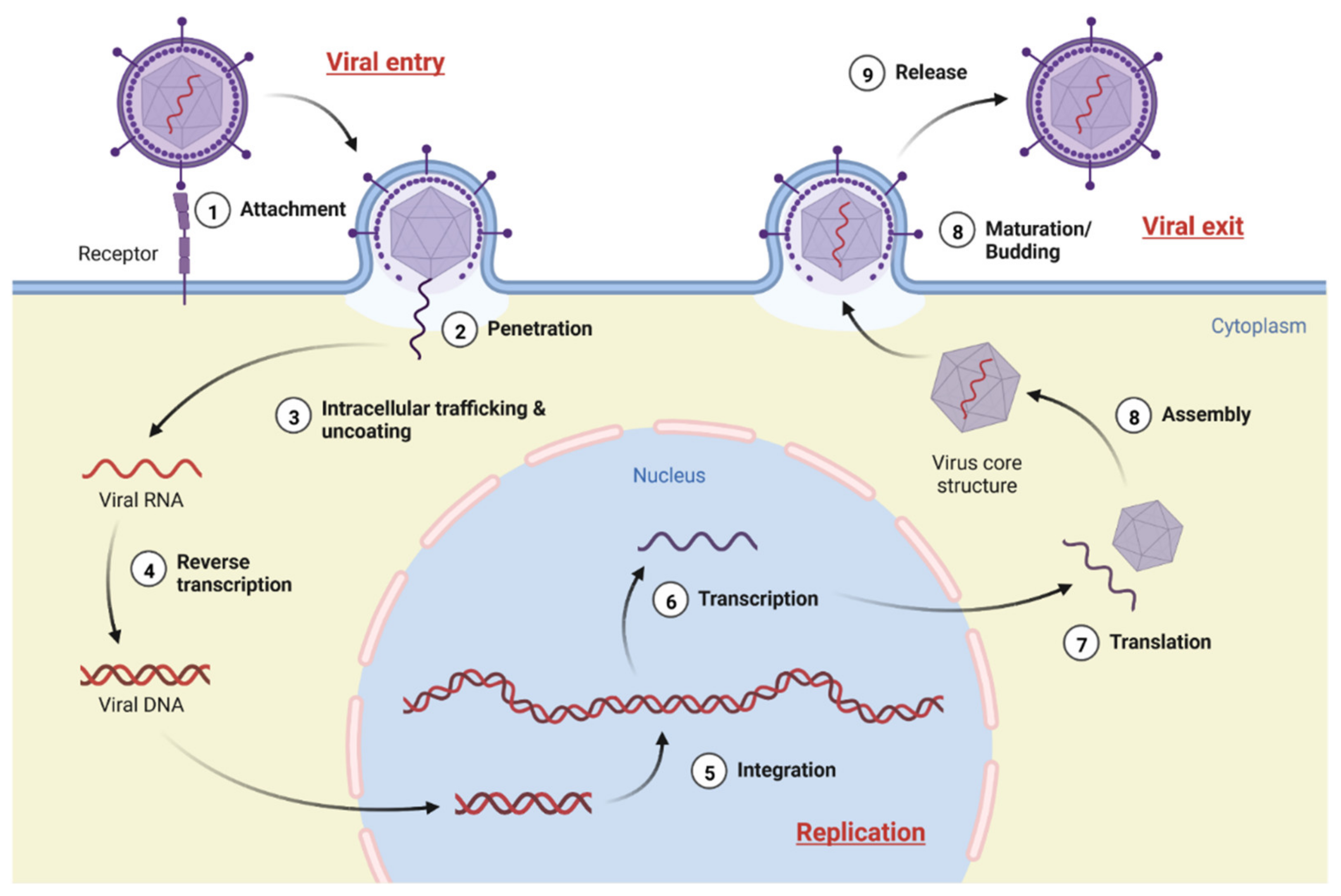
8. Viruses in Their Biological Make-Up and Possible Life Cycle Target
8.1. Viral Entry
8.1.1. Mechanism
8.1.2. Possible Target of Viral Entry
8.2. Genome Replication
8.2.1. Mechanism
8.2.2. Possible Target
8.3. Viral Exit
8.3.1. Assembly and Maturation
8.3.2. Release
9. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Crowley, D.; Avramovic, G.; Cullen, W.; Farrell, C.; Halpin, A.; Keevans, M.; Laird, E.; McHugh, T.; McKiernan, S.; Miggin, S.J. New hepatitis C virus infection, re-infection and associated risk behaviour in male Irish prisoners: A cohort study, 2019. Arch. Public Health 2021, 79, 97. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Dávila, J.D.; Allen, A.; Haklay, M.; Tacoli, C.; Fèvre, E.M. Does urbanization make emergence of zoonosis more likely? Evidence, myths and gaps. Environ. Urban. 2019, 31, 443–460. [Google Scholar] [CrossRef] [PubMed]
- Spinella, C.; Mio, A.M. Simulation of the impact of people mobility, vaccination rate, and virus variants on the evolution of COVID-19 outbreak in Italy. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Delaugerre, C.; Foissac, F.; Abdoul, H.; Masson, G.; Choupeaux, L.; Dufour, E.; Gastli, N.; Delarue, S.M.; Néré, M.L.; Minier, M. Prevention of SARS-CoV-2 transmission during a large, live, indoor gathering (SPRING): A non-inferiority, randomised, controlled trial. Lancet Infect. Dis. 2022, 22, 341–348. [Google Scholar] [CrossRef]
- Zell, R.; Krumbholz, A.; Wutzler, P. Impact of global warming on viral diseases: What is the evidence? Curr. Opin. Biotech. 2008, 19, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.-M.; Wu, G. Engineering. Trends in global warming and evolution of polymerase basic protein 2 family from influenza A virus. J. Biomed. Sci. 2009, 2, 458. [Google Scholar]
- Frediansyah, A.; Tiwari, R.; Sharun, K.; Dhama, K.; Harapan, H. Antivirals for COVID-19: A critical review. Clin. Epidemiol. Glob. Health 2021, 9, 90–98. [Google Scholar] [CrossRef]
- Smith, K.F.; Sax, D.F.; Gaines, S.D.; Guernier, V.; Guégan, J.-F.J.E. Globalization of human infectious disease. Ecology 2007, 88, 1903–1910. [Google Scholar] [CrossRef]
- Chaitanya, K. Structure and Organization of Virus Genomes. In Genome and Genomics; Springer: Cham, Switzerland, 2019; pp. 1–30. [Google Scholar]
- Worldometers. COVID-19 Coronavirus Pandemic. Available online: https://www.worldometers.info/coronavirus/ (accessed on 21 May 2022).
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Zhang, L.; An, R.; Wang, J.; Sun, N.; Zhang, S.; Hu, J.; Kuai, J. Exploring novel bioactive compounds from marine microbes. Curr. Opin. Microbiol. 2005, 8, 276–281. [Google Scholar] [CrossRef]
- Haruna, A.; Yahaya, S.M. Recent advances in the chemistry of bioactive compounds from plants and soil microbes: A review. Chem. Afr. 2021, 4, 231–248. [Google Scholar] [CrossRef]
- Firn, R.D.; Jones, C.G. Natural products—A simple model to explain chemical diversity. Nat. Prod. Rep. 2003, 20, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Henrich, C.J.; Beutler, J.A. Matching the power of high throughput screening to the chemical diversity of natural products. Nat. Prod. Rep. 2013, 30, 1284–1298. [Google Scholar] [CrossRef] [PubMed]
- Lautie, E.; Russo, O.; Ducrot, P.; Boutin, J.A. Unraveling plant natural chemical diversity for drug discovery purposes. Front. Pharmacol. 2020, 11, 397. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef]
- Corcoran, O.; Spraul, M. LC–NMR–MS in drug discovery. Drug Discov. Today 2003, 8, 624–631. [Google Scholar] [CrossRef]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef]
- Nishiuchi, K.; Ohashi, H.; Nishioka, K.; Yamasaki, M.; Furuta, M.; Mashiko, T.; Tomoshige, S.; Ohgane, K.; Kamisuki, S.; Watashi, K.J. Synthesis and Antiviral Activities of Neoechinulin B and Its Derivatives. J. Nat. Prod. 2021, 85, 284–291. [Google Scholar] [CrossRef]
- Cullen, B.R. Human immunodeficiency virus as a prototypic complex retrovirus. J. Virol. 1991, 65, 1053. [Google Scholar] [CrossRef]
- Foley, J.F.; Yu, C.-R.; Solow, R.; Yacobucci, M.; Peden, K.W.; Farber, J.M. Roles for CXC chemokine ligands 10 and 11 in recruiting CD4+ T cells to HIV-1-infected monocyte-derived macrophages, dendritic cells, and lymph nodes. J. Immunol. 2005, 174, 4892–4900. [Google Scholar] [CrossRef]
- Montarroyos, U.R.; Miranda-Filho, D.B.; César, C.C.; Souza, W.V.; Lacerda, H.R.; de Fátima Pessoa Militão Albuquerque, M.; Aguiar, M.F.; de Alencar Ximenes, R.A. Factors related to changes in CD4+ T-cell counts over time in patients living with HIV/AIDS: A multilevel analysis. PLoS ONE 2014, 9, e84276. [Google Scholar] [CrossRef] [PubMed]
- UNAIDS. 38 million people are living with HIV around the world. 2020. Available online: https://www.unaids.org/en/resources/infographics/people-living-with-hiv-around-the-world (accessed on 1 February 2022).
