Synthetic Perturbations in IL6 Biological Circuit Induces Dynamical Cellular Response
Abstract
:1. Introduction
2. Results
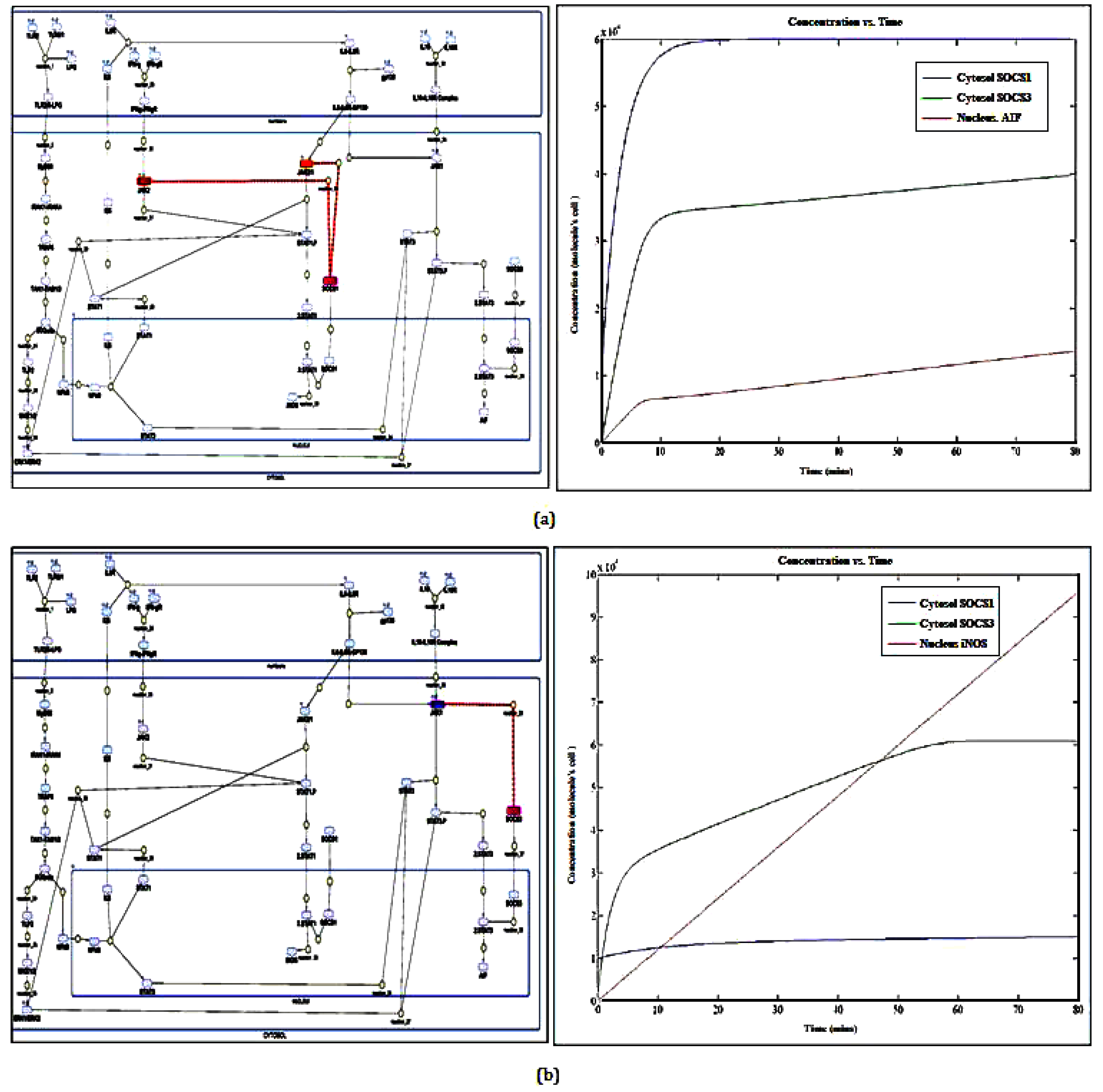
2.1. SOCS1/SOCS3 Differential Expression Governing Macrophage Polarization
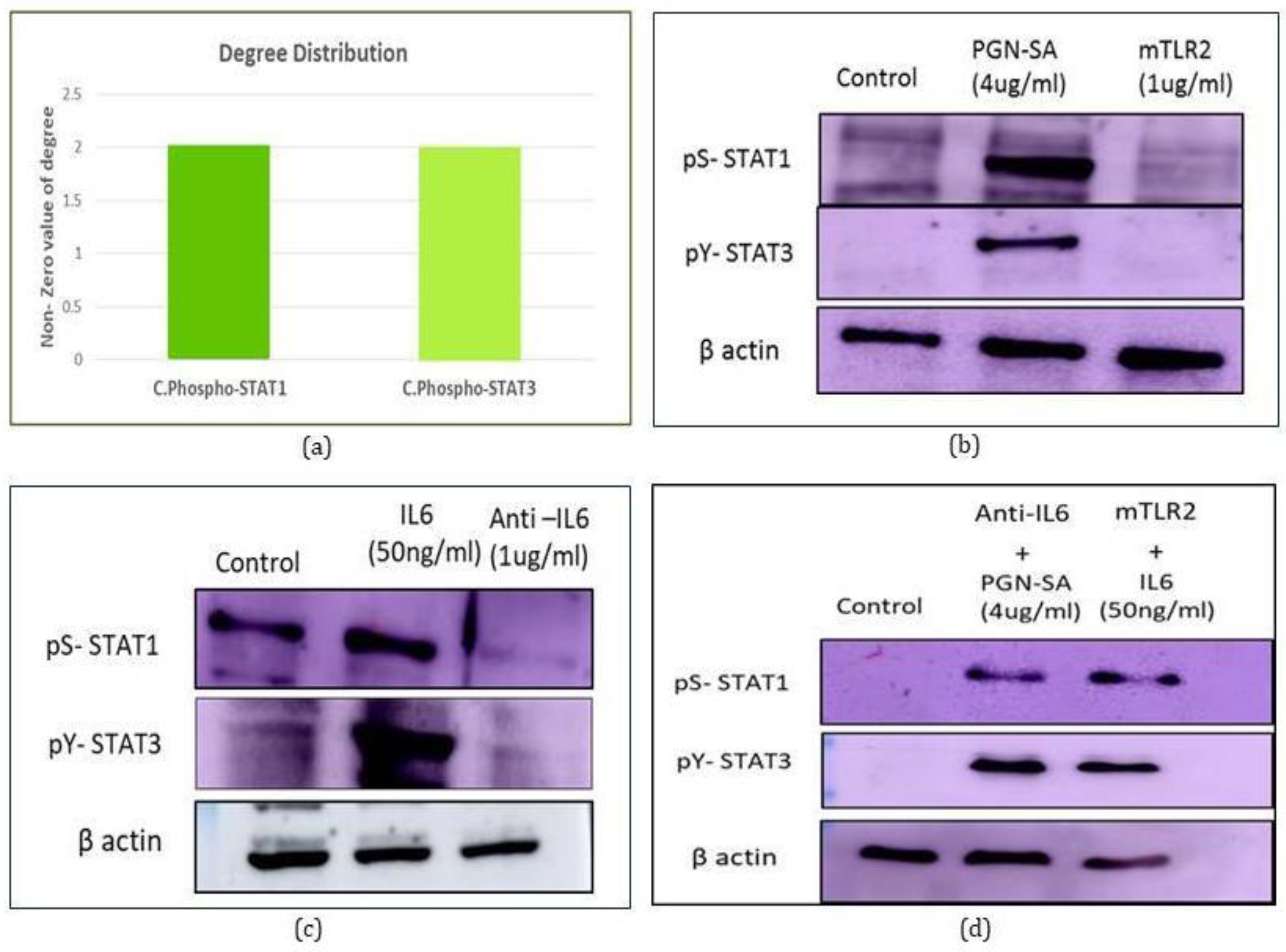
2.2. Systems Study Reveals Phosphorylated STAT1 and STAT3 as Cross Talk
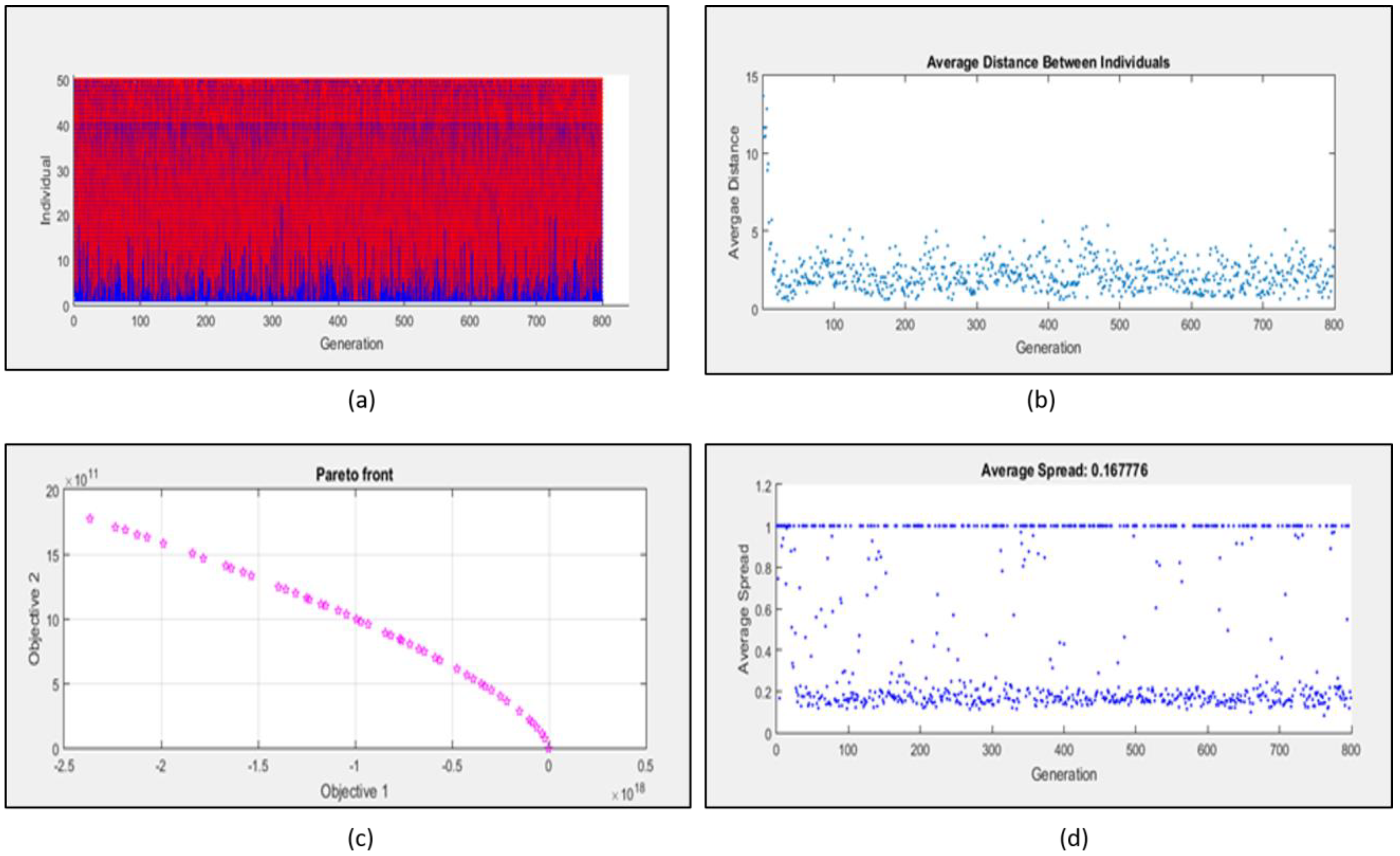
2.3. Multi Objective Optimization of Mathematical Model
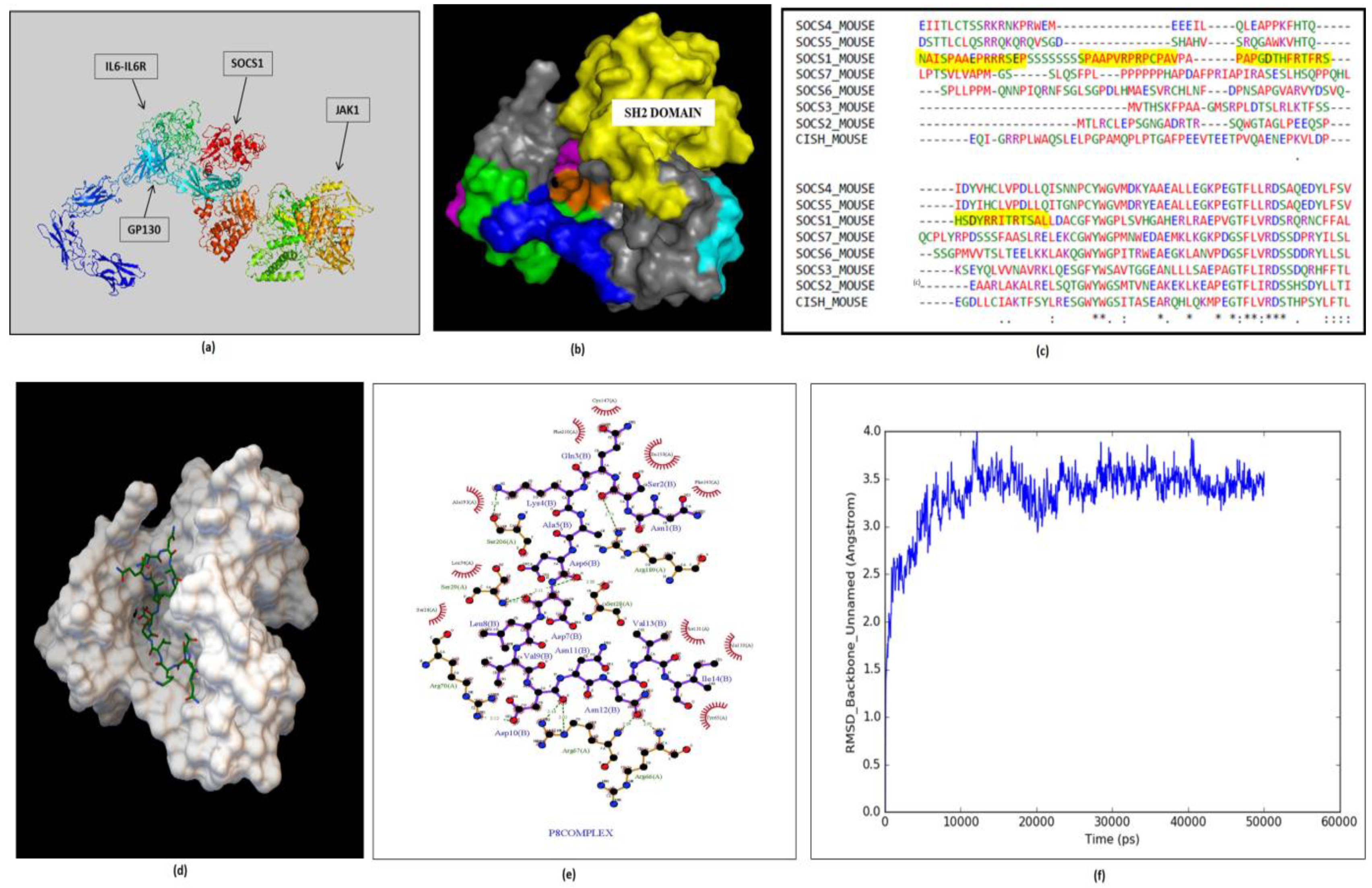
2.4. SOCS1 as a Target for Therapeutic Intervention
2.5. Peptide Design, Docking and MD Simulation of Selected Complex
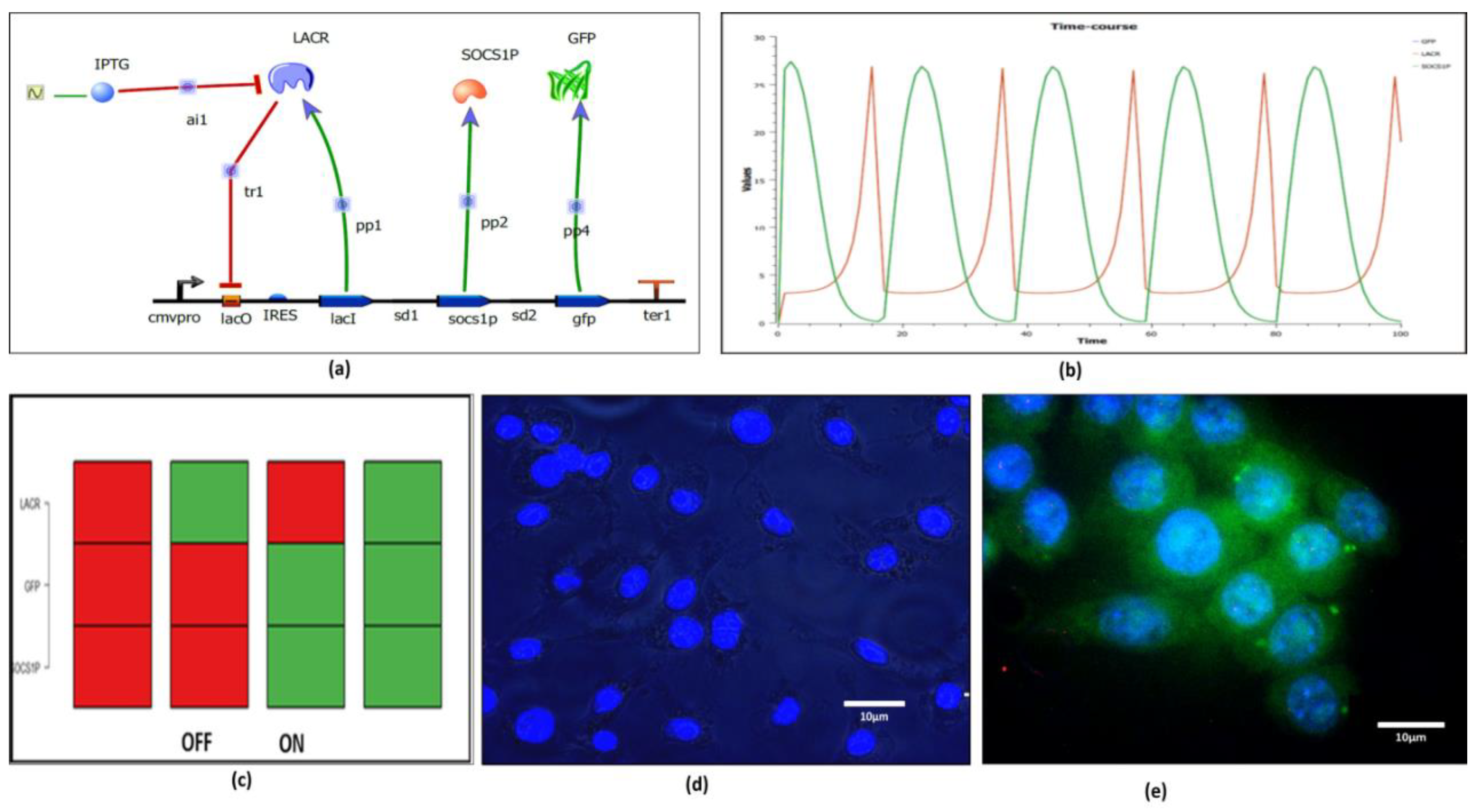
2.6. Systems Driven Synthetic Biological Circuit Design
2.7. In Vitro Validation
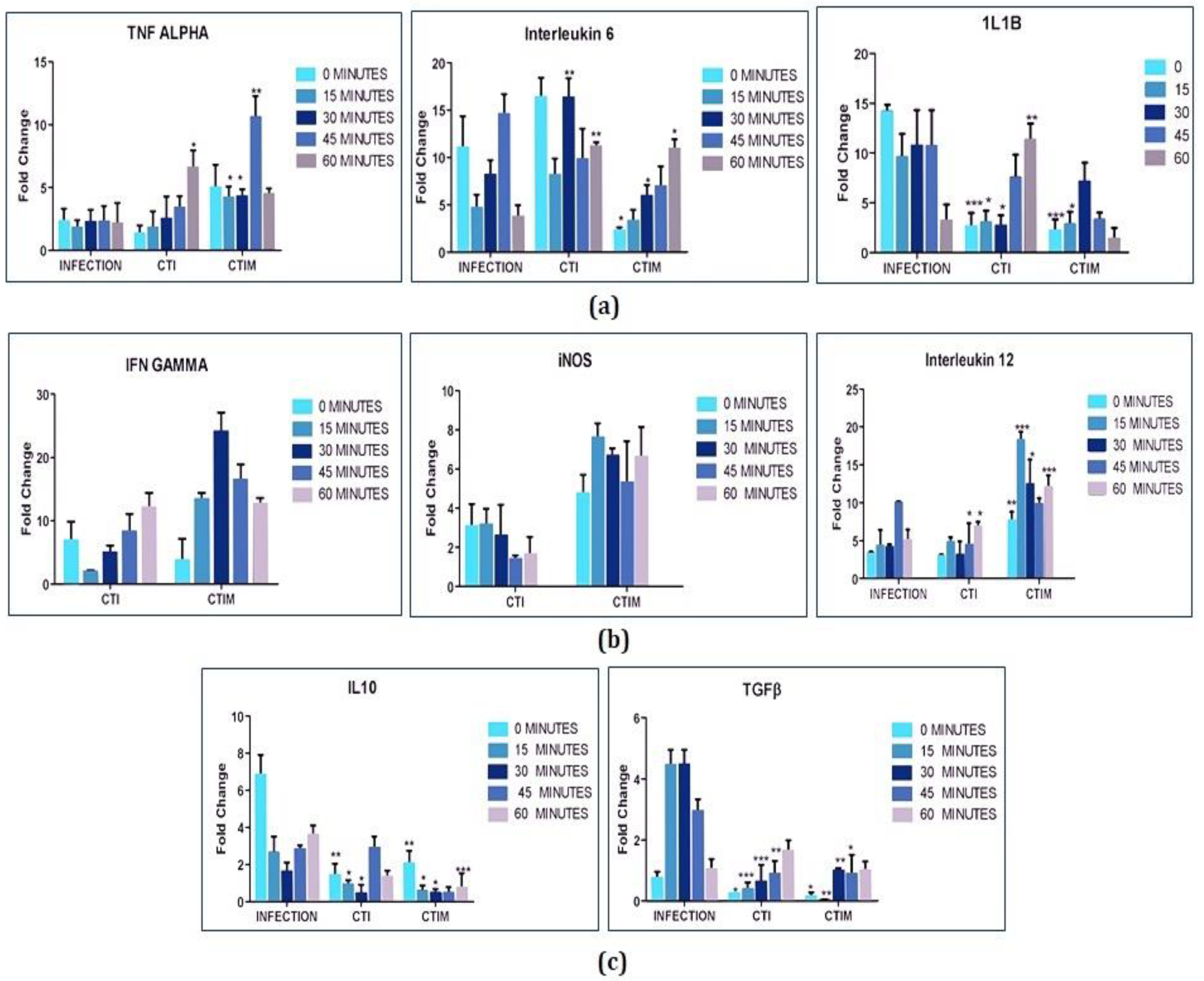
2.7.1. Cytokine Profiling
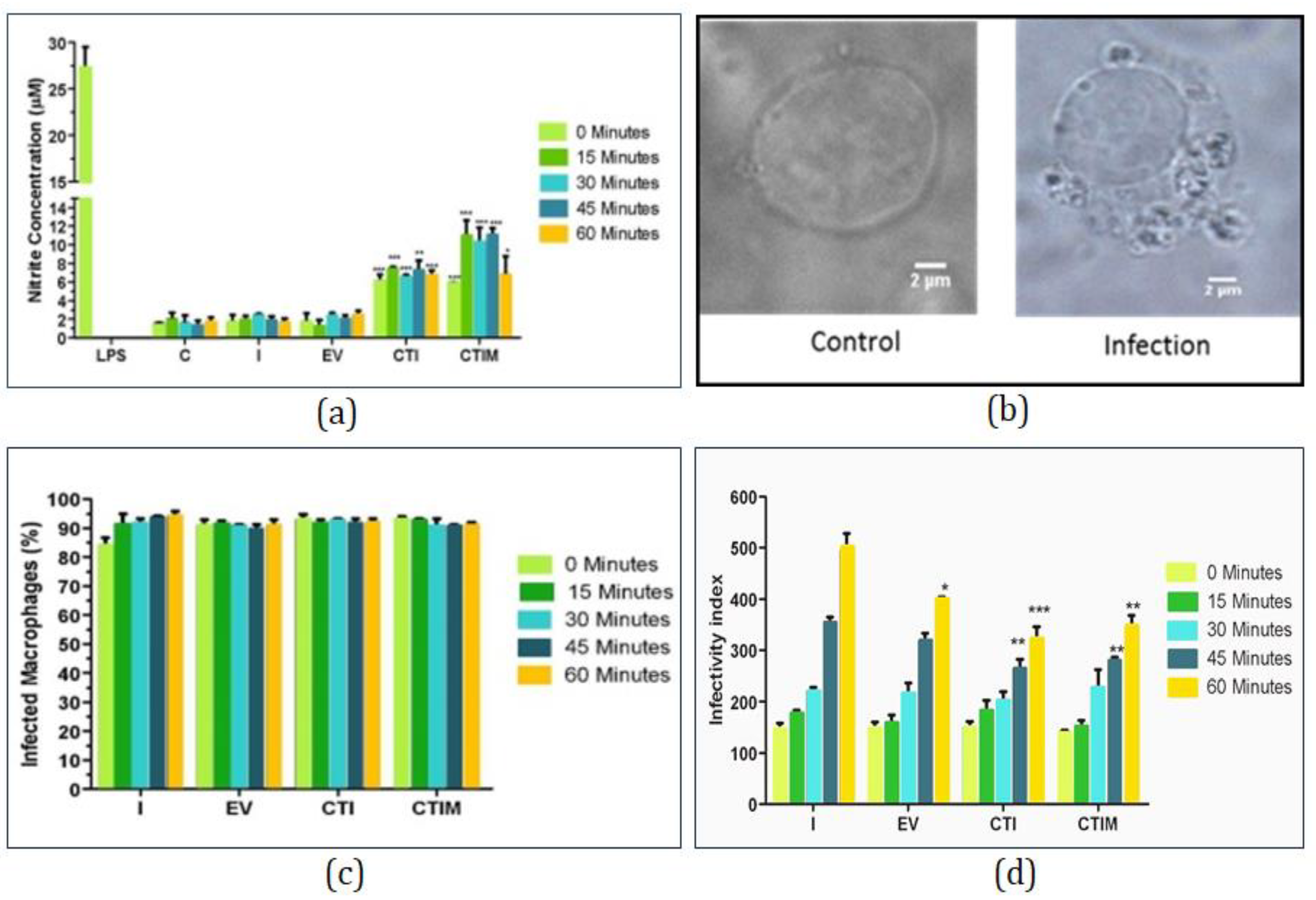
2.7.2. Nitrite Estimation
2.7.3. Parasite Load Assay
3. Discussion
4. Materials and Methods
4.1. In Silico
4.1.1. Reconstruction and Analysis of IL6 Mathematical Model
4.1.2. Multi Objective Optimization and Evolvability
4.1.3. Target Identification and Protein-Protein Docking
4.1.4. Peptide Design, Docking and MD Simulations
4.1.5. Synthetic Circuit Design and Quasipotential Landscape
4.2. In Vitro
4.2.1. Reagents, Antibodies, Probes and Constructs
4.2.2. Macrophage and Parasite Infection
