Unified Nanotechnology Format: One Way to Store Them All
Abstract
:1. Introduction
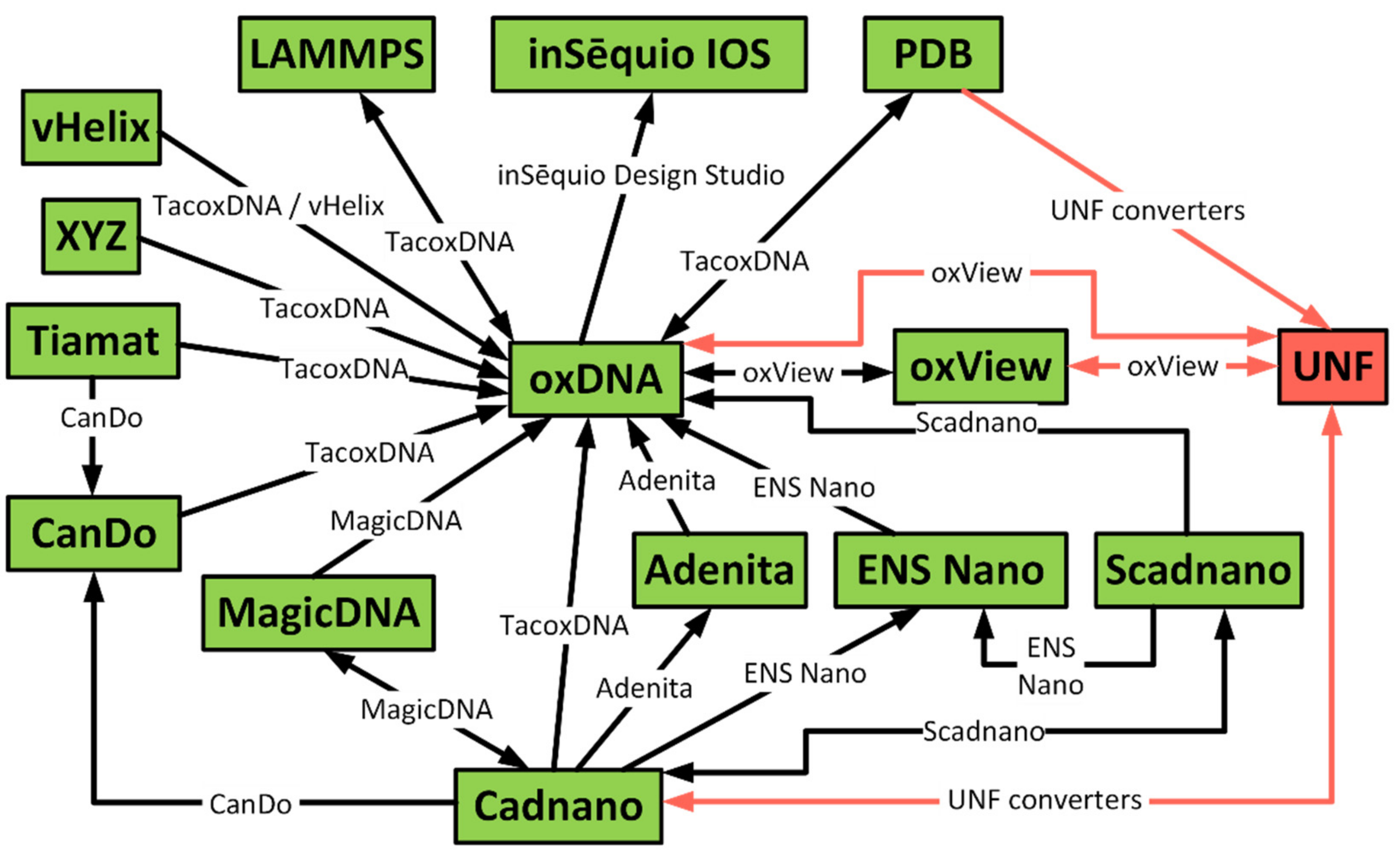
2. State of the Art
3. Format Description
3.1. Overall Goal
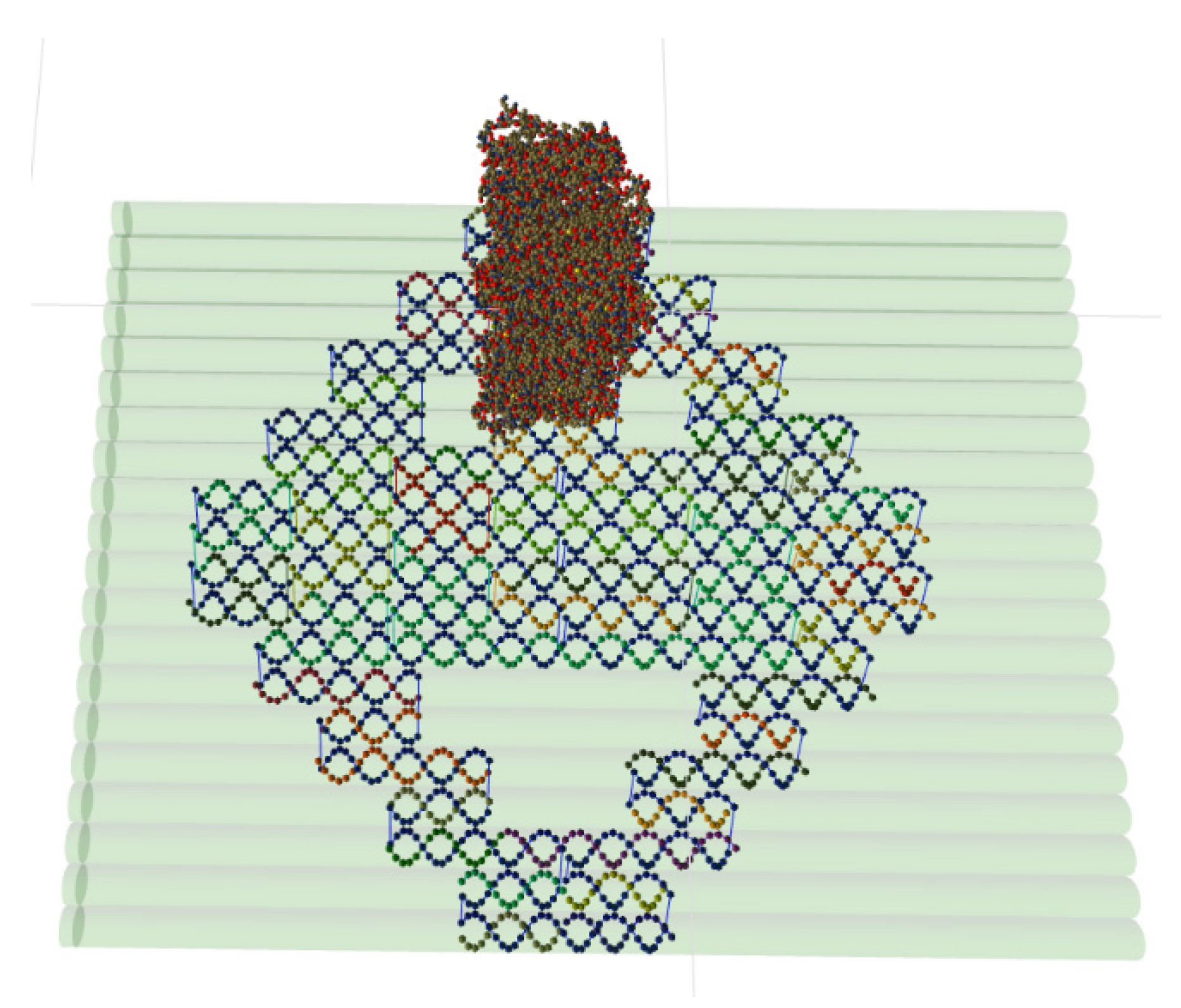
- Due to the popularity of the multilayer DNA origami technique, the format should be able to explicitly store designs of such lattice-constrained nanostructures. However, at the same time, it must also support the storing of the free-form DNA nanostructures, allowing for the description of arbitrarily shaped designs and simulation outcomes.
- The format should be viable for DNA-protein and RNA-protein nanotechnology engineering by storing coarse-grained representations of protein structures.
- For conversion from fully atomistic models to coarse-grained ones and vice versa, the format should have some way of referencing the original source data from crystallography, NMR, cryoEM, and all-atom simulation experiments.
