Chemical Profiling, Bioactivity Evaluation and the Discovery of a Novel Biopigment Produced by Penicillium purpurogenum CBS 113139
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Pre-Culture Media
2.2. Shake Flask Fermentations
2.3. Pigment Production at Optimum Conditions
2.4. pH Stability
2.5. Analytical Methods
2.6. Purification of Pigments
2.7. High-Resolution Mass Spectrometry (HRMS) Analysis
2.8. NMR Spectroscopy
2.9. Antioxidant Capacity
2.10. Cytotoxicity Assay
3. Results and Discussion
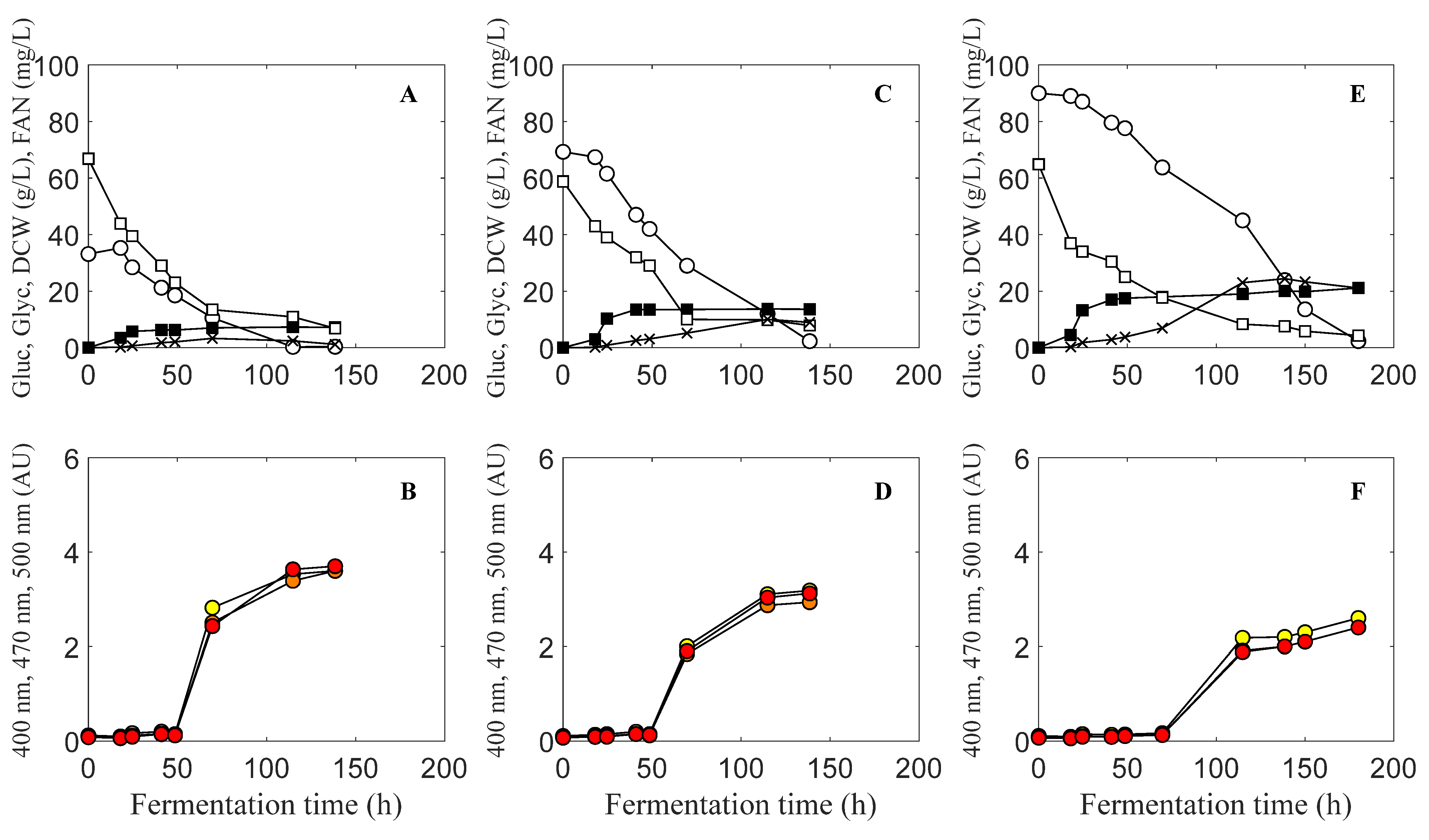
3.1. Optimization of Initial Glucose Concentration and Carbon to Nitrogen Ratio
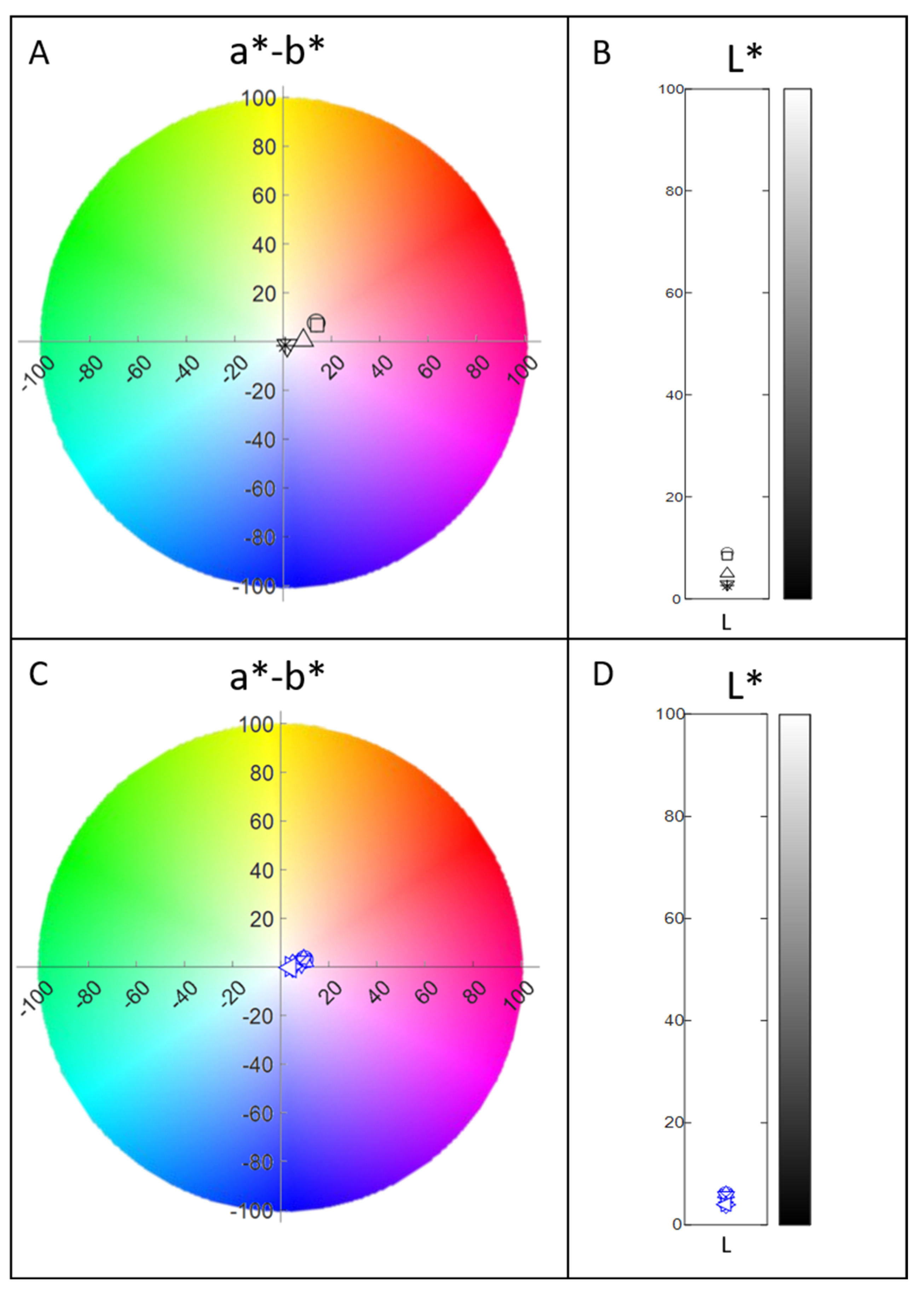
3.2. pH Stability
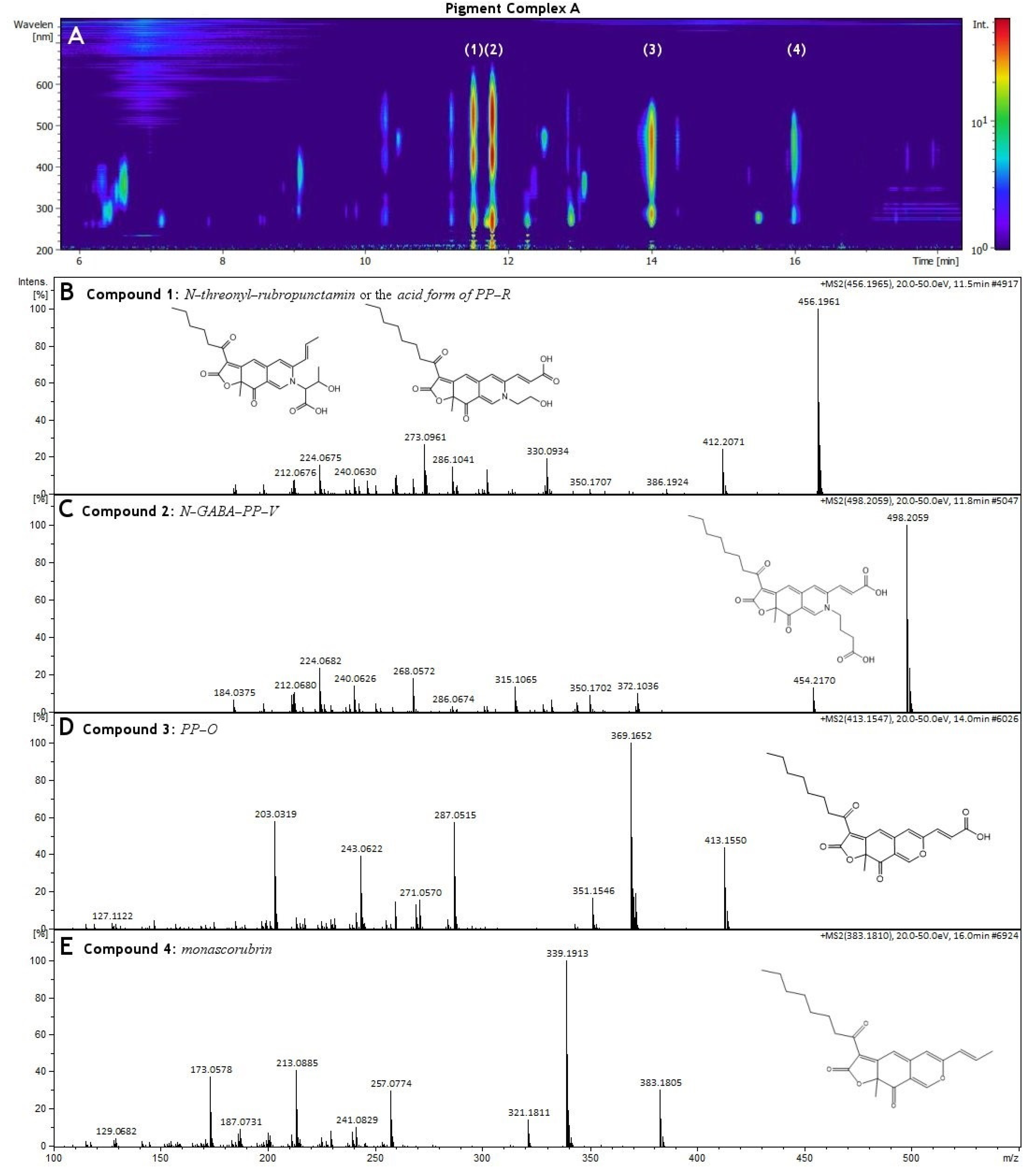
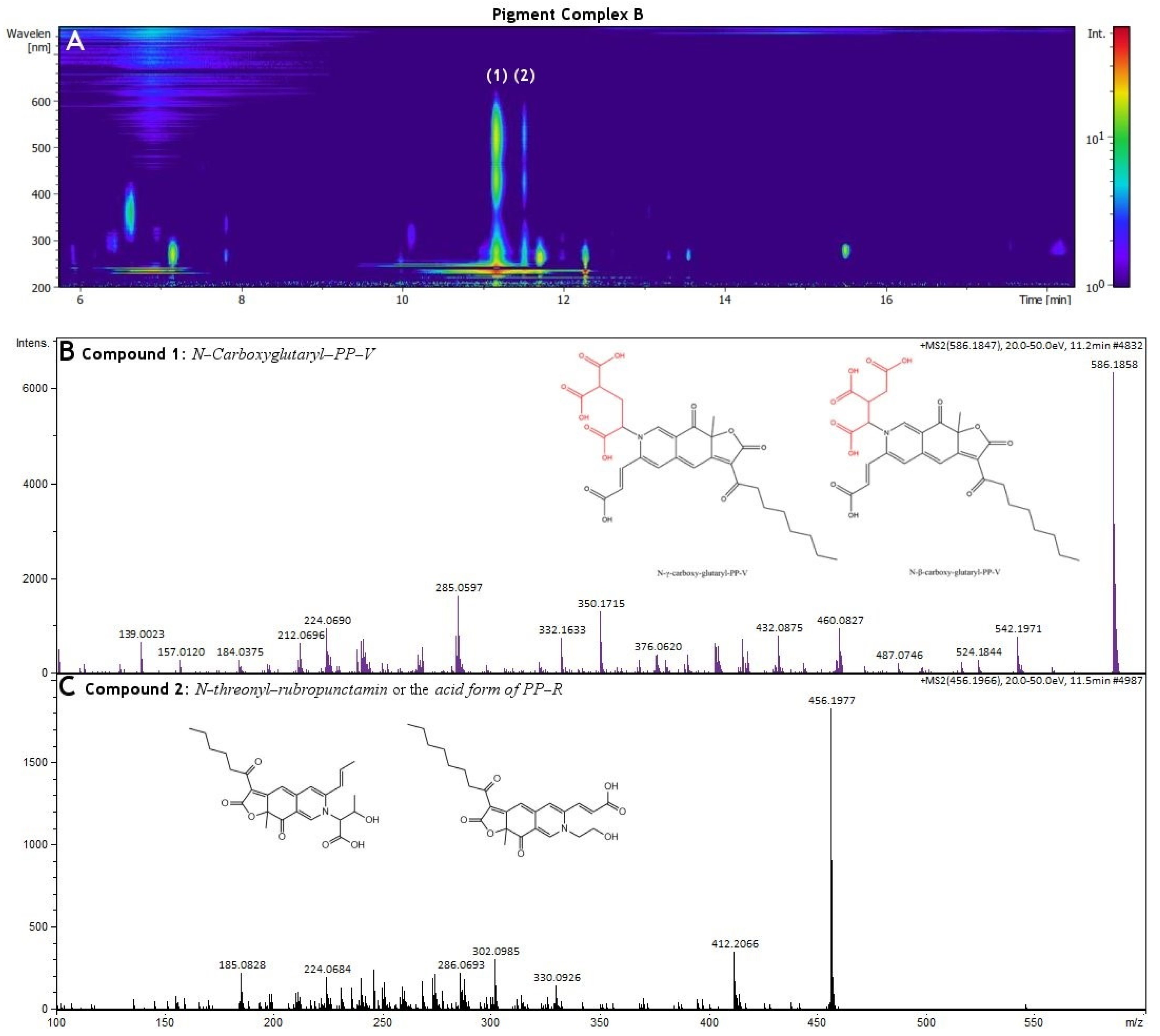
3.3. HR-MS and NMR Analysis of Biopigments
3.4. Antioxidant Capacity (DPPH Test)
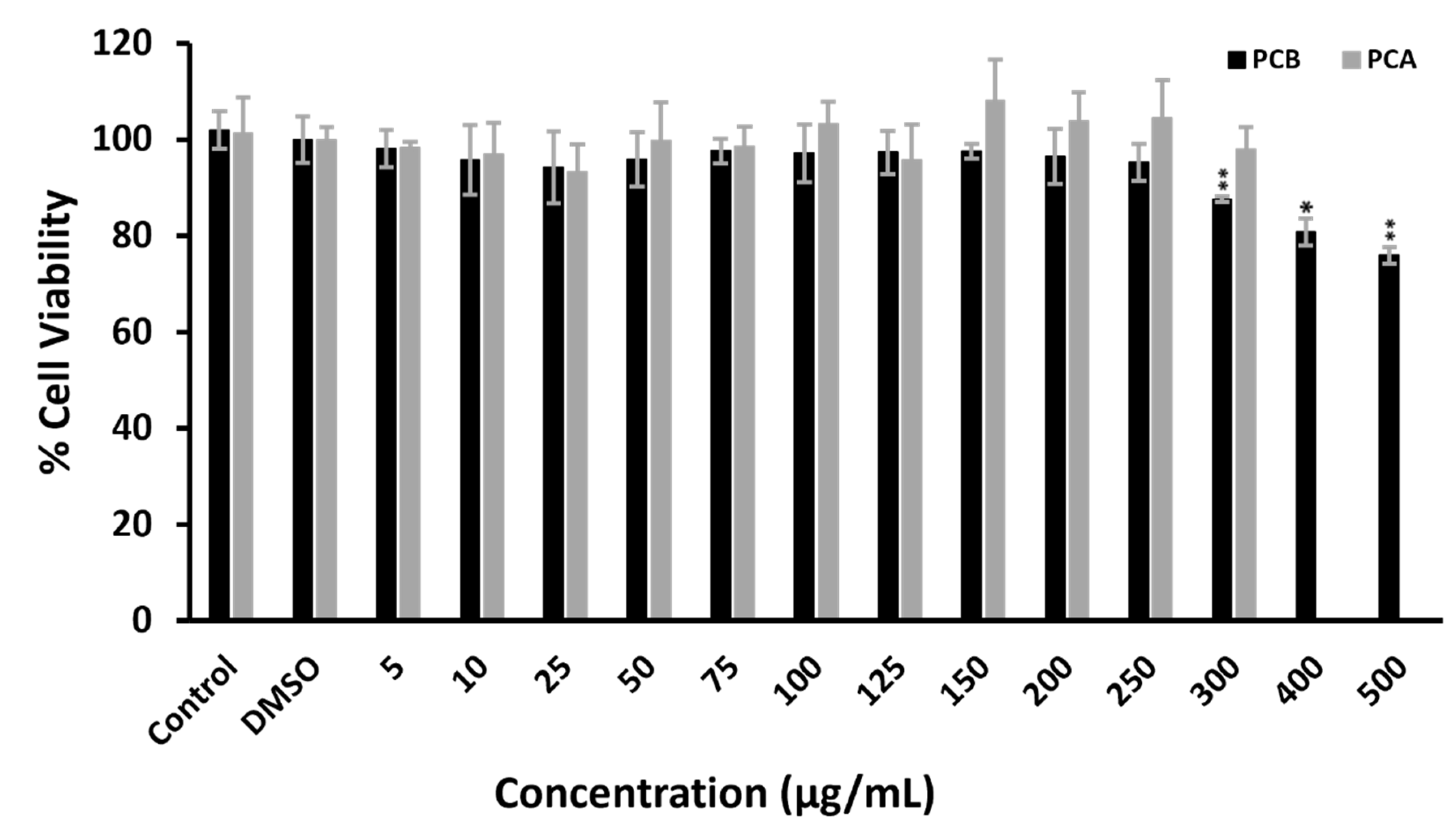
3.5. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Mapari, S.A.S.; Hansen, M.; Meyer, A.S.; Thrane, U. Computerized Screening for Novel Producers of Monascus-like Food Pigments in Penicillium Species. J. Agric. Food Chem. 2008, 56, 9981–9989. [Google Scholar] [CrossRef] [PubMed]