- Zhang, Y.; Tian, R.; Liu, S.; Chen, X.; Liu, X.; Che, Y.J.B. Alachalasins A–G, new cytochalasins from the fungus Stachybotrys charatum. Bioorg. Med. Chem. 2008, 16, 2627–2634. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tian, R.; Liu, S.; Chen, X.; Liu, X.; Che, Y. Corrigendum to "Alachalasins A–G, new cytochalasins from the fungus Stachybotrys charatum". Bioorg. Med. Chem. 2009, 1, 428. [Google Scholar] [CrossRef]
- Liu, L.; Liu, S.; Niu, S.; Guo, L.; Chen, X.; Che, Y. Isoprenylated chromone derivatives from the plant endophytic fungus Pestalotiopsis fici. J. Nat. Prod. 2009, 72, 1482–1486. [Google Scholar] [CrossRef]
- Liu, L.; Liu, S.; Chen, X.; Guo, L.; Che, Y. Pestalofones A–E, bioactive cyclohexanone derivatives from the plant endophytic fungus Pestalotiopsis fici. Bioorg. Med. Chem. 2009, 17, 606–613. [Google Scholar] [CrossRef]
- Guo, H.; Sun, B.; Gao, H.; Chen, X.; Liu, S.; Yao, X.; Liu, X.; Che, Y. Diketopiperazines from the Cordyceps-colonizing fungus Epicoccum nigrum. J. Nat. Prod. 2009, 72, 2115–2119. [Google Scholar] [CrossRef]
- Zou, X.; Liu, S.; Zheng, Z.; Zhang, H.; Chen, X.; Liu, X.; Li, E. Two New Imidazolone-Containing Alkaloids and Further Metabolites from the Ascomycete Fungus Tricladium sp. Chem. Biodivers. 2011, 8, 1914–1920. [Google Scholar] [CrossRef]
- Chen, C.; Zhu, H.; Wang, J.; Yang, J.; Li, X.N.; Wang, J.; Chen, K.; Wang, Y.; Luo, Z.; Yao, G. Armochaetoglobins K–R, Anti-HIV Pyrrole-Based Cytochalasans from Chaetomium globosum TW1-1. Eur. J. Org. Chem. 2015, 2015, 3086–3094. [Google Scholar] [CrossRef]
- Ma, X.; Li, L.; Zhu, T.; Ba, M.; Li, G.; Gu, Q.; Guo, Y.; Li, D. Phenylspirodrimanes with anti-HIV activity from the sponge-derived fungus Stachybotrys chartarum MXH-X73. J. Nat. Prod. 2013, 76, 2298–2306. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, J.; Shen, Y.; Tan, Z.; Zhang, M.; Chen, R.; Zhao, J.; Zhang, D.; Yu, L.; Dai, J. Stachybotrysams A–E, prenylated isoindolinone derivatives with anti-HIV activity from the fungus Stachybotrys chartarum. Phytochem. Lett. 2017, 20, 289–294. [Google Scholar] [CrossRef]
- Li, Y.; Liu, D.; Cen, S.; Proksch, P.; Lin, W. Isoindolinone-type alkaloids from the sponge-derived fungus Stachybotrys chartarum. Tetrahedron 2014, 70, 7010–7015. [Google Scholar] [CrossRef]
- Zhou, X.; Fang, W.; Tan, S.; Lin, X.; Xun, T.; Yang, B.; Liu, S.; Liu, Y. Aspernigrins with anti-HIV-1 activities from the marine-derived fungus Aspergillus niger SCSIO Jcsw6F30. Bioorg. Med. Chem. Lett. 2016, 26, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Liu, D.; Shao, Z.; Proksch, P.; Lin, W. Eutypellazines A–M, thiodiketopiperazine-type alkaloids from deep sea derived fungus Eutypella sp. MCCC 3A00281. RSC Adv. 2017, 7, 33580–33590. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, D.; Proksch, P.; Zhou, D.; Lin, W. Truncateols OV, further isoprenylated cyclohexanols from the sponge-associated fungus Truncatella angustata with antiviral activities. Phytochemistry 2018, 155, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Yang, B.; Liu, J.; Xun, T.; Liu, Y.; Zhou, X. Penicillixanthone A, a marine-derived dual-coreceptor antagonist as anti-HIV-1 agent. Nat. Prod. Res. 2019, 33, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.Q.; Li, Y.H.; Fan, Z.W.; Yan, W.L.; He, Z.H.; Zhong, T.H.; Gai, Y.B.; Yang, X.W. Anti-HIV Compounds from the Deep-Sea-Derived Fungus Chaetomium globosum. Chem. Biodivers. 2022, 19, e202100804. [Google Scholar] [CrossRef]
- Yang, Z.; Ding, J.; Ding, K.; Chen, D.; Cen, S.; Ge, M. Phomonaphthalenone A: A novel dihydronaphthalenone with anti-HIV activity from Phomopsis sp. HCCB04730. Phytochem. Lett. 2013, 6, 257–260. [Google Scholar] [CrossRef]
- Bashyal, B.P.; Wellensiek, B.P.; Ramakrishnan, R.; Faeth, S.H.; Ahmad, N.; Gunatilaka, A.L. Altertoxins with potent anti-HIV activity from Alternaria tenuissima QUE1Se, a fungal endophyte of Quercus emoryi. Bioorg. Med. Chem. 2014, 22, 6112–6116. [Google Scholar] [CrossRef]
- Ding, J.; Zhao, J.; Yang, Z.; Ma, L.; Mi, Z.; Wu, Y.; Guo, J.; Zhou, J.; Li, X.; Guo, Y.J.V. Microbial natural product alternariol 5-O-methyl ether inhibits HIV-1 integration by blocking nuclear import of the pre-integration complex. Viruses 2017, 9, 105. [Google Scholar] [CrossRef]
- Zhao, J.-L.; Zhang, M.; Liu, J.-M.; Tan, Z.; Chen, R.-D.; Xie, K.-B.; Dai, J.-G. Bioactive steroids and sorbicillinoids isolated from the endophytic fungus Trichoderma sp. Xy24. J. Asian. Nat. Prod. Res. 2017, 19, 1028–1035. [Google Scholar] [CrossRef]
- Yang, Z.-J.; Zhang, Y.-F.; Wu, K.; Xu, Y.-X.; Meng, X.-G.; Jiang, Z.-T.; Ge, M.; Shao, L. New azaphilones, phomopsones AC with biological activities from an endophytic fungus Phomopsis sp. CGMCC No. 5416. Fitoterapia 2020, 145, 104573. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, M.; Chen, R.; Xie, K.; Chen, D.; Si, S.; Dai, J.J. Three new compounds from endophytic fungus Periconia sp. F-31. Chin. Pharm. Sci. 2020, 29, 244–251. [Google Scholar]
- Pang, X.; Zhao, J.-Y.; Fang, X.-M.; Zhang, T.; Zhang, D.-W.; Liu, H.-Y.; Su, J.; Cen, S.; Yu, L.-Y. Metabolites from the plant endophytic fungus Aspergillus sp. CPCC 400735 and their anti-HIV activities. J. Nat. Prod. 2017, 80, 2595–2601. [Google Scholar] [CrossRef]
- Lianeras, J.; Riveiro-Barciela, M.; Rando-Segura, A.; Marcos-Fosch, C.; Roade, L.; Velázquez, F.; Rodríguez-Frías, F.; Esteban, R.; Buti, M. Etiologies and features of acute viral hepatitis in Spain. Clin. Gastroenterol. Hepatol. 2021, 19, 1030–1037. [Google Scholar] [CrossRef]
- Frediansyah, A.; Sallam, M.; Yufika, A.; Sharun, K.; Iqhrammullah, M.; Chandran, D.; Mamada, S.S.; Sallam, D.E.; Khader, Y.; Lemu, Y.K. Acute severe hepatitis of unknown etiology in children: A mini-review. Narra J. 2022, 2, 1–11. [Google Scholar] [CrossRef]
- Bandiera, S.; Bian, C.B.; Hoshida, Y.; Baumert, T.F.; Zeisel, M.B. Chronic hepatitis C virus infection and pathogenesis of hepatocellular carcinoma. Curr. Opin. Virol. 2016, 20, 99–105. [Google Scholar] [CrossRef]
- Seto, W.K.; Lai, C.L.; Yuen, M.F. Acute-on-chronic liver failure in chronic hepatitis B. J. Gastroenterol. Hepatol. 2012, 27, 662–669. [Google Scholar] [CrossRef]
- Lavanchy, D. Chronic viral hepatitis as a public health issue in the world. Best Pract. Gastroenterol. 2008, 22, 991–1008. [Google Scholar] [CrossRef]
- WHO. Hepatitis C. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (accessed on 23 April 2022).
- Nakamoto, S.; Kanda, T.; Wu, S.; Shirasawa, H.; Yokosuka, O. Hepatitis C virus NS5A inhibitors and drug resistance mutations. World J. Gastroenterol. 2014, 20, 2902. [Google Scholar] [CrossRef]
- Shih, I.-H.; Vliegen, I.; Peng, B.; Yang, H.; Hebner, C.; Paeshuyse, J.; Pürstinger, G.; Fenaux, M.; Tian, Y.; Mabery, E. Mechanistic characterization of GS-9190 (Tegobuvir), a novel nonnucleoside inhibitor of hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 2011, 55, 4196–4203. [Google Scholar] [CrossRef]
- Lamarre, D.; Anderson, P.C.; Bailey, M.; Beaulieu, P.; Bolger, G.; Bonneau, P.; Bös, M.; Cameron, D.R.; Cartier, M.; Cordingley, M.G. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 2003, 426, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Geddawy, A.; Ibrahim, Y.F.; Elbahie, N.M.; Ibrahim, M.A. Direct acting anti-hepatitis C virus drugs: Clinical pharmacology and future direction. J. Trans. Intern. Med. 2017, 5, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Marchelli, R.; Dossena, A.; Pochini, A.; Dradi, E. The structures of five new didehydropeptides related to neoechinulin, isolated from Aspergillus amstelodami. J. Chem. Soc. Perkin Trans. 1977, 7, 713–717. [Google Scholar] [CrossRef]
- Nakajima, S.; Watashi, K.; Ohashi, H.; Kamisuki, S.; Izaguirre-Carbonell, J.; Kwon, A.T.-J.; Suzuki, H.; Kataoka, M.; Tsukuda, S.; Okada, M. Fungus-derived neoechinulin B as a novel antagonist of liver X receptor, identified by chemical genetics using a hepatitis C virus cell culture system. J. Virol. 2016, 90, 9058–9074. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.C.F.; Wong, J.H.; Pan, W.L.; Chan, Y.S.; Yin, C.M.; Dan, X.L.; Wang, H.X.; Fang, E.F.; Lam, S.K.; Ngai, P.H.K. Antifungal and antiviral products of marine organisms. Appl. Microbiol. Biotechnol. 2014, 98, 3475–3494. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.; Rodriguez, A.; Taglialatela-Scafati, O.; Fusetani, N. Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2003, 11, 2510–2573. [Google Scholar] [CrossRef]
- Singh, R.P.; Kumari, P.; Reddy, C. Antimicrobial compounds from seaweeds-associated bacteria and fungi. Appl. Microbiol. Biotechnol. 2015, 99, 1571–1586. [Google Scholar] [CrossRef]
- Li, J.; Hu, Y.; Hao, X.; Tan, J.; Li, F.; Qiao, X.; Chen, S.; Xiao, C.; Chen, M.; Peng, Z. Raistrickindole A, an anti-HCV oxazinoindole alkaloid from Penicillium raistrickii IMB17-034. J. Nat. Prod. 2019, 82, 1391–1395. [Google Scholar] [CrossRef]
- Li, B.; Li, L.; Peng, Z.; Liu, D.; Si, L.; Wang, J.; Yuan, B.; Huang, J.; Proksch, P.; Lin, W. Harzianoic acids A and B, new natural scaffolds with inhibitory effects against hepatitis C virus. Bioorg. Med. Chem. 2019, 27, 560–567. [Google Scholar] [CrossRef]
- Nishikori, S.; Takemoto, K.; Kamisuki, S.; Nakajima, S.; Kuramochi, K.; Tsukuda, S.; Iwamoto, M.; Katayama, Y.; Suzuki, T.; Kobayashi, S. Anti-hepatitis C virus natural product from a fungus, Penicillium herquei. J. Nat. Prod. 2016, 79, 442–446. [Google Scholar] [CrossRef]
- Ahmed, E.; Rateb, M.; El-Kassem, A.; Hawas, U.W. Anti-HCV protease of diketopiperazines produced by the Red Sea sponge-associated fungus Aspergillus versicolor. Appl. Biochem. Microbiol. 2017, 53, 101–106. [Google Scholar] [CrossRef]
- Hawas, U.W.; El-Halawany, A.M.; Ahmede, E.F. Hepatitis C virus NS3-NS4A protease inhibitors from the endophytic Penicillium chrysogenum isolated from the red alga Liagora viscida. Z. Nat. C 2013, 68, 355–366. [Google Scholar]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef]