- Related to the previous point, the format should support references to other types of molecules to allow the creation of more complex molecular scenes, possibly including all-atom structures together with coarse-grained ones.
- To facilitate the implementation of the format in various tools, it must be well defined, with a clear and properly explained terminology. Furthermore, the documentation of its structure should be easily available.
- The UNF file itself should be human-readable to allow for quick changes using a simple text editor in case of need. At the same time, it must be easy to process from the perspective of software developers.
- Ideally, the format should reuse well-defined concepts and terms from the existing DNA nanotechnology file formats and software applications to make the transition from other ways of data storage easier.
- Finally, due to the nature of the goal summarized at the beginning of this chapter, it is expected that the format will gradually evolve to meet the needs of its potential end-users. Therefore, it should be open for extension, making it possible to shape it in the future without a need for a complete rewrite.
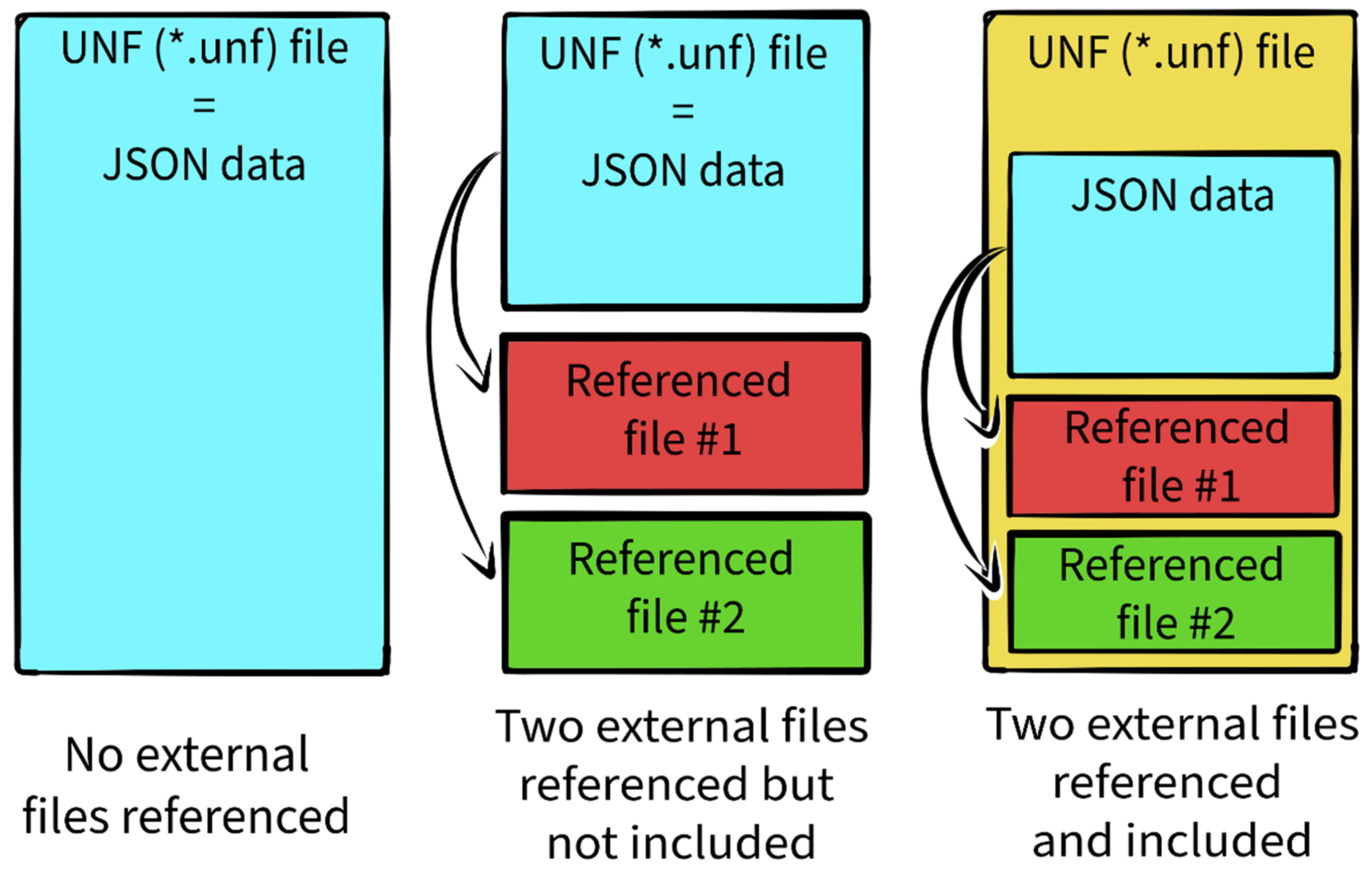
3.2. UNF File Structure
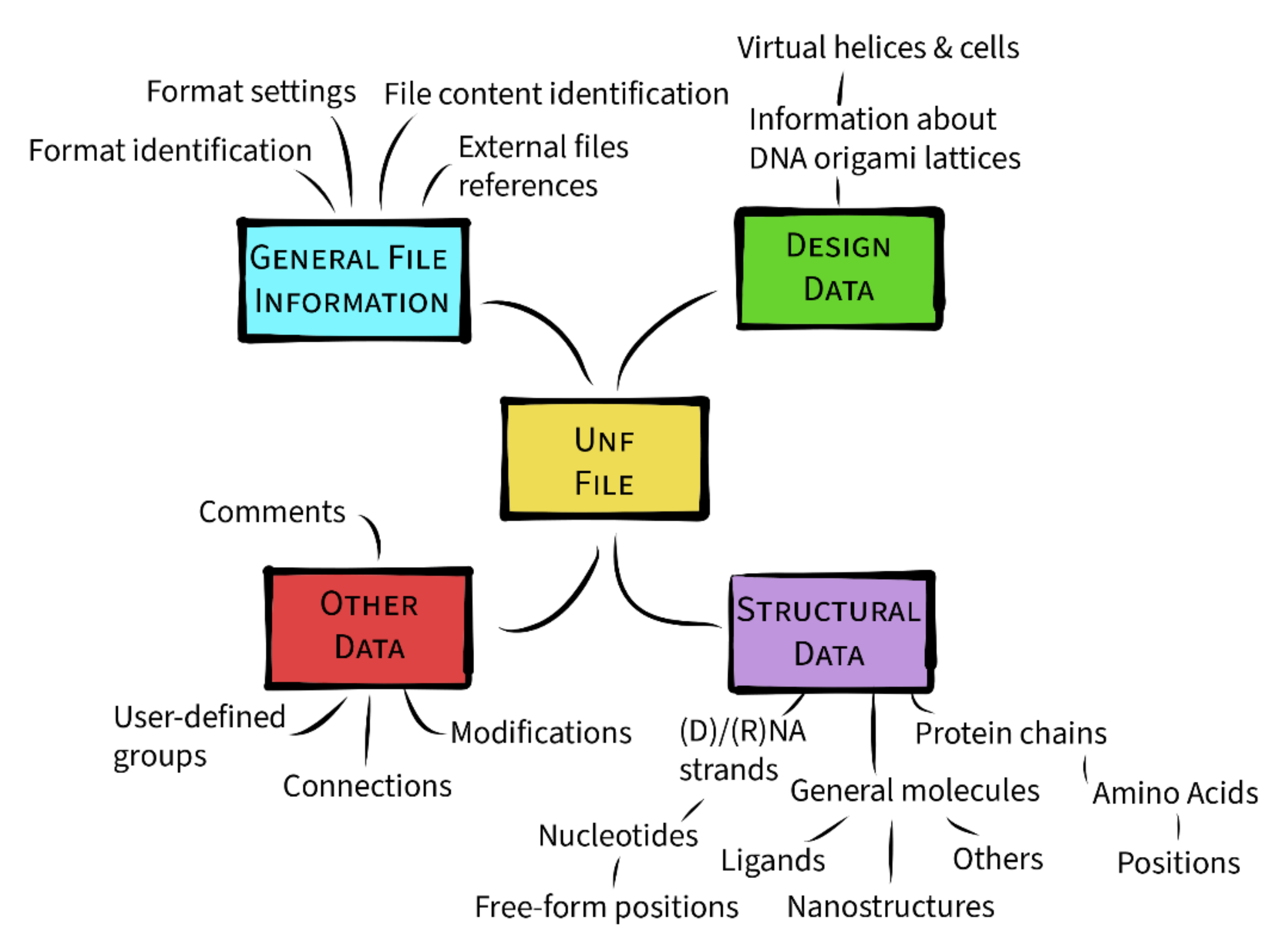
