BSA Interaction, Molecular Docking, and Antibacterial Activity of Zinc(II) Complexes Containing the Sterically Demanding Biomimetic N3S2 Ligand: The Effect of Structure Flexibility
Abstract
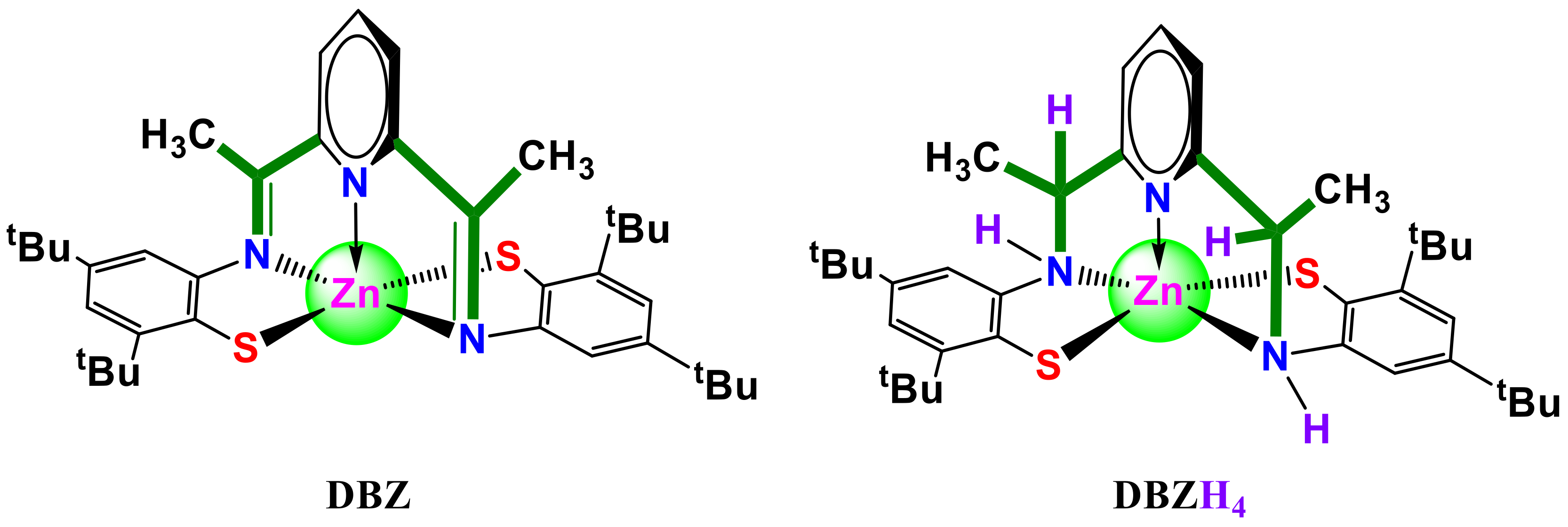
1. Introduction
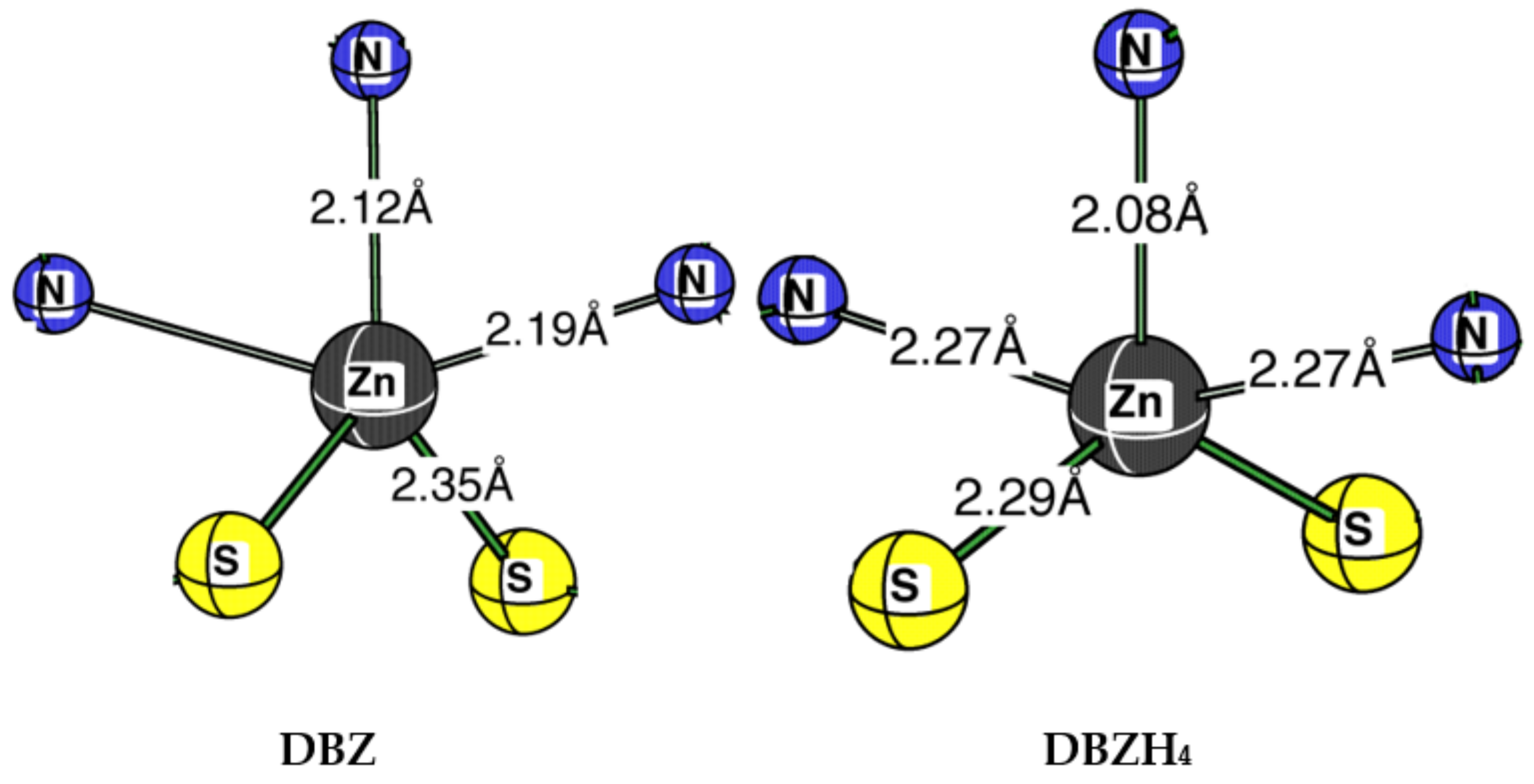
2. Results and Discussion
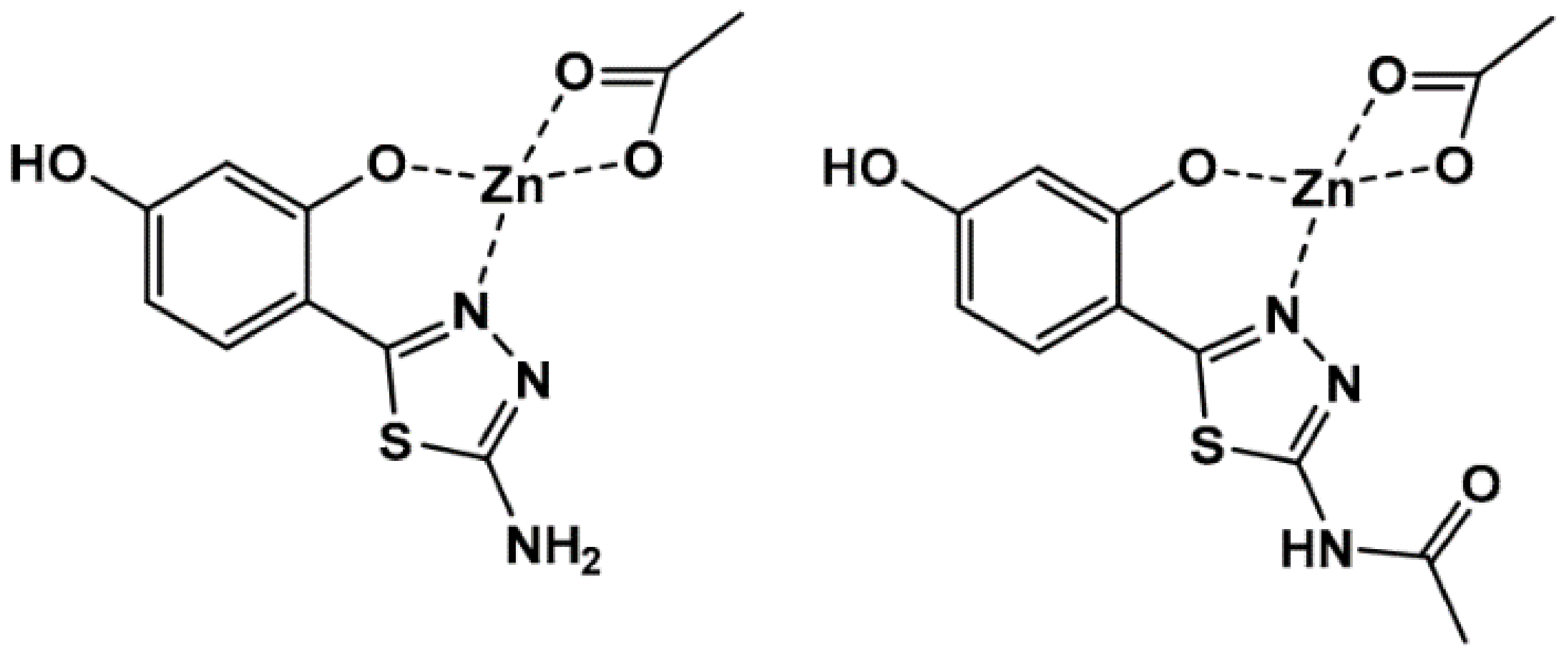
2.1. Interaction of BSA with DBZ and DBZH4
2.1.1. Emission Studies
2.1.2. Absorption Studies
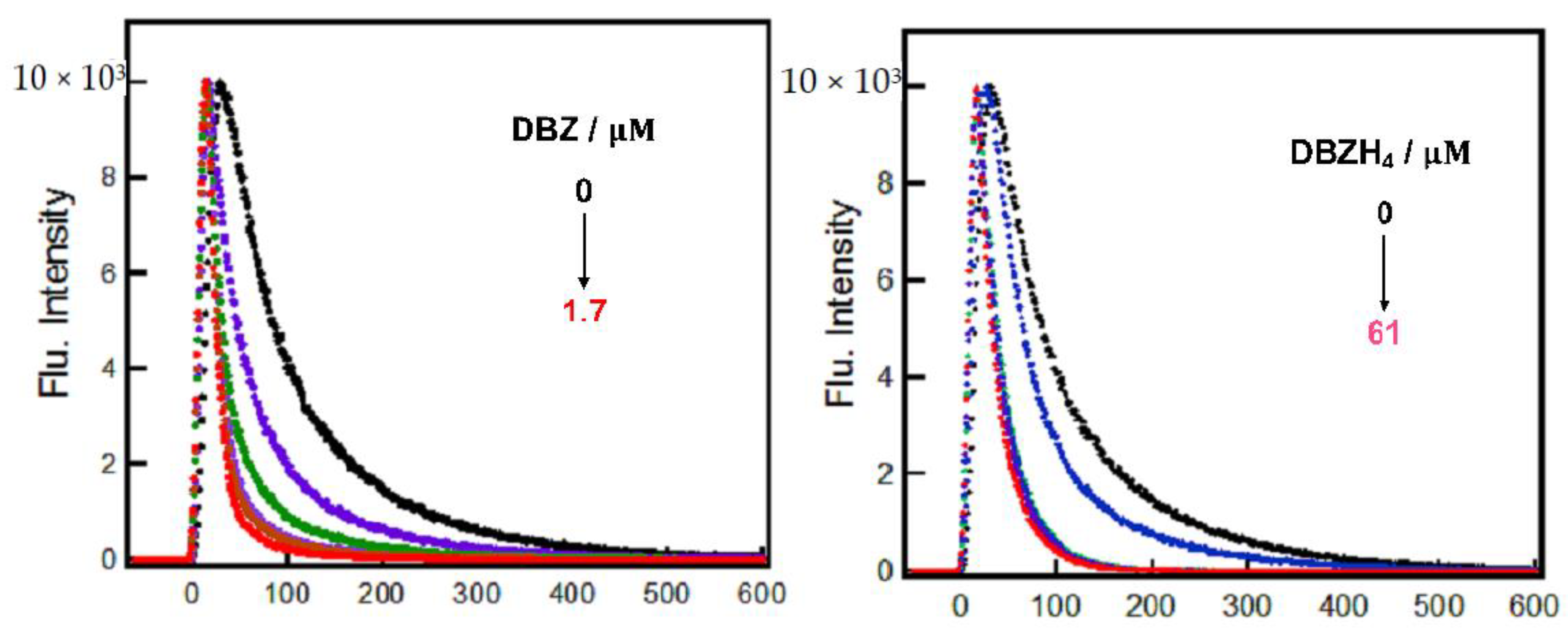
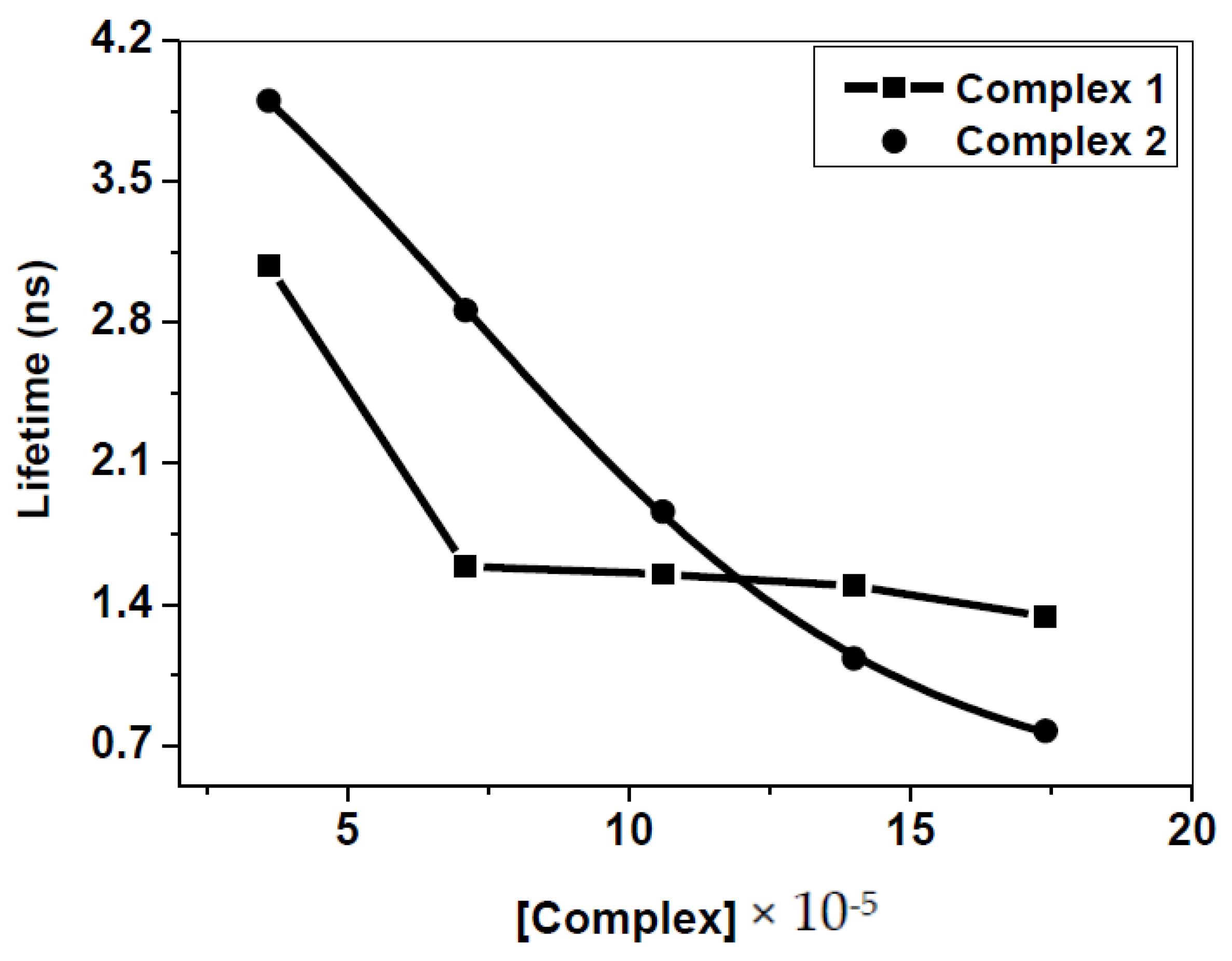
2.1.3. Time-Resolved Studies
2.1.4. Determination of Thermodynamic Parameters
2.1.5. Cyclic Voltammetry Studies
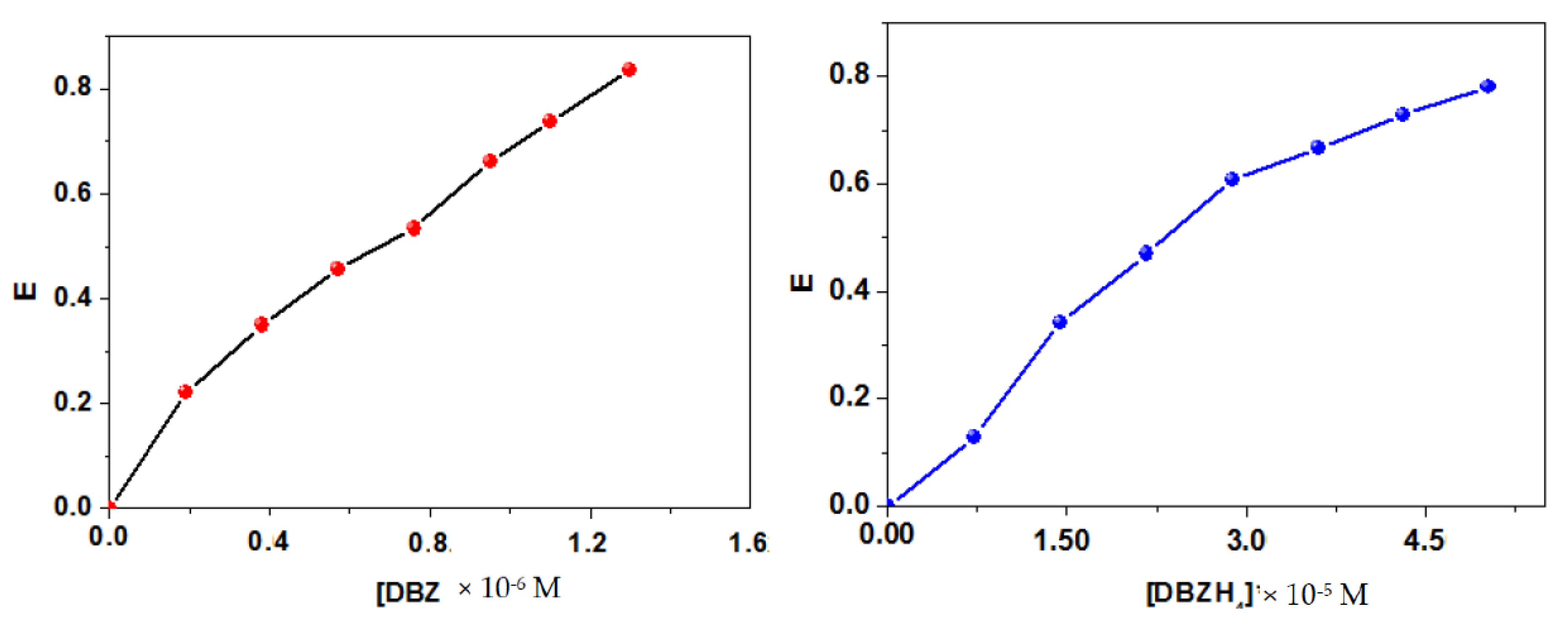
2.2. Energy Transfer from BSA to DBZ and DBZH4 Complexes
2.3. Antibacterial Activity
2.4. Protein Docking
3. Experimental Section
3.1. Materials
3.2. Instrumentation
3.3. Antibacterial Activity
3.4. Molecular Docking
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Declaration of Interest Statement
References
- Carter, D.C.; Ho, J.X. Structure of serum albumin. Adv. Protein Chem. 1994, 45, 153–203. [Google Scholar]
- Olson, R.E.; Christ, D.D. Chapter 33. Plasma Protein Binding of Drugs. Annu. Rep. Med. Chem. 1996, 31, 327–337. [Google Scholar]
- Naik, K.M.; Kolli, D.B.; Nandibewoor, S.T. Elucidation of binding mechanism of hydroxyurea on serum albumins by different spectroscopic studies. SpringerPlus 2014, 3, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Ashoka, S.; Seetharamappa, J.; Kandagal, P.B.; Shaikh, S.M.T. Investigation of the interaction between trazodone hydrochloride and bovine serum albumin. J. Lumin. 2006, 121, 179–186. [Google Scholar] [CrossRef]