4.2.3. Transfection of Macrophages
4.2.4. mRNA Isolation, RT PCR and Real Time PCR
4.2.5. Western Blotting
Cross Talk Validation
SOCS1/SOCS3 Validation
4.2.6. Parasite Load Assay
4.2.7. Estimation of NO Production
4.2.8. Animal Maintenance
4.2.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIF | Anti-inflammatory factor |
| CMV | Cytomegalovirus |
| CL | Cutaneous leishmaniasis |
| CT | Comparative threshold |
| CTI | Control + Transfected + Infected |
| CTIM | Control + Transfected + Infected + Miltefosine |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DSM | Diseased State Model |
| EDTA | Ethylenediaminetetraacetic acid |
| EV | Empty Vector |
| GFP | Green Fluorescent Protein |
| HSM | Healthy State Model |
| IFNγ | Interferon gamma |
| IPTG | Isopropyl β-d-1-thiogalactopyranoside |
| iGEM | International Genetically Engineered Machine |
| iNOS | Inducible nitric oxide synthase |
| IL6 | Interleukin 6 |
| IL1β | Interleukin 1 beta |
| IL10 | Interleukin 10 |
| JAK/STAT | Janus kinase/signal transducer and activator of transcription |
| LACR | Lactose Repressor |
| LPS | Lipopolysaccharide |
| MOGA | Multi-objective genetic algorithm |
| ODE | Ordinary differential equation |
| PBC | Periodic boundary condition |
| PCA | Principal component analysis |
| PFA | Paraformaldehyde |
| PGN-SA | Peptidoglycan from Staphylococcus aureus |