3.3. Data Hierarchy
3.3.1. General File Information
3.3.2. Design Data
3.3.3. Structural Data
3.3.4. Other Data
4. Converters from Existing Formats
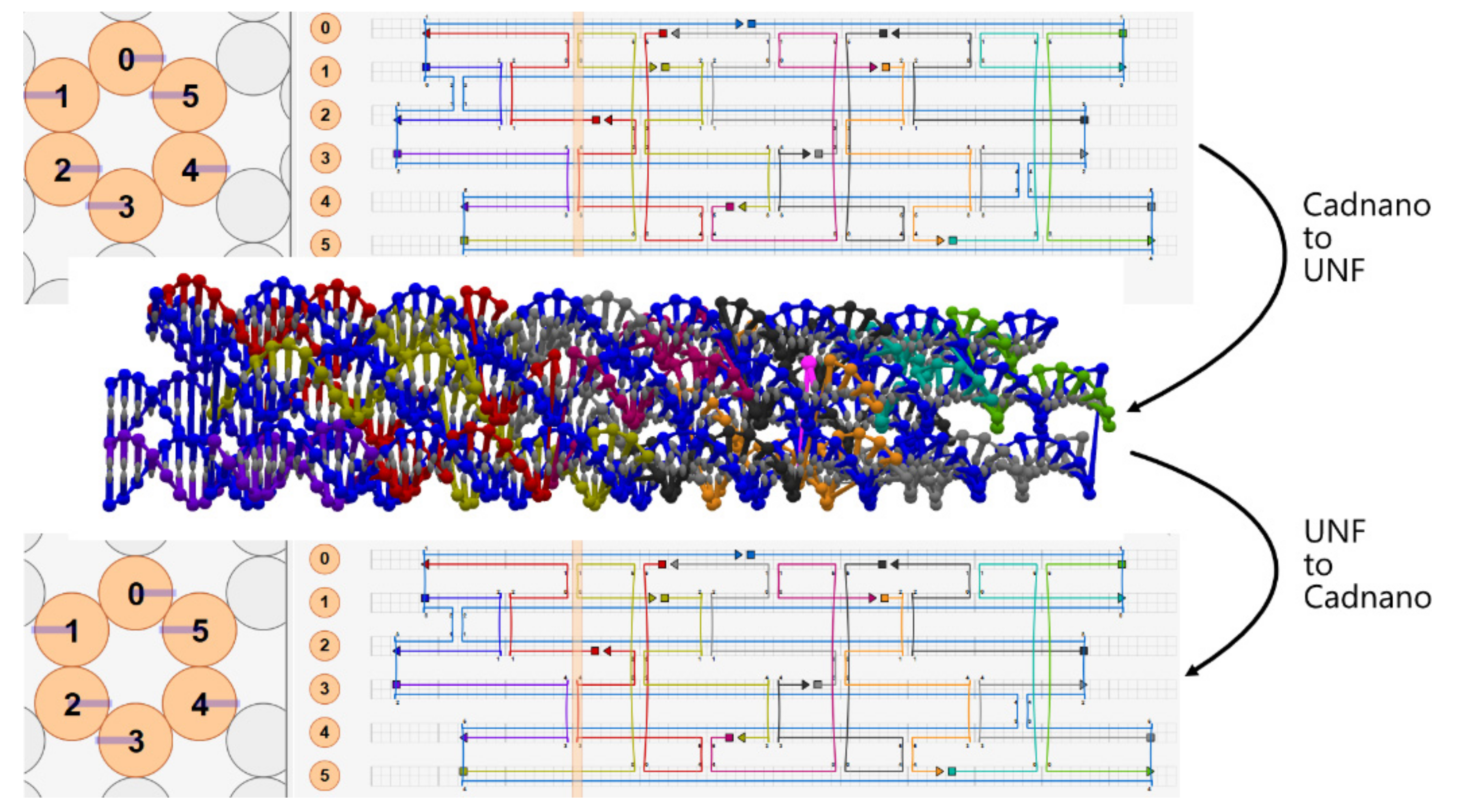
4.1. Cadnano ⇄ UNF Converter
4.2. PDB → UNF Converter
4.3. oxDNA/oxView ⇄ UNF
5. Use Cases
5.1. Multi-Component Designs
5.2. Multilayer DNA Origami Structures and All-Atom Molecules
5.3. Coarse-Grained DNA-Protein Hybrids
5.4. Coarse-Grained RNA Structures
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Appendix A
- The official UNF repository is accessible via the following link: https://github.com/barisicgroup/unf (accessed on 15 November 2021).