- Gao, D.; Tian, Y.; Liang, F.; Jin, D.; Chen, Y.; Zhang, H.; Yu, A. Investigation on the pH-dependent binding of Eosin Y and bovine serum albumin by spectral methods. J. Lumin. 2007, 127, 515–522. [Google Scholar] [CrossRef]
- Zhou, B.; Qi, Z.; Xiao, Q.; Dong, J.X.; Zhang, Y.Z.; Liu, Y. Interaction of loratadine with serum albumins studied by fluorescence quenching method. J. Biochem. Biophys. Meth 2007, 70, 743–747. [Google Scholar] [CrossRef]
- Hongwei, Z.; Min, G.; Zhaoxia, Z.; Wenfeng, W.; Guozhong, W. Spectroscopic studies on the interaction between riboflavin and albumins. Spectrochim. Acta Part A 2006, 65, 811–817. [Google Scholar]
- Yin, Y.B.; Wang, Y.N.; Ma, J.B. Aggregation of two carboxylic derivatives of porphyrin and their affinity to bovine serum albumin. Spectrochim. Acta Part A 2006, 64, 1032–1038. [Google Scholar] [CrossRef]
- Hu, Y.J.; Liu, Y.; Zhao, R.M.; Dong, J.X.; Qu, S.S. Spectroscopic studies on the interaction between methylene blue and bovine serum albumin. J. Photochem. Photobiol. A Chem. 2006, 179, 324–329. [Google Scholar] [CrossRef]
- Hansen, U.K. Molecular aspects of ligand binding to serum albumin. Pharmacol. Rev. 1981, 33, 17–35. [Google Scholar]
- Tesmer, M.; Shu, M.; Vahrenkamp, H. Sulfur-rich zinc chemistry: New tris(thioimidazolyl)hydroborate ligands and their zinc complex chemistry related to the structure and function of alcohol dehydrogenase. Inorg. Chem. 2001, 40, 4022–4029. [Google Scholar] [CrossRef]
- Bonnet, D.; Leducn, P.; Bill, E.; Chottard, G.; Mansuy, D.; Artaud, I. CoII Complexes with Mixed Amino-N and Thiolato-S Donor Sets—Structural Characterization and Electronic Properties of a Stable Bis(μ-thiolato)-Bridged Binuclear CoII Complex. Eur. J. Inorg. Chem. 2001, 1449–1456. [Google Scholar] [CrossRef]