- Dufossé, L.; Fouillaud, M.; Caro, Y.; Mapari, S.A.; Sutthiwong, N. Filamentous fungi are large-scale producers of pigments and colorants for the food industry. Curr. Opin. Biotechnol. 2014, 26, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.G.; Puttananjaiah, M.H.; Peddha, M.S.; Dhale, M.A. Safety efficacy and chemical profiling of water-soluble Talaromyces purpureogenus CFRM02 pigment. Food Chem. 2020, 310, 125869. [Google Scholar] [CrossRef] [PubMed]
- Morales-Oyervides, L.; Oliveira, J.; Sousa-Gallagher, M.; Méndez-Zavala, A.; Montañez, J. Effect of heat exposure on the colour intensity of red pigments produced by Penicillium purpurogenum GH2. J. Food Eng. 2015, 164, 21–29. [Google Scholar] [CrossRef]
- Dufossé, L. Red colourants from filamentous fungi: Are they ready for the food industry? J. Food Compos. Anal. 2018, 69, 156–161. [Google Scholar] [CrossRef]
- Kantifedaki, A.; Kachrimanidou, V.; Mallouchos, A.; Papanikolaou, S.; Koutinas, A. Orange processing waste valorisation for the production of bio-based pigments using the fungal strains Monascus purpureus and Penicillium purpurogenum. J. Clean. Prod. 2018, 185, 882–890. [Google Scholar] [CrossRef] [Green Version]