- Khaldi, N.; Seifuddin, F.T.; Turner, G.; Haft, D.; Nierman, W.C.; Wolfe, K.H.; Fedorova, N.D. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010, 47, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; Huimi Wang, H.; Davies Frs, J. Antibiotics as signalling molecules. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1195–1200. [Google Scholar] [CrossRef] [PubMed]
- El-Gendy, M.M.A.A.; Yahya, S.M.; Hamed, A.R.; Soltan, M.M.; El-Bondkly, A.M.A. Phylogenetic analysis and biological evaluation of marine endophytic fungi derived from Red Sea sponge Hyrtios erectus. Appl. Biochem. Biotechnol. 2018, 185, 755–777. [Google Scholar] [CrossRef]
- El-Kassem, L.A.; Hawas, U.W.; El-Souda, S.; Ahmed, E.F.; El-Khateeb, W.; Fayad, W. Anti-HCV protease potential of endophytic fungi and cytotoxic activity. Biocatal. Agric. Biotechnol. 2019, 19, 101170. [Google Scholar] [CrossRef]
- Lok, A.S.; McMahon, B.J. Chronic hepatitis B. Hepatology 2007, 346, 1682–1683. [Google Scholar] [CrossRef]
- Maynard, J.E. Hepatitis B: Global importance and need for control. Vaccine 1990, 8, S18–S20. [Google Scholar] [CrossRef]
- Yuan, B.H.; Li, R.H.; Huo, R.R.; Li, M.J.; Papatheodoridis, G.; Zhong, J.H. Lower risk of hepatocellular carcinoma with tenofovir than entecavir treatment in subsets of chronic hepatitis B patients: An updated meta-analysis. J. Gastroenterol. Hepatol. 2022, 37, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.-L.; Zhang, L.-M.; Chen, Y.-P.; Zi, S.-H.; Xiang, H.; Zhao, D.-K.; Shen, Y. Two new compounds from an endophytic fungus Alternaria solani. J. Asian Nat. Prod. 2012, 14, 1144–1148. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Qin, S.; Gao, H.; Zhu, G.; Wang, W.; Zhu, W.; Wang, Y. An anti-HBV anthraquinone from aciduric fungus Penicillium sp. OUCMDZ-4736 under low pH stress. Extremophiles 2018, 22, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, H.; Kamisuki, S.; Kaneko, M.; Yamaguchi, Y.; Takeuchi, T.; Watashi, K.; Sugawara, F. Isolation and structure of vanitaracin A, a novel anti-hepatitis B virus compound from Talaromyces sp. Bioorg. Med. Chem. Lett. 2015, 25, 4325–4328. [Google Scholar] [CrossRef]
- Kaneko, M.; Watashi, K.; Kamisuki, S.; Matsunaga, H.; Iwamoto, M.; Kawai, F.; Ohashi, H.; Tsukuda, S.; Shimura, S.; Suzuki, R. A novel tricyclic polyketide, vanitaracin A, specifically inhibits the entry of hepatitis B and D viruses by targeting sodium taurocholate cotransporting polypeptide. J. Virol. 2015, 89, 11945–11953. [Google Scholar] [CrossRef]
- Isaka, M.; Kittakoop, P.; Kirtikara, K.; Hywel-Jones, N.L.; Thebtaranonth, Y. Bioactive substances from insect pathogenic fungi. Acc. Chem. Res. 2005, 38, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Kuephadungphan, W.; Phongpaichit, S.; Luangsa-ard, J.J.; Rukachaisirikul, V. Antimicrobial activity of invertebrate-pathogenic fungi in the genera Akanthomyces and Gibellula. Mycoscience 2014, 55, 127–133. [Google Scholar] [CrossRef]
- Wagenaar, M.M.; Gibson, D.M.; Clardy, J. Akanthomycin, a New Antibiotic Pyridone from the Entomopathogenic Fungus Akanthomyces gracilis. Org. Lett. 2002, 4, 671–673. [Google Scholar] [CrossRef]
- Dong, C.; Yu, J.; Zhu, Y.; Dong, C. Inhibition of hepatitis B virus gene expression & replication by crude destruxins from Metarhizium anisopliae var. dcjhyium. Indian J. Med. Res. 2013, 138, 969. [Google Scholar]
- Chen, H.-C.; Chou, C.-K.; Sun, C.-M.; Yeh, S.F. Suppressive effects of destruxin B on hepatitis B virus surface antigen gene expression in human hepatoma cells. Antivir. Res. 1997, 34, 137–144. [Google Scholar] [CrossRef]
- Whitley, R.J. Herpes simplex virus infection. Semin. Pediatr. Infect. Dis. 2002, 13, 6–11. [Google Scholar] [CrossRef]
- Arduino, P.G.; Porter, S.R. Herpes Simplex Virus Type 1 infection: Overview on relevant clinico-pathological features. J. Oral Pathol. Med. 2008, 37, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Pebody, R.; Andrews, N.; Brown, D.; Gopal, R.; De Melker, H.; François, G.; Gatcheva, N.; Hellenbrand, W.; Jokinen, S.; Klavs, I. The seroepidemiology of herpes simplex virus type 1 and 2 in Europe. Sex. Transm. Infect. 2004, 80, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Casrouge, A.; Zhang, S.-Y.; Eidenschenk, C.; Jouanguy, E.; Puel, A.; Yang, K.; Alcais, A.; Picard, C.; Mahfoufi, N.; Nicolas, N. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 2006, 314, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Sarangi, P.P.; Kim, B.; Kurt-Jones, E.; Rouse, B.T. Innate recognition network driving herpes simplex virus-induced corneal immunopathology: Role of the toll pathway in early inflammatory events in stromal keratitis. J. Virol. 2007, 81, 11128–11138. [Google Scholar] [CrossRef]
- James, C.; Harfouche, M.; Welton, N.J.; Turner, K.M.; Abu-Raddad, L.J.; Gottlieb, S.L.; Looker, K.J. Herpes simplex virus: Global infection prevalence and incidence estimates, 2016. Bull. World Health Org. 2020, 98, 315. [Google Scholar] [CrossRef]
- Pottage, J., Jr.; Kessler, H. Herpes simplex virus resistance to acyclovir: Clinical relevance. Infect. Agents Dis. 1995, 4, 115–124. [Google Scholar]
- Bacon, T.H.; Levin, M.J.; Leary, J.J.; Sarisky, R.T.; Sutton, D. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin. Microbiol. Rev. 2003, 16, 114–128. [Google Scholar] [CrossRef]
- Corey, L.; Wald, A.; Patel, R.; Sacks, S.L.; Tyring, S.K.; Warren, T.; Douglas, J.M., Jr.; Paavonen, J.; Morrow, R.A.; Beutner, K.R. Once-daily valacyclovir to reduce the risk of transmission of genital herpes. NEJM 2004, 350, 11–20. [Google Scholar] [CrossRef]
- Moghadamtousi, S.Z.; Nikzad, S.; Kadir, H.A.; Abubakar, S.; Zandi, K. Potential antiviral agents from marine fungi: An overview. Mar. Drugs 2015, 13, 4520–4538. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, K.; Wang, W.; Zhang, G.; Zhu, T.; Che, Q.; Gu, Q.; Li, D. Amphiepicoccins A–J: Epipolythiodioxopiperazines from the fish-gill-derived fungus Epicoccum nigrum HDN17-88. J. Nat. Prod. 2020, 83, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jia, J.; Wang, L.; Li, F.; Wang, Y.; Jiang, Y.; Song, X.; Qin, S.; Zheng, K.; Ye, J. Anti-HSV-1 activity of Aspergillipeptide D, a cyclic pentapeptide isolated from fungus Aspergillus sp. SCSIO 41501. Virol. J. 2020, 17, 41. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Nong, X.-H.; Ren, Z.; Wang, J.; Liang, X.; Wang, L.; Qi, S.-H. Antiviral peptides from marine gorgonian-derived fungus Aspergillus sp. SCSIO 41501. Tetrahedron Lett. 2017, 58, 1151–1155. [Google Scholar] [CrossRef]
- Huang, Z.; Nong, X.; Ren, Z.; Wang, J.; Zhang, X.; Qi, S. Anti-HSV-1, antioxidant and antifouling phenolic compounds from the deep-sea-derived fungus Aspergillus versicolor SCSIO 41502. Bioorg. Med. Chem. Lett. 2017, 27, 787–791. [Google Scholar] [CrossRef]
- Sun, Y.-L.; Wang, J.; Wang, Y.-F.; Zhang, X.-Y.; Nong, X.-H.; Chen, M.-Y.; Xu, X.-Y.; Qi, S.-H. Cytotoxic and antiviral tetramic acid derivatives from the deep-sea-derived fungus Trichobotrys effuse DFFSCS021. Tetrahedron 2015, 71, 9328–9332. [Google Scholar] [CrossRef]
- Nong, X.-H.; Wang, Y.-F.; Zhang, X.-Y.; Zhou, M.-P.; Xu, X.-Y.; Qi, S.-H. Territrem and butyrolactone derivatives from a marine-derived fungus Aspergillus terreus. Mar. Drugs 2014, 12, 6113–6124. [Google Scholar] [CrossRef]
- Rowley, D.C.; Kelly, S.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Halovirs A–E, new antiviral agents from a marine-derived fungus of the genus Scytalidium. Bioorg. Med. Chem. 2003, 11, 4263–4274. [Google Scholar] [CrossRef]
- Shushni, M.A.; Singh, R.; Mentel, R.; Lindequist, U. Balticolid: A new 12-membered macrolide with antiviral activity from an ascomycetous fungus of marine origin. Mar. Drugs 2011, 9, 844–851. [Google Scholar] [CrossRef]
- Gill, E.E.; Franco, O.L.; Hancock, R.E. Antibiotic adjuvants: Diverse strategies for controlling drug-resistant pathogens. Chem. Biol. Drug Des. 2015, 85, 56–78. [Google Scholar] [CrossRef]
- Ateba, J.E.; Toghueo, R.M.; Awantu, A.F.; Mba’ning, B.M.; Gohlke, S.; Sahal, D.; Rodrigues-Filho, E.; Tsamo, E.; Boyom, F.F.; Sewald, N. Antiplasmodial properties and cytotoxicity of endophytic fungi from Symphonia globulifera (Clusiaceae). J. Fungi 2018, 4, 70. [Google Scholar] [CrossRef]
- Manganyi, M.; Regnier, T.; Kumar, A.; Bezuidenhout, C.; Ateba, C. Biodiversity and antibacterial screening of endophytic fungi isolated from Pelargonium sidoides. S. Afr. J. Bot. 2018, 116, 192–199. [Google Scholar] [CrossRef]
- Selim, K.A.; Elkhateeb, W.A.; Tawila, A.M.; El-Beih, A.A.; Abdel-Rahman, T.M.; El-Diwany, A.I.; Ahmed, E.F. Antiviral and antioxidant potential of fungal endophytes of Egyptian medicinal plants. Fermentation 2018, 4, 49. [Google Scholar] [CrossRef]
- Bunyapaiboonsri, T.; Yoiprommarat, S.; Srikitikulchai, P.; Srichomthong, K.; Lumyong, S. Oblongolides from the endophytic fungus Phomopsis sp. BCC 9789. J. Nat. Prod. 2010, 73, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, S.-J.; Li, J.-J.; Liang, Z.-Z.; Zhao, C.-Q. Novel natural products from extremophilic fungi. Mar. Drugs 2018, 16, 194. [Google Scholar] [CrossRef] [PubMed]
- Arena, A.; Maugeri, T.L.; Pavone, B.; Iannello, D.; Gugliandolo, C.; Bisignano, G. Antiviral and immunoregulatory effect of a novel exopolysaccharide from a marine thermotolerant Bacillus licheniformis. Int. Immunopharmacol. 2006, 6, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Arena, A.; Gugliandolo, C.; Stassi, G.; Pavone, B.; Iannello, D.; Bisignano, G.; Maugeri, T.L. An exopolysaccharide produced by Geobacillus thermodenitrificans strain B3-72: Antiviral activity on immunocompetent cells. Immunol. Lett. 2009, 123, 132–137. [Google Scholar] [CrossRef]