- Brand, U.; Burth, R.; Vahrenkamp, H. Design of Trigonal-Bipyramidal ZnN3S2 Complexes. J. Inorg. Chem. 1996, 35, 1083–1086. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, Y.; Ding, Y.; Povey, M.; York, D. Investigation into the antibacterial behaviour of suspensions of ZnO nanoparticles (ZnO nanofluids). J. Nanoparticle Res. 2007, 9, 479–489. [Google Scholar] [CrossRef]
- Maret, W. Zinc and Sulfur: A Critical Biological Partnership. Biochemistry 2004, 43, 3301–3309. [Google Scholar] [CrossRef]
- Shaban, S.Y.; Puchta, R.; van Eldik, R. Five-coordinate Zinc(II) Complexes Containing Sterically Demanding Bio-mimetic N3S2 Ligands. Syntheses, Characterization and DFT Calculations. Z. Naturforsch. 2010, 65, 251–257. [Google Scholar] [CrossRef]
- Soliman, E.; L-Khouly, M.E.; Ramadan, A.; L-Mehasseb, I.E.; Shaban, S.Y. Energy-transfer versus electron-transfer reactions for the light-harvesting phthalocyanine/dithiolato-bisimino zinc system. J. Coord. Chem. 2020, 68, 2054–2064. [Google Scholar] [CrossRef]
- Shaban, N.Z.; Yehia, S.A.; Shoueir, K.R.; Saleh, S.R.; Awad, D.; Shaban, S.Y. Design, DNA binding and kinetic stuies, antibaterial and cytotoxic activities of stable dithiophenolatotitanium(IV)-chitosan Nanocomposite. J. Mol. Liq. 2019, 287, 111002–111011. [Google Scholar] [CrossRef]
- El-Shamy, H.; Shaban, S.Y.; El-Mehasseb, I.; El-Kemary, M.A.; van Eldik, R. Probing the interaction of iron complex containing N3S2 macrocyclic ligand with bovine serum albumin using spectroscopic techniques. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 228, 117811–117815. [Google Scholar] [CrossRef]