| PEI | Polyethylenimine |
| RPMI | Roswell Park Memorial Institute |
| RMSD | Root-mean-square deviation |
| RMSF | Root mean square fluctuation |
| SH2 | Src Homology 2 |
| SOCS | Suppressor Of Cytokine Signaling |
| TIP3P | Transferable intermolecular potential with 3-point |
| TLR | Toll-like Receptors |
References
- Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 2 May 2021).
- Lainson, R. The Neotropical Leishmania species: A brief historical review of their discovery, ecology and taxonomy. Rev. Pan-Amazônica Saúde 2010, 1. [Google Scholar] [CrossRef]
- de Veer, M.J.; Curtis, J.M.; Baldwin, T.M.; DiDonato, J.A.; Sexton, A.; McConville, M.J.; Handman, E.; Schofield, L. MyD88 is essential for clearance of Leishmania major: Possible role for lipophosphoglycan and Toll-like receptor 2 signaling. Eur. J. Immunol. 2003, 33, 2822–2831. [Google Scholar] [CrossRef]
- Castellano, L.R.; Filho, D.C.; Argiro, L.; Dessein, H.; Prata, A.; Dessein, A.; Rodrigues, V. Th1/Th2 immune responses are associated with active cutaneous leishmaniasis and clinical cure is associated with strong interferon-γ production. Hum. Immunol. 2009, 70, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Tomiotto-Pellissier, F.; Bortoleti, B.T.D.S.; Assolini, J.P.; Gonçalves, M.D.; Carloto, A.C.M.; Miranda-Sapla, M.M.; Conchon-Costa, I.; Bordignon, J.; Pavanelli, W.R. Macrophage Polarization in Leishmaniasis: Broadening Horizons. Front. Immunol. 2018, 9, 2529. [Google Scholar] [CrossRef] [Green Version]