- The repository of the oxView application is available using this link: https://github.com/sulcgroup/oxdna-viewer (accessed on 15 November 2021).
References
- Sun, L.; Yu, L.; Shen, W. DNA nanotechnology and its applications in biomedical research. J. Biomed. Nanotechnol. 2014, 10, 2350–2370. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.S.L.; Shiu, S.C.-C.; Godonoga, M.; Cheung, Y.-W.; Liang, S.; Dirkzwager, R.M.; Kinghorn, A.B.; Fraser, L.A.; Heddle, J.G.; Tanner, J.A. An aptamer-enabled DNA nanobox for protein sensing. Nanomedicine 2018, 14, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.-Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef] [Green Version]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Högberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef] [Green Version]
- Weizmann, Y.; Andersen, E.S. RNA nanotechnology—The knots and folds of RNA nanoparticle engineering. MRS Bull. 2017, 42, 930–935. [Google Scholar] [CrossRef]
- Hernandez-Garcia, A. Strategies to Build Hybrid Protein-DNA Nanostructures. Nanomaterials 2021, 11, 1332. [Google Scholar] [CrossRef]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef] [Green Version]
- De Llano, E.; Miao, H.; Ahmadi, Y.; Wilson, A.J.; Beeby, M.; Viola, I.; Barisic, I. Adenita: Interactive 3D modelling and visualization of DNA nanostructures. Nucleic Acids Res. 2020, 48, 8269–8275. [Google Scholar] [CrossRef]
- Šulc, P.; Romano, F.; Ouldridge, T.E.; Rovigatti, L.; Doye, J.P.K.; Louis, A.A. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 2012, 137, 135101. [Google Scholar] [CrossRef]
- Doye, J.P.K.; Fowler, H.; Prešern, D.; Bohlin, J.; Rovigatti, L.; Romano, F.; Šulc, P.; Wong, C.K.; Louis, A.A.; Schreck, J.S.; et al. The oxDNA Coarse-Grained Model as a Tool to Simulate DNA Origami. 2020. Available online: https://arxiv.org/pdf/2004.05052 (accessed on 15 November 2021).
- Suma, A.; Poppleton, E.; Matthies, M.; Šulc, P.; Romano, F.; Louis, A.A.; Doye, J.P.K.; Micheletti, C.; Rovigatti, L. TacoxDNA: A user-friendly web server for simulations of complex DNA structures, from single strands to origami. J. Comput. Chem. 2019, 40, 2586–2595. [Google Scholar] [CrossRef] [PubMed]
- Dalby, A.; Nourse, J.G.; Hounshell, W.D.; Gushurst, A.K.I.; Grier, D.L.; Leland, B.A.; Laufer, J. Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. J. Chem. Inf. Comput. Sci. 1992, 32, 244–255. [Google Scholar] [CrossRef]
- XYZ (Format)—Open Babel. Available online: http://openbabel.org/wiki/XYZ_%28format%29 (accessed on 15 October 2021).
- Atomic Coordinate Entry Format Version 3.3. Available online: https://www.wwpdb.org/documentation/file-format-content/format33/v3.3.html (accessed on 15 October 2021).
- Bourne, P.E.; Berman, H.M.; McMahon, B.; Watenpaugh, K.D.; Westbrook, J.D.; Fitzgerald, P.M. [30] Macromolecular crystallographic information file. In Macromolecular Crystallography Part B; Elsevier: Amsterdam, The Netherlands, 1997; pp. 571–590. ISBN 9780121821784. [Google Scholar]
- Fitzgerald, P.M.D.; Berman, H.; Bourne, P.; McMahon, B.; Watenpaugh, K.; Westbrook, J. The mmCIF dictionary: Community review and final approval. Acta Cryst. Sect. A 1996, 52, C575. [Google Scholar] [CrossRef] [Green Version]
- Jewett, A.I.; Stelter, D.; Lambert, J.; Saladi, S.M.; Roscioni, O.M.; Ricci, M.; Autin, L.; Maritan, M.; Bashusqeh, S.M.; Keyes, T.; et al. Moltemplate: A Tool for Coarse-Grained Modeling of Complex Biological Matter and Soft Condensed Matter Physics. J. Mol. Biol. 2021, 433, 166841. [Google Scholar] [CrossRef]
- LAMMPS Molecular Dynamics Simulator. Available online: https://www.lammps.org/ (accessed on 15 October 2021).