- Frisvad, J.; Yilmaz, N.; Thrane, U.; Rasmussen, K.B.; Houbraken, J.; Samson, R.A. Talaromyces atroroseus, a New Species Efficiently Producing Industrially Relevant Red Pigments. PLoS ONE 2013, 8, e84102. [Google Scholar] [CrossRef] [Green Version]
- Venkatachalam, M.; Magalon, H.; Dufossé, L.; Fouillaud, M. Production of pigments from the tropical marine-derived fungi Talaromyces albobiverticillius: New resources for natural red-colored metabolites. J. Food Compos. Anal. 2018, 70, 35–48. [Google Scholar] [CrossRef]
- Pisareva, E.I.; Kujumdzieva, A.V. Influence of Carbon and Nitrogen Sources on Growth and Pigment Production by Monascus Pilosus C1Strain. Biotechnol. Biotechnol. Equip. 2010, 24, 501–506. [Google Scholar] [CrossRef]
- Yam, K.L.; Papadakis, S.E. A simple digital imaging method for measuring and analyzing color of food surfaces. J. Food Eng. 2004, 61, 137–142. [Google Scholar] [CrossRef]
- Lie, S. The Ebc-Ninhydrin Method for Determination of Free Alpha Amino Nitrogen. J. Inst. Brew. 1973, 79, 37–41. [Google Scholar] [CrossRef]
- Firuzi, O.; Miri, R.; Asadollahi, M.; Eslami, S.; Jassbi, A.R. Cytotoxic, Antioxidant and Antimicrobial Activities and Phenolic Contents of Eleven Salvia Species from Iran. Iran. J. Pharm. Res 2013, 12, 801–810. [Google Scholar] [PubMed]
