NMR-Based Metabolomic Analyses to Identify the Effect of Harvesting Frequencies on the Leaf Metabolite Profile of a Moringa oleifera Cultivar Grown in an Open Hydroponic System
Abstract
1. Introduction
2. Results and Discussion
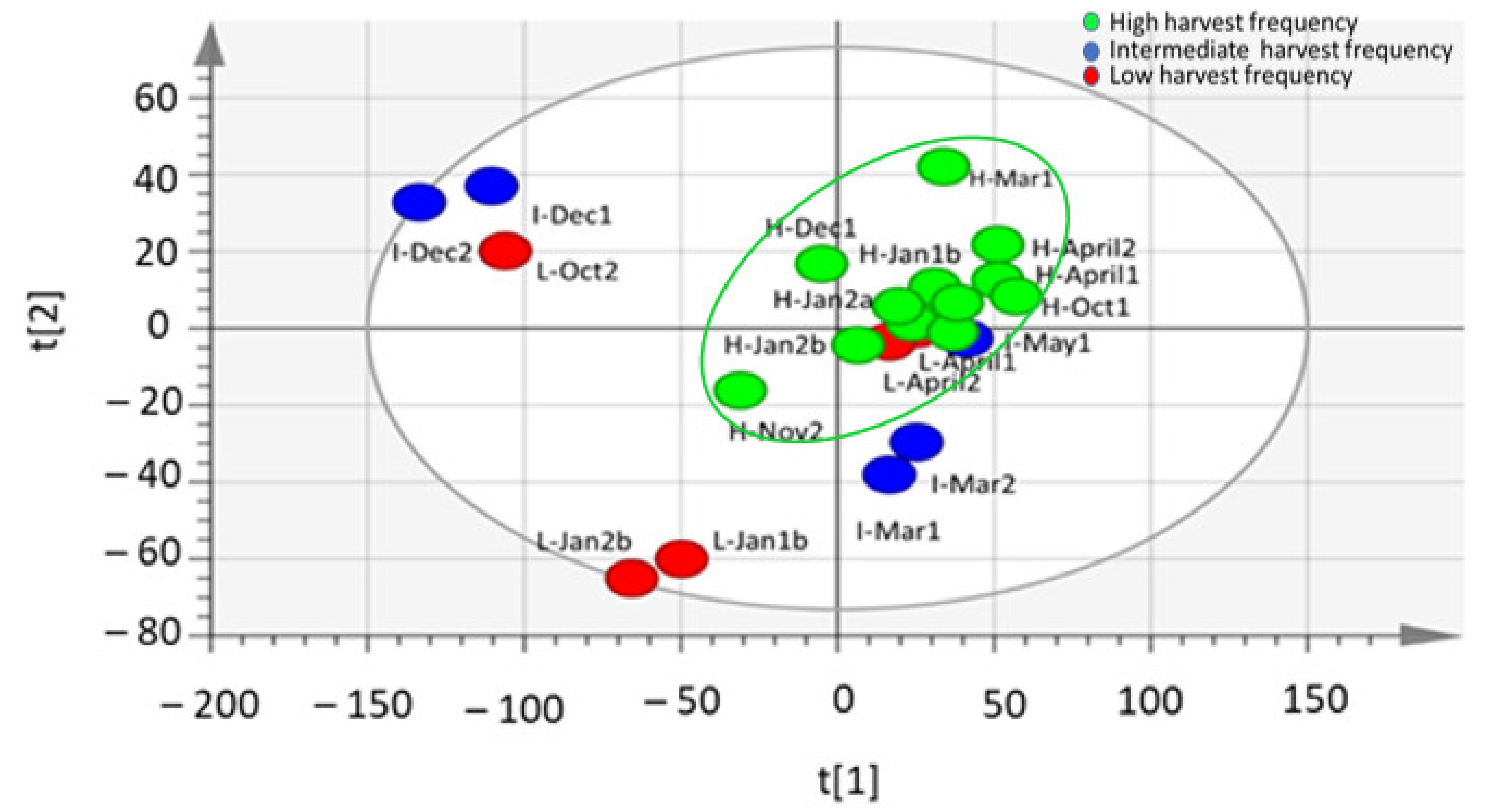
2.1. Changes in the Metabolite Profile
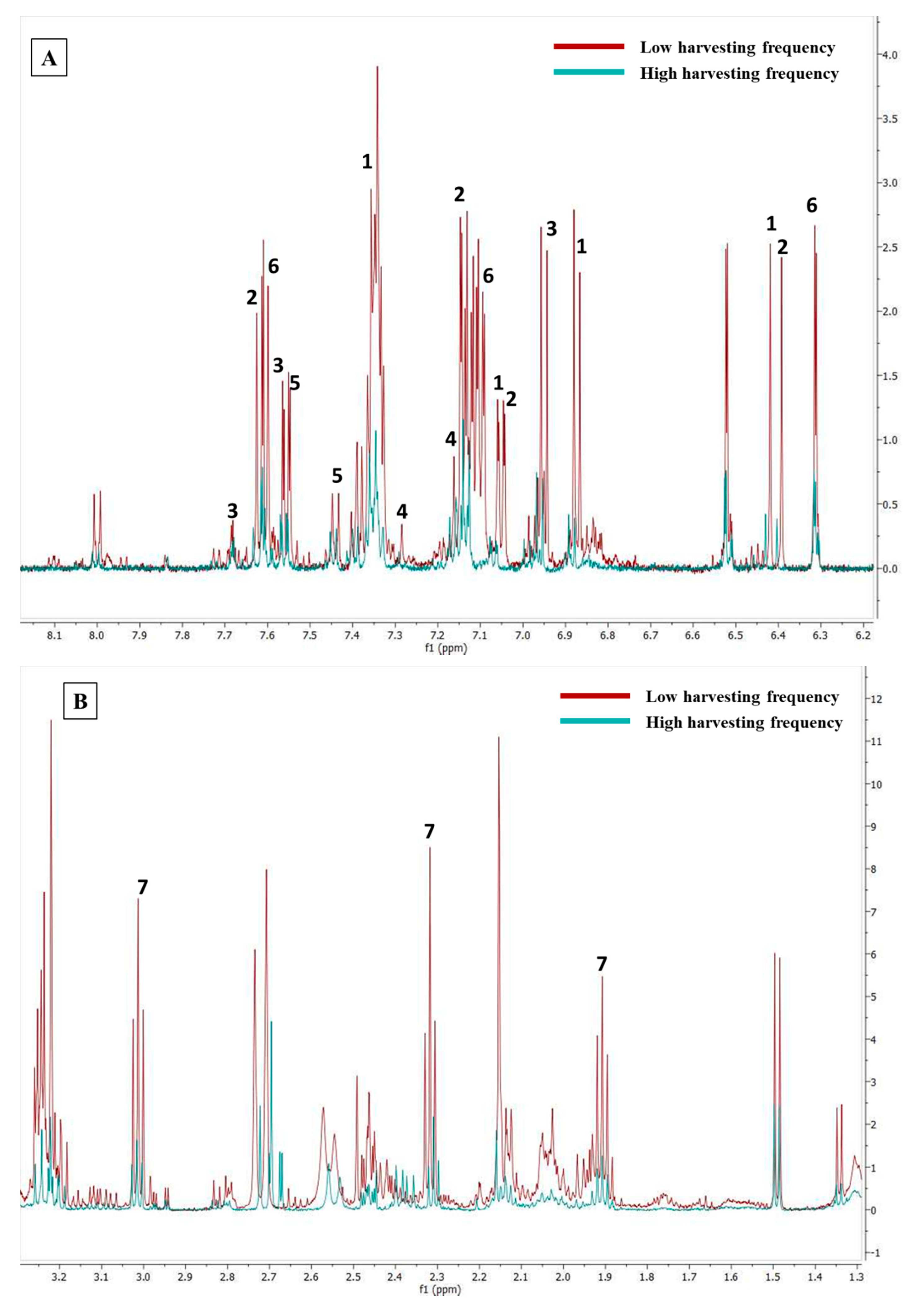
2.2. Targeted Metabolites That Were Affected by Changes in the Metabolic Profile
3. Materials and Methods
3.1. Hydroponic Planting, Maintenance, and Leaf Harvesting
3.2. Preparation of Extract for Metabolomics Analysis
3.3. Data Acquisition, Processing, and 1H-NMR Spectra Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Anwar, F.; Latif, S.; Ashraf, M.; Gilani, A.H. Moringa oleifera: A food plant with multiple medicinal uses. Phyther. Res. 2007, 21, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Siddhuraju, P.; Becker, K. Antioxidant properties of various solvent extracts of total phenolic constituents from three different agroclimatic origins of drumstick tree (Moringa oleifera Lam.) leaves. J. Agric. Food Chem. 2003, 51, 2144–2155. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.L. Soluble extract from Moringa oleifera leaves with a new anticancer activity. PLoS ONE 2014, 9, e95492. [Google Scholar] [CrossRef]
- Guevara, A.P.; Vargas, C.; Sakurai, H.; Fujiwara, Y.; Hashimoto, K.; Maoka, T.; Kozuka, M.; Ito, Y.; Tokuda, H.; Nishino, H. An antitumor promoter from Moringa oleifera Lam. Mutat. Res. Toxicol. Environ. Mutagen. 1999, 440, 181–188. [Google Scholar] [CrossRef]
- Jaja-Chimedza, A.; Graf, B.L.; Simmler, C.; Kim, Y.; Kuhn, P.; Pauli, G.F.; Raskin, I. Biochemical characterization and anti-inflammatory properties of an isothiocyanate-enriched moringa (Moringa oleifera) seed extract. PLoS ONE 2017, 12, e0182658. [Google Scholar] [CrossRef]
- Faizi, S.; Siddiqui, B.S.; Saleem, R.; Siddiqui, S.; Aftab, K.; Gilani, A.H. Isolation and structure elucidation of new nitrile and mustard oil glycosides from Moringa oleifera and their effect on blood pressure. J. Nat. Prod. 1994, 57, 1256–1261. [Google Scholar] [CrossRef]