- Elshami, F.I.; Ramadan, A.; Ibrahim, M.M.; Elmehasseb, I.M.; Al-Juaid, S.; Shaban, S.Y. Metformin Containing Nickel (II) Complexes: Synthesis, Structural Characterization, Binding and Kinetic Interactions with BSA, Antibacterial and in-vitro Cytotoxicity Studies. Appl. Organometal. Chem. 2020, 34, 5437–5454. [Google Scholar] [CrossRef]
- Shaban, S.Y.; El-Kemary, M.A.; Samir, G.; Elbaradei, H. Synthesis, characterization, antibacterial activities testing and the interaction of DNA with ciprofloxacin and its La(III)-based complex. J. Chin. Adv. Mater. Soc. 2018, 6, 123–133. [Google Scholar] [CrossRef]
- Shaban, S.Y.; El-Shafai, N.; Mansour, H.; van Eldik, R. Iron(II) complexes containing the 2,6-bis-iminopyridyl moiety. Synthesis, characterization, reactivity, and DNA binding. J. Coord. Chem. 2015, 68, 2054–2064. [Google Scholar] [CrossRef][Green Version]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 7, 1349–1356. [Google Scholar] [CrossRef]
- Uhlenbrock, S.; Wagner, R.; Krebs, B. 2,4-Diferrocenyl-1,3-dithiadiphosphetane 2,4-disulfide; structure and reactions with catechols and [PtCl2(PR3)2](R = Et or Bun). J. Chem. Soc. Dalton Trans. 1996, 18, 3731–3736. [Google Scholar] [CrossRef]
- Sharma, A.; Schulman, S.G. Introduction to Fluorescence Spectroscopy; Wiley Press: New York, NY, USA, 1999. [Google Scholar]
- Sulkowska, A. Interaction of drugs with bovine and human serum albumin. J. Mol. Struct. 2002, 614, 227–232. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principle of Fluorescence Spectroscopy; Springer: New York, NY, USA, 1999. [Google Scholar]
- Wang, R.; Chai, Y.; Wang, R. Study of the interaction between bovine serum albumin and analogs of Biphenyldicarboxylate by spectrofluorimetry. Spectrochim. Acta A 2012, 96, 324–331. [Google Scholar] [CrossRef]