- Bougouffa, S.; Radovanovic, A.; Essack, M.; Bajic, V.B. DEOP: A database on osmoprotectants and associated pathways. Database 2014, 2014, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Osmotic Stress Signaling and Osmoadaptation in Yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caro, Y.; Anamale, L.; Fouillaud, M.; Laurent, P.; Petit, T.; Dufossé, L. Natural hydroxyanthraquinoid pigments as potent food grade colorants: An overview. Nat. Prod. Bioprospect. 2012, 2, 174–193. [Google Scholar] [CrossRef]
- Said, F.B.M.; Brooks, J.; Chisti, Y. Optimal C:N ratio for the production of red pigments by Monascus ruber. World J. Microbiol. Biotechnol. 2014, 30, 2471–2479. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.-B.; Eksriwong, T.; Chauvatcharin, S.; Zhong, J.-J. Optimization of carbon source and carbon/nitrogen ratio for cordycepin production by submerged cultivation of medicinal mushroom Cordyceps militaris. Process. Biochem. 2005, 40, 1667–1672. [Google Scholar] [CrossRef]
- Lebeau, J.; Petit, T.; Fouillaud, M.; Dufossé, L.; Caro, Y. Alternative Extraction and Characterization of Nitrogen-Containing Azaphilone Red Pigments and Ergosterol Derivatives from the Marine-Derived Fungal Talaromyces sp. 30570 Strain with Industrial Relevance. Microorganisms 2020, 8, 1920. [Google Scholar] [CrossRef]
- Pimenta, L.; Gomes, D.; Cardoso, P.; Takahashi, J. Recent Findings in Azaphilone Pigments. J. Fungi 2021, 7, 541. [Google Scholar] [CrossRef]
- Morales-Oyervides, L.; Ruiz-Sánchez, J.P.; Oliveira, J.C.; Sousa-Gallagher, M.J.; Zavala, A.M.; Giuffrida, D.; Dufossé, L.; Montañez, J. Biotechnological approaches for the production of natural colorants by Talaromyces/Penicillium: A review. Biotechnol. Adv. 2020, 43, 107601. [Google Scholar] [CrossRef]
- Koli, S.H.; Suryawanshi, R.K.; Mohite, B.V.; Patil, S.V. Prospective of Monascus Pigments as an Additive to Commercial Sunscreens. Nat. Prod. Commun. 2019, 14, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Hsu, Y.-W.; Hsu, L.-C.; Liang, Y.-H.; Kuo, Y.-H.; Pan, T.-M. Monaphilones A−C, Three New Antiproliferative Azaphilone Derivatives from Monascus purpureus NTU 568. J. Agric. Food Chem. 2010, 58, 8211–8216. [Google Scholar] [CrossRef] [PubMed]
