Allicin, the Odor of Freshly Crushed Garlic: A Review of Recent Progress in Understanding Allicin’s Effects on Cells §
Abstract
1. Introduction
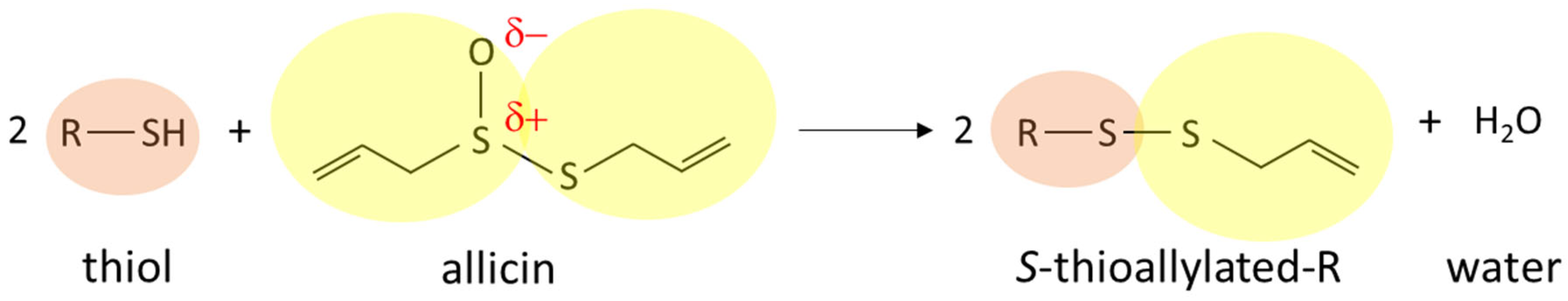
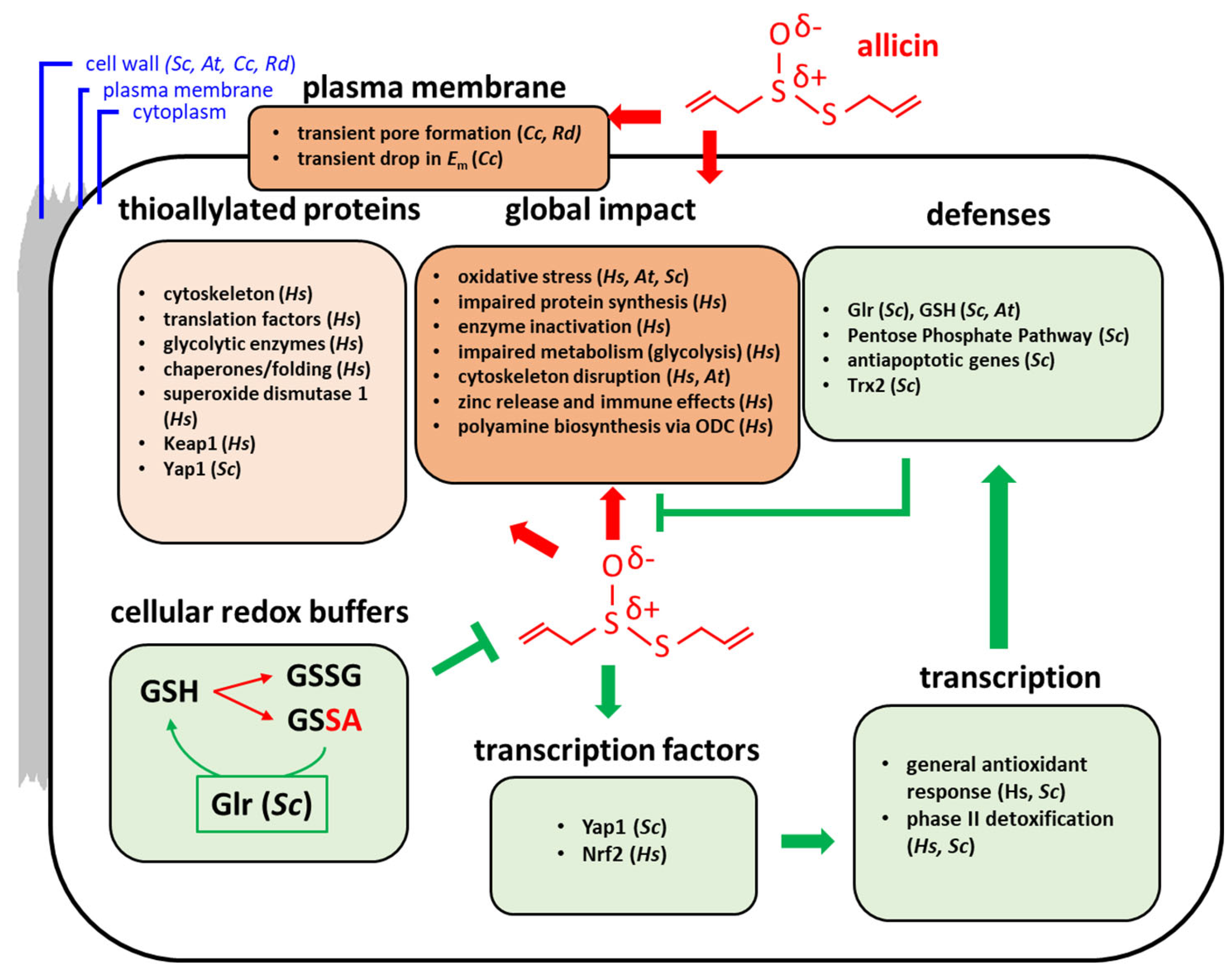
2. Allicin and Oxidative Stress in Cells and Organisms
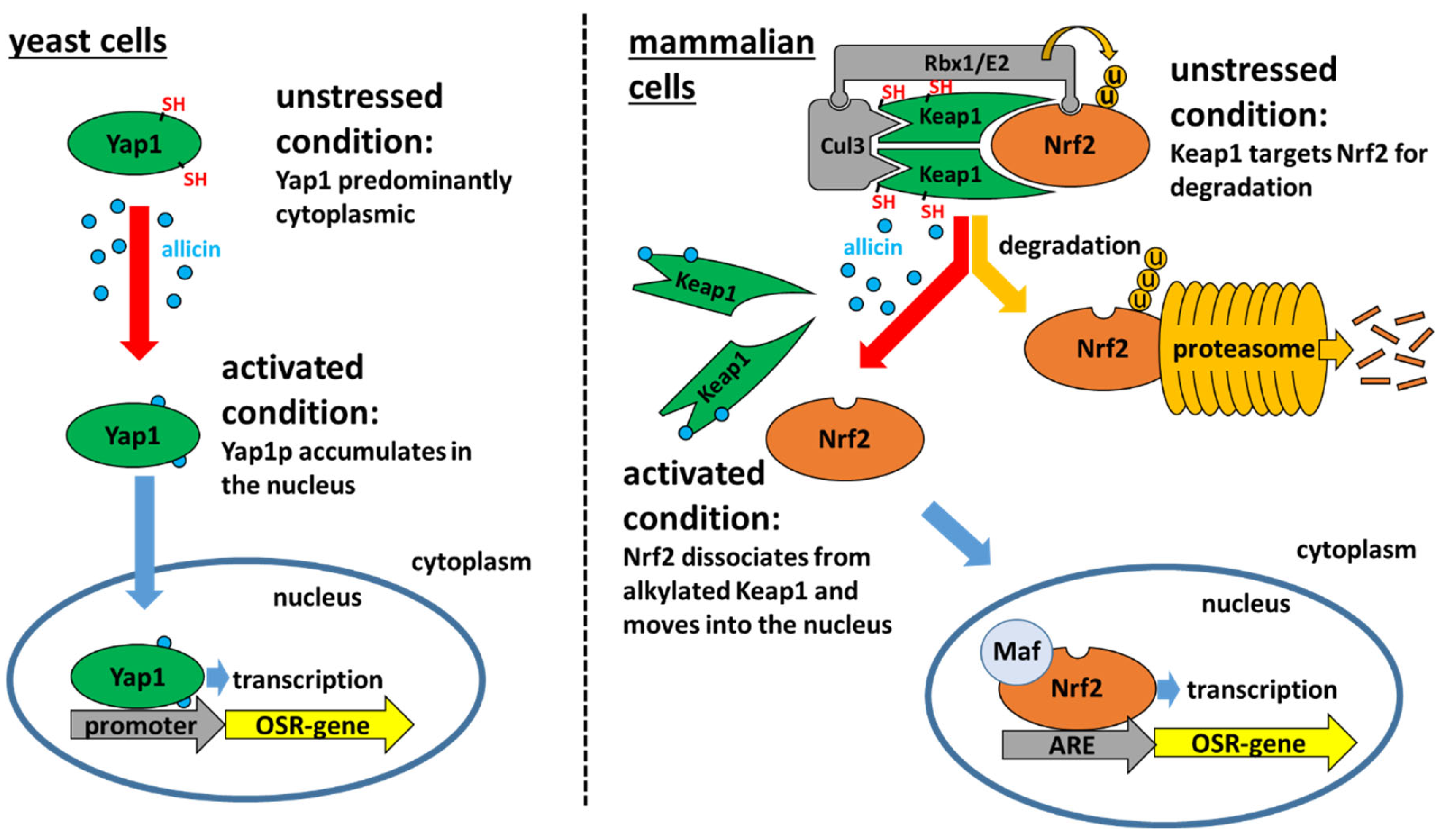
2.1. The Yeast Yap1 Oxidative Stress Sensor System
2.2. The Mammalian Keap1-Nrf2/ARE Oxidative Stress Sensor System
2.3. Bacterial Redox Sensors
2.4. The Central Role of GSH Metabolism in Allicin Resistance
2.4.1. Plants (Arabidopsis thaliana)
2.4.2. Yeast (Saccharomyces cerevisiae)
2.4.3. Glutathione in Gram-Negative Bacteria (E. coli, Pseudomonas spp.) and Bacillithiol in Gram-Positive Firmicutes (B. subtilis, S. aureus)
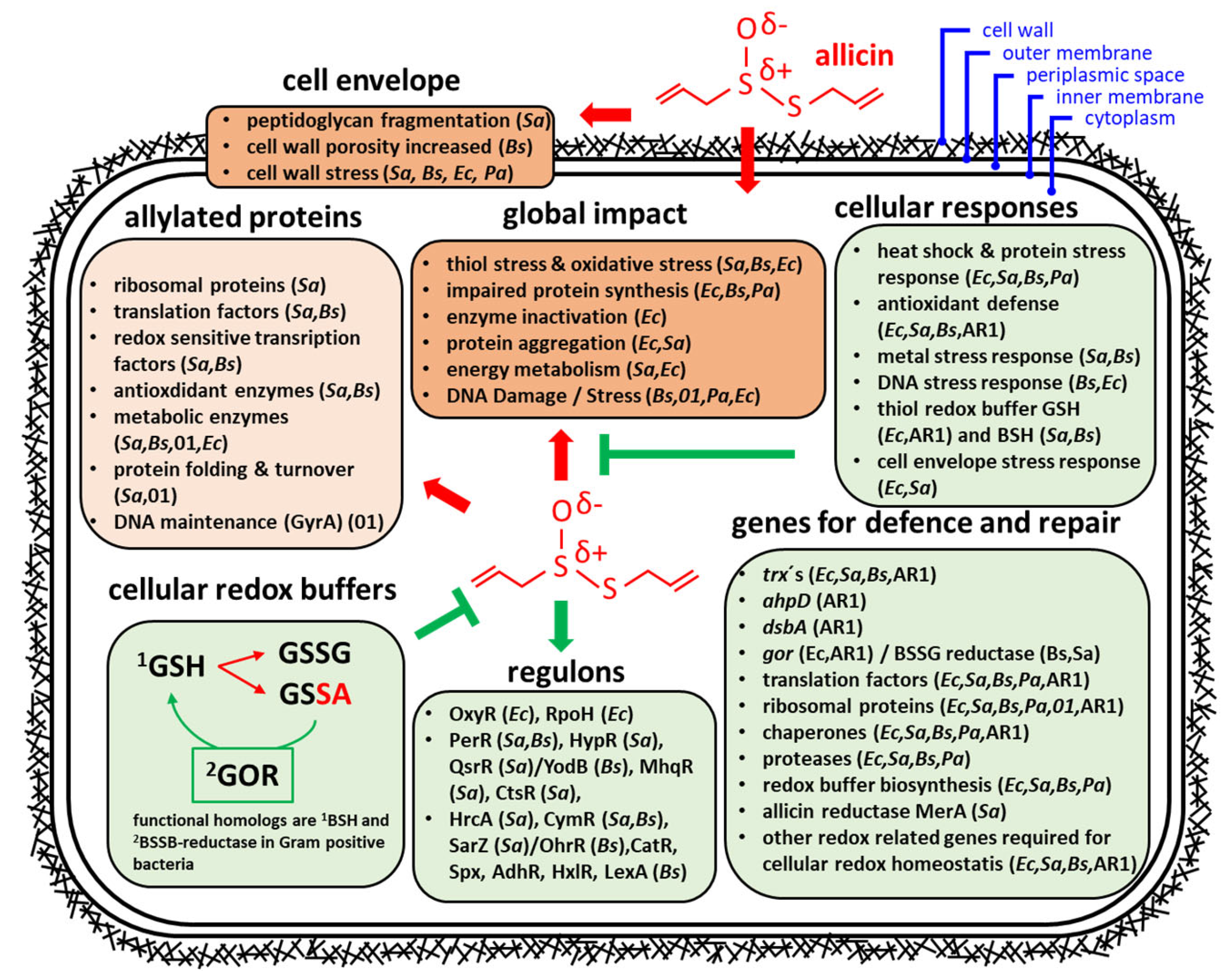