- Topala, T.; Bodoki, A.; Operean, L.; Oprean, R. Bovine Serum Albumin Interactions with Metal Complexes. Clujul. Med. 2014, 87, 215–219. [Google Scholar] [CrossRef]
- Gok, E.; Ozturk, C.; Akbay, N. Interaction of Thyroxine with 7 Hydroxycoumarin: A Fluorescence Quenching Study. J. Fluoresc. 2008, 18, 781–785. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Green, R.J.; Frazier, R.A. Interaction of flavonoids with bovine serum albumin: A fluorescence quenching study. J. Agric. Food Chem. 2005, 53, 158–163. [Google Scholar] [CrossRef]
- Xie, M.X.; Long, M.; Liu, Y. Characterization of the interaction between human serum albumin and morin. Biophys. Acta 2006, 1760, 1184–1191. [Google Scholar] [CrossRef]
- Ross, D.P.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Liua, Y.; Chen, M.; Luoa, Z. Investigation on the site-selective binding of bovine serum albumin by erlotinib hydrochloride. J. Biomol. Struct. Dyn. 2013, 31, 1160–1174. [Google Scholar] [CrossRef]
- Hu, Y.-H.; Liu, Y.; Xiao, X.-H. Investigation of the interaction between Berberine and human serum albumin. Biomacromolecules 2009, 10, 517–521. [Google Scholar] [CrossRef]
- Carter, M.T.; Bard, A.J. Voltammetric studies of the interaction of tris(1,10-phenanthroline)cobalt(III) with DNA. J. Am. Chem. Soc. 1987, 24, 7528–7530. [Google Scholar] [CrossRef]
- Förster, T. Studies on the Binding Mechanism of VB1 and VB9 with Trypsin. Modern Quantum Chemistry; Academic Press: New York, NY, USA, 1996. [Google Scholar]
- Cyril, L.; Earl, J.K.; Sperry, W.M. Biochemists Handbook; E & FN Epon Led. Press: London, UK, 1961. [Google Scholar]
- Valeur, B.; Brochon, J.C. New Trends in Fluorescence Spectroscopy; Springer: Berlin, Germany, 2001. [Google Scholar]