- Molnar, I.; Gibson, D.M.; Krasnoff, S.B. Secondary metabolites from entomopathogenic Hypocrealean fungi. Nat. Prod. Rep. 2010, 27, 1241–1275. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, L. Beauvericin, a bioactive compound produced by fungi: A short review. Molecules 2012, 17, 2367–2377. [Google Scholar] [CrossRef]
- Kornsakulkarn, J.; Thongpanchang, C.; Lapanun, S.; Srichomthong, K. Isocoumarin glucosides from the scale insect fungus Torrubiella tenuis BCC 12732. J. Nat. Prod. 2009, 72, 1341–1343. [Google Scholar] [CrossRef]
- Bunyapaiboonsri, T.; Yoiprommarat, S.; Intereya, K.; Kocharin, K. New diphenyl ethers from the insect pathogenic fungus Cordyceps sp. BCC 1861. Chem. Pharmaceut. Bull. 2007, 55, 304–307. [Google Scholar] [CrossRef]
- Sekurova, O.N.; Schneider, O.; Zotchev, S.B. Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering. Microb. Biotechnol. 2019, 12, 828–844. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.-S.; Ngo, D.-H.; Van Ta, Q.; Kim, S.-K. Marine organisms as a therapeutic source against herpes simplex virus infection. Eur. J. Pharm. Sci. 2011, 44, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ni, S.; Wu, L.; Wang, Y.; Wang, Y.; Tao, P.; He, W.; Wang, X. Novel 4, 5-Dihydro-thiazinogeldanamycin in a gdmP Mutant Strain of Streptomyces hygroscopicus 17997. Biosci. Biotechnol. Biochem. 2011, 75, 2042–2045. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Férir, G.; Petrova, M.I.; Andrei, G.; Huskens, D.; Hoorelbeke, B.; Snoeck, R.; Vanderleyden, J.; Balzarini, J.; Bartoschek, S.; Brönstrup, M. The lantibiotic peptide labyrinthopeptin A1 demonstrates broad anti-HIV and anti-HSV activity with potential for microbicidal applications. PLoS ONE 2013, 8, e64010. [Google Scholar] [CrossRef] [PubMed]
- Lopes, V.R.; Ramos, V.; Martins, A.; Sousa, M.; Welker, M.; Antunes, A.; Vasconcelos, V.M. Phylogenetic, chemical and morphological diversity of cyanobacteria from Portuguese temperate estuaries. Mar. Environ. Res. 2012, 73, 7–16. [Google Scholar] [CrossRef]
- Bunyapaiboonsri, T.; Yoiprommarat, S.; Khonsanit, A.; Komwijit, S. Phenolic glycosides from the filamentous fungus Acremonium sp. BCC 14080. J. Nat. Prod. 2008, 71, 891–894. [Google Scholar] [CrossRef]
- Pittayakhajonwut, P.; Suvannakad, R.; Thienhirun, S.; Prabpai, S.; Kongsaeree, P.; Tanticharoen, M. An anti-herpes simplex virus-type 1 agent from Xylaria mellisii (BCC 1005). Tetrahedron Lett. 2005, 46, 1341–1344. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.; Lin, Y.; Wang, Y. Exocellular polysaccharides extracted from mangrove fungus Paecilomyces Lilacinuson present anti-HSV-1 activity in mice. J. Virol. Methods 2021, 297, 114246. [Google Scholar] [CrossRef]
- De Vries, E.; Du, W.; Guo, H.; de Haan, C.A. Influenza A virus hemagglutinin–neuraminidase–receptor balance: Preserving virus motility. Trends Microbiol. 2020, 28, 57–67. [Google Scholar] [CrossRef]
- Kumara, T.S.R.; Chen, G.-W. A Numbering Scheme for Influenza A Virus Neuraminidase (NA) Subtypes. TANET2019 2019, 1188–1193. [Google Scholar] [CrossRef]
- Bellino, S.; Bella, A.; Puzelli, S.; Di Martino, A.; Facchini, M.; Punzo, O.; Pezzotti, P.; Castrucci, M.R.; The InfluNet Study Group. Moderate influenza vaccine effectiveness against A (H1N1) pdm09 virus, and low effectiveness against A (H3N2) subtype, 2018/19 season in Italy. Expert Rev. Vaccines 2019, 18, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, F.; Liu, Y.; Liu, Y.; Li, K.; Yang, X.; Liu, S.; Zhou, X.; Yang, J. Spirostaphylotrichin X from a marine-derived fungus as an anti-influenza agent targeting RNA polymerase PB2. J. Nat. Prod. 2018, 81, 2722–2730. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Sun, X.; Yu, G.; Wang, W.; Zhu, T.; Gu, Q.; Li, D. Cladosins A–E, hybrid polyketides from a deep-sea-derived fungus, Cladosporium sphaerospermum. J. Nat. Prod. 2014, 77, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, B.; Qin, Y.; Karthik, L.; Zhu, G.; Hou, C.; Jiang, L.; Liu, M.; Ye, X.; Liu, M. A new abyssomicin polyketide with anti-influenza A virus activity from a marine-derived Verrucosispora sp. MS100137. Appl. Microbiol. Biotechnol. 2020, 104, 1533–1543. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Wang, W.; Fu, P.; Liu, P.; Zhu, W. Anti-influenza virus polyketides from the acid-tolerant fungus Penicillium purpurogenum JS03-21. J. Nat. Prod. 2011, 74, 2014–2018. [Google Scholar] [CrossRef]
- Luo, X.; Yang, J.; Chen, F.; Lin, X.; Chen, C.; Zhou, X.; Liu, S.; Liu, Y. Structurally diverse polyketides from the mangrove-derived fungus Diaporthe sp. SCSIO 41011 with their anti-influenza A virus activities. Front. Chem. 2018, 6, 282. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, Y.; Liu, P.; Fu, P.; Zhu, T.; Wang, W.; Zhu, W. Indole-diterpenoids with anti-H1N1 activity from the aciduric fungus Penicillium camemberti OUCMDZ-1492. J. Nat. Prod. 2013, 76, 1328–1336. [Google Scholar] [CrossRef]
- Peng, J.; Lin, T.; Wang, W.; Xin, Z.; Zhu, T.; Gu, Q.; Li, D. Antiviral alkaloids produced by the mangrove-derived fungus Cladosporium sp. PJX-41. J. Nat. Prod. 2013, 76, 1133–1140. [Google Scholar] [CrossRef]
- Rodrigo-Muñoz, J.; Sastre, B.; Cañas, J.; Gil-Martínez, M.; Redondo, N.; Del Pozo, V.; de Enfermedades Respiratorias, C. Eosinophil response against classical and emerging respiratory viruses: COVID-19. J. Investig. Allergol. Clin. Immunol. 2020, 31, 94–107. [Google Scholar] [CrossRef]
- Falsey, A.R.; Cunningham, C.K.; Barker, W.H.; Kouides, R.W.; Yuen, J.B.; Menegus, M.; Weiner, L.B.; Bonville, C.A.; Betts, R.F. Respiratory syncytial virus and influenza A infections in the hospitalized elderly. J. Infect. Dis. 1995, 172, 389–394. [Google Scholar] [CrossRef]
- Leung, A.K.; Kellner, J.D.; Davies, H.D. Respiratory syncytial virus bronchiolitis. J. Nat. Med. Assoc. 2005, 97, 1708. [Google Scholar]
- Feltes, T.F.; Cabalka, A.K.; Meissner, H.C.; Piazza, F.M.; Carlin, D.A.; Top, F.H., Jr.; Connor, E.M.; Sondheimer, H.M.; Cardiac Synagis Study Group. Palivizumab prophylaxis reduces hospitalization due to respiratory syncytial virus in young children with hemodynamically significant congenital heart disease. J. Pediatr. 2003, 143, 532–540. [Google Scholar] [CrossRef]
- Hall, C.B.; McBride, J.T.; Walsh, E.E.; Bell, D.M.; Gala, C.L.; Hildreth, S.; Ten Eyck, L.G.; Hall, W.J. Aerosolized ribavirin treatment of infants with respiratory syncytial viral infection: A randomized double-blind study. NEJM 1983, 308, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Shao, C.-L.; Meng, H.; She, Z.-G.; Wang, C.-Y. Anti-respiratory syncytial virus prenylated dihydroquinolone derivatives from the gorgonian-derived fungus Aspergillus sp. XS-20090B15. J. Nat. Prod. 2014, 77, 2720–2724. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.J.; Burleson, S.L.; Crosby, J.C.; Heimann, M.A.; Pigott, D.C. Coronavirus disease 2019: International public health considerations. J. Am. Coll. Emerg. Phys. Open 2020, 1, 70–77. [Google Scholar] [CrossRef]
- Rodriguez-Morales, A.J.; Cardona-Ospina, J.A.; Gutiérrez-Ocampo, E.; Villamizar-Peña, R.; Holguin-Rivera, Y.; Escalera-Antezana, J.P.; Alvarado-Arnez, L.E.; Bonilla-Aldana, D.K.; Franco-Paredes, C.; Henao-Martinez, A.F. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med. Infect. Dis. 2020, 34, 101623. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Chen, Y.; Qin, Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020, 92, 568–576. [Google Scholar] [CrossRef]
- Mutiawati, E.; Fahriani, M.; Mamada, S.S.; Fajar, J.K.; Frediansyah, A.; Maliga, H.A.; Ilmawan, M.; Emran, T.B.; Ophinni, Y.; Ichsan, I. Anosmia and dysgeusia in SARS-CoV-2 infection: Incidence and effects on COVID-19 severity and mortality, and the possible pathobiology mechanisms-a systematic review and meta-analysis. F1000Research 2021, 10, 40. [Google Scholar] [CrossRef]
- Syahrul, S.; Maliga, H.A.; Ilmawan, M.; Fahriani, M.; Mamada, S.S.; Fajar, J.K.; Frediansyah, A.; Syahrul, F.N.; Imran, I.; Haris, S. Hemorrhagic and ischemic stroke in patients with coronavirus disease 2019: Incidence, risk factors, and pathogenesis-a systematic review and meta-analysis. F1000Research 2021, 10, 34. [Google Scholar] [CrossRef]
- Ledford, H.; Cyranoski, D.; Van Noorden, R. The UK has approved a COVID vaccine—Here’s what scientists now want to know. Nature 2020, 588, 205–206. [Google Scholar] [CrossRef]
- Nainu, F.; Abidin, R.S.; Bahar, M.A.; Frediansyah, A.; Emran, T.B.; Rabaan, A.A.; Dhama, K.; Harapan, H. SARS-CoV-2 reinfection and implications for vaccine development. Vaccines Immunother. 2020, 16, 3061–3073. [Google Scholar] [CrossRef] [PubMed]
- Fahriani, M.; Ilmawan, M.; Fajar, J.K.; Maliga, H.A.; Frediansyah, A.; Masyeni, S.; Yusuf, H.; Nainu, F.; Rosiello, F.; Sirinam, S. Persistence of long COVID symptoms in COVID-19 survivors worldwide and its potential pathogenesis-a systematic review and meta-analysis. Narra J. 2021, 1, e36. [Google Scholar] [CrossRef]
- Frediansyah, A.; Nainu, F.; Dhama, K.; Mudatsir, M.; Harapan, H. Remdesivir and its antiviral activity against COVID-19: A systematic review. Clin. Epidemiol. Glob. Health 2020, 9, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Masyeni, S.; Iqhrammullah, M.; Frediansyah, A.; Nainu, F.; Tallei, T.; Emran, T.B.; Ophinni, Y.; Dhama, K.; Harapan, H. Molnupiravir: A lethal mutagenic drug against rapidly mutating severe acute respiratory syndrome coronavirus 2—A narrative review. J. Med. Virol. 2022, 94, 3006–3016. [Google Scholar] [CrossRef]
- Mudatsir, M.; Yufika, A.; Nainu, F.; Frediansyah, A.; Megawati, D.; Pranata, A.; Mahdani, W.; Ichsan, I.; Dhama, K.; Harapan, H. Antiviral Activity of Ivermectin Against SARS-CoV-2: An Old-Fashioned Dog with a New Trick—A Literature Review. Sci. Pharm. 2020, 88, 36. [Google Scholar] [CrossRef]
- FDA. Coronavirus (COVID-19) Drugs. 2022. Available online: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs (accessed on 4 April 2022).