- Al-Asmari, A.K.; Albalawi, S.M.; Athar, M.T.; Khan, A.Q.; Al-Shahrani, H.; Islam, M. Moringa oleifera as an anti-cancer agent against breast and colorectal cancer cell lines. PLoS ONE 2015, 10, e0135814. [Google Scholar] [CrossRef]
- Sreelatha, S.; Jeyachitra, A.; Padma, P.R. Antiproliferation and induction of apoptosis by Moringa oleifera leaf extract on human cancer cells. Food Chem. Toxicol. 2011, 49, 1270–1275. [Google Scholar] [CrossRef]
- Pangastuti, A.; Amin, I.F.; Amin, A.Z.; Amin, M. Natural bioactive compound from Moringa oleifera against cancer based on in silico screening. J. Teknol. Sci. Eng. 2016, 78, 5. [Google Scholar]
- Vongsak, B.; Sithisarn, P.; Gritsanapan, W. Simultaneous determination of crypto-chlorogenic acid, isoquercetin, and astragalin contents in Moringa oleifera leaf extracts by TLC-densitometric method. Evid. Based Complement. Altern. Med. 2013. [Google Scholar] [CrossRef]
- Leone, A.; Fiorillo, G.; Criscuoli, F.; Ravasenghi, S.; Santagostini, L.; Fico, G.; Spadafranca, A.; Battezzati, A.; Schiraldi, A.; Pozzi, F.; et al. Nutritional characterization and phenolic profiling of Moringa oleifera leaves grown in chad, sahrawi refugee camps, and Haiti. Int. J. Mol. Sci. 2015, 16, 18923–18937. [Google Scholar] [CrossRef] [PubMed]
- Atawodi, S.E.; Atawodi, J.C.; Idakwo, G.A.; Pfundstein, B.; Haubner, R.; Wurtele, G.; Bartsch, H.; Owen, W.R. Evaluation of the polyphenol content and antioxidant properties of methanol extracts of the leaves, stem, and root barks of Moringa oleifera Lam. J. Med. Food 2010, 13, 710–716. [Google Scholar] [CrossRef]
- Awodele, O.; Oreagba, I.A.; Odoma, S.; da Silva, J.A.T.; Osunkalu, V.O. Toxicological evaluation of the aqueous leaf extract of Moringa oleifera Lam.(Moringaceae). J. Ethnopharmacol. 2012, 139, 330–336. [Google Scholar] [CrossRef]
- Craker, L.E.; Gardner, Z. Sustaining the harvest: Challenges in MAP production and markets. Acta Hortic. 2003, 2, 25–30. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Int. J. Plant Chem. Biochem. Tech. 2010, 21, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Verpoorte, R.; Contin, A.; Memelink, J. Biotechnology for the production of plant secondary metabolites. Phytochem. Rev. 2002, 1, 13–25. [Google Scholar] [CrossRef]