- Deepa, S.; Mishra, A.K. Fluorescence spectroscopic study of serum albumin–bromadiolone interaction: Fluorimetric determination of bromadiolone. J. Pharm. Biomed. Anal. 2005, 38, 556–563. [Google Scholar] [CrossRef]
- Kathiravan, A.; Paramaguru, G.; Renganathan, R. Study on the binding of colloidal zinc oxide nanoparticles with bovine serum albumin. J. Mol. Struct. 2009, 934, 129–137. [Google Scholar] [CrossRef]
- Karcz, D.; Matwijczuk, A.; Kami´nski, D.; Creaven, B.; Ciszkowicz, E.; Lecka-Szlachta, K.; Starzak, K. Structural Features of 1,3,4-Thiadiazole-Derived Ligands and Their Zn(II) and Cu(II) Complexes Which Demonstrate Synergistic Antibacterial Effects with Kanamycin. Int. J. Mol. Sci. 2020, 21, 5735. [Google Scholar] [CrossRef]
- Sharma, A.K.; Chandra, S. Complexation of nitrogen and sulphur donor Schiff’s base ligand to Cr(III) and Ni(II) metal ions: Synthesis, spectroscopic and antipathogenic studies. Spectrochim. Acta A 2011, 78, 337. [Google Scholar] [CrossRef]
- Dey, D.; Maiti, C.; Maiti, S.; Dhara, D. Interaction between Calf Thymus DNA and Cationic Bottle-Brush Copolymers: Equilibrium and Stopped-Flow Kinetic Studies. Phys. Chem. Chem. Phys. 2015, 17, 2366–2377. [Google Scholar] [CrossRef]
- Chung, Y.-C.; Su, Y.P.; Chen, C.C.; Jia, G.; Wang, H.L.; Wu, J.G.; Lin, J.G. Relationship between antibacterial activity of chitosan and surface characteristics of cell wall. Acta Pharmacol. Sin. 2004, 25, 932. [Google Scholar]
- Xue, Z.-X.; Yang, G.-P.; Zhang, Z.-P.; He, B.-L. Application of chitosan microspheres as carriers of LH-RH analogue TX46. React. Funct. Polym. 2006, 66, 893–901. [Google Scholar] [CrossRef]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin Acta Crystallogr. Sect. D Struct. Biol. 2012, 68, 1278–1289. [Google Scholar]