- WHO. WHO recommends Two New Drugs to Treat COVID-19. 2022. Available online: https://www.who.int/news/item/14-01-2022-who-recommends-two-new-drugs-to-treat-covid-19 (accessed on 13 April 2022).
- ElNaggar, M.H.; Abdelwahab, G.M.; Kutkat, O.; GabAllah, M.; Ali, M.A.; El-Metwally, M.E.; Sayed, A.M.; Abdelmohsen, U.R.; Khalil, A.T. Aurasperone A Inhibits SARS CoV-2 In Vitro: An Integrated In Vitro and In Silico Study. Mar. Drugs 2022, 20, 179. [Google Scholar] [CrossRef]
- Alhadrami, H.A.; Burgio, G.; Thissera, B.; Orfali, R.; Jiffri, S.E.; Yaseen, M.; Sayed, A.M.; Rateb, M.E. Neoechinulin A as a promising SARS-CoV-2 Mpro inhibitor: In vitro and in silico study showing the ability of simulations in discerning active from inactive enzyme inhibitors. Mar. Drugs 2022, 20, 163. [Google Scholar] [CrossRef]
- Liang, X.-X.; Zhang, X.-J.; Zhao, Y.-X.; Feng, J.; Zeng, J.-C.; Shi, Q.-Q.; Kaunda, J.S.; Li, X.-L.; Wang, W.-G.; Xiao, W.-L. Aspulvins A–H, Aspulvinone Analogues with SARS-CoV-2 Mpro Inhibitory and Anti-inflammatory Activities from an Endophytic Cladosporium sp. J. Nat. Prod. 2022, 85, 878–887. [Google Scholar] [CrossRef]
- Fayed, M.A.; El-Behairy, M.F.; Abdallah, I.A.; Abdel-Bar, H.M.; Elimam, H.; Mostafa, A.; Moatasim, Y.; Abouzid, K.A.; Elshaier, Y.A. Structure-and ligand-based in silico studies towards the repurposing of marine bioactive compounds to target SARS-CoV-2. Arab. J. Chem. 2021, 14, 103092. [Google Scholar] [CrossRef]
- Ebrahimi, K.S.; Ansari, M.; Moghaddam, M.S.H.; Ebrahimi, Z.; Shahlaei, M.; Moradi, S. In silico investigation on the inhibitory effect of fungal secondary metabolites on RNA dependent RNA polymerase of SARS-CoV-II: A docking and molecular dynamic simulation study. Comp. Biol. Med. 2021, 135, 104613. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Mostafa, A.; Mostafa, Y.A.; Abou-Ghadir, O.F.; Abdelazeem, A.H.; Gouda, A.M.; Kutkat, O.; Shama, N.M.A.; Shehata, M.; Gomaa, H.A. Discovery of novel oxazole-based macrocycles as anti-coronaviral agents targeting SARS-CoV-2 main protease. Bioorg. Chem. 2021, 116, 105363. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P.; Prangishvili, D. The origin of viruses. Res. Microbiol. 2009, 160, 466–472. [Google Scholar] [CrossRef]
- Helenius, A. Virus entry: Looking back and moving forward. J. Mol. Biol. 2018, 430, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Drakesmith, H.; Prentice, A. Viral infection and iron metabolism. Nat. Rev. Microbiol. 2008, 6, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.W.; Chang, K.-M. Hepatitis C virus: Virology and life cycle. Clin. Mol. Hepatol. 2013, 19, 17. [Google Scholar] [CrossRef]
- Weiss, R.A. Thirty years on: HIV receptor gymnastics and the prevention of infection. BMC Biol. 2013, 11, 1–5. [Google Scholar] [CrossRef]
- Wilen, C.B.; Tilton, J.C.; Doms, R.W. Molecular mechanisms of HIV entry. In Viral Molecular Machines; Springer: Cham, Switzerland, 2012; pp. 223–242. [Google Scholar]
- Altan-Bonnet, N. Extracellular vesicles are the Trojan horses of viral infection. Curr. Opin. Microbiol. 2016, 32, 77–81. [Google Scholar] [CrossRef]
- Vanheule, V.; Vervaeke, P.; Mortier, A.; Noppen, S.; Gouwy, M.; Snoeck, R.; Andrei, G.; Van Damme, J.; Liekens, S.; Proost, P. Basic chemokine-derived glycosaminoglycan binding peptides exert antiviral properties against dengue virus serotype 2, herpes simplex virus-1 and respiratory syncytial virus. Biochem. Pharmacol. 2016, 100, 73–85. [Google Scholar] [CrossRef]
- Meredith, L.W.; Wilson, G.K.; Fletcher, N.F.; McKeating, J.A. Hepatitis C virus entry: Beyond receptors. Rev. Med. Virol. 2012, 22, 182–193. [Google Scholar] [CrossRef]
- Gao, H.; Shi, W.; Freund, L.B. Mechanics of receptor-mediated endocytosis. Proc. Nat. Acad. Sci. USA 2005, 102, 9469–9474. [Google Scholar] [CrossRef]
- Neil, S.J.D.; Eastman, S.W.; Jouvenet, N.; Bieniasz, P.D. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006, 2, e39. [Google Scholar] [CrossRef] [PubMed]
- Pelkmans, L.; Helenius, A. Insider information: What viruses tell us about endocytosis. Curr. Opin Cell Biol. 2003, 15, 414–422. [Google Scholar] [CrossRef]
- Permanyer, M.; Ballana, E.; Esté, J.A. Endocytosis of HIV: Anything goes. Trends Microbiol. 2010, 18, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Weissenhorn, W.; Dessen, A.; Calder, L.; Harrison, S.; Skehel, J.; Wiley, D. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 1999, 16, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Seth, P. Mechanism of adenovirus-mediated endosome lysis: Role of the intact adenovirus capsid structure. Biochem. Biophys. Res. Commun. 1994, 205, 1318–1324. [Google Scholar] [CrossRef]
- Helle, F.; Dubuisson, J. Hepatitis C virus entry into host cells. Cell. Mol. Life Sci. 2008, 65, 100–112. [Google Scholar] [CrossRef]
- Sieczkarski, S.B.; Whittaker, G.R. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J. Virol. 2002, 76, 10455–10464. [Google Scholar] [CrossRef]
- Leopold, P.L.; Pfister, K.K. Viral strategies for intracellular trafficking: Motors and microtubules. Traffic 2006, 7, 516–523. [Google Scholar] [CrossRef]
- Den Boon, J.A.; Diaz, A.; Ahlquist, P. Cytoplasmic viral replication complexes. Cell Host Microbe 2010, 8, 77–85. [Google Scholar] [CrossRef]
- Kobiler, O.; Drayman, N.; Butin-Israeli, V.; Oppenheim, A. Virus strategies for passing the nuclear envelope barrier. Nucleus 2012, 3, 526–539. [Google Scholar] [CrossRef]
- Morrison, L.A.; DeLassus, G.S. Breach of the nuclear lamina during assembly of herpes simplex viruses. Nucleus 2011, 2, 137–147. [Google Scholar] [CrossRef]
- Sodeik, B.; Ebersold, M.W.; Helenius, A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997, 136, 1007–1021. [Google Scholar] [CrossRef]
- Siddiqa, A.; Broniarczyk, J.; Banks, L. Papillomaviruses and endocytic trafficking. Int. J. Mol. Sci. 2018, 19, 2619. [Google Scholar] [CrossRef] [PubMed]
- Meunier, B. Hybrid Molecules with a Dual Mode of Action: Dream or Reality? Acc. Chem. Res.. 2008, 41, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.N.Y.; Nislow, C.; Emili, A. Recent advances and method development for drug target identification. Trends Pharmacol. Sci. 2010, 31, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Wittine, K.; Saftić, L.; Peršurić, Ž.; Kraljević Pavelić, S. Novel antiretroviral structures from marine organisms. Molecules 2019, 24, 3486. [Google Scholar] [CrossRef]
- Grande, F.; Occhiuzzi, M.A.; Rizzuti, B.; Ioele, G.; De Luca, M.; Tucci, P.; Svicher, V.; Aquaro, S.; Garofalo, A. CCR5/CXCR4 dual antagonism for the improvement of HIV infection therapy. Molecules 2019, 24, 550. [Google Scholar] [CrossRef]
- Chen, X.; Si, L.; Liu, D.; Proksch, P.; Zhang, L.; Zhou, D.; Lin, W. Neoechinulin B and its analogues as potential entry inhibitors of influenza viruses, targeting viral hemagglutinin. Eur. J. Med. Chem. 2015, 93, 182–195. [Google Scholar] [CrossRef]
- Copeland, A.M.; Newcomb, W.W.; Brown, J.C. Herpes simplex virus replication: Roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 2009, 83, 1660–1668. [Google Scholar] [CrossRef]
- Penin, F.; Dubuisson, J.; Rey, F.A.; Moradpour, D.; Pawlotsky, J.M. Structural biology of hepatitis C virus. Hepatology 2004, 39, 5–19. [Google Scholar] [CrossRef]
- Rosenberg, S. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 2001, 313, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Suzuki, T.; Ishii, K.; Matsuura, Y.; Miyamura, T. Processing and functions of Hepatitis C virus proteins. Intervirology 1999, 42, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Tong, K.; Yao, R.; Zhou, Y.; Lin, H.; Du, L.; Jin, Y.; Cao, L.; Tan, J.; Zhang, X.-D. Genome-wide analysis of protein–protein interactions and involvement of viral proteins in SARS-CoV-2 replication. Cell Biosci. 2021, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bartenschlager, R.; Lohmann, V.; Penin, F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 2013, 11, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Moradpour, D.; Penin, F.; Rice, C.M. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007, 5, 453–463. [Google Scholar] [CrossRef]
- Li, X.-D.; Sun, L.; Seth, R.B.; Pineda, G.; Chen, Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Nat. Acad. Sci. USA 2005, 102, 17717–17722. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Klupp, B.G.; Granzow, H. Herpesvirus assembly: A tale of two membranes. Curr. Opin. Microbiol. 2006, 9, 423–429. [Google Scholar] [CrossRef]
- Sugimoto, K.; Uema, M.; Sagara, H.; Tanaka, M.; Sata, T.; Hashimoto, Y.; Kawaguchi, Y. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 2008, 82, 5198–5211. [Google Scholar] [CrossRef]
- Johnson, J.E. Virus particle maturation: Insights into elegantly programmed nanomachines. Curr. Opin. Struct. Biol. 2010, 20, 210–216. [Google Scholar] [CrossRef]
- Konvalinka, J.; Kräusslich, H.-G.; Müller, B. Retroviral proteases and their roles in virion maturation. Virology 2015, 479, 403–417. [Google Scholar] [CrossRef]
- Van der Grein, S.G.; Defourny, K.A.; Slot, E.F. Intricate relationships between naked viruses and extracellular vesicles in the crosstalk between pathogen and host. Semin. Immunopathol. 2018, 40, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Perlmutter, J.D.; Hagan, M.F. Mechanisms of virus assembly. Ann. Rev. Phys. Chem. 2015, 66, 217. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Nishiyama, T.; Primadharsini, P.P.; Okamoto, H. Characterization of the quasi-enveloped hepatitis E virus particles released by the cellular exosomal pathway. J. Virol. 2017, 91, e00822-17. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Bae, H. The diversity of fungal genome. Biol. Proc. Online 2015, 17, 8. [Google Scholar] [CrossRef]
- Mahoney, M.; Damalanka, V.C.; Tartell, M.A.; Chung, D.H.; Lourenço, A.L.; Pwee, D.; Mayer Bridwell, A.E.; Hoffmann, M.; Voss, J.; Karmakar, P. A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proc. Nat. Acad. Sci. USA 2021, 118, e2108728118. [Google Scholar] [CrossRef]

| Compound Name [Ref.] | Compound Type | Microbial Strain | Strain Origin/Host | Viral Target | IC50/EC50/ED50 | Target Inhibition |
|---|---|---|---|---|---|---|
| alachalasin A [25] | alkaloid | Podospora vesticola XJ03-56-1 | glacier | HIV-1 | EC50 = 8.01 μM | ND |
| pestalofone A [28] | terpenoid | Pestalotiopsis fici W106-1 | plant endophyte | HIV-1 | EC50 = 90.4 μM | ND |
| pestalofone B [28] | terpenoid | P. fici W106-1 | plant endophyte | HIV-1 | EC50 = 64.0 μM | ND |
| pestalofone E [28] | terpenoid | P. fici W106-2 | plant endophyte | HIV-1 | EC50 = 93.7 μM | ND |
| pestaloficiol G [28] | terpenoid | P. fici W106-3 | plant endophyte | HIV-1 | EC50 = 89.2 μM | ND |
| pestaloficiol H [28] | terpenoid | P. fici W106-4 | plant endophyte | HIV-1 | EC50 = 89.2 μM | ND |
| pestaloficiol J [28] | terpenoid | P. fici W106-5 | plant endophyte | HIV-1 | EC50 = 8 μM | ND |
| pestaloficiol K [28] | terpenoid | P. fici W106-6 | plant endophyte | HIV-1 | EC50 = 78.2 μM | ND |
| epicoccin G [95] | alkaloid | Epicoccum nigrum XZC04-CS-302 | Cordyceps sinensis fungus | HIV-1 | EC50 = 13.5 μM | ND |
| epicoccin H [95] | alkaloid | E. nigrum XZC04-CS-302 | C. sinensis | HIV-1 | EC50 = 42.2 μM | ND |
| diphenylalazine A [95] | peptide | E. nigrum XZC04-CS-302 | C. sinensis | HIV-1 | EC50 = 27.9 μM | ND |
| bacillamide B [30] | peptide | Tricladium sp. No. 2520 | soil in which C. sinensis grow | HIV-1 | EC50 = 24.8 μM | ND |
| armochaetoglobin K [31] | alkaloid | Chaetomium globosum TW 1-1 | Armadillidium vulgare insect | HIV-1 | EC50 = 1.23 μM | ND |
| armochaetoglobin L [31] | alkaloid | C. globosum TW 1-1 | A. vulgare insect | HIV-1 | EC50 = 0.48 μM | ND |
| armochaetoglobin M [31] | alkaloid | C. globosum TW 1-1 | A. vulgare insect | HIV-1 | EC50 = 0.55μM | ND |
| armochaetoglobin N [31] | alkaloid | C. globosum TW 1-1 | A. vulgare insect | HIV-1 | EC50 = 0.25 μM | ND |
| armochaetoglobin O [31] | alkaloid | C. globosum TW 1-1 | A. vulgare insect | HIV-1 | EC50 = 0.61 μM | ND |
| armochaetoglobin P [31] | alkaloid | C. globosum TW 1-1 | A. vulgare insect | HIV-1 | EC50 = 0.68 μM | ND |
| armochaetoglobin Q [31] | alkaloid | C. globosum TW 1-1 | A. vulgare insect | HIV-1 | EC50 = 0.31 μM | ND |
| armochaetoglobin R [31] | alkaloid | C. globosum TW 1-1 | A. vulgare insect | HIV-1 | EC50 = 0.34 μM | ND |
| stachybotrin D [32] | terpenoid | Stachybotrys chartarum MXH-X73 | Xestospongia testudinaris sponge | HIV-1 | EC50 = 8.4 μM | replication |
| stachybotrysam A [33] | alkaloid | S. chartarum CGMCC 3.5365. | ND | HIV-1 | EC50 = 9.3 μM | ND |
| stachybotrysam B [33] | alkaloid | S. chartarum CGMCC 3.5365. | ND | HIV-1 | EC50 = 1.0 μM | ND |
| stachybotrysam C [33] | alkaloid | S. chartarum CGMCC 3.5365. | ND | HIV-1 | EC50 = 9.6 μM | ND |
| chartarutine B [34] | alkaloid | S. chartarum WGC-25C-6 | Niphates sp. sponge | HIV-1 | IC50 = 4.90 μM | ND |
| chartarutine G [34] | alkaloid | S. chartarum WGC-25C-6 | Niphates sp. sponge | HIV-1 | IC50 = 5.57 μM | ND |
| chartarutine H [34] | alkaloid | S. chartarum WGC-25C-6 | Niphates sp. sponge | HIV-1 | IC50 = 5.58 μM | ND |
| malformin C [35] | peptide | Aspergillus niger SCSIO Jcsw6F30 | marine | HIV-1 | IC50 = 1.4 μM | entry |
| aspernigrin C [181] | alkaloid | A. niger SCSIO Jcsw6F30 | marine | HIV-1 | IC50 = 4.7 μM | entry |
| eutypellazine E [36] | alkaloid | Eutypella sp. MCCC 3A00281 | deep sea sediment | HIV-1 | IC50 = 3.2 μM | ND |
| truncateol O [37] | terpenoid | Truncatella angustata XSB-01-43 | Amphimedon sp. sponge | HIV-1 and H1N1 | IC50 = 39.0 μM (HIV) and 30.4 μM (H1N1) | ND |
| truncateol P [37] | terpenoid | T. angustata XSB-01-43 | Amphimedon sp. sponge | HIV-1 | IC50 = 16.1 μM | ND |
| penicillixanthone A [38] | polyketide | Aspergillus fumigatus | jellyfish | HIV-1 | IC50 = 0.26 μM | entry |
| DTM [39] | polyketide | C. globosum | deep sea sediment | HIV-1 | 75.1% at 20 μg/mL | ND |
| epicoccone B [39] | polyketide | C. globosum | deep sea sediment | HIV-1 | 88.4% at 20 μg/mL | ND |
| xylariol [39] | polyketide | C. globosum | deep sea sediment | HIV-1 | 70.2% at 20 μg/mL | ND |
| phomonaphthalenone A [40] | polyketide | Phomopsis sp. HCCB04730 | Stephania japonica-plant endophyte | HIV-1 | IC50: 11.6 μg/mL | ND |
| bostrycoidin [40] | polyketide | Phomopsis sp. HCCB04730 | S. japonica plant endophyte | HIV-1 | IC50: 9.4 μg/mL | ND |
| altertoxin I [41] | phenalene | Alternaria tenuissima QUE1Se | Quercus emoryi plant endophyte | HIV-1 | IC50: 1.42 μM | ND |