- Lucchesini, M.; Mensuali-Sodi, A. Plant Tissue Culture—An Opportunity for the Production of Nutraceuticals. In Bio-Farms for Nutraceuticals; Springer: Boston, MA, USA, 2010; pp. 185–202. [Google Scholar]
- Iqbal, S.; Bhanger, M.I. Effect of season and production location on antioxidant activity of Moringa oleifera leaves grown in Pakistan. J. Food Compos. Anal. 2006, 19, 544–551. [Google Scholar] [CrossRef]
- Almela, L.; Sánchez-Munoz, B.; Fernández-López, J.A.; Roca, M.J.; Rabe, V. Liquid chromatograpic–mass spectrometric analysis of phenolics and free radical scavenging activity of rosemary extract from different raw material. J. Chromatogr. A 2006, 1120, 221–229. [Google Scholar] [CrossRef]
- Jorquera-Fontena, E.; Alberdi, M.; Franck, N. Pruning severity affects yield, fruit load and fruit and leaf traits of’Brigitta’blueberry. J. Soil. Sci. Plant. Nutr. 2014, 14, 855–868. [Google Scholar]
- Kim, S.W.; Ban, S.H.; Jeong, S.C.; Chung, H.J.; Ko, S.M.; Yoo, O.J.; Liu, J.R. Genetic discrimination between Catharanthus roseus cultivars by metabolic fingerprinting using 1H NMR spectra of aromatic compounds. Biotechnol. Bioprocess. Eng. 2007, 12, 646. [Google Scholar] [CrossRef]
- Palama, T.L.; Menard, P.; Fock, I.; Choi, Y.H.; Bourdon, E.; Govinden-Soulange, J.; Bahut, M.; Payet, B.; Verpoorte, R.; Kodja, H.; et al. Shoot differentiation from protocorm callus cultures of Vanilla planifolia (Orchidaceae): Proteomic and metabolic responses at early stage. BMC Plant. Biol. 2010, 10, 82. [Google Scholar] [CrossRef]
- Mahmud, I.; Chowdhury, K.; Boroujerdi, A. Tissue-specific metabolic profile study of Moringa oleifera L. using nuclear magnetic resonance spectroscopy. Plant. Tissue Cult. Biotechnol. 2014, 24, 77. [Google Scholar] [CrossRef]
- Mahmud, I.; Thapaliya, M.; Boroujerdi, A.; Chowdhury, K. NMR-based metabolomics study of the biochemical relationship between sugarcane callus tissues and their respective nutrient culture media. Anal. Bioanal. Chem. 2014, 406, 5997–6005. [Google Scholar] [CrossRef]
- Gilani, A.H.; Aftab, K.; Suria, A.; Siddiqui, S.; Salem, R.; Siddiqui, B.S.; Faizi, S. Pharmacological studies on hypotensive and spasmolytic activities of pure compounds from Moringa oleifera. Phyther. Res. 1994, 8, 87–91. [Google Scholar] [CrossRef]
- Maiyo, F.C.; Moodley, R.; Singh, M. Cytotoxicity, antioxidant and apoptosis studies of quercetin-3-O glucoside and 4-(β-D-glucopyranosyl-1→ 4-α-L-rhamnopyranosyloxy)-benzyl isothiocyanate from Moringa oleifera. Anti Cancer Agents Med. Chem. 2016, 16, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.R.; Vijayakumar, M.; Mathela, C.S.; Rao, C.V. In Vitro and In Vivo antioxidant properties of different fractions of Moringa oleifera leaves. Food Chem Toxicol. 2009, 47, 2196–2201. [Google Scholar] [CrossRef] [PubMed]
- Sabino, A.R.; Tavares, S.S.; Riffel, A.; Li, J.V.; Oliveira, D.J.A.; Feres, C.I.M.A.; Henrique, L.; Oliveira, J.S.; Correia, G.D.S.; Sabino, A.R.; et al. 1H NMR metabolomic approach reveals chlorogenic acid as a response of sugarcane induced by exposure to Diatraea saccharalis. Ind Crops Prod. 2019, 140, 111651. [Google Scholar] [CrossRef]
- Maurya, A.; Gupta, S.; Srivastava, S.K. Preparative isolation of bioactive nitrile glycoside “Niazirin” from the fruits of Moringa oleifera using fast centrifugal partition chromatography. Sep. Sci Technol. 2011, 46, 1195–1199. [Google Scholar] [CrossRef]
- Aboul-Ela, M.A.; Abdul-Ghani, M.M.; El-Fiky, F.K.; El-Lakany, A.M.; Mekky, H.M.; Ghazy, N.M. Chemical constituens of Cirsium syriacum and Cichorium Intybus (Asteraceae) growing in Egypt. Alex. J. Pharm Sci. 2002, 16, 152–156. [Google Scholar]
- Jang, J.; Kim, H.P.; Park, H. Structure and antiinflammatory activity relationships of wogonin derivatives. Arch. Pharm Res. 2005, 28, 877. [Google Scholar] [CrossRef]
- Huang, S.T.; Wang, C.Y.; Yang, R.C.; Chu, C.J.; Wu, H.T.; Pang, J.H.S. Wogonin, an active compound in Scutellaria baicalensis, induces apoptosis and reduces telomerase activity in the HL-60 leukemia cells. Phytomedicine 2010, 17, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Calviño, E.; García-Pérez, A.; Herráez, A.; Diez, J.C. Human acute promyelocytic leukemia NB4 cells are sensitive to esculetin through induction of an apoptotic mechanism. Chem. Biol. Interact. 2014, 220, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.Y.; Tsai, Y.Y.; Wang, C.J.; Lin, W.L.; Tseng, T.H. Induction of apoptosis by esculetin in human leukemia cells. Eur. J. Pharmacol. 2001, 416, 25–32. [Google Scholar] [CrossRef]
- Ali, F.T.; Hassan, N.S.; Abdrabou, R.R. Hepatoprotective and antiproliferative activity of moringinine, chlorogenic acid and quercetin. Int. J. Res. Med. Sci. 2016, 4, 1147–1153. [Google Scholar] [CrossRef][Green Version]
- Farah, A.; de Paulis, T.; Trugo, L.C.; Martin, P.R. Effect of roasting on the formation of chlorogenic acid lactones in coffee. J. Agric. Food Chem. 2005, 53, 1505–1513. [Google Scholar] [CrossRef]
- De Sotillo, D.V.R.; Hadley, M. Chlorogenic acid modifies plasma and liver concentrations of: Cholesterol, triacylglycerol, and minerals in (fa/fa) Zucker rats. J. Nutr. Biochem. 2002, 13, 717–726. [Google Scholar] [CrossRef]
- Cho, A.-S.; Jeon, S.M.; Kim, M.J.; Yeo, J.; Seo, K.-I.; Choi, M.S.; Lee, M.-K. Chlorogenic acid exhibits anti-obesity property and improves lipid metabolism in high-fat diet-induced-obese mice. Food Chem. Toxicol. 2010, 48, 937–943. [Google Scholar] [CrossRef]
- Singh, R.S.G.; Negi, P.S.; Radha, C. Phenolic composition, antioxidant and antimicrobial activities of free and bound phenolic extracts of Moringa oleifera seed flour. J. Funct. Foods. 2013, 5, 1883–1891. [Google Scholar] [CrossRef]
- Doss, H.M.; Dey, C.; Sudandiradoss, C.; Rasool, M.K. Targeting inflammatory mediators with ferulic acid, a dietary polyphenol, for the suppression of monosodium urate crystal-induced inflammation in rats. Life. Sci. 2016, 148, 201–210. [Google Scholar] [CrossRef]
- Nile, S.H.; Ko, E.Y.; Kim, D.H.; Keum, Y.-S. Screening of ferulic acid related compounds as inhibitors of xanthine oxidase and cyclooxygenase-2 with anti-inflammatory activity. Rev. Bras. Farmacogn. 2016, 26, 50–55. [Google Scholar] [CrossRef]
- Balasubashini, M.S.; Rukkumani, R.; Viswanathan, P.; Menon, V.P. Ferulic acid alleviates lipid peroxidation in diabetic rats. Phyther. Res. 2004, 18, 310–314. [Google Scholar] [CrossRef]
- Anson, N.M.; van den Berg, R.; Havenaar, R.; Bast, A.; Haenen, G.R.M.M. Bioavailability of ferulic acid is determined by its bioaccessibility. J. Cereal Sci. 2009, 49, 296–300. [Google Scholar] [CrossRef]
- Saleem, A.; Saleem, M.; Akhtar, M.F. Antioxidant, anti-inflammatory and antiarthritic potential of Moringa oleifera Lam: An ethnomedicinal plant of Moringaceae family. S. Afr. J. Bot. 2020, 128, 246–256. [Google Scholar] [CrossRef]
- Ma, Y.Q.; Ye, X.Q.; Fang, Z.X.; Chen, J.C.; Xu, G.H.; Liu, D.H. Phenolic compounds and antioxidant activity of extracts from ultrasonic treatment of Satsuma mandarin (Citrus unshiu Marc.) peels. J. Agric. Food Chem. 2008, 56, 5682–5690. [Google Scholar] [CrossRef] [PubMed]
- Merkl, R.; Hradkova, I.; Filip, V.; ŠMIdRkal, J. Antimicrobial and antioxidant properties of phenolic acids alkyl esters. Czech J. Food Sci. 2010, 28, 275–279. [Google Scholar] [CrossRef]
- Ingale, S.P.; Gandhi, F.P. Effect of aqueous extract of Moringa oleifera leaves on pharmacological models of epilepsy and anxiety in mice. Int. J. Epilepsy 2016, 3, 12–19. [Google Scholar] [CrossRef]
- Bakre, A.G.; Aderibigbe, A.O.; Ademowo, O.G. Studies on neuropharmacological profile of ethanol extract of Moringa oleifera leaves in mice. J. Ethnopharmacol. 2013, 149, 783–789. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Darko, E.; Heydarizadeh, P.; Schoefs, B.; Sabzalian, M.R. Photosynthesis under artificial light: The shift in primary and secondary metabolism. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130243. [Google Scholar] [CrossRef]
- Leone, A.; Spada, A.; Battezzati, A.; Schiraldi, A.; Aristil, J.; Bertoli, S. Cultivation, genetic, ethnopharmacology, phytochemistry and pharmacology of Moringa oleifera leaves: An overview. Int. J. Mol. Sci. 2015, 16, 12791–12835. [Google Scholar] [CrossRef]
- Singh, B.N.; Singh, B.R.; Singh, R.L.; Prakash, D.; Dhakarey, R.; Upadhyay, G.; Singh, H.B. Oxidative DNA damage protective activity, antioxidant and anti-quorum sensing potentials of Moringa oleifera. Food Chem. Toxicol. 2009, 47, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Birošová, L.; Mikulášová, M.; Vaverková, Š. Antimutagenic effect of phenolic acids. Biomed. Pap. 2005, 149, 489–491. [Google Scholar] [CrossRef]
- Perkins, N.D. Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 2007, 8, 49–62. [Google Scholar] [CrossRef]
- Balasubramanian, T.; Karthikeyan, M.; Muhammed Anees, K.P.; Kadeeja, C.P.; Jaseela, K. Antidiabetic and antioxidant potentials of amaranthus hybridus in streptozotocin-induced diabetic rats. J. Diet. Suppl. 2017, 14, 395–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, H.; Yang, T.; Ye, Y.; Shan, J.; Yin, Z.; Luo, L. Chlorogenic acid protects mice against lipopolysaccharide-induced acute lung injury. Injury 2010, 41, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Mediani, A.; Abas, F.; Khatib, A.; Maulidiani, H.; Shaari, K.; Choi, Y.H.; Lajis, N.H. 1H-NMR-based metabolomics approach to understanding the drying effects on the phytochemicals in Cosmos caudatus. Food Res. Int. 2012, 49, 763–770. [Google Scholar] [CrossRef]

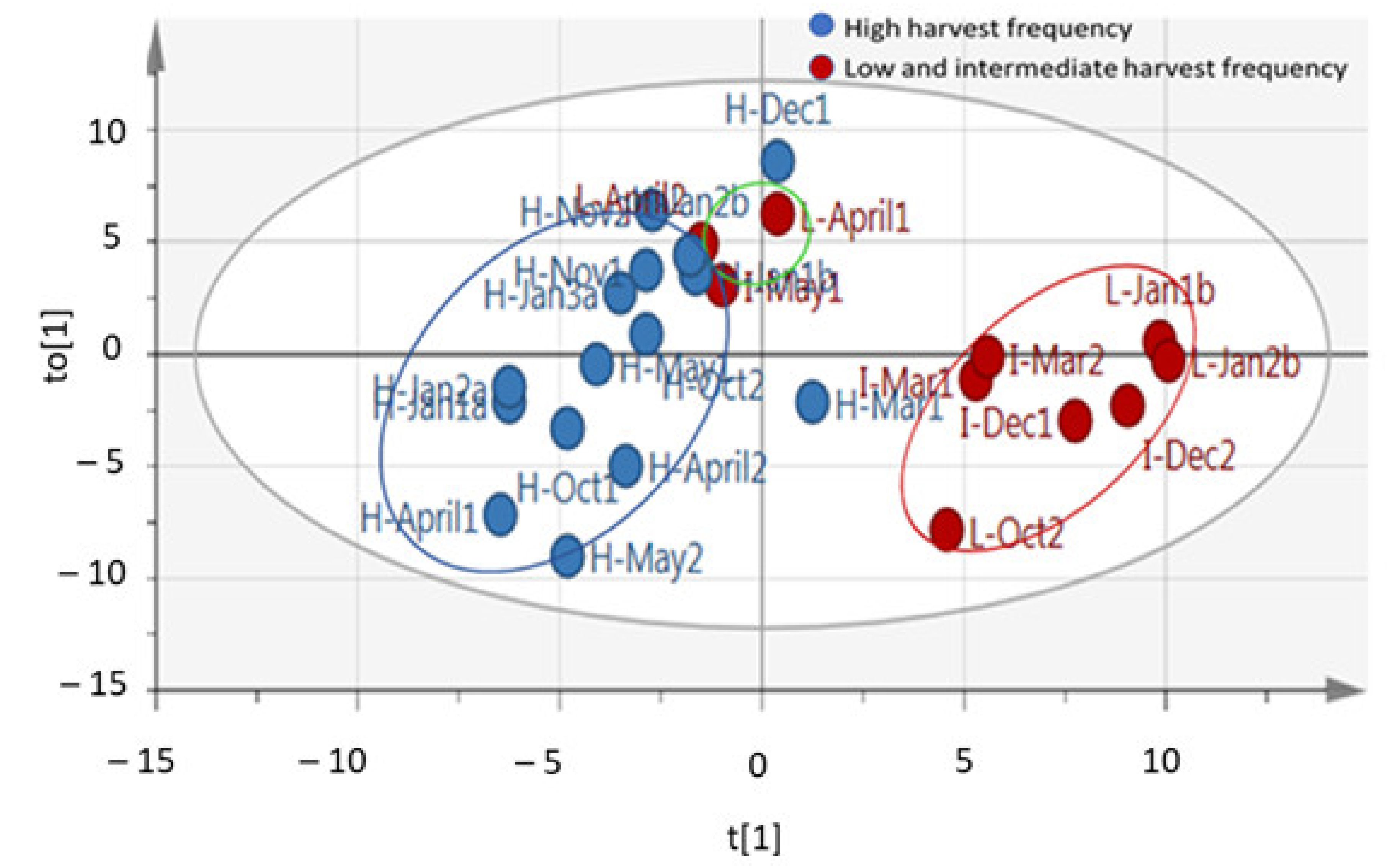

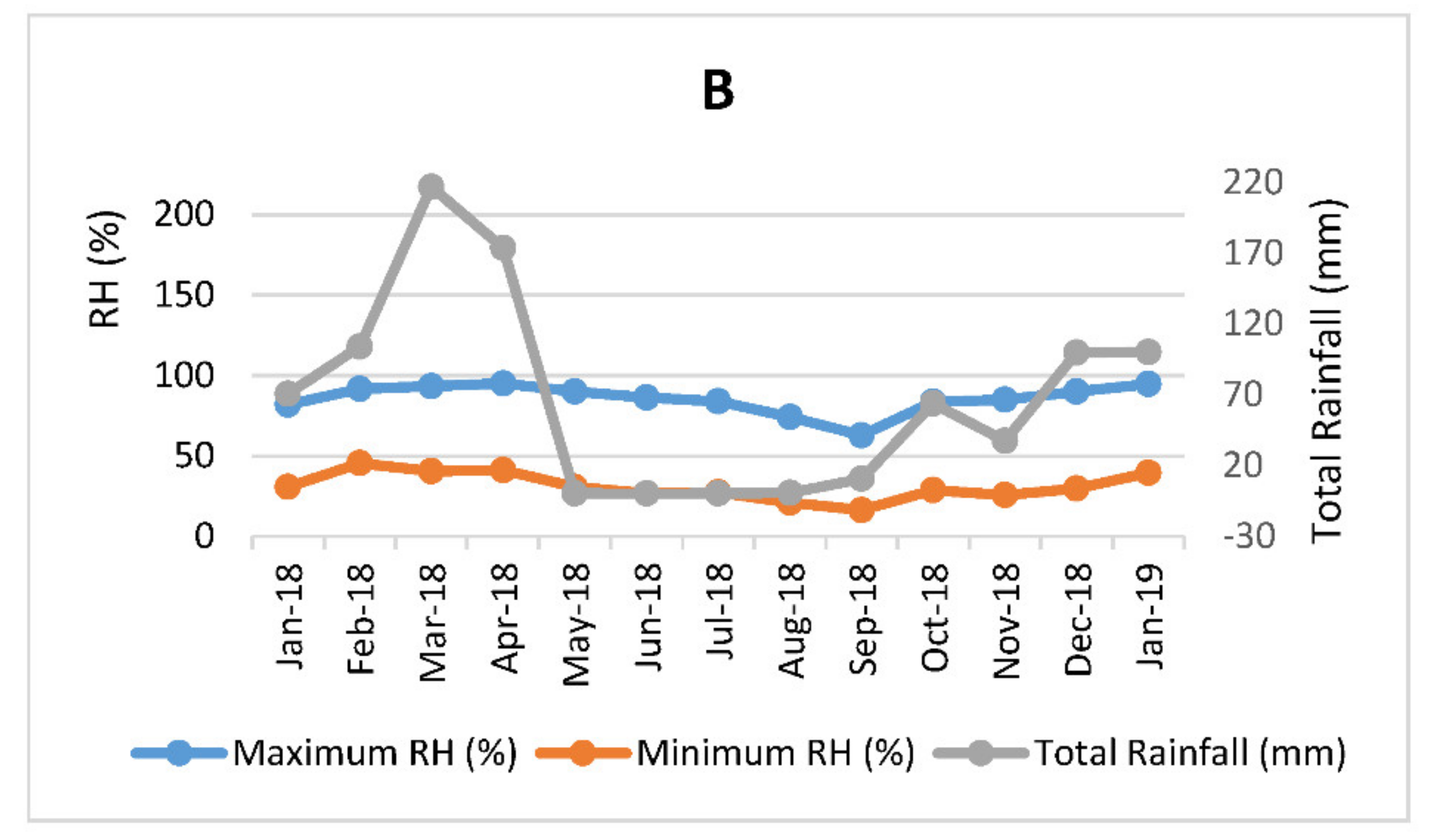
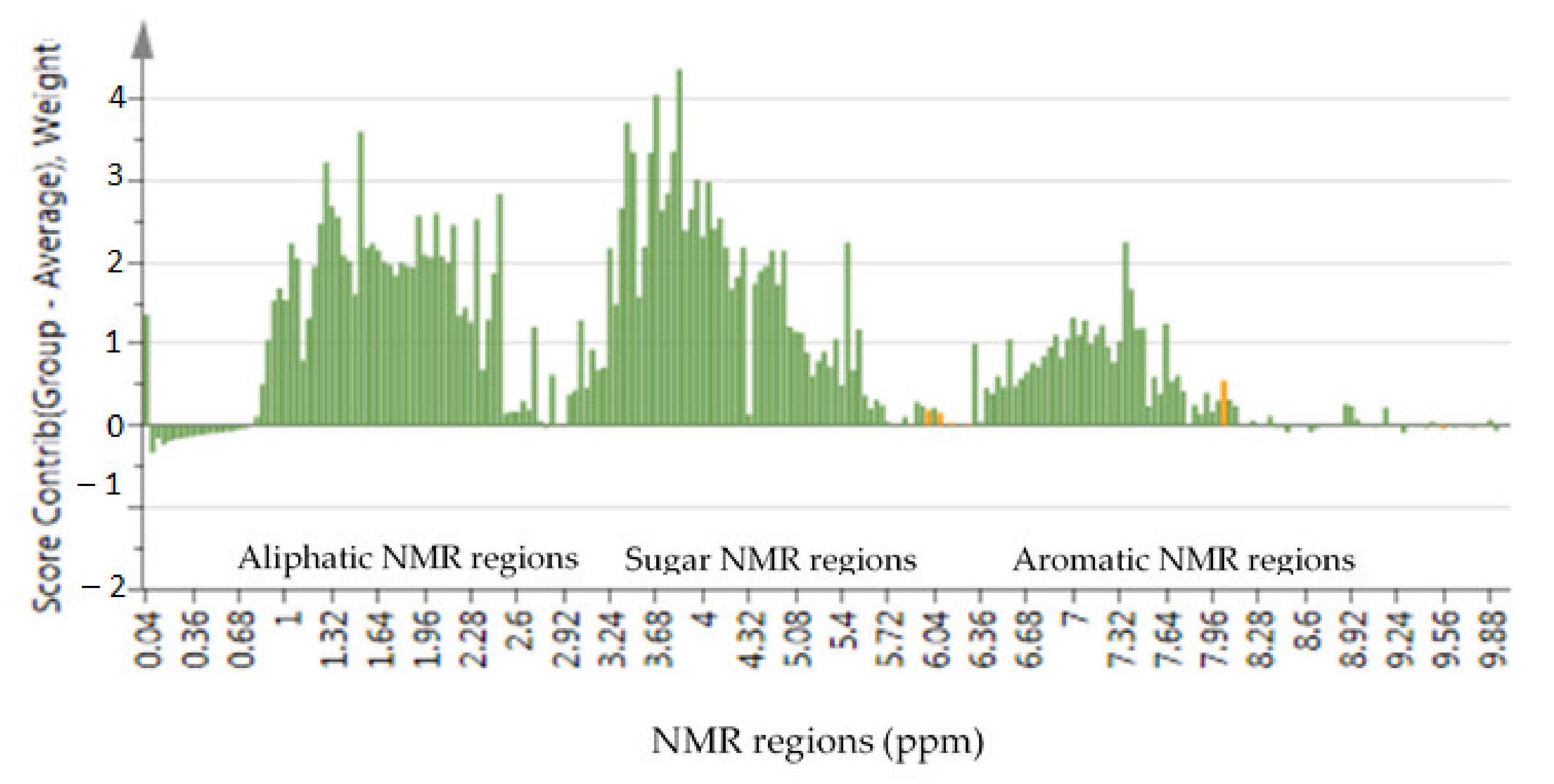
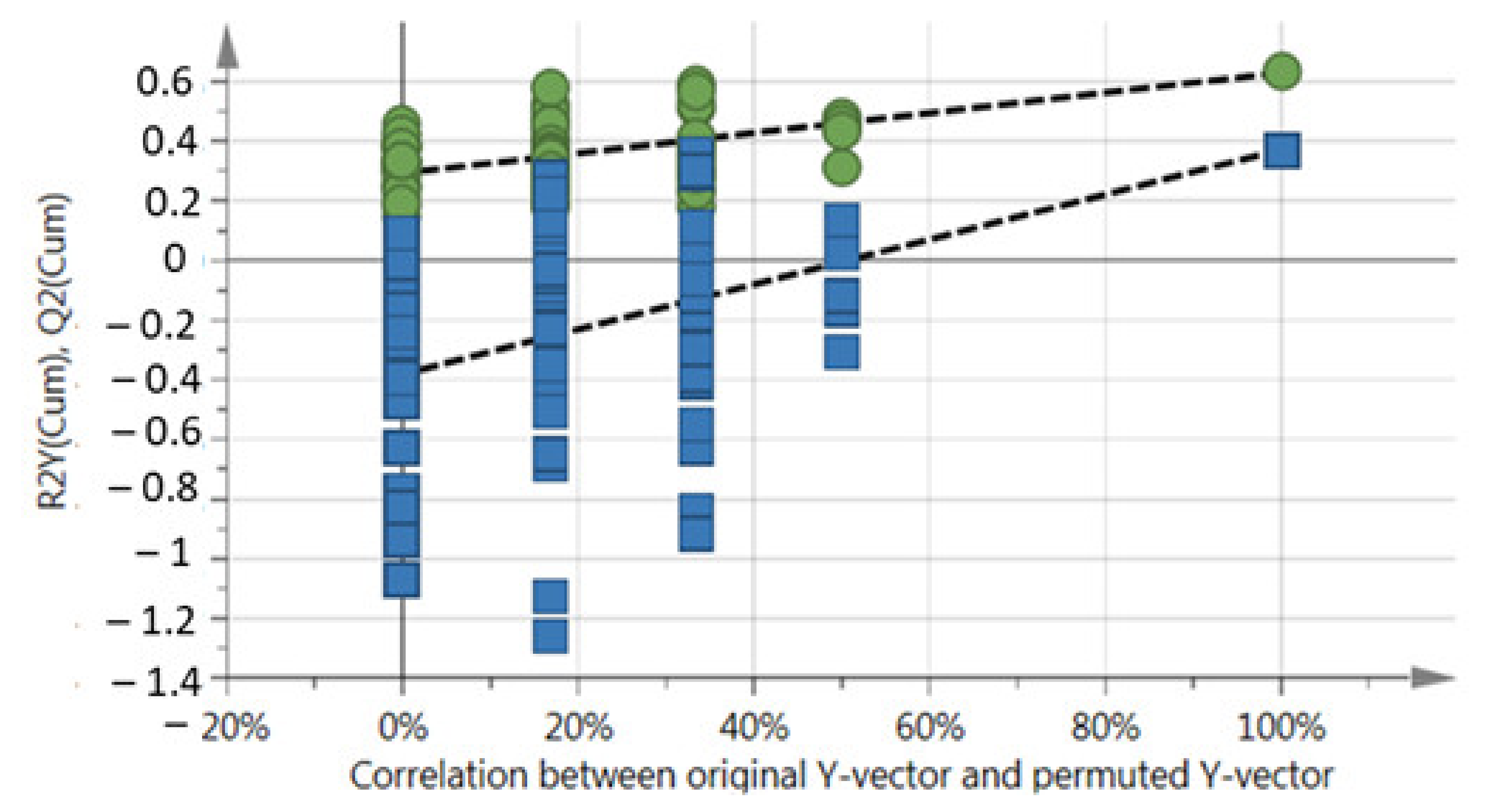

| Harvest Date | Harvest Frequency | Sample Labels |
|---|---|---|
| January 2018 | High | H-Jan1a H-Jan2a H-Jan3a |
| Intermediate | I-Jan1a * I-Jan2a * | |
| Low | L-Jan1a * L-Jan2a * | |
| February 2018 | High | H-Feb1 ** H-Feb2 ** |
| March 2018 | High | H-Mar1 H-Mar2 ** |
| Intermediate | I-Mar1 I-Mar2 | |
| April 2018 | High | H-April1 H-April2 |
| Low | L-April1 L-Apri2 | |
| May 2018 | High | H-May1 H-May2 |
| Intermediate | I-May1 I-May2 ** | |
| June 2018 | Excluded from metabolomics analysis as the leaf quality was affected by the winter season | |
| July 2018 | ||
| August 2018 | ||
| September 2018 | ||
| October 2018 | High | H-Oct1 H-Oct2 |
| Intermediate | I-Oct1 ** I-Oct2 ** | |
| Low | L-Oct1 ** L-Oct2 | |
| November 2018 | High | H-Nov1 H-Nov2 |
| December 2018 | High | H-Dec1 H-Dec2 ** |
| Intermediate | I-Dec1 I-Dec2 | |
| January 2019 | High | H-Jan1b H-Jan2b |
| Low | L-Jan1b L-Jan2b | |
| Compounds Name | Chemical Shift Peaks (ppm) | Identification Level * | Associated Medicinal Properties | References |
|---|---|---|---|---|
| Niazirin | δ 7.16, 7.26 | Level 2 | Anticancer activities | [4,6] |
| Wogonin | δ 6.32, 7.09, 7.59 | Level 2 | Anti-inflammatory | [33] |
| Esculetin | δ 7.42,7.55 | Level 2 | Antioxidant, anti-inflammatory | [34] |
| Chlorogenic acid | δ 6.410, 7.05, 7.15, 7.64 | Level 3 | Anti-inflammatory, anti-oxidants, antispasmodic, anti-cancer, anti-obesity | [35,36,37,38,39] |
| Ferulic acid | δ 6.38, 6.86, 7.05, 7.33 | Level 2 | Anti-oxidants, anti-microbial, anti-inflammatory, anti-diabetic | [28,40,41,42,43,44] |
| Vanillic acid | δ 6.94, 7.55, 7.65 | Level 2 | Anti-inflammatory, anti-oxidants, anti-cancer, anti-sickling, anti-microbial | [36,45,46,47] |
| Gamma-aminobutyric Acid (GABA) | δ 1.9, 2.3, 3.0 | Level 3 | Anti-epileptic, anti-anxiety | [48,49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Managa, L.R.; du Toit, E.S.; Prinsloo, G. NMR-Based Metabolomic Analyses to Identify the Effect of Harvesting Frequencies on the Leaf Metabolite Profile of a Moringa oleifera Cultivar Grown in an Open Hydroponic System. Molecules 2021, 26, 2298. https://doi.org/10.3390/molecules26082298
Managa LR, du Toit ES, Prinsloo G. NMR-Based Metabolomic Analyses to Identify the Effect of Harvesting Frequencies on the Leaf Metabolite Profile of a Moringa oleifera Cultivar Grown in an Open Hydroponic System. Molecules. 2021; 26(8):2298. https://doi.org/10.3390/molecules26082298
Chicago/Turabian StyleManaga, Lavhelesani R., Elsa S. du Toit, and Gerhard Prinsloo. 2021. "NMR-Based Metabolomic Analyses to Identify the Effect of Harvesting Frequencies on the Leaf Metabolite Profile of a Moringa oleifera Cultivar Grown in an Open Hydroponic System" Molecules 26, no. 8: 2298. https://doi.org/10.3390/molecules26082298
APA StyleManaga, L. R., du Toit, E. S., & Prinsloo, G. (2021). NMR-Based Metabolomic Analyses to Identify the Effect of Harvesting Frequencies on the Leaf Metabolite Profile of a Moringa oleifera Cultivar Grown in an Open Hydroponic System. Molecules, 26(8), 2298. https://doi.org/10.3390/molecules26082298