- Heyneman, D. Immunology of leishmaniasis. Bull. World Health Organ. 1971, 44, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Singh, S. Immunobiology of leishmaniasis. Indian J. Exp. Biol. 2009, 47, 412–423. [Google Scholar] [PubMed]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 2011, 1813, 878–888. [Google Scholar] [CrossRef] [Green Version]
- Fernando, M.R.; Reyes, J.L.; Iannuzzi, J.; Leung, G.; McKay, D.M. The pro-inflammatory cytokine, interleukin-6, enhances the polarization of alternatively activated macrophages. PLoS ONE 2014, 9, e94188. [Google Scholar] [CrossRef]
- Wilson, H.M. SOCS Proteins in Macrophage Polarization and Function. Front. Immunol. 2014, 5, 357. [Google Scholar] [CrossRef] [Green Version]
- Soni, B.; Saha, B.; Singh, S. Systems cues governing IL6 signaling in leishmaniasis. Cytokine 2018, 106, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Whyte, C.S.; Bishop, E.T.; Rückerl, D.; Gaspar-Pereira, S.; Barker, R.N.; Allen, J.E.; Rees, A.J.; Wilson, H.M. Suppressor of cytokine signaling (SOCS)1 is a key determinant of differential macrophage activation and function. J. Leukoc. Biol. 2011, 90, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Ryo, A.; Tsurutani, N.; Ohba, K.; Kimura, R.; Komano, J.; Nishi, M.; Soeda, H.; Hattori, S.; Perrem, K.; Yamamoto, M.; et al. SOCS1 is an inducible host factor during HIV-1 infection and regulates the intracellular trafficking and stability of HIV-1 Gag. Proc. Natl. Acad. Sci. USA 2008, 105, 294–299. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, C.M.; Dabelic, R.; Bedoya, S.K.; Larkin, J.; Johnson, H.M. A SOCS1/3 Antagonist Peptide Protects Mice Against Lethal Infection with Influenza A Virus. Front. Immunol. 2015, 6, 574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrakar, P.; Parmar, N.; Descoteaux, A.; Kar, S. Differential Induction of SOCS Isoforms by Leishmania donovani Impairs Macrophage–T Cell Cross-Talk and Host Defense. J. Immunol. 2020, 204, 596–610. [Google Scholar] [CrossRef]
- Dittrich, A.; Quaiser, T.; Khouri, C.; Görtz, D.; Mönnigmann, M.; Schaper, F. Model-driven experimental analysis of the function of SHP-2 in IL-6-induced Jak/STAT signaling. Mol. Biosyst. 2012, 8, 2119–2134. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ebrahim, A.; Lloyd, C.J.; Saunders, M.A.; Palsson, B.O. DynamicME: Dynamic simulation and refinement of integrated models of metabolism and protein expression. BMC Syst. Biol. 2019, 13, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahl, S.K.; Kremling, A. A Comparison of Deterministic and Stochastic Modeling Approaches for Biochemical Reaction Systems: On Fixed Points, Means, and Modes. Front. Genet. 