- Su, N.-W.; Lin, Y.-L.; Lee, M.-H.; Ho, C.-Y. Ankaflavin from Monascus-Fermented Red Rice Exhibits Selective Cytotoxic Effect and Induces Cell Death on Hep G2 Cells. J. Agric. Food Chem. 2005, 53, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Akihisa, T.; Tokuda, H.; Yasukawa, K.; Ukiya, M.; Kiyota, A.; Sakamoto, N.; Suzuki, T.; Tanabe, A.N.; Nishino, H. Azaphilones, Furanoisophthalides, and Amino Acids from the Extracts of Monascus pilosus—Fermented Rice (Red-Mold Rice) and Their Chemopreventive Effects. J. Agric. Food Chem. 2005, 53, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.-C.; Liao, V.H.-C.; Pan, T.-M. Monascin from red mold dioscorea as a novel antidiabetic and antioxidative stress agent in rats and Caenorhabditis elegans. Free Radic. Biol. Med. 2012, 52, 109–117. [Google Scholar] [CrossRef]
 ). Absorbance at 470 nm (
). Absorbance at 470 nm (  ). Absorbance at 500 nm (
). Absorbance at 500 nm (  ).
).
 ). Absorbance at 470 nm (
). Absorbance at 470 nm (  ). Absorbance at 500 nm (
). Absorbance at 500 nm (  ).
).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsiailanis, A.D.; Pateraki, C.; Kyriazou, M.; Chatzigiannis, C.M.; Chatziathanasiadou, M.; Parisis, N.; Mandala, I.; Tzakos, A.G.; Koutinas, A. Chemical Profiling, Bioactivity Evaluation and the Discovery of a Novel Biopigment Produced by Penicillium purpurogenum CBS 113139. Molecules 2022, 27, 69. https://doi.org/10.3390/molecules27010069