- Plimpton, S. Fast Parallel Algorithms for Short-Range Molecular Dynamics. J. Comput. Phys. 1995, 117, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roel-Touris, J.; Don, C.G.; Honorato, R.V.; Rodrigues, J.P.G.L.M.; Bonvin, A.M.J.J. Less Is More: Coarse-Grained Integrative Modeling of Large Biomolecular Assemblies with HADDOCK. J. Chem. Theory Comput. 2019, 15, 6358–6367. [Google Scholar] [CrossRef]
- Honorato, R.V.; Roel-Touris, J.; Bonvin, A.M.J.J. MARTINI-Based Protein-DNA Coarse-Grained HADDOCKing. Front. Mol. Biosci. 2019, 6, 102. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [Green Version]
- Gamini, R.; Chandler, D. Residue-Based Coarse Graining Using MARTINI Force Field in NAMD; University of Illinois at Urbana-Champaign, Computational Biophysics Workshop: Urbana, IL, USA, 2013. [Google Scholar]
- Doty, D.; Lee, B.L.; Stérin, T. scadnano: A Browser-Based, Scriptable Tool for Designing DNA Nanostructures. In 26th International Conference on DNA Computing and Molecular Programming (DNA 26); Geary, C., Matthew, P.J., Eds.; Schloss Dagstuhl-Leibniz-Zentrum für Informatik: Dagstuhl, Germany, 2020; pp. 9:1–9:17. ISBN 978-3-95977-163-4. [Google Scholar]
- Williams, S.; Lund, K.; Lin, C.; Wonka, P.; Lindsay, S.; Yan, H. Tiamat: A Three-Dimensional Editing Tool for Complex DNA Structures. In DNA Computing; Goel, A., Simmel, F.C., Sosík, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 90–101. ISBN 978-3-642-03075-8. [Google Scholar]
- Parabon NanoLabs. The Parabon™ inSēquio™ Design Studio: The Quintessential Application for Designing DNA-Based Nanostructures. Available online: https://parabon-nanolabs.com/therapeutics/insequio.html (accessed on 30 November 2021).
- Huang, C.-M.; Kucinic, A.; Johnson, J.A.; Su, H.-J.; Castro, C.E. Integrated computer-aided engineering and design for DNA assemblies. Nat. Mater. 2021, 20, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- University of Oxford. Documentation—OxDNA. Available online: https://dna.physics.ox.ac.uk/index.php/Documentation#Configuration_and_topology_files (accessed on 15 October 2021).