- Ashbrook, J.D.; Spector, A.A.; Santos, E.C.; Fletcher, J.E. Structures of bovine, equine and leporine serum albumin. J. Biol.Chem. 1975, 250, 2333–2338. [Google Scholar] [CrossRef]
- NCCL. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 5th ed.; NCCLS: Wayne, PA, USA, 2000; ISBN 1-56238-394-9. [Google Scholar]
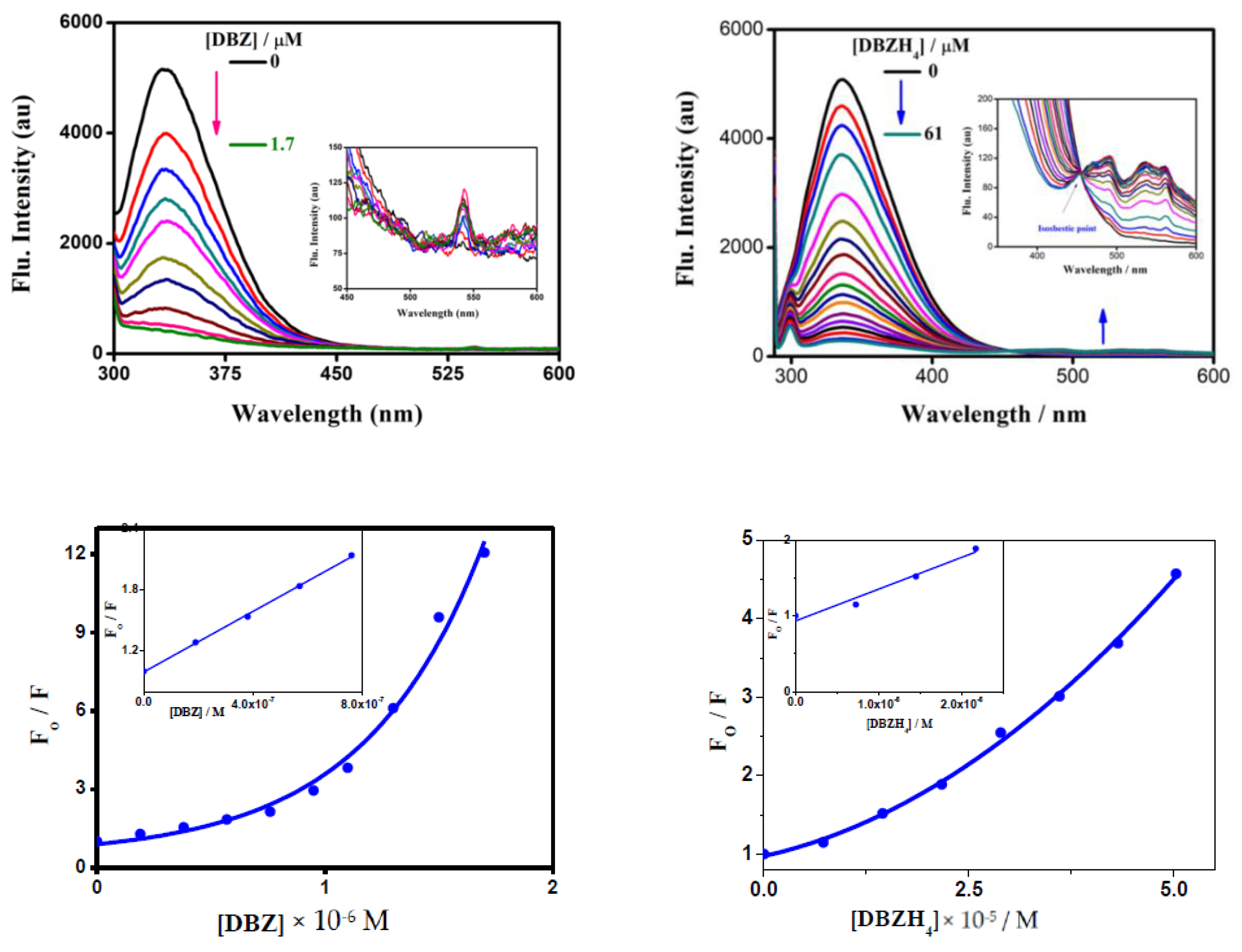
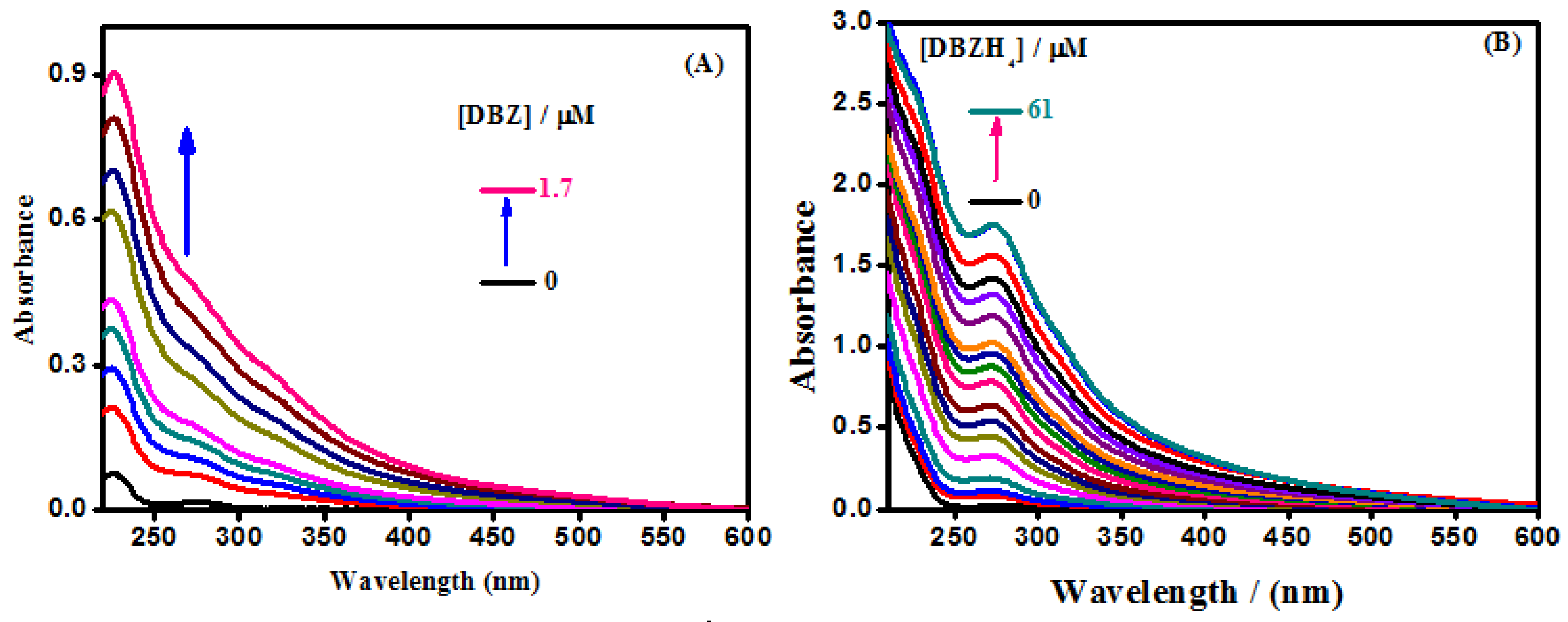



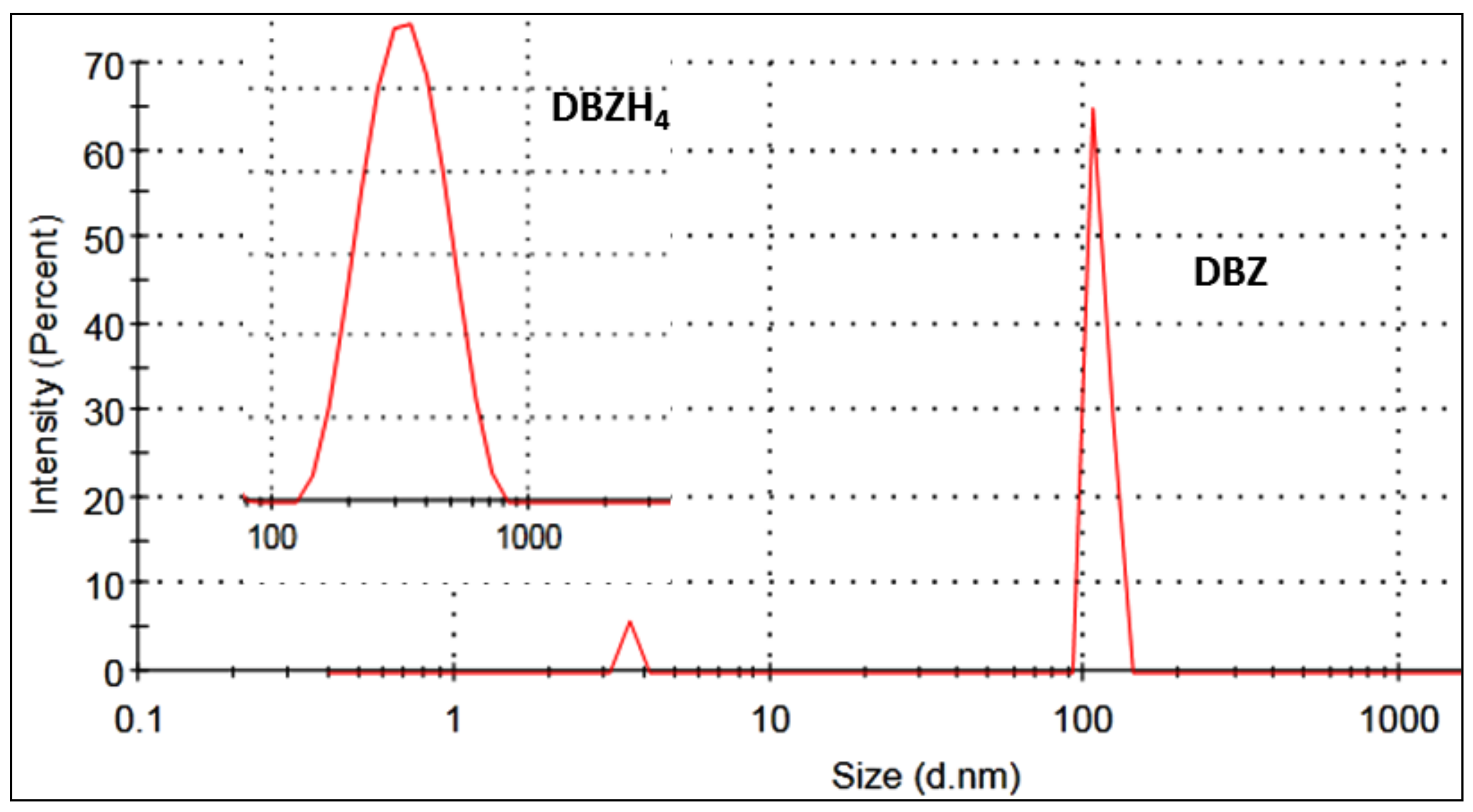
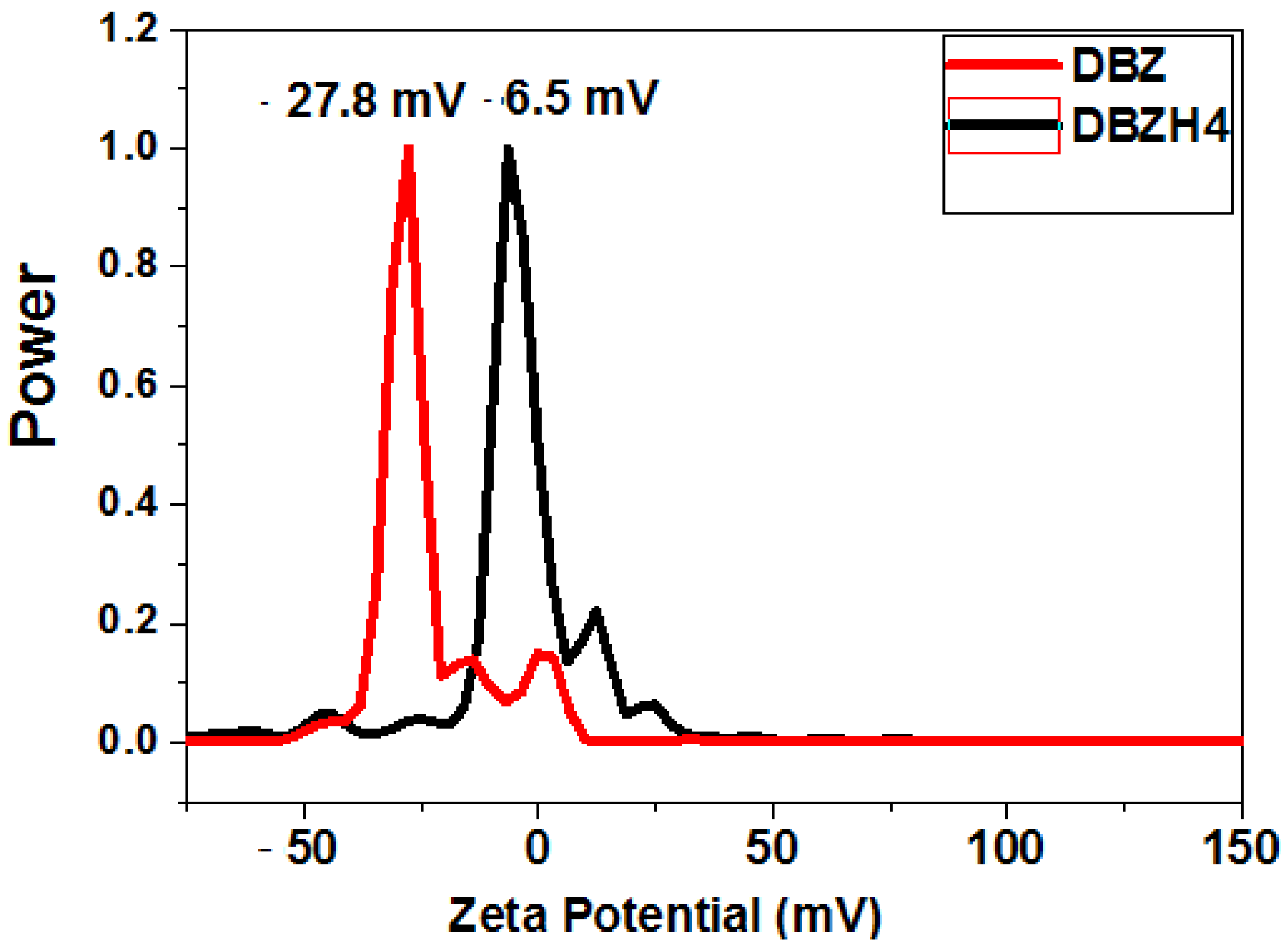
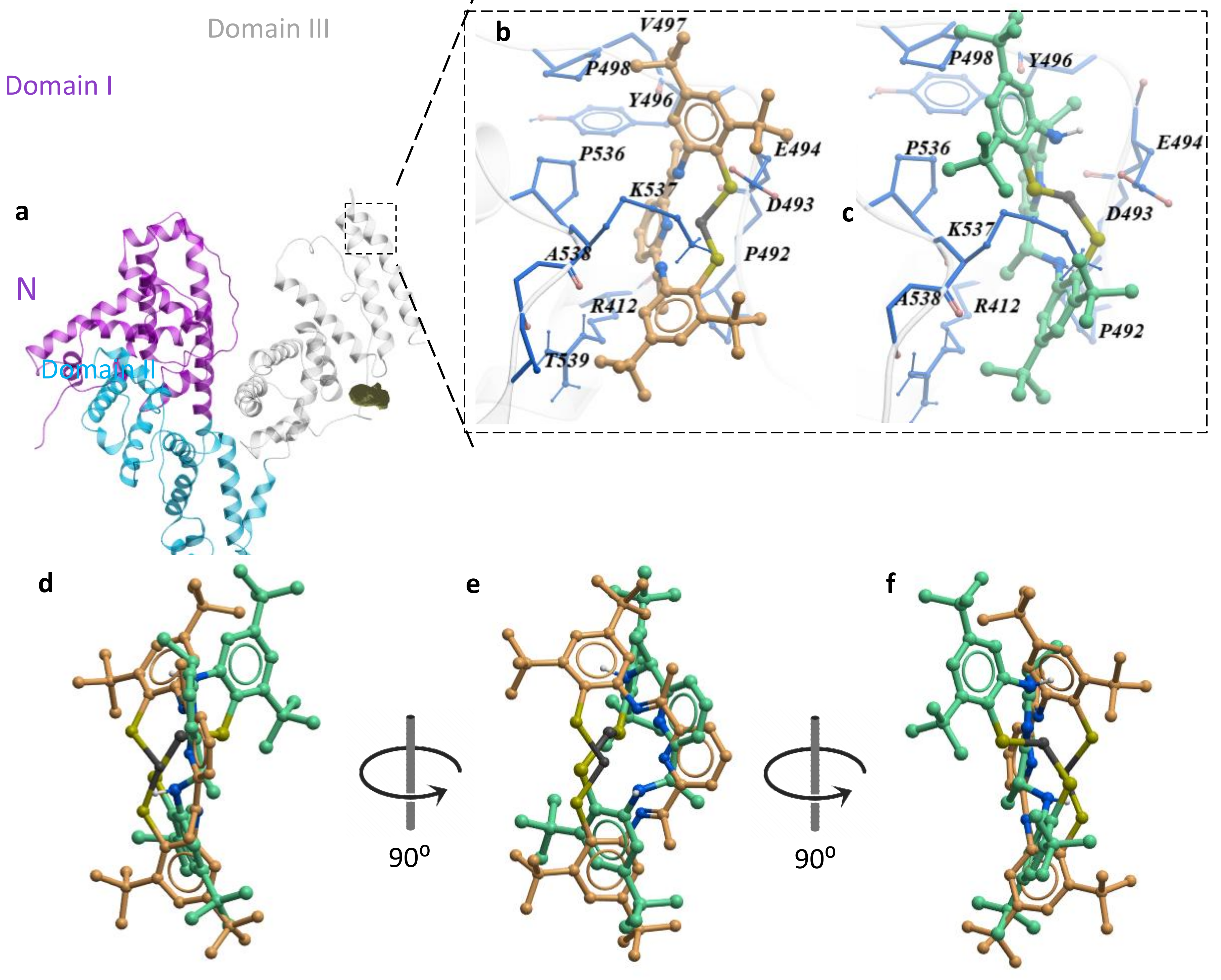
| Compounds | T (K) | Ksv × (105 M−1) | Kq × (1014 M−1 s−1) |
|---|---|---|---|
| DBZ | 288 | 19.0 | 4.15 |
| 298 | 24.7 | 5.40 | |
| 318 | 27.3 | 5.97 | |
| DBZH4 | 288 | 0.69 | 0.150 |
| 298 | 0.70 | 0.153 | |
| 318 | 0.71 | 0.155 |
| T (K) | KA × 105 (M−1) | n | ΔHº (kJ mol−1) | ΔGº (kJ mol−1) | ΔSº (J mol−1 K−1) | |
|---|---|---|---|---|---|---|
| DBZ | 288 | 9.9 | 1.12 | 42 ± 10 | −33.2 ± 0.08 | 261 ± 35 |
| 298 | 22.8 | 1.32 | −35.8 ± 0.4 | |||
| 308 | 30.7 | 1.45 | −38.4 ± 0.8 | |||
| DBZH4 | 288 | 0.13 | 1.22 | 65 ± 2 | −22.6 ± 0.3 | 304 ± 6 |
| 298 | 0.31 | 1.40 | −25.6 ± 0.2 | |||
| 308 | 0.76 | 1.42 | −28.6 ± 0.2 |
| J (10−13 L mol−1 cm3) | R0 (nm) | r (nm) | E (%) | ||
|---|---|---|---|---|---|
| DBZ–BSA | 79.6 | 9.0 | 2.51 | 3.21 | 0.19 |
| DBZH4–BSA | 90.0 | 5.4 | 2.31 | 3.67 | 0.06 |
| Microorganisms | DBZ | DBZH4 | ||
|---|---|---|---|---|
| MIC | MBC | MIC | MBC | |
| Gram negative | ||||
| Escherichia coli | 25.0 | 50.0 | 0 | 0 |
| Pseudomonas aeruginosa | 12.5 | 25.0 | 15.0 | 30.0 |
| Gram positive | ||||
| Enterococcus | 25.0 | 50.0 | 30.0 | 60.0 |
| Staphylococcus aureus | 12.5 | 25.0 | 15.0 | 30.0 |
| Parameters | DBZ | DBZH4 |
|---|---|---|
| Docking score | −12.63 | −9.76 |