Tsiailanis AD, Pateraki C, Kyriazou M, Chatzigiannis CM, Chatziathanasiadou M, Parisis N, Mandala I, Tzakos AG, Koutinas A. Chemical Profiling, Bioactivity Evaluation and the Discovery of a Novel Biopigment Produced by Penicillium purpurogenum CBS 113139. Molecules. 2022; 27(1):69. https://doi.org/10.3390/molecules27010069
Chicago/Turabian StyleTsiailanis, Antonis D., Chrysanthi Pateraki, Mary Kyriazou, Christos M. Chatzigiannis, Maria Chatziathanasiadou, Nikolaos Parisis, Ioanna Mandala, Andreas G. Tzakos, and Apostolis Koutinas. 2022. "Chemical Profiling, Bioactivity Evaluation and the Discovery of a Novel Biopigment Produced by Penicillium purpurogenum CBS 113139" Molecules 27, no. 1: 69. https://doi.org/10.3390/molecules27010069
APA StyleTsiailanis, A. D., Pateraki, C., Kyriazou, M., Chatzigiannis, C. M., Chatziathanasiadou, M., Parisis, N., Mandala, I., Tzakos, A. G., & Koutinas, A. (2022). Chemical Profiling, Bioactivity Evaluation and the Discovery of a Novel Biopigment Produced by Penicillium purpurogenum CBS 113139. Molecules, 27(1), 69. https://doi.org/10.3390/molecules27010069