- Procyk, J.; Poppleton, E.; Šulc, P. Coarse-grained nucleic acid-protein model for hybrid nanotechnology. Soft Matter 2021, 17, 3586–3593. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-N.; Kilchherr, F.; Dietz, H.; Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012, 40, 2862–2868. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Lee, J.G.; Yun, G.; Lee, C.; Kim, Y.-J.; Kim, K.S.; Kim, T.H.; Kim, D.-N. Rapid Computational Analysis of DNA Origami Assemblies at Near-Atomic Resolution. ACS Nano 2021, 15, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Poppleton, E.; Bohlin, J.; Matthies, M.; Sharma, S.; Zhang, F.; Šulc, P. Design, optimization and analysis of large DNA and RNA nanostructures through interactive visualization, editing and molecular simulation. Nucleic Acids Res. 2020, 48, e72. [Google Scholar] [CrossRef]
- Fernandez-Castanon, J.; Bomboi, F.; Rovigatti, L.; Zanatta, M.; Paciaroni, A.; Comez, L.; Porcar, L.; Jafta, C.J.; Fadda, G.C.; Bellini, T.; et al. Small-angle neutron scattering and molecular dynamics structural study of gelling DNA nanostars. J. Chem. Phys. 2016, 145, 84910. [Google Scholar] [CrossRef] [Green Version]
- Snodin, B.E.K.; Randisi, F.; Mosayebi, M.; Šulc, P.; Schreck, J.S.; Romano, F.; Ouldridge, T.E.; Tsukanov, R.; Nir, E.; Louis, A.A.; et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J. Chem. Phys. 2015, 142, 234901. [Google Scholar] [CrossRef] [Green Version]
- Zhan, P.; Urban, M.J.; Both, S.; Duan, X.; Kuzyk, A.; Weiss, T.; Liu, N. DNA-assembled nanoarchitectures with multiple components in regulated and coordinated motion. Sci. Adv. 2019, 5, eaax6023. [Google Scholar] [CrossRef] [Green Version]
- Marras, A.E.; Zhou, L.; Kolliopoulos, V.; Su, H.-J.; Castro, C.E. Directing folding pathways for multi-component DNA origami nanostructures with complex topology. New J. Phys. 2016, 18, 55005. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.A.; Dehankar, A.; Robbins, A.; Kabtiyal, P.; Jergens, E.; Ho Lee, K.; Johnston-Halperin, E.; Poirier, M.; Castro, C.E.; Winter, J.O. The path towards functional nanoparticle-DNA origami composites. Mater. Sci. Eng. R Rep. 2019, 138, 153–209. [Google Scholar] [CrossRef]
- Ahmadi, Y.; Nord, A.L.; Wilson, A.J.; Hütter, C.; Schroeder, F.; Beeby, M.; Barišić, I. The Brownian and Flow-Driven Rotational Dynamics of a Multicomponent DNA Origami-Based Rotor. Small 2020, 16, e2001855. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.J.; Skaletsky, E.; McPherson, A. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 1998, 275, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Converting caDNAno Design JSON File to All-Atom PDB File|The Aksimentiev Group. Available online: https://bionano.physics.illinois.edu/sites/default/files/smileyFace.json (accessed on 2 November 2021).
- Nagamura, R.; Fukuda, M.; Kawamoto, A.; Matoba, K.; Dohmae, N.; Ishitani, R.; Takagi, J.; Nureki, O. Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2. Acta Crystallogr. F Struct. Biol. Commun. 2019, 75, 348–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, A.N.-S.; Bradley, P.; Cernadas, R.A.; Bogdanove, A.J.; Stoddard, B.L. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 2012, 335, 716–719. [Google Scholar] [CrossRef] [Green Version]
- Xue, H.; Yao, T.; Cao, M.; Zhu, G.; Li, Y.; Yuan, G.; Chen, Y.; Lei, M.; Huang, J. Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature 2019, 573, 445–449. [Google Scholar] [CrossRef]
- Zuo, X.; Wang, J.; Foster, T.R.; Schwieters, C.D.; Tiede, D.M. Rigid-Body Refinement of the Tetraloop-Receptor RNA Complex. Available online: https://www.rcsb.org/structure/2JYH (accessed on 21 December 2021).
- Grau, F.C.; Jaeger, J.; Groher, F.; Suess, B.; Muller, Y.A. The complex formed between a synthetic RNA aptamer and the transcription repressor TetR is a structural and functional twin of the operator DNA-TetR regulator complex. Nucleic Acids Res. 2020, 48, 3366–3378. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuťák, D.; Poppleton, E.; Miao, H.; Šulc, P.; Barišić, I. Unified Nanotechnology Format: One Way to Store Them All. Molecules 2022, 27, 63. https://doi.org/10.3390/molecules27010063
Kuťák D, Poppleton E, Miao H, Šulc P, Barišić I. Unified Nanotechnology Format: One Way to Store Them All. Molecules. 2022; 27(1):63. https://doi.org/10.3390/molecules27010063
Chicago/Turabian StyleKuťák, David, Erik Poppleton, Haichao Miao, Petr Šulc, and Ivan Barišić. 2022. "Unified Nanotechnology Format: One Way to Store Them All" Molecules 27, no. 1: 63. https://doi.org/10.3390/molecules27010063
APA StyleKuťák, D., Poppleton, E., Miao, H., Šulc, P., & Barišić, I. (2022). Unified Nanotechnology Format: One Way to Store Them All. Molecules, 27(1), 63. https://doi.org/10.3390/molecules27010063