| Hydrophobic binding energy (Kcal/mol) | −7.59 | −7.86 |
| Van der Waals binding energy (Kcal/mol) | −19.46 | −18.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soliman, E.; Ibrahim, M.M.; El-Khouly, M.E.; El-Mehasseb, I.; Ramadan, A.E.-M.M.; Mahfouz, M.E.; Shaban, S.Y.; van Eldik, R. BSA Interaction, Molecular Docking, and Antibacterial Activity of Zinc(II) Complexes Containing the Sterically Demanding Biomimetic N3S2 Ligand: The Effect of Structure Flexibility. Molecules 2022, 27, 3543. https://doi.org/10.3390/molecules27113543
Soliman E, Ibrahim MM, El-Khouly ME, El-Mehasseb I, Ramadan AE-MM, Mahfouz ME, Shaban SY, van Eldik R. BSA Interaction, Molecular Docking, and Antibacterial Activity of Zinc(II) Complexes Containing the Sterically Demanding Biomimetic N3S2 Ligand: The Effect of Structure Flexibility. Molecules. 2022; 27(11):3543. https://doi.org/10.3390/molecules27113543
Chicago/Turabian StyleSoliman, Eman, Mohamed M. Ibrahim, Mohamed E. El-Khouly, Ibrahim El-Mehasseb, Abd El-Motaleb M. Ramadan, Magdy E. Mahfouz, Shaban Y. Shaban, and Rudi van Eldik. 2022. "BSA Interaction, Molecular Docking, and Antibacterial Activity of Zinc(II) Complexes Containing the Sterically Demanding Biomimetic N3S2 Ligand: The Effect of Structure Flexibility" Molecules 27, no. 11: 3543. https://doi.org/10.3390/molecules27113543
APA StyleSoliman, E., Ibrahim, M. M., El-Khouly, M. E., El-Mehasseb, I., Ramadan, A. E.-M. M., Mahfouz, M. E., Shaban, S. Y., & van Eldik, R. (2022). BSA Interaction, Molecular Docking, and Antibacterial Activity of Zinc(II) Complexes Containing the Sterically Demanding Biomimetic N3S2 Ligand: The Effect of Structure Flexibility. Molecules, 27(11), 3543. https://doi.org/10.3390/molecules27113543