3. Transcriptomic and Proteomic Studies
3.1. Effects of Allicin on the Transcriptome and Proteome in Bacteria
3.2. Effects of Allicin on the Proteome of Human Cells (Jurkat T Lymphocytes)
4. Allicin in Medicine
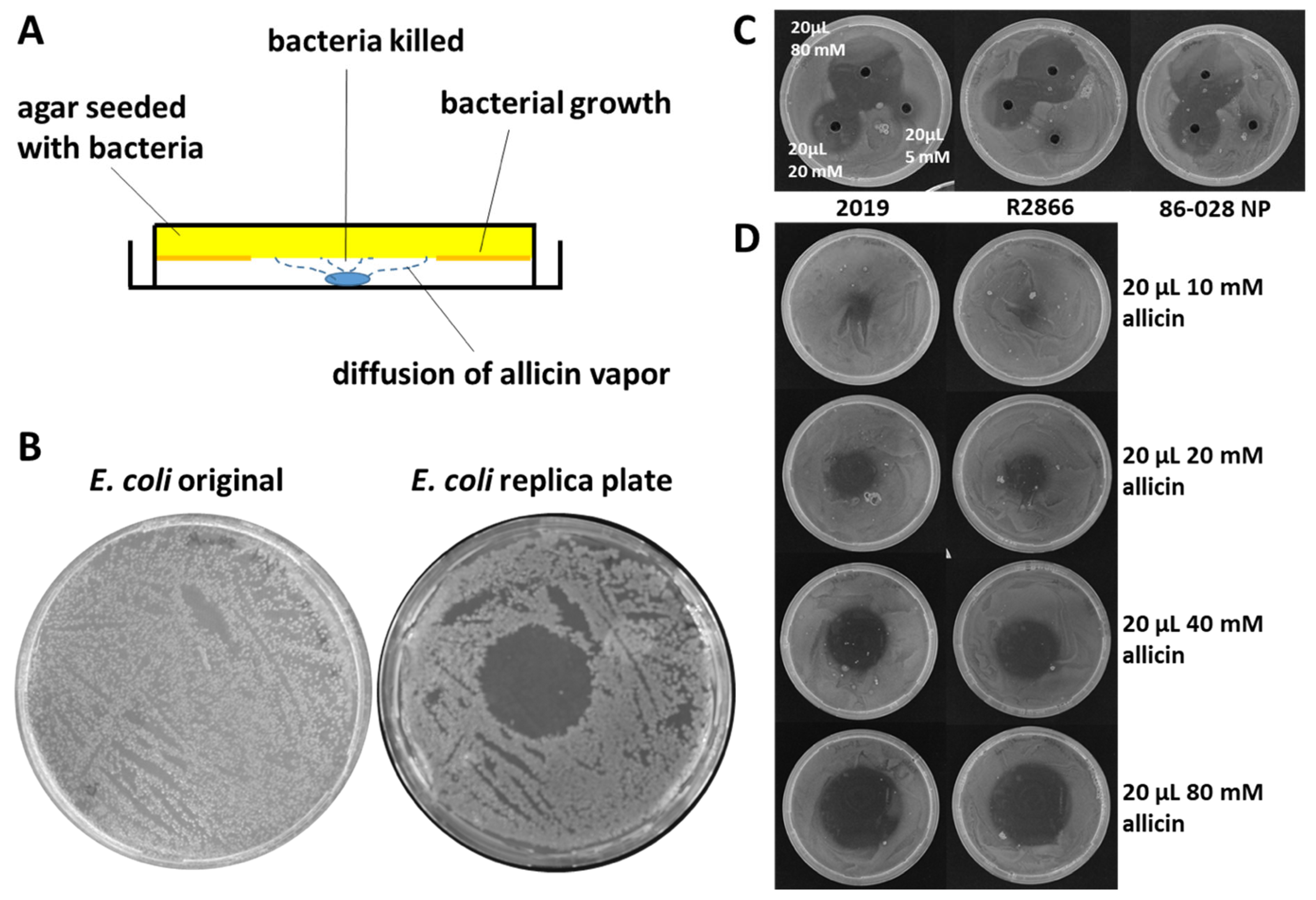
4.1. Allicin Activity against Respiratory Tract Pathogens
4.2. Allicin vs. Cancer
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cavallito, C.J.; Bailey, H.J. Allicin, the antibacterial principle of Allium sativum I. Isolation, physical properties and antibacterial action. J. Am. Chem. Soc. 1944, 66, 1950–1951. [Google Scholar] [CrossRef]
- Cavallito, C.J.; Buck, J.S.; Suter, C.M. Allicin, the antibacterial principle of Allium sativum II. Determination of the chemical structure. J. Am. Chem. Soc. 1944, 66, 1952–1954. [Google Scholar] [CrossRef]