2016, 7, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smole, A.; Lainšček, D.; Bezeljak, U.; Horvat, S.; Jerala, R. A Synthetic Mammalian Therapeutic Gene Circuit for Sensing and Suppressing Inflammation. Mol. Ther. 2017, 25, 102–119. [Google Scholar] [CrossRef] [Green Version]
- Ye, H.; Xie, M.; Xue, S.; Hamri, G.C.E.; Yin, J.; Zulewski, H.; Fussenegger, M. Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat. Biomed. Eng. 2017, 1, 0005. [Google Scholar] [CrossRef]
- Kis, Z.; Pereira, H.S.; Homma, T.; Pedrigi, R.M.; Krams, R. Mammalian synthetic biology: Emerging medical applications. J. R. Soc. Interface 2015, 12, 20141000. [Google Scholar] [CrossRef] [Green Version]
- Ye, H.; Fussenegger, M. Synthetic therapeutic gene circuits in mammalian cells. FEBS Lett. 2014, 588, 2537–2544. [Google Scholar] [CrossRef] [PubMed]
- Wroblewska, L.; Kitada, T.; Endo, K.; Siciliano, V.; Stillo, B.; Saito, H.; Weiss, R. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015, 33, 839–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, N.; Ninfa, A.J. Synthetic networks: Oscillators and toggle switches for escherichia coli. Methods Mol. Biol. 2012, 813, 287–300. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Leibier, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Hishitani, Y.; Ogata, A. Monoclonal antibodies in rheumatoid arthritis: Comparative effectiveness of tocilizumab with tumor necrosis factor inhibitors. Biol. Targets Ther. 2014, 8, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsume, A.; Saito, H.; Yamada, Y.; Yorozu, K.; Ueda, O.; Akamatsu, K.-I.; Nishimoto, N.; Kishimoto, T.; Yoshizaki, K.; Ohsugi, Y. Anti-interleukin 6 (IL-6) receptor antibody suppresses Castleman’s disease like symptoms emerged in IL-6 transgenic mice. Cytokine 2002, 20, 304–311. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Ogata, A.; Kishimoto, T. A new era for the treatment of inflammatory autoimmune diseases by interleukin-6 blockade strategy. Semin. Immunol. 2014, 26, 88–96. [Google Scholar] [CrossRef]
- Ulhaq, Z.S.; Soraya, G.V. Interleukin-6 as a potential biomarker of COVID-19 progression. Med. Mal. Infect. 2020, 50, 382–383. [Google Scholar] [CrossRef]
- Wang, C.; Fei, D.; Li, X.; Zhao, M.; Yu, K. IL-6 may be a good biomarker for earlier detection of COVID-19 progression. Intensive Care Med. 2020, 46, 1475–1476. [Google Scholar] [CrossRef]
- Nct Anti-il6 Treatment of Serious COVID-19 Disease with Threatening Respiratory Failure. 2020. Available online: https://clinicaltrials.gov/show/NCT04322773 (accessed on 27 May 2021).
- Biran, N.; Ip, A.; Ahn, J.; Go, R.C.; Wang, S.; Mathura, S.; Sinclaire, B.A.; Bednarz, U.; Marafelias, M.; Hansen, E.; et al. Tocilizumab among patients with COVID-19 in the intensive care unit: A multicentre observational study. Lancet Rheumatol. 2020, 2, e603–e612. [Google Scholar] [CrossRef]
- Xu, X.; Qi, L.S. A CRISPR–dCas Toolbox for Genetic Engineering and Synthetic Biology. J. Mol. Biol. 2019, 431, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, M.; Moore, R.T.; Rajamani, S.; Panchal, R.G. Bacterial genome engineering and synthetic biology: Combating pathogens. BMC Microbiol. 2016, 16, 258. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Batra, H.; Dong, J.; Chen, C.; Rao, V.B.; Tao, P. Genetic Engineering of Bacteriophages Against Infectious Diseases. Front. Microbiol. 2019, 10, 954. [Google Scholar] [CrossRef]
- Contreras-Llano, L.E.; Tan, C. High-throughput screening of biomolecules using cell-free gene expression systems. Synth. Biol. (Oxf. Engl.) 2018, 3, ysy012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.; Shin, J.-Y.; Leijten, J.; Jeon, O.; Camci-Unal, G.; Dikina, A.D.; Brinegar, K.; Ghaemmaghami, A.M.; Alsberg, E.; Khademhosseini, A. High-throughput approaches for screening and analysis of cell behaviors. Biomaterials 2018, 153, 85–101. [Google Scholar] [CrossRef]
- Courbet, A.; Renard, E.; Molina, F. Bringing next-generation diagnostics to the clinic through synthetic biology. EMBO Mol. Med. 2016, 8, 987–991. [Google Scholar] [CrossRef] [Green Version]
- Slomovic, S.; Pardee, K.; Collins, J.J. Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl. Acad. Sci. USA 2015, 112, 14429–14435. [Google Scholar] [CrossRef] [Green Version]
- Geraldi, A.; Giri-Rachman, E.A. Synthetic biology-based portable in vitro diagnostic platforms. Alex. J. Med. 2018, 54, 423–428. [Google Scholar] [CrossRef] [Green Version]
- Trosset, J.-Y.; Carbonell, P. Synthetic biology for pharmaceutical drug discovery. Drug Des. Dev. Ther. 2015, 9, 6285–6302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunes, S.F.; Hamers, C.; Ratinier, M.; Shaw, A.; Brunet, S.; Hudelet, P.; Palmarini, M. A Synthetic Biology Approach for a Vaccine Platform against Known and Newly Emerging Serotypes of Bluetongue Virus. J. Virol. 2014, 88, 12222–12232. [Google Scholar] [CrossRef] [Green Version]
- Deb, K.; Pratap, A.; Agarwal, S.; Meyarivan, T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 2002, 6, 182–197. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Singh, H.; Tuknait, A.; Chaudhary, K.; Singh, B.; Kumaran, S.; Raghava, G.P.S. PEPstrMOD: Structure prediction of peptides containing natural, non-natural and modified residues. Biol. Direct 2015, 10, 73. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Arthur, J. Olson AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. Ligplot: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Bowers, K.J.; Sacerdoti, F.D.; Salmon, J.K.; Shan, Y.; Shaw, D.E.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the SC ′06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 1–17 November 2006; p. 84.
- Chandran, D.; Bergmann, F.T.; Sauro, H.M. TinkerCell: Modular CAD tool for synthetic biology. J. Biol. Eng. 2009, 3, 19. [Google Scholar] [CrossRef]
- Müssel, C.; Hopfensitz, M.; Kestler, H.A. BoolNet-an R package for generation, reconstruction and analysis of Boolean networks. Bioinformatics 2010, 26, 1378–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrissey, E.R.; Juárez, M.A.; Denby, K.J.; Burroughs, N.J.; Ideker, T. On reverse engineering of gene interaction networks using time course data with repeated measurements. Bioinformatics 2011, 26, 2305–2312. [Google Scholar] [CrossRef]
- Morrissey, E. GRENITS: Gene Regulatory Network Inference Using Time Series (R Package Version 1.24.0). Bioinformatics 2012, 1, 1–5. [Google Scholar]
- Bhattacharya, S.; Zhang, Q.; Andersen, M.E. A deterministic map of Waddington’s epigenetic landscape for cell fate specification. BMC Syst. Biol. 2011, 5, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kébaïer, C.; Louzir, H.; Chenik, M.; Ben Salah, A.; Dellagi, K. Heterogeneity of wild Leishmania major isolates in experimental murine pathogenicity and specific immune response. Infect. Immun. 2001, 69, 4906–4915. [Google Scholar] [CrossRef] [Green Version]

| S.No. | Reactions | Flux (Molecules/s) | PCA Score |
|---|---|---|---|
| 1. | TLR2/6-LPG -> MyD88 | 207.4748058 | 0.95 |
| 2. | JAK1 + STAT3{CYTOSOL} -> STAT3.P | 666.8136585 | 0.73 |
| 3. | SOCS1{NUCLEUS} -> SOCS1{CYTOSOL} | 15,045.06 | 0.9 |
| 4. | SOCS3{NUCLEUS} -> SOCS3{CYTOSOL} | 4999.92 | 0.81 |
| S.No. | Biological Parts | Accession ID |
|---|---|---|
| 1. | CMV promoter + RBS | BBa_I712004 |
| 2. | LacR | BBa_K731500 |
| 3. | GFP+ Terminator | BBa_K259006 |
| 4. | Spacer DNA | BBa_K1123011 |
| 5. | pcDNA 3.1 (Backbone) | BBa_K3030004 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soni, B.; Singh, S. Synthetic Perturbations in IL6 Biological Circuit Induces Dynamical Cellular Response. Molecules 2022, 27, 124. https://doi.org/10.3390/molecules27010124
Soni B, Singh S. Synthetic Perturbations in IL6 Biological Circuit Induces Dynamical Cellular Response. Molecules. 2022; 27(1):124. https://doi.org/10.3390/molecules27010124
Chicago/Turabian StyleSoni, Bhavnita, and Shailza Singh. 2022. "Synthetic Perturbations in IL6 Biological Circuit Induces Dynamical Cellular Response" Molecules 27, no. 1: 124. https://doi.org/10.3390/molecules27010124
APA StyleSoni, B., & Singh, S. (2022). Synthetic Perturbations in IL6 Biological Circuit Induces Dynamical Cellular Response. Molecules, 27(1), 124. https://doi.org/10.3390/molecules27010124