| altertoxin II [41] | phenalene | A. tenuissima QUE1Se | Q. emoryi plant endophyte | HIV-1 | IC50: 0.21 μM | ND |
| altertoxin III [41] | phenalene | A. tenuissima QUE1Se | Q. emoryi plant endophyte | HIV-1 | IC50: 0.29 μM | ND |
| alternariol 5-O-methyl ether [42] | phenolic | Colletotrichum sp | plant endophyte | HIV-1 | EC50: 30.9 μM | replication |
| ergokonin A [43] | terpenoid | Trichoderma sp. Xy24 | Xylocarpus granatum plant endophyte | HIV-1 | IC50: 22.3 μM | ND |
| ergokonin B [43] | terpenoid | Trichoderma sp. Xy24 | X. granatum plant endophyte | HIV-1 | IC50: 1.9 μM | ND |
| sorrentanone [43] | terpenoid | Trichoderma sp. Xy24 | X. granatum plant endophyte | HIV-1 | IC50: 4.7 μM | ND |
| cerevisterol [43] | terpenoid | Trichoderma sp. Xy24 | X. granatum plant endophyte | HIV-1 | IC50: 9.3 μM | ND |
| phomopsone B [44] | alkaloid | Phomopsis sp. CGMCC 5416 | Achyranthes bidentata plant endophyte | HIV-1 | IC50: 7.6 μmol/L | ND |
| phomopsone C [44] | alkaloid | Phomopsis sp. CGMCC 5416 | A. bidentata plant endophyte | HIV-1 | IC50: 0.5 μmol/L | ND |
| pericochlorosin B [45] | polyketide | Periconia sp. F-31 | plant endophyte | HIV-1 | IC50: 2.2 μM | ND |
| asperphenalenone A [46] | alkaloid | Aspergillus sp. | Kadsura longipedunculata plant endophyte | HIV-1 | IC50: 4.5 μM | ND |
| asperphenalenone D [46] | alkaloid | Aspergillus sp. | K. longipedunculata plant endophyte | HIV-1 | IC50: 2.4 μM | ND |
| cytochalasin Z8 [46] | alkaloid | Aspergillus sp. | K. longipedunculata plant endophyte | HIV-1 | IC50: 9.2 μM | ND |
| epicocconigrone A [46] | alkaloid | Aspergillus sp. | K. longipedunculata plant endophyte | HIV-1 | IC50: 6.6 μM | ND |
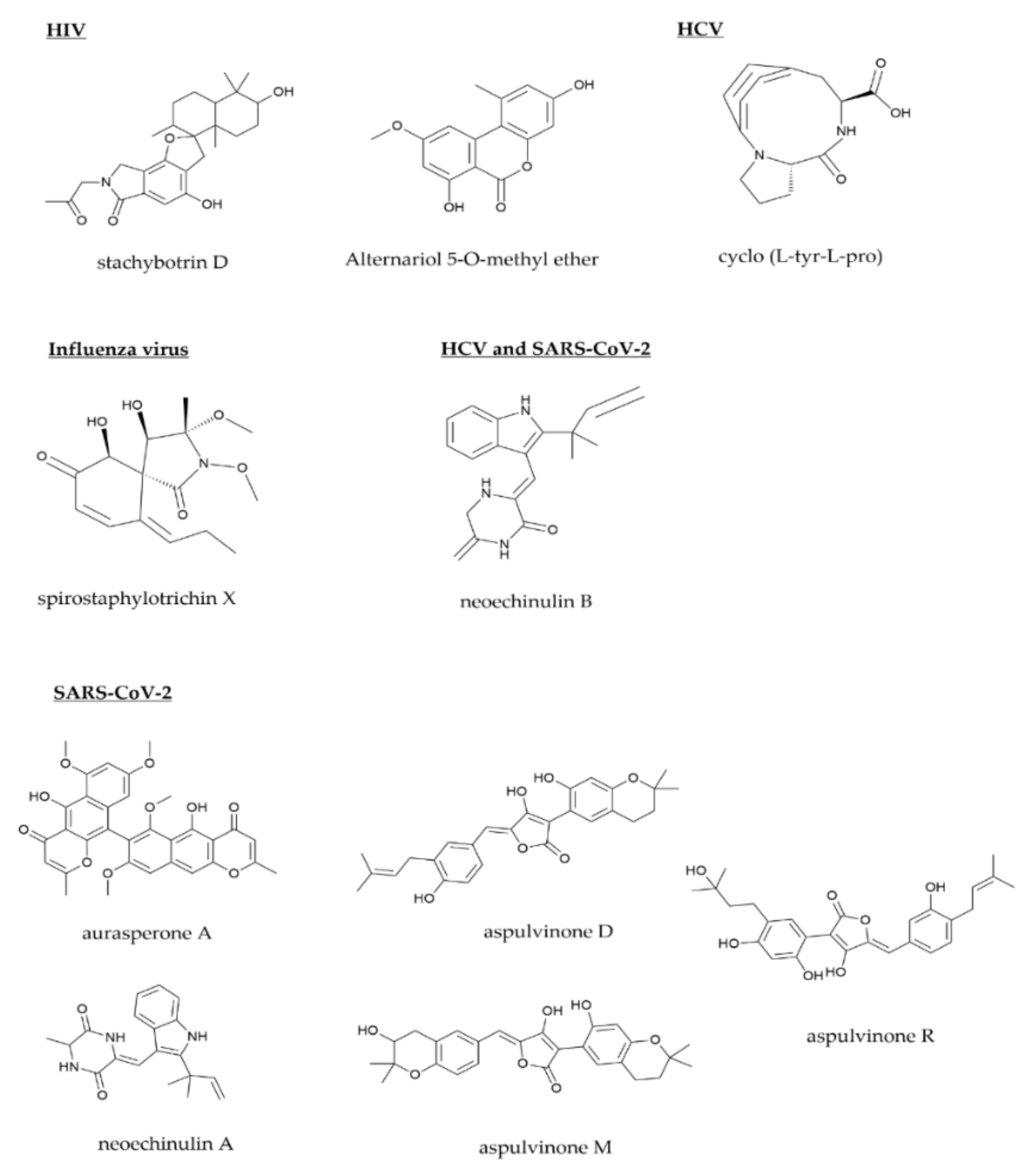
| neoechinulin B/NeoB [57,153,185] | alkaloid | Aspergillus amstelodami | ND | HCV and SARS-CoV-2 | IC50: 5.5 μM (HCV) and 32.9 μM (SARS-CoV-2) | replication |
| Eurotium rubrum F33 | marine sediment | H1N1 | IC50; 7 μM | entry | ||
| raistrickindole A [62] | alkaloid | Penicillium raistrickii IMB17-034 | mangrove sediment | HCV | EC50: 5.7 μM | ND |
| raistrickin [62] | alkaloid | P. raistrickii IMB17-035 | mangrove sediment | HCV | EC50: 7.0 μM | ND |
| sclerotigenin [62] | alkaloid | P. raistrickii IMB17-036 | mangrove sediment | HCV | EC50: 5.8 μM | ND |
| harzianoic acid A [43] | terpenoid | Trichoderma harzianum LZDX-32-08 | Xestospongia testudinaria sponge | HCV | IC50: 5.5 μM | entry |
| harzianoic acid B [43] | terpenoid | T. harzianum LZDX-32-08 | X. testudinaria sponge | HCV | IC50: 42.9 μM | entry |
| peniciherquamide C [64] | peptide | Penicillium herquei P14190 | seaweed | HCV | IC50: 5.1 μM | ND |
| cyclo (L-Tyr-L-Pro) [65] | peptide | Aspergillus versicolor | Spongia officinalis sponge | HCV | IC50: 8.2 μg/mL | replication |
| 7-dehydroxyl-zinniol [76] | alkaloid | Alternia solani | Aconitum transsectum plant endophyte | HBV | IC50: 0.38 mM | ND |
| THA [77] | polyketide | Penicillium sp. OUCMDZ-4736 | mangrove sediment | HBV | IC50: 4.63 μM | ND |
| MDMX [77] | polyketide | Penicillium sp. OUCMDZ-4736 | mangrove sediment | HBV | IC50: 11.35 μM | ND |
| vanitaracin A [78] | polyketide | Talaromyces sp. | sand | HBV | IC50: 10.58 μM | entry |
| destruxin A [83] | peptide | Metarhizium anisopliae var. dcjhyium | Odontoternes formosanus termite | HBV | IC50: 1.2 μg/mL (mix A+B+E) | ND |
| destruxin B [83] | peptide | M. anisopliae var. dcjhyium; | O. formosanus termite | HBV | IC50: 1.2 μg/mL (mix A+B+E) | ND |
| destruxin E [83] | peptide | M. anisopliae var. dcjhyium | O. formosanus termite | HBV | IC50: 1.2 μg/mL (mix A+B+E) | ND |
| amphiepicoccin A [95] | alkaloid | Epicoccum nigrum HDN17-88 | Amphilophus sp. fish gill | HSV-2 | IC50: 70 μM | ND |
| amphiepicoccin C [95] | alkaloid | E. nigrum HDN17-88 | Amphilophus sp. fish gill | HSV-2 | IC50: 64 μM | ND |
| amphiepicoccin F [95] | alkaloid | E. nigrum HDN17-88 | Amphilophus sp. fish gill | HSV-2 | IC50: 29 μM | ND |
| aspergillipeptide D [96] | peptide | Aspergillus sp. SCSIO 41501 | gorgonian coral | HSV-1 | IC50: 7.93 μM | entry |
| aspergilol H [98] | polyketide | Aspergillus versicolor SCSIO 41501 | deep sea sediment | HSV-1 | EC50 = 4.68 μM | ND |
| aspergilol I [98] | polyketide | A. versicolor SCSIO 41503 | deep sea sediment | HSV-1 | IC50 = 6.25 μM | ND |
| coccoquinone A [98] | polyketide | A. versicolor SCSIO 41504 | deep sea sediment | HSV-1 | IC50 = 3.12 μM | ND |
| trichobotrysin A [99] | alkaloid | Trichobotrys effuse DFFSCS021 | deep sea sediment | HSV-1 | IC50 = 3.08 μM | ND |
| trichobotrysin B [99] | alkaloid | Trichobotrys effuse DFFSCS021 | deep sea sediment | HSV-1 | IC50 = 9.37 μM | ND |
| trichobotrysin D [99] | alkaloid | Trichobotrys effuse DFFSCS021 | deep sea sediment | HSV-1 | IC50 = 3.12 μM | ND |
| 11a-dehydroxyisoterreulactone A [100] | terpenoid | Aspergillus terreus SCSGAF0162 | gorgonian corals Echinogorgia aurantiaca | HSV-1 | IC50 = 16.4 μg/mL | ND |
| arisugacin A [100] | terpenoid | Aspergillus terreus SCSGAF0162 | gorgonian corals E. aurantiaca | HSV-1 | IC50 = 6.34 μg/mL | ND |
| isobutyrolactone II [100] | terpenoid | Aspergillus terreus SCSGAF0162 | gorgonian corals E. aurantiaca | HSV-1 | IC50 = 21.8 μg/mL | ND |
| aspernolide A [100] | terpenoid | Aspergillus terreus SCSGAF0162 | gorgonian corals E. aurantiaca | HSV-1 | IC50 = 28.9 μg/mL | ND |
| halovir A [101] | peptide | Scytalidium sp. | NI | HSV-1 and HSV-2 | ED50 = 1.1 μM (HSV-1) and 0.28 (HSV-2) | ND |
| halovir B [101] | peptide | Scytalidium sp. | NI | HSV-1 | ED50 = 3.5 μM | ND |
| halovir C [101] | peptide | Scytalidium sp. | NI | HSV-1 | ED50 = 2.2 μM | ND |
| halovir D [101] | peptide | Scytalidium sp. | NI | HSV-1 | ED50 = 2.0 μM | ND |
| halovir E [101] | peptide | Scytalidium sp. | NI | HSV-1 | ED50 = 3.1 μM | ND |
| balticolid [102] | polyketide | Ascomycetous fungus | driftwood | HSV-1 | IC50 = 0.45 μM | ND |
| alternariol [106] | phenolic | Pleospora tarda | Ephedra aphylla endphyte | HSV-1 | IC50 = 13.5 μM | ND |
| alternariol-(9)-methyl ether [106] | phenolic | Pleospora tarda | E. aphylla endophyte | HSV-1 | IC50 = 21.3 μM | ND |
| oblongolide Z [107] | polyketide | Phomopsis sp. BCC 9789 | Musa acuminata endophyte | HSV-1 | IC50: 14 μM | ND |
| DHI [113] | phenolic | Torrubiella tenuis BCC 12732 | Homoptera scale insect | HSV-1 | IC50: 50 μg/mL | ND |
| cordyol C [114] | polyketide | Cordyceps sp. BCC 1861 | Homoptera-cicada nymph | HSV-1 | IC50: 1.3 μg/mL | ND |
| DTD [117] | polyketide | Streptomyces hygroscopicus 17997 | GdmP mutant | HSV-1 | IC50: 0.252 μgmol/L | ND |
| labyrinthopeptin A1/LabyA1 [118] | peptide | Actinomadura namibiensis DSM 6313 | desert soil | HSV-1 and HSV-2 | EC50 = 0.56 μM (HSV-1) and 0.32 μM (HSV-2) | entry |
| HIV-1 and HIV-2 | EC50 = 2.0 μM (HIV-1) and 1.9 μM (HIV-2) | entry | ||||
| monogalactopyranose [120] | polyphenol | Acremonium sp. BCC 14080 | palm leaf | HSV | IC50: 7.2 μM | ND |
| mellisol [121] | polyketide | Xylaria mellisii BCC 1005 | NI | HSV | IC50: 10.5 μg/mL | ND |
| DOG [121] | polyketide | Xylaria mellisii BCC 1005 | NI | HSV | IC50: 8.4 μg/mL | ND |