- Lawson, L.D.; Wood, S.G.; Hughes, B.G. HPLC analysis of allicin and other thiosulfinates in garlic clove homogenates. Planta Med. 1991, 57, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.E.; Collin, H.A. Presence of alliinase in isolated vacuoles and of alkyl cysteine sulphoxides in the cytoplasm of bulbs of onion (Allium cepa). Plant Sci. Lett. 1981, 22, 169–176. [Google Scholar] [CrossRef]
- Block, E.; Naganathan, S.; Putman, D.; Zhao, S.H. Allium chemistry: HPLC Analysis of Thiosulfinates from Onion, Garlic, Wild Garlic (Ramsons), Leek, Scallion, Shallot, Elephant (Great-Headed) Garlic, Chive, and Chinese Chive. Uniquely High Allyl to Methyl Ratios in Some Garlic Samples. J. Agric. Food Chem. 1992, 40, 2418–2430. [Google Scholar] [CrossRef]
- Yamazaki, M.; Sugiyama, M.; Saito, K. Intercellular Localization of Cysteine Synthase and Alliinase in Bundle Sheaths of Allium Plants. Plant Biotechnol. 2002, 19, 7–10. [Google Scholar] [CrossRef]
- Block, E. Fifty years of smelling sulfur. J. Sulfur Chem. 2013, 34, 158–207. [Google Scholar] [CrossRef]
- Lawson, L.D.; Hughes, B.G. Characterization of the formation of allicin and other thiosulfinates from garlic. Planta Med. 1992, 58, 345–350. [Google Scholar] [CrossRef]
- Albrecht, F.; Leontiev, R.; Jacob, C.; Slusarenko, A.J. An Optimized Facile Procedure to synthesize and purify Allicin. Molecules 2017, 22, 770. [Google Scholar] [CrossRef]
- Fowler, F.G.; Fowler, H.W. The Concise Oxford Dictionary of Current English, 5th ed.; Oxford Clarendon Press: Oxford, UK, 1964. [Google Scholar]
- Block, E.; Batista, V.S.; Matsunami, H.; Zhuang, H.; Ahmed, L. The role of metals in mammalian olfaction of low molecular weight organosulfur compounds. Nat. Prod. Rep. 2017, 34, 529–597. [Google Scholar] [CrossRef]
- Hile, A.G.; Shan, Z.; Zhang, S.Z.; Block, E. Aversion of European Starlings (Sturnus vulgaris) to Garlic Oil Treated Granules: Garlic Oil as an Avian Repellent. Garlic Oil Analysis by Nuclear Magnetic Resonance Spectroscopy. Agric. Food Chem. 2004, 52, 2192–2196. [Google Scholar] [CrossRef] [PubMed]
- Bautista, D.M.; Movahed, P.; Hinman, A.; Axelsson, H.E.; Sterner, O.; Högestätt, E.D.; Julius, D.; Jordt, S.-E.; Zygmunt, P.M. Pungent products from garlic activate the sensory ion channel TRPA1. Proc. Natl. Acad. Sci. USA 2005, 102, 12248–12252. [Google Scholar] [CