A New Pyrimidine Schiff Base with Selective Activities against Enterococcus faecalis and Gastric Adenocarcinoma
Abstract
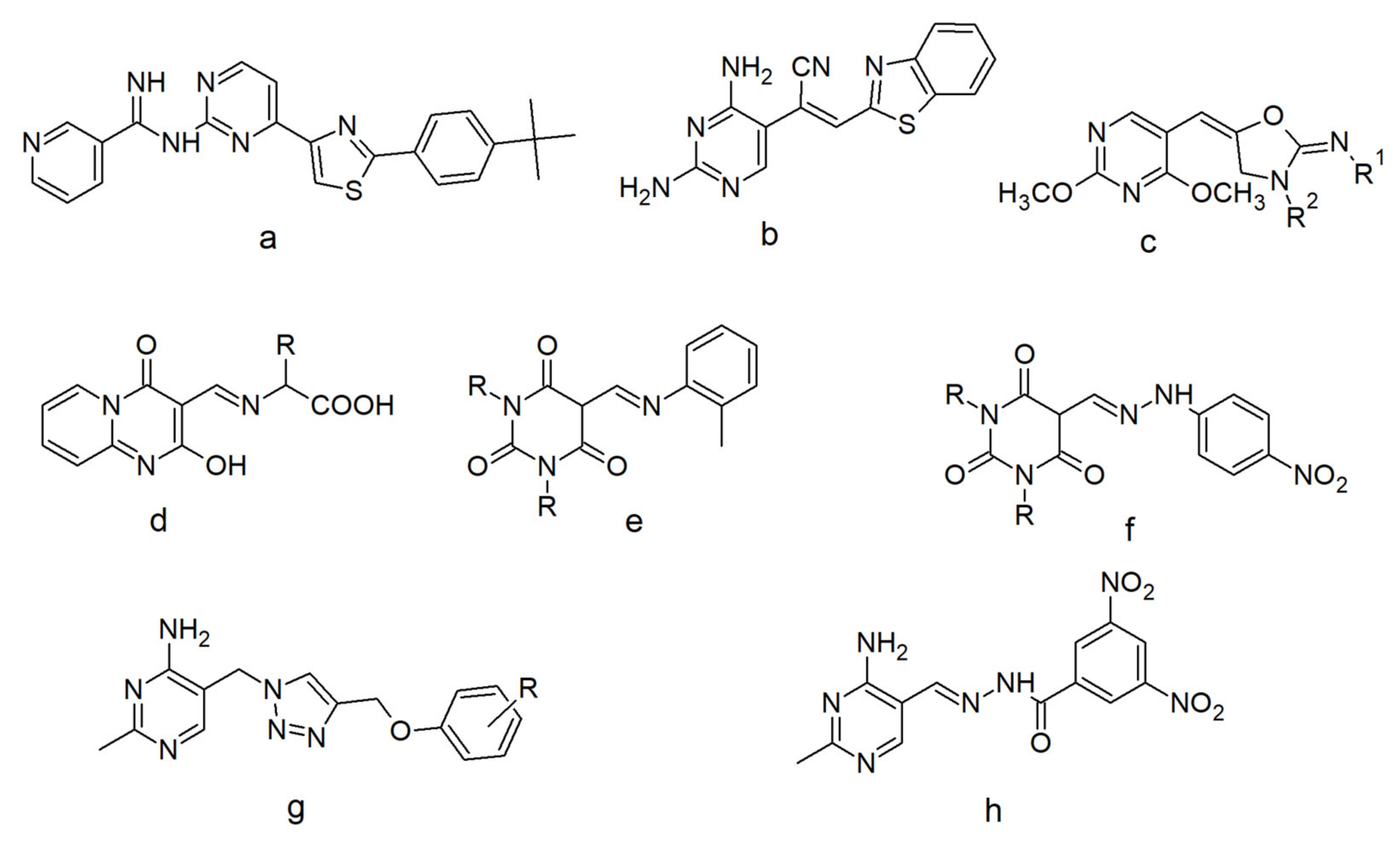
1. Introduction
2. Results
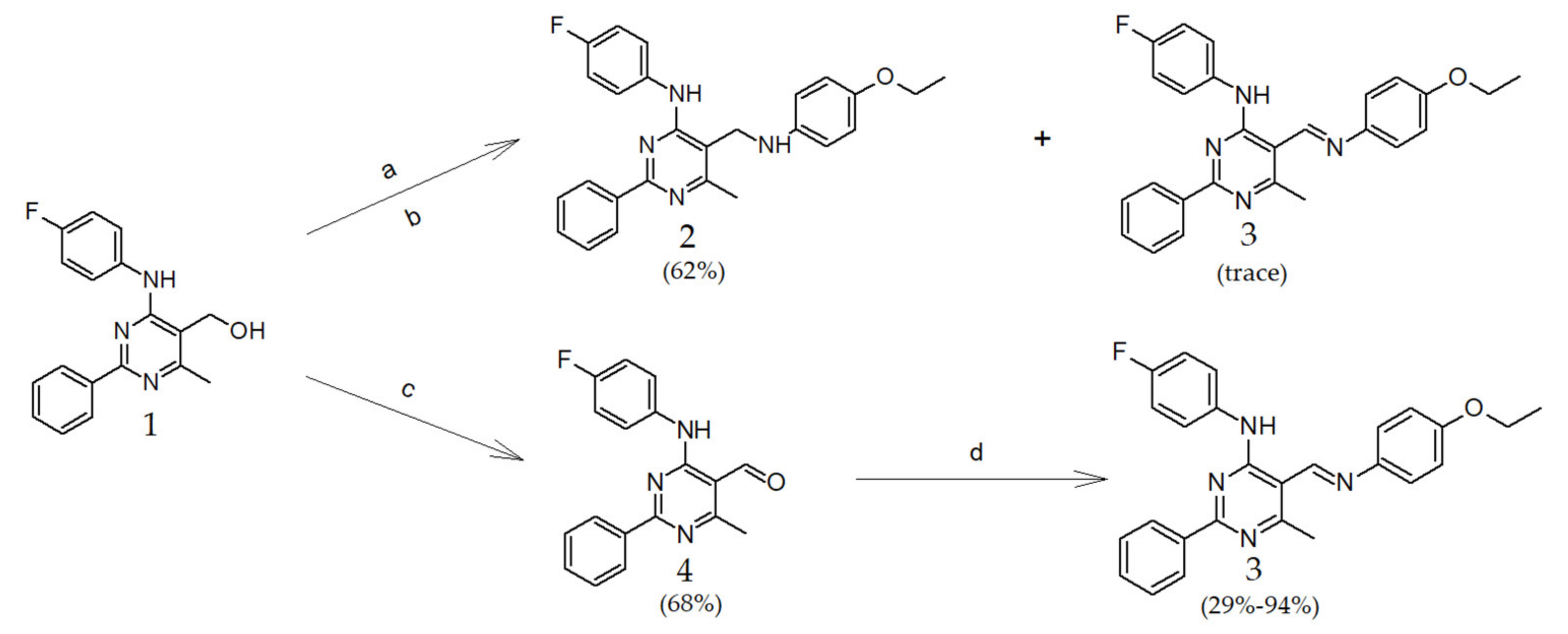
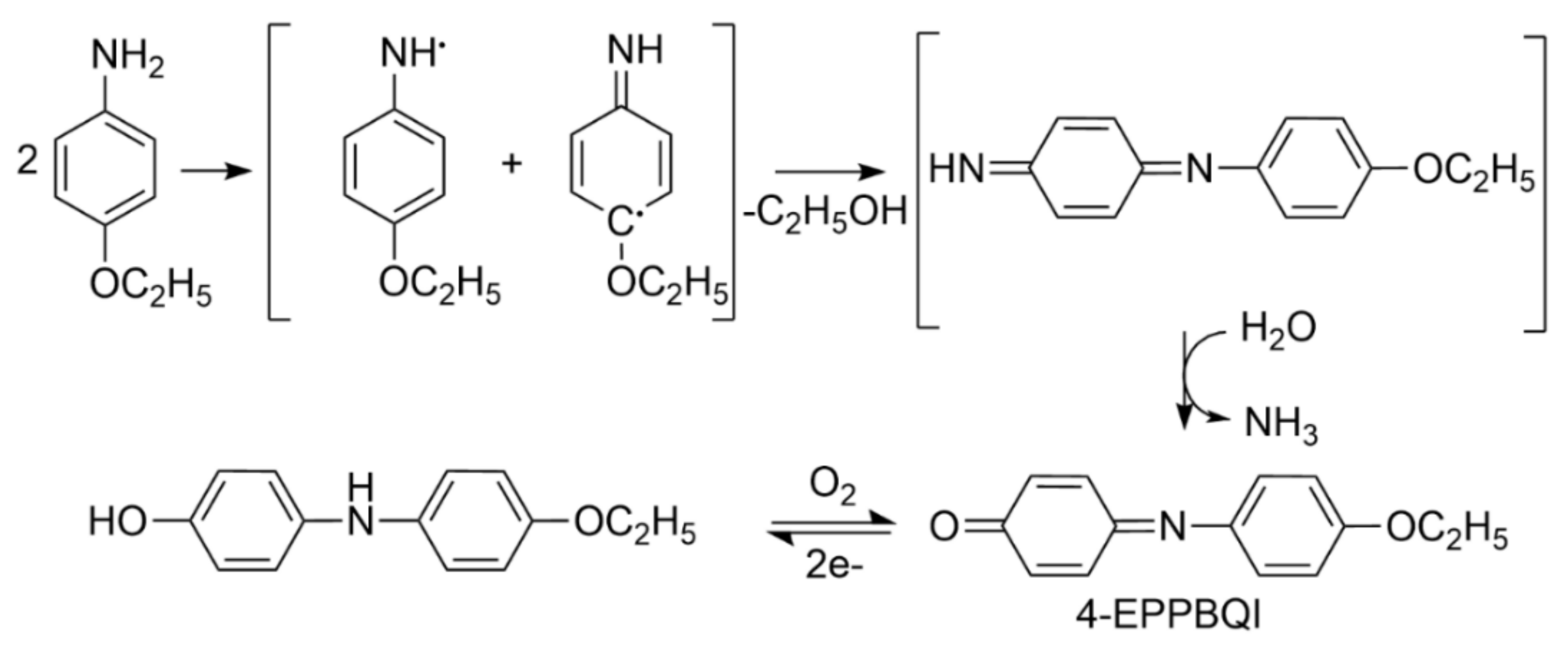
2.1. Chemistry
2.2. X-ray Structural Studies
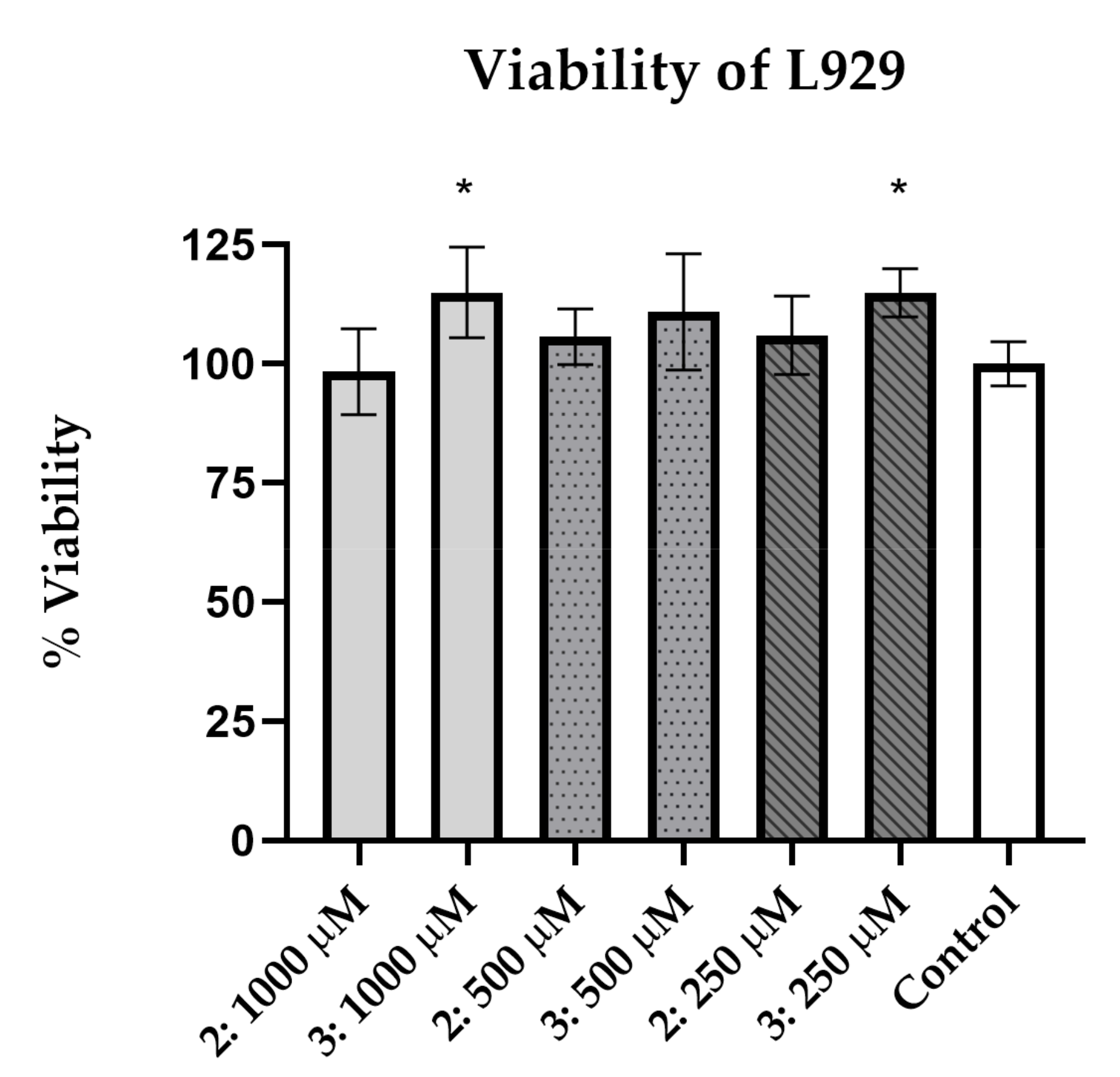
2.3. Cells and Cytotoxicity Assay
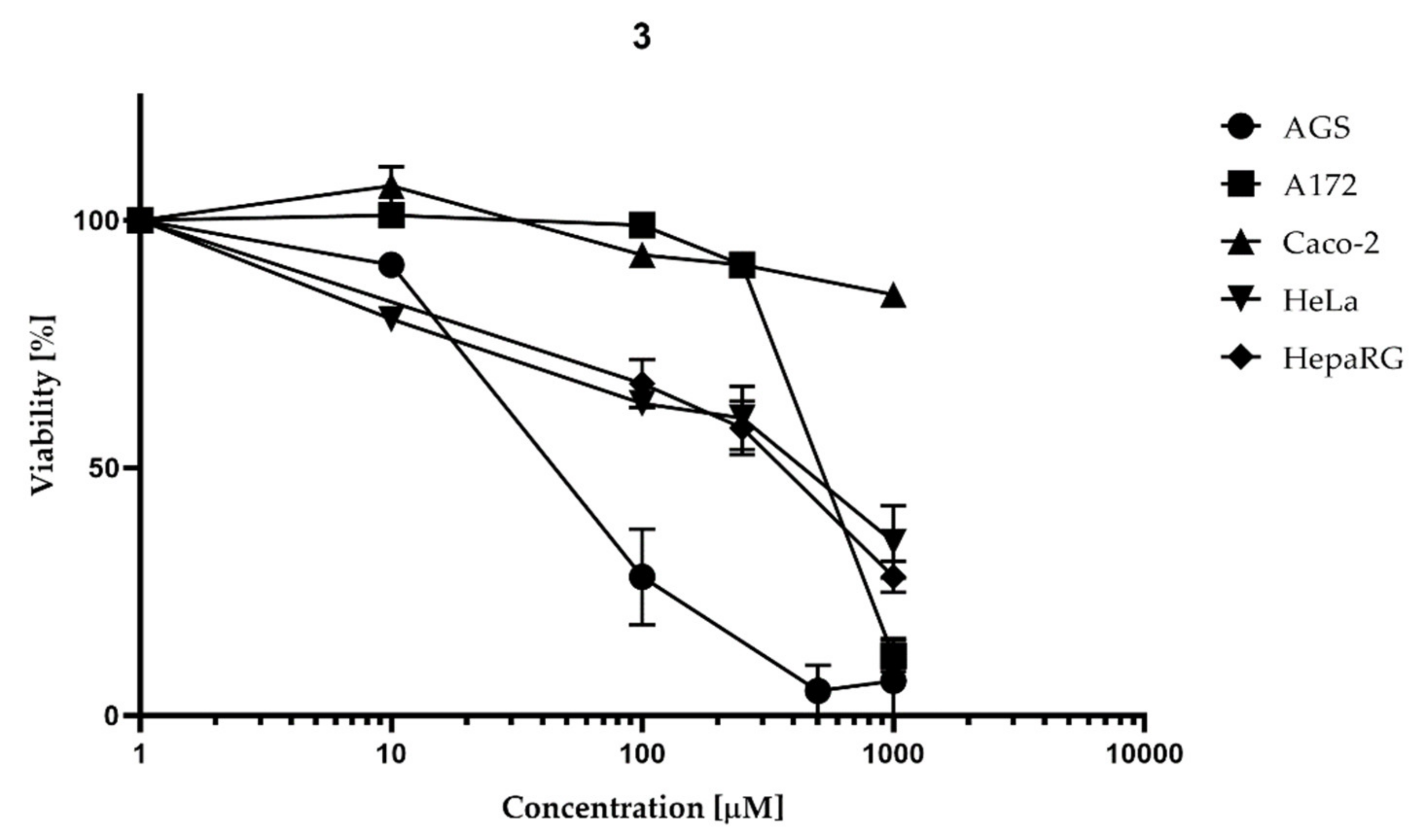
2.3.1. Proliferation Inhibition Caused by 2 and 3
2.3.2. Morphological Changes Induced by Imine 3
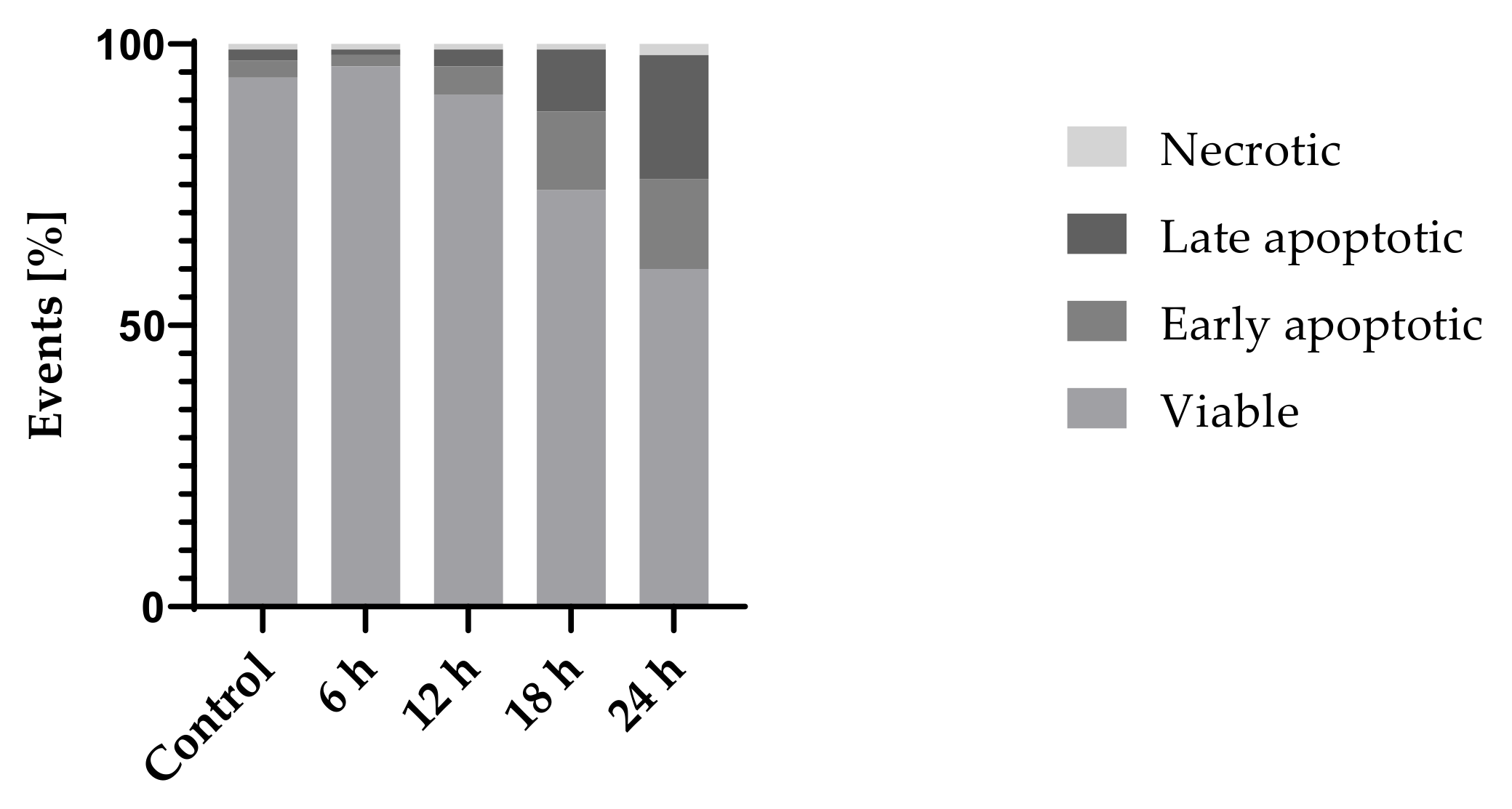
2.3.3. Cell Death
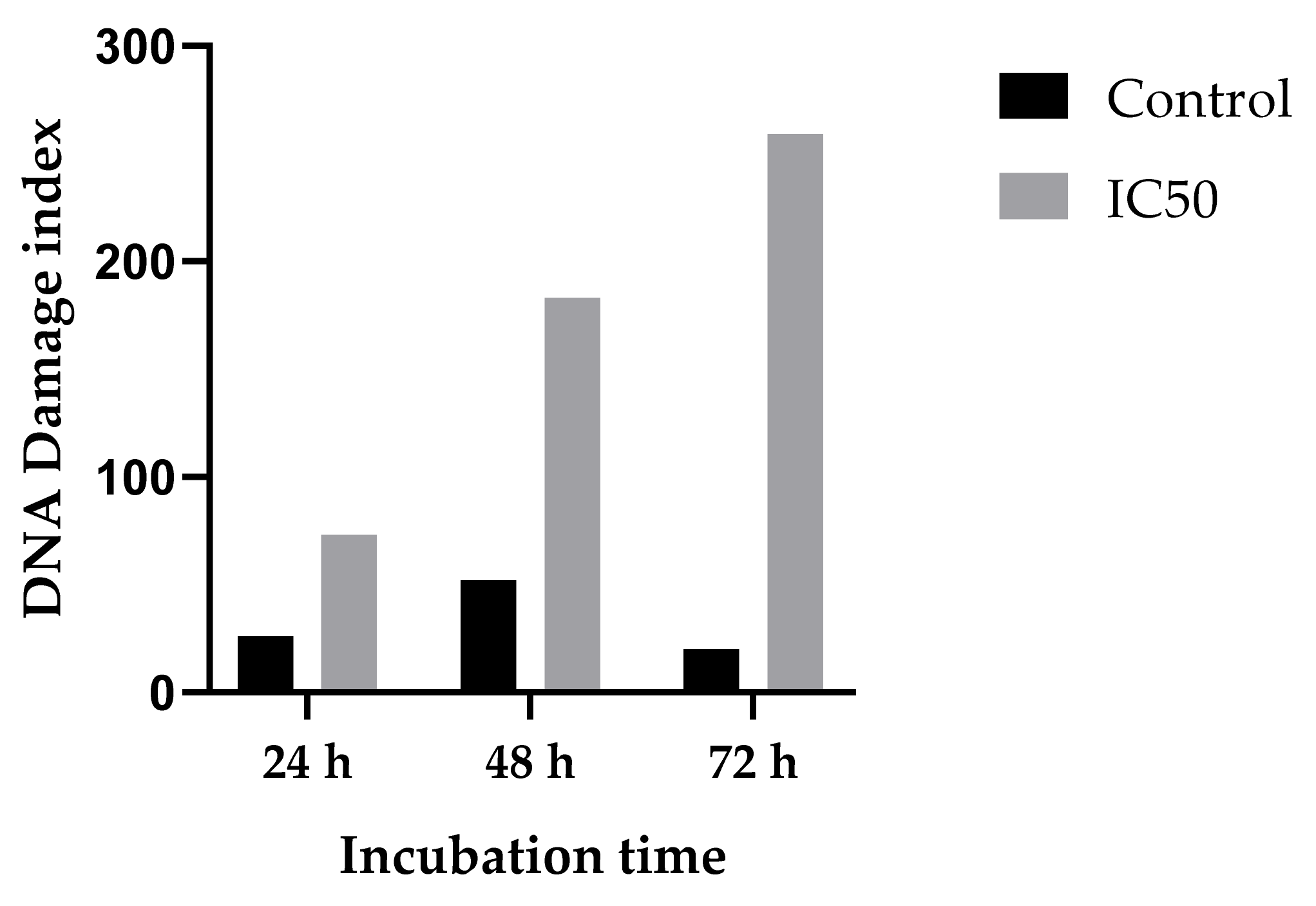
2.3.4. Genotoxicity/DNA Damage
2.4. Antimicrobial Activity Assay
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.2. X-ray Structural Studies
4.3. Cells and Cytotoxicity Assay
4.3.1. Chemicals
4.3.2. Cell Cultures and Reagent Preparation
4.3.3. Neutral Red Cell Viability Screening
4.3.4. Flow Cytometry
4.3.5. Microscopic Observation
4.3.6. Data Processing and Statistical Analysis
4.3.7. Annexin V/PI Staining
4.3.8. Comet Assay
4.4. Antimicrobial Activity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H.E. Lactic acid bacteria: From starter cultures to producers of chemicals. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- Godfree, A.F.; Kay, D.; Wyer, M.D. Faecal streptococci as indicators of faecal contamination in water. J. Appl. Microbiol. 1997, 83, 110–119. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Stiles, M.E.; Schleifer, K.H.; Holzapfel, W.H. Enterococci in foods—a conundrum for food safety. Int. J. Food Microbiol. 2003, 88, 105–122. [Google Scholar] [CrossRef]
- Giraffa, G.; Carminati, D.; Neviani, E. Enterococci Isolated from Dairy Products: A Review of Risks and Potential Technological Use. J. Food Prot. 1997, 60, 732–738. [Google Scholar] [CrossRef]
- Zirakzadeh, A.; Patel, R. Vancomycin-Resistant Enterococci: Colonization, Infection, Detection, and Treatment. Mayo Clin. Proc. 2006, 81, 529–536. [Google Scholar] [CrossRef]
- Linden, P.K. Optimizing therapy for vancomycin-resistant enterococci (VRE). Semin Respir Crit Care Med. 2007, 28, 632–645. [Google Scholar] [CrossRef]
- Huycke, M.M.; Joyce, W.; Wack, M.F. Augmented Production of Extracellular Superoxide by Blood Isolates of Enterococcus faecalis. J. Infect. Dis. 1996, 173, 743–745. [Google Scholar] [CrossRef]
- Huycke, M.M.; Abrams, V.; Moore, D.R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 2002, 23, 529–536. [Google Scholar] [CrossRef]
- Wang, X.; Huycke, M.M. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 2007, 132, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis Induces Aneuploidy and Tetraploidy in Colonic Epithelial Cells through a Bystander Effect. Cancer Res. 2008, 68, 9909–9917. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Allen, T.D.; Yang, Y.; Moore, D.R.; Huycke, M.M. Cyclooxygenase-2 Generates the Endogenous Mutagen trans-4-Hydroxy-2-nonenal in Enterococcus faecalis–Infected Macrophages. Cancer Prev. Res. 2013, 6, 206–216. [Google Scholar] [CrossRef]
- Balamurugan, R.; Rajendiran, E.; George, S.; Samuel, G.V.; Ramakrishna, B.S. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 2008, 23, 1298–1303. [Google Scholar] [CrossRef]
- Friis–Hansen, L.; Rieneck, K.; Nilsson, H.; Wadström, T.; Rehfeld, J.F. Gastric Inflammation, Metaplasia, and Tumor Development in Gastrin–Deficient Mice. Gastroenterology 2006, 131, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Dicksved, J.; Lindberg, M.; Rosenquist, M.; Enroth, H.; Jansson, J.K.; Engstrand, L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J. Med. Microbiol. 2009, 58, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Strickertsson, J.A.B.; Desler, C.; Martin-Bertelsen, T.; Machado, A.M.D.; Wadstrøm, T.; Winther, O.; Rasmussen, L.J.; Friis-Hansen, L. Enterococcus faecalis Infection Causes Inflammation, Intracellular Oxphos-Independent ROS Production, and DNA Damage in Human Gastric Cancer Cells. PLoS ONE 2013, 8, e63147. [Google Scholar] [CrossRef]
- Strickertsson, J.A.B.; Rasmussen, L.J.; Friis-Hansen, L. Enterococcus faecalis Infection and Reactive Oxygen Species Down-Regulates the miR-17-92 Cluster in Gastric Adenocarcinoma Cell Culture. Genes 2014, 5, 726. [Google Scholar] [CrossRef]
- Kumar, S.; Narasimhan, B. Therapeutic potential of heterocyclic pyrimidine scaffolds. Chem. Cent. J. 2018, 12, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Ma, S. Recent Development of Pyrimidine-Containing Antimicrobial Agents. ChemMedChem 2020, 15, 1875–1886. [Google Scholar] [CrossRef]
- Kotb, A.; Abutaleb, N.S.; Seleem, M.A.; Hagras, M.; Mohammad, H.; Bayoumi, A.; Ghiaty, A.; Seleem, M.N.; Mayhoub, A.S. Phenylthiazoles with tert-Butyl side chain: Metabolically stable with anti-biofilm activity. Eur. J. Med. Chem. 2018, 151, 110–120. [Google Scholar] [CrossRef]
- AlNeyadi, S.S.; Salem, A.A.; Ghattas, M.A.; Atatreh, N.; Abdou, I.M. Antibacterial activity and mechanism of action of the benzazole acrylonitrile-based compounds: In vitro, spectroscopic, and docking studies. Eur. J. Med. Chem. 2017, 136, 270–282. [Google Scholar] [CrossRef]
- Romeo, R.; Chiacchio, M.A.; Campisi, A.; Monciino, G.; Veltri, L.; Iannazzo, D.; Broggini, G.; Giofre, S.V. Synthesis and Biological Evaluation of Pyrimidine-oxazolidin-2-arylimino Hybrid Molecules as Antibacterial Agents. Molecules 2018, 23, 1754. [Google Scholar] [CrossRef]
- Alwan, S.M. Synthesis and Preliminary Antimicrobial Activity of New Schiff Bases of Pyrido [1,2-A] Pyrimidine Derivatives with Certain Amino Acids. Med. Chem. 2014, 4, 635–639. [Google Scholar] [CrossRef]
- Neumann, D.M.; Cammarata, A.; Backes, G.; Palmer, G.E.; Jursic, B.S. Synthesis and antifungal activity of substituted 2,4,6-pyrimidinetrione carbaldehyde hydrazones. Bioorg. Med. Chem. 2014, 22, 813–826. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Wang, W.; Zhou, Y.; Xia, Q.; Ren, Y.; Feng, J.; Peng, H.; He, H.; Feng, L. Rational design, synthesis and biological evaluation of 1,3,4-oxadiazole pyrimidine derivatives as novel pyruvate dehydrogenase complex E1 inhibitors. Bioorg. Med. Chem. 2016, 24, 1879–1888. [Google Scholar] [CrossRef]
- He, H.; Xia, H.; Xia, Q.; Ren, Y.; He, H. Design and optimization of N-acylhydrazone pyrimidine derivatives as E. coli PDHc E1 inhibitors: Structure-activity relationship analysis, biological evaluation and molecular docking study. Bioorg. Med. Chem. 2017, 25, 5652–5661. [Google Scholar] [CrossRef]
- Parikh, K.S.; Vyas, S.P. Synthesis and Spectral Studies of some Novel Schiff Base derived with Pyrimidines. J. Chem. Pharm. Res. 2012, 4, 2109–2111. [Google Scholar]
- Fırıncı, E. Pyrimidine-2,4,6-trione copper(II) complexes and their catalytic activities in the peroxidative oxidation of cyclohexane. J. Mol. Struct. 2019, 1193, 125–130. [Google Scholar] [CrossRef]
- Schiff, H. Mittheilungen aus dem Universitätslaboratorium in Pisa: Eine neue Reihe organischer Basen. Justus Liebigs Ann. Chem. 1864, 131, 118–119. [Google Scholar] [CrossRef]
- Stacy, G.W.; Day, R.I.; Morath, R.J. Schiff Bases and Related Substances. II. Reactions of Thiols with N-Benzylideneaniline and N-Benzylideneanthranilic Acid1. J. Am. Chem. Soc. 1955, 77, 3869–3873. [Google Scholar] [CrossRef]
- Thomas, A.B.; Tupe, P.N.; Badhe, R.V.; Nanda, R.K.; Kothapalli, L.P.; Paradkar, O.D.; Sharma, P.A.; Deshpande, A.D. Green route synthesis of Schiff’s bases of isonicotinic acid hydrazide. Green Chem. Lett. Rev. 2009, 2, 23–27. [Google Scholar] [CrossRef]
- Chakraborti, A.K.; Bhagat, S.; Rudrawar, S. Magnesium perchlorate as an efficient catalyst for the synthesis of imines and phenylhydrazones. Tetrahedron Lett. 2004, 45, 7641–7644. [Google Scholar] [CrossRef]
- Vass, A.; Dudás, J.; Varma, R.S. Solvent-free synthesis of N-sulfonylimines using microwave irradiation. Tetrahedron Lett. 1999, 40, 4951–4954. [Google Scholar] [CrossRef]
- Avaji, P.G.; Vinod Kumar, C.H.; Patil, S.A.; Shivananda, K.N.; Nagaraju, C. Synthesis, spectral characterization, in-vitro microbiological evaluation and cytotoxic activities of novel macrocyclic bis hydrazone. Eur. J. Med. Chem. 2009, 44, 3552–3559. [Google Scholar] [CrossRef]
- Miri, R.; Razzaghi-asl, N.; Mohammadi, M.K. QM study and conformational analysis of an isatin Schiff base as a potential cytotoxic agent. J. Mol. Model. 2013, 19, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.M.M.; Azad, M.A.K.; Jesmin, M.; Ahsan, S.; Rahman, M.M.; Khanam, J.A.; Islam, M.N.; Shahriar, S.M.S. In vivo anticancer activity of vanillin semicarbazone. Asian Pac. J. Trop. Biomed. 2012, 2, 438–442. [Google Scholar] [CrossRef]
- Sondhi, S.M.; Singh, N.; Kumar, A.; Lozach, O.; Meijer, L. Synthesis, anti-inflammatory, analgesic and kinase (CDK-1, CDK-5 and GSK-3) inhibition activity evaluation of benzimidazole/benzoxazole derivatives and some Schiff’s bases. Bioorg. Med. Chem. 2006, 14, 3758–3765. [Google Scholar] [CrossRef]
- Chinnasamy, R.P.; Sundararajan, R.; Govindaraj, S. Synthesis, characterization, and analgesic activity of novel schiff base of isatin derivatives. J. Adv. Pharm. Technol. Res. 2010, 1, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Cieplik, J.; Raginia, G.; Pluta, J.; Gubrynowicz, O.; Bryndal, I.; Lis, T. Synthesis and antibacterial properties of 1,2,3-aryl-1,2,3,4-tetrahydropyrimido[4,5-d]pyrimidine derivatives. Acta Pol. Pharm. 2008, 65, 427–434. [Google Scholar] [PubMed]
- Cieplik, J.; Stolarczyk, M.; Pluta, J.; Gubrynowicz, O.; Bryndal, I.; Lis, T.; Mikulewicz, M. Synthesis and antibacterial properties of pyrimidine derivatives. Acta Pol. Pharm. 2015, 72, 53–64. [Google Scholar] [PubMed]
- Wang, B.; Zhao, B.; Chen, Z.S.; Pang, L.P.; Zhao, Y.D.; Guo, Q.; Zhang, X.H.; Liu, Y.; Liu, G.Y.; Zhang, H.; et al. Exploration of 1,2,3-triazole-pyrimidine hybrids as potent reversal agents against ABCB1-mediated multidrug resistance. Eur. J. Med. Chem. 2018, 143, 1535–1542. [Google Scholar] [CrossRef] [PubMed]
- Cieplik, J.; Pluta, J.; Gubrynowicz, O. Synthesis and antibacterial activity of 1,3-diarylpyrimido[4,5-d]pyrimidines. Boll. Chim. Farm. 2003, 142, 146–150. [Google Scholar]
- Cieplik, J.; Stolarczyk, M.; Pluta, J.; Gubrynowicz, O.; Bryndal, I.; Lis, T.; Mikulewicz, M. Synthesis and antibacterial properties of pyrimidine derivatives. Acta Pol. Pharm. 2011, 68, 57–65. [Google Scholar] [PubMed]
- Cieplik, J.; Pluta, J.; Gubrynowicz, O. Synthesis and antimicrobial properties of 3-sulfonyl-1,2,3,4-tetrahydropyrimido [4,5-d] pyrimidines. Boll. Chim. Farm. 2004, 143, 321–328. [Google Scholar] [PubMed]
- Corey, E.J.; Suggs, J.W. Pyridinium chlorochromate. An efficient reagent for oxidation of primary and secondary alcohols to carbonyl compounds. Tetrahedron Lett. 1975, 16, 2647–2650. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.-L. Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew. Chem. Int. Ed. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Cieplik, J.; Pluta, J.; Bryndal, I.; Lis, T. Two polymorphic forms of N-(4-chlorophenyl)-5-[(4-chlorophenyl)aminomethyl]-6-methyl-2-phenylpyrimidin-4-amine. Acta Crystallogr. C 2006, 62, 259–261. [Google Scholar] [CrossRef]
- Cieplik, J.; Pluta, J.; Bryndal, I.; Lis, T. N-(2-Fluorophenyl)-5-[(4-methoxyphenyl)aminomethyl]-6-methyl-2-phenylpyrimidin-4-amine. Acta Crystallogr. E 2011, 67, 3162. [Google Scholar] [CrossRef]
- Cieplik, J.; Stolarczyk, M.; Bryndal, I.; Lis, T. 5-[(4-Ethoxyanilino)methyl]-N-(2-fluorophenyl)-6-methyl-2-phenylpyrimidin-4-amine. Acta Crystallogr. E 2012, 68, 1729–1730. [Google Scholar] [CrossRef]
- Vermes, I.; Haanen, C.; Steffens-Nakken, H.; Reutellingsperger, C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 1995, 184, 39–51. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 10. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 13 May 2020).
- Lavilla Lerma, L.; Benomar, N.; Valenzuela, A.S.; Muñoz, M.D.C.C.; Gálvez, A.; Abriouel, H. Role of EfrAB efflux pump in biocide tolerance and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from traditional fermented foods and the effect of EDTA as EfrAB inhibitor. Food Microbiol. 2014, 44, 249–257. [Google Scholar] [CrossRef]
- Selleck, E.M.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- International Organization for Standardization ISO 20776-1:2019. Available online: https://www.iso.org/cms/render/live/en/sites/isoorg/contents/data/standard/07/04/70464.html (accessed on 13 May 2020).
- Gabrielson, J.; Hart, M.; Jarelöv, A.; Kühn, I.; McKenzie, D.; Möllby, R. Evaluation of redox indicators and the use of digital scanners and spectrophotometer for quantification of microbial growth in microplates. J. Microbiol. Methods 2002, 50, 63–73. [Google Scholar] [CrossRef]
- Francisco, F.L.; Saviano, A.M.; Pinto, T.D.J.A.; Lourenço, F.R. Development, optimization and validation of a rapid colorimetric microplate bioassay for neomycin sulfate in pharmaceutical drug products. J. Microbiol. Methods 2014, 103, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Sabaeifard, P.; Abdi-Ali, A.; Soudi, M.R.; Dinarvand, R. Optimization of tetrazolium salt assay for Pseudomonas aeruginosa biofilm using microtiter plate method. J. Microbiol. Methods 2014, 105, 134–140. [Google Scholar] [CrossRef]
- Feely, W.; Lehn, W.; Boekelheide, V. Communications—Alkaline Decomposition of Quaternary Salts of Amine Oxides. J. Org. Chem. 1957, 22, 1135. [Google Scholar] [CrossRef]
- Mukaiyama, S.; Inanaga, J.; Yamaguchi, M. 4-Dimethylaminopyridine N-Oxide as an Efficient Oxidizing Agent for Alkyl Halides. Bull. Chem. Soc. Jpn. 1981, 54, 2221–2222. [Google Scholar] [CrossRef]
- Chen, D.X.; Ho, C.M.; Wu, Q.Y.R.; Wu, P.R.; Wong, F.M.; Wu, W. Convenient oxidation of benzylic and allylic halides to aldehydes and ketones. Tetrahedron Lett. 2008, 49, 4147–4148. [Google Scholar] [CrossRef]
- Franzen, V.; Otto, S. Eine neue Methode zur Darstellung von Carbonylverbindungen. Chem. Ber. 1961, 94, 1360–1363. [Google Scholar] [CrossRef]
- Suzuki, S.; Onishi, T.; Fujita, Y.; Misawa, H.; Otera, J. A Convenient Method for Conversion of Allylic Chlorides to α,β-Unsaturated Aldehydes. Bull. Chem. Soc. Jpn. 1986, 59, 3287–3288. [Google Scholar] [CrossRef]
- Larsson, R.; Ross, D.; Nordenskjöld, M.; Lindeke, B.; Olsson, L.-I.; Moldéus, P. Reactive products formed by peroxidase catalyzed oxidation of p-phenetidine. Chem. Biol. Interact. 1984, 52, 1–14. [Google Scholar] [CrossRef]
- Porubek, D.; Rundgren, M.; Larsson, R.; Albano, E.; Ross, D.; Nelson, S.D.; Moldéus, P. Quinone Imines as Biological Reactive Intermediates. In Biological Reactive Intermediates III: Mechanisms of Action in Animal Models and Human Disease, 1st ed.; Kocsis, J.J., Jollow, D.J., Witmer, C.M., Nelson, J.O., Snyder, R., Eds.; Springer: Boston, MA, USA, 1986; pp. 631–644. ISBN 978-1-4684-5134-4. [Google Scholar] [CrossRef]
- Mitchell, J.R.; Jollow, D.J.; Potter, W.Z.; Gillette, J.R.; Brodie, B.B. Acetaminophen-induced hepatic necrosis. IV. Protective role of glutathione. J. Pharmacol. Exp. Ther. 1973, 187, 211–217. [Google Scholar] [PubMed]
- Moore, M.; Thor, H.; Moore, G.; Nelson, S.; Moldéus, P.; Orrenius, S. The toxicity of acetaminophen and N-acetyl-p-benzoquinone imine in isolated hepatocytes is associated with thiol depletion and increased cytosolic Ca2+. J. Biol. Chem. 1985, 260, 13035–13040. [Google Scholar] [CrossRef]
- Rosen, G.M.; Singletary, W.V.; Rauckman, E.J.; Killenberg, P.G. Acetaminophen hepatotoxicity: An alternative mechanism. Biochem. Pharmacol. 1983, 32, 2053–2059. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. B 2016, 72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- CrysAlis PRO; Agilent Technologies Ltd.: Yarnton, Oxfordshire, UK, 2014.
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, K. DIAMOND; Crystal Impact GbR: Bonn, Germany, 1999. [Google Scholar]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Rotman, B.; Papermaster, B.W. Membrane properties of living mammalian cells as studied by enzymatic hydrolysis of fluorogenic esters. Proc. Natl. Acad. Sci. USA 1966, 55, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.H.; Senft, J.A. An improved method to determine cell viability by simultaneous staining with fluorescein diacetate-propidium iodide. J. Histochem. Cytochem. 2017, 33, 77–79. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; CLSI: Wayne, PA, USA, 2018; ISBN 1-56238-837-1. [Google Scholar]
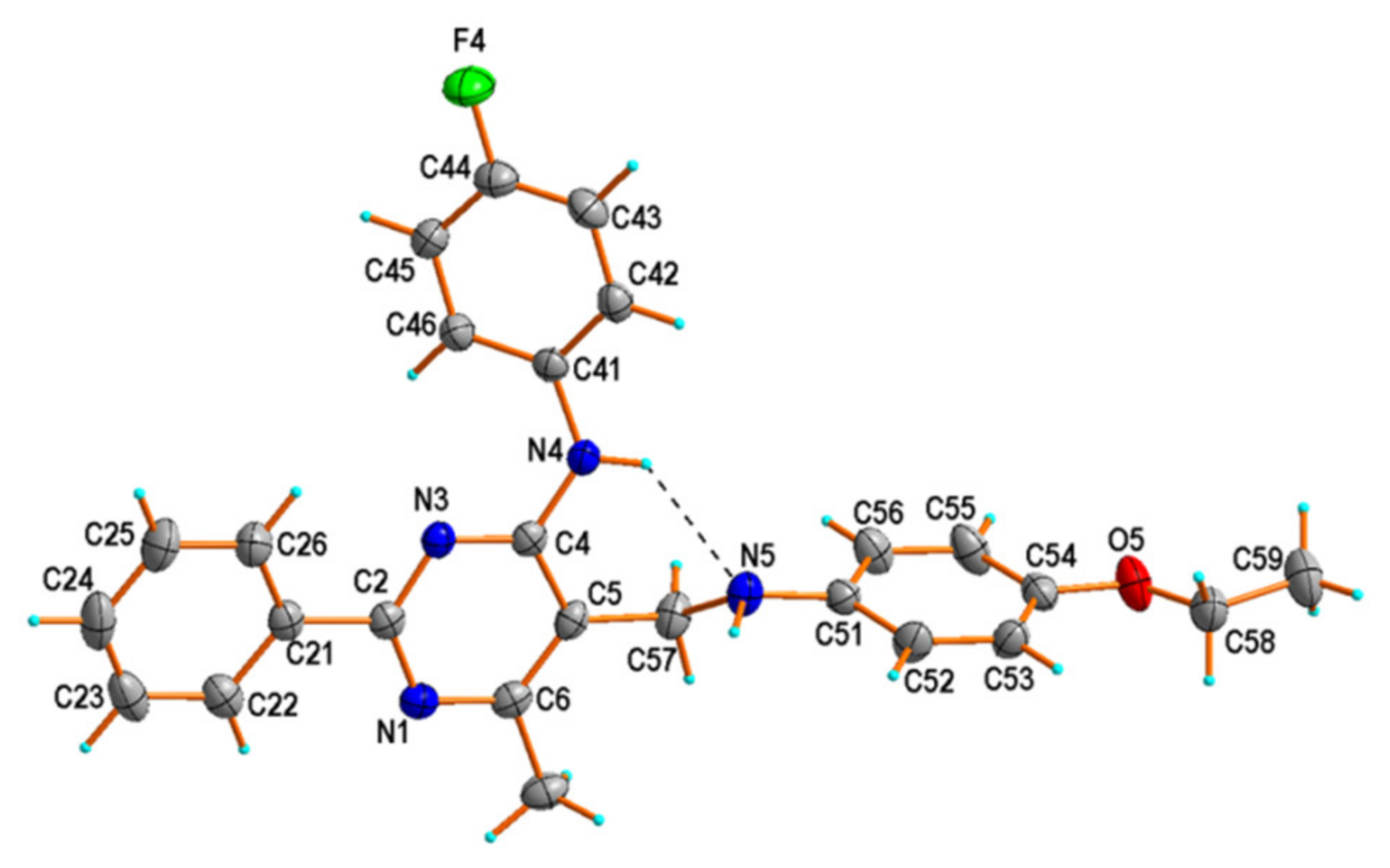
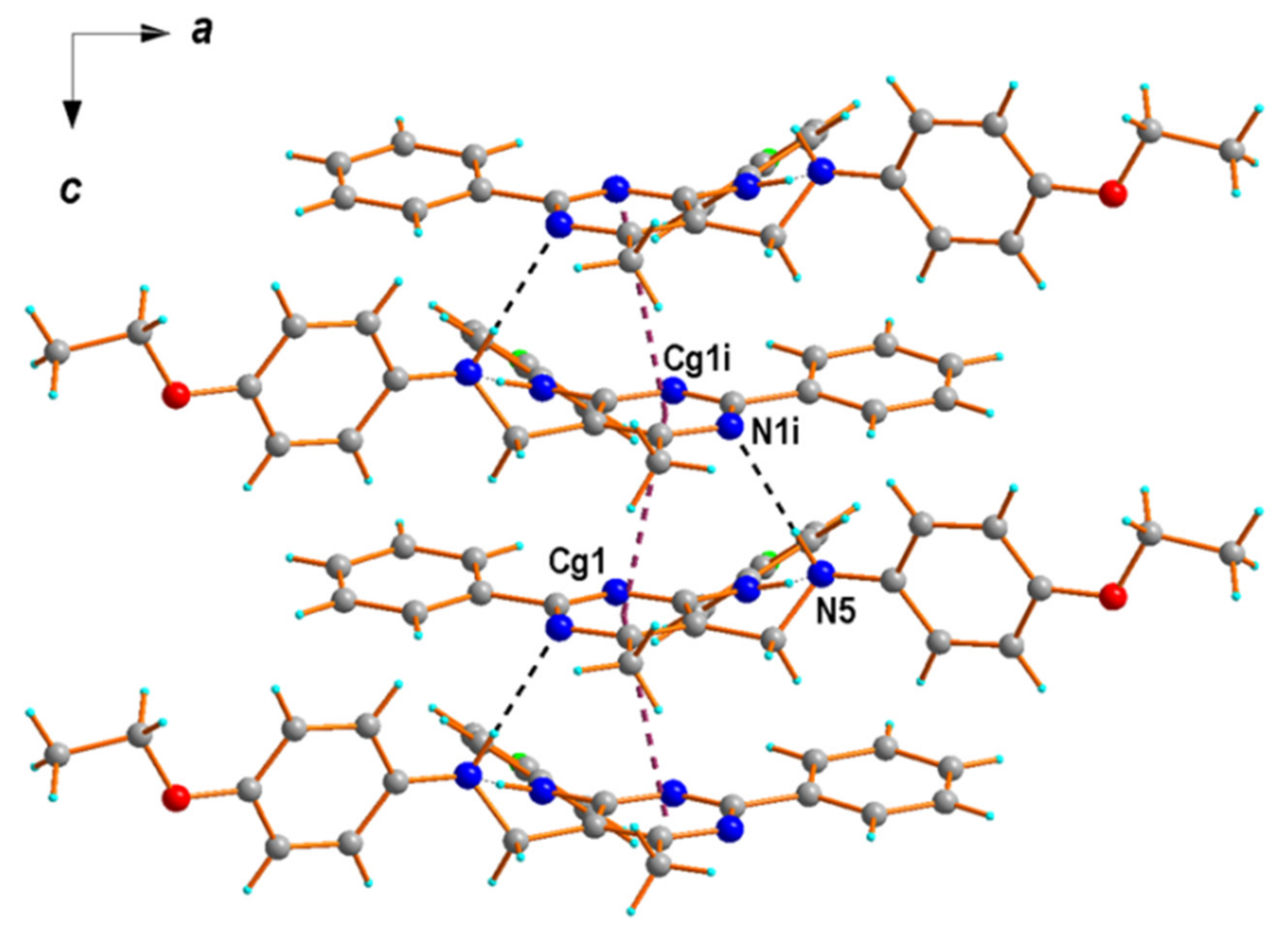





| D—H⋯A | d(D—H) | d(H⋯A) | d(D⋯A) | <(D—H⋯A) |
|---|---|---|---|---|
| Compound 2 | ||||
| N4—H4⋯N5 | 0.860 (17) | 2.277 (17) | 2.9365 (18) | 133.6 (14) |
| N5—H5⋯N1i | 0.93 (2) | 2.32 (2) | 3.1358 (18) | 145.4 (14) |
| C46—H46⋯N3 | 0.95 | 2.40 | 2.8894 (18) | 111 |
| C43—H43⋯O5ii | 0.95 | 2.59 | 3.496 (2) | 159 |
| C61—H611⋯F4iii | 0.98 | 2.54 | 3.277 (2) | 132 |
| Compound 3 | ||||
| N4—H4⋯N5 | 1.00 (2) | 1.75 (2) | 2.664 (2) | 150.4 (17) |
| C46—H46⋯N3 | 0.95 | 2.35 | 2.918 (2) | 118 |
| C25—H25⋯O5i | 0.95 | 2.53 | 3.452 (3) | 165 |
| C24—H24⋯F4ii | 0.95 | 2.63 | 3.572 (2) | 171 |
| C53—H53⋯F4iii | 0.95 | 2.64 | 3.298 (2) | 127 |
| Symmetry codes: 2: (i) – x + 1/2, y, z − 1/2; (ii) – x + 1, − y + 1, z − 1/2; (iii) – x + 1/2, y + 1, z + 1/2; 3: (i) x + 1, y, z − 1; (ii) – x + 1, y − 1/2, − z + 1/2; (iii) − x, y − 1/2, − z + 3/2. | ||||
| Strain | Enterococcus faecalis ATCC 29212 | Streptococcus pneumonia ATCC 49619 | Staphylococcus aureus ATCC 43300 | Escherichia coli ATCC 25922 | Klebsiella pneumonia ATCC 13883 | Pseudomonas aeruginosa ATCC 27853 | Candida albicans ATCC 10231 |
|---|---|---|---|---|---|---|---|
| MBC (ug/mL) | |||||||
| 2 | 1024 | n/d | n/d | n/d | n/d | n/d | n/d |
| 3 | 32 | n/d | n/d | n/d | n/d | n/d | n/d |
| MIC (ug/mL) | |||||||
| 2 | 512 | n/d | n/d | n/d | n/d | n/d | n/d |
| 3 | 16 | n/d | n/d | n/d | n/d | n/d | n/d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stolarczyk, M.; Wolska, A.; Mikołajczyk, A.; Bryndal, I.; Cieplik, J.; Lis, T.; Matera-Witkiewicz, A. A New Pyrimidine Schiff Base with Selective Activities against Enterococcus faecalis and Gastric Adenocarcinoma. Molecules 2021, 26, 2296. https://doi.org/10.3390/molecules26082296
Stolarczyk M, Wolska A, Mikołajczyk A, Bryndal I, Cieplik J, Lis T, Matera-Witkiewicz A. A New Pyrimidine Schiff Base with Selective Activities against Enterococcus faecalis and Gastric Adenocarcinoma. Molecules. 2021; 26(8):2296. https://doi.org/10.3390/molecules26082296
Chicago/Turabian StyleStolarczyk, Marcin, Aleksandra Wolska, Aleksandra Mikołajczyk, Iwona Bryndal, Jerzy Cieplik, Tadeusz Lis, and Agnieszka Matera-Witkiewicz. 2021. "A New Pyrimidine Schiff Base with Selective Activities against Enterococcus faecalis and Gastric Adenocarcinoma" Molecules 26, no. 8: 2296. https://doi.org/10.3390/molecules26082296
APA StyleStolarczyk, M., Wolska, A., Mikołajczyk, A., Bryndal, I., Cieplik, J., Lis, T., & Matera-Witkiewicz, A. (2021). A New Pyrimidine Schiff Base with Selective Activities against Enterococcus faecalis and Gastric Adenocarcinoma. Molecules, 26(8), 2296. https://doi.org/10.3390/molecules26082296