| spirostaphylotrichin X [126] | polyketide | Cochliobolus lunatus SCSIO41401 | marine algae | H1N1 and H3N2 | IC50: 1.6 μM (H1N1) and 4.1 μM (H3N2) | replication |
| cladosin C [127] | polyketide | Cladosporium sphaerospermum 2005-01-E3 | deep sea sludge | H1N1 | IC50: 276 μM | ND |
| abyssomicin Y [118] | polyketide | Verrucosispora sp. MS100137 | deep sea sediment | H1N1 | inhibition rate: 97.9% | ND |
| purpurquinone B [129] | polyketide | Penicillium purpurogenum JS03-21 | acidic red soil | H1N1 | IC50: 61.3 μM | ND |
| purpurquinone C [129] | polyketide | Penicillium purpurogenum JS03-22 | acidic red soil | H1N1 | IC50: 64 μM | ND |
| purpurester A [129] | polyketide | Penicillium purpurogenum JS03-23 | acidic red soil | H1N1 | IC50: 85.3 μM | ND |
| TAN-931 [129] | polyketide | Penicillium purpurogenum JS03-24 | acidic red soil | H1N1 | IC50: 58.6 μM | ND |
| pestalotiopsone B [130] | polyketide | Diaporthe sp. SCSIO 41011 | Rhizophora stylosa mangrove endophte | H1N1 and H3N2 | IC50: 2.56 μM (H1N1) and 6.76 μM (H3N2) | ND |
| pestalotiopsone F [130] | polyketide | Diaporthe sp. SCSIO 41012 | R. stylosa mangrove endophte | H1N1 and H3N2 | IC50: 21.8 μM (H1N1) and 6.17 μM (H3N2) | ND |
| DMXC [130] | polyketide | Diaporthe sp. SCSIO 41013 | R. stylosa mangrove endophte | H1N1 and H3N2 | IC50: 9.4 μM (H1N1) and 5.12 μM (H3N2) | ND |
| 5-chloroisorotiorin [130] | polyketide | Diaporthe sp. SCSIO 41014 | R. stylosa mangrove endophte | H1N1 and H3N2 | IC50: 2.53 μM (H1N1) and 10.1 μM (H3N2) | ND |
| 3-deoxo-4b-deoxypaxilline [131] | alkaloid | Penicillium camemberti | mangrove sediment | H1N1 | IC50: 28.3 μM | ND |
| DCA [131] | alkaloid | P. camemberti OUCMDZ-1492 | mangrove sediment | H1N1 | IC50: 38.9 μM | ND |
| DPT [131] | alkaloid | P. camemberti OUCMDZ-1492 | mangrove sediment | H1N1 | IC50: 32.2 μM | ND |
| 9,10-diisopentenylpaxilline | alkaloid | P. camemberti OUCMDZ-1492 | mangrove sediment | H1N1 | IC50: 73.3 μM | ND |
| TTD [131] | alkaloid | P. camemberti OUCMDZ-1492 | mangrove sediment | H1N1 | IC50: 34.1 μM | ND |
| emindole SB [131] | alkaloid | P. camemberti OUCMDZ-1492 | mangrove sediment | H1N1 | IC50: 26.2 μM | ND |
| 21-isopentenylpaxilline [131] | alkaloid | P. camemberti OUCMDZ-1492 | mangrove sediment | H1N1 | IC50: 6.6 μM | ND |
| paspaline [131] | alkaloid | P. camemberti OUCMDZ-1492 | mangrove sediment | H1N1 | IC50: 77.9 μM | ND |
| paxilline [131] | alkaloid | P. camemberti OUCMDZ-1492 | mangrove sediment | H1N1 | IC50: 17.7 μM | ND |
| (14S)-oxoglyantrypine [132] | alkaloid | Cladosporium sp. PJX-41 | mangrove sediment | H1N1 | IC50: 85 μM | ND |
| norquinadoline A [132] | alkaloid | Cladosporium sp. PJX-42 | mangrove sediment | H1N1 | IC50: 82 μM | ND |
| deoxynortryptoquivaline [132] | alkaloid | Cladosporium sp. PJX-43 | mangrove sediment | H1N1 | IC50: 85 μM | ND |
| deoxytryptoquivaline [132] | alkaloid | Cladosporium sp. PJX-44 | mangrove sediment | H1N1 | IC50: 85 μM | ND |
| tryptoquivaline [132] | alkaloid | Cladosporium sp. PJX-45 | mangrove sediment | H1N1 | IC50: 89 μM | ND |
| quinadoline B [132] | alkaloid | Cladosporium sp. PJX-46 | mangrove sediment | H1N1 | IC50: 82 μM | ND |
| 22-O-(N-Me-l-valyl)-21-epi-aflaquinolone B [138] | alkaloid | Aspergillus sp strain XS-2009 | Muricella abnormaliz gorgonian | RSV | IC50: 0.042 μM | ND |
| aflaquinolone D [138] | alkaloid | Aspergillus sp strain XS-2009 | M. abnormaliz gorgonian | RSV | IC50: 6.6 μM | ND |
| aurasperone A [152] | polyphenol | Aspergillus niger No.LC582533 | Phallusia nigra tunicate | SARS-CoV-2 | IC50: 12.25 μM | replication |
| neoechinulin A [153] | alkaloid | Aspergillus fumigatus MR2012 | marine sediment | SARS-CoV-2 | IC50: 0.47 μM | replication |
| aspulvinone D [154] | polyphenol | Cladosporium sp. 7951 | Paris polyphylla endophyte | SARS-CoV-2 | IC50: 10.3 μM | replication |
| aspulvinone M [154] | polyphenol | Cladosporium sp. 7951 | P. polyphylla endophyte | SARS-CoV-2 | IC50: 9.4 μM | replication |
| aspulvinone R [154] | polyphenol | Cladosporium sp. 7952 | P. polyphylla endophyte | SARS-CoV-2 | IC50: 7.7 μM | replication |
| HIV | HCV | HBV | Influenza Virus | HSV | RSV | SARS-CoV-2 | |
|---|---|---|---|---|---|---|---|
| Taxonomy | |||||||
| Family | Retroviridae | Flaviridae | Hepadnaviridae | Orthomyxoviridae | Herpesviridae | Paramyxoviridae | Coronaviridae |
| Genus | Lentivirus | Hepacivirus | Orthohepadnavirus | Alphainfluenzavirus | Simplexvirus | Orthopneumovirus | Betacoronavirus |
| Type | positive-strand RNA | positive-strand RNA | partially double-stranded DNA | negative-strand RNA | double-stranded DNA | negative- strand RNA | positive-strand RNA |
| Viral structure | |||||||
| Genome size | 9.2 kb | ±3 kB | 3.2 kb | 0.89–2.3 kb | 125 kb | 15.2 kb, | ±29.9 kB |
| Core shape and diameter | cone-shaped and 145 nm | spherical and 40-80 nm | spherical or filamentous and 42 nm | spherical or pleomorphic and 80–120 nm | spherical and 155-240 nm | filamentous and 130 nm | spherical or ellipsoidal and 108 nm |
| Envelope glycoprotein | SU (gp120) and TM (gp41) | E1/E2 heterodimers, p7 | LHBs, MHBs (preS1 and preS2) and SHBs | HA, NA | gD, gH-gL, gB, and additional gK, gC-gG, gE/gI, gN, gM, UL45 | glycoprotein (G) and the fusion (F) glycoprotein | CoV envelope (E) |
| Non-structural protein | Gag-pol | NS2, NS3, NS4A, NS4B, NS5A, and NS5B | HBeAg and HBx | PA-X, PB1-F2, PB1-N40, PA-N155, PA-N182, M42 and NS3 | - | NS1 and NS2 | NSP 1 to NSP 16 |
| Host | |||||||
| Receptor (coreceptor) | CD4 receptor; CXCR4, and CCR5 coreceptors | CD81, Claudin 1, Occludin; LXRs | NTCP | SA | gD receptor: nectin-1, HVEM, 3-OS HS;NatasagB receptor: PILRα, MAG, NMHC-IIA | CX3CR1, nucleolin, EGFR, IGF1R, ICAM-1 | ACE2 |
| Attachment factor | DC-SIGN, L-SIGN | SR-B1, LDL | HSPG | - | HSPG | HSPG | L-SIGN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frediansyah, A.; Sofyantoro, F.; Alhumaid, S.; Al Mutair, A.; Albayat, H.; Altaweil, H.I.; Al-Afghani, H.M.; AlRamadhan, A.A.; AlGhazal, M.R.; Turkistani, S.A.; et al. Microbial Natural Products with Antiviral Activities, Including Anti-SARS-CoV-2: A Review. Molecules 2022, 27, 4305. https://doi.org/10.3390/molecules27134305
Frediansyah A, Sofyantoro F, Alhumaid S, Al Mutair A, Albayat H, Altaweil HI, Al-Afghani HM, AlRamadhan AA, AlGhazal MR, Turkistani SA, et al. Microbial Natural Products with Antiviral Activities, Including Anti-SARS-CoV-2: A Review. Molecules. 2022; 27(13):4305. https://doi.org/10.3390/molecules27134305
Chicago/Turabian StyleFrediansyah, Andri, Fajar Sofyantoro, Saad Alhumaid, Abbas Al Mutair, Hawra Albayat, Hayyan I. Altaweil, Hani M. Al-Afghani, Abdullah A. AlRamadhan, Mariam R. AlGhazal, Safaa A. Turkistani, and et al. 2022. "Microbial Natural Products with Antiviral Activities, Including Anti-SARS-CoV-2: A Review" Molecules 27, no. 13: 4305. https://doi.org/10.3390/molecules27134305
APA StyleFrediansyah, A., Sofyantoro, F., Alhumaid, S., Al Mutair, A., Albayat, H., Altaweil, H. I., Al-Afghani, H. M., AlRamadhan, A. A., AlGhazal, M. R., Turkistani, S. A., Abuzaid, A. A., & Rabaan, A. A. (2022). Microbial Natural Products with Antiviral Activities, Including Anti-SARS-CoV-2: A Review. Molecules, 27(13), 4305. https://doi.org/10.3390/molecules27134305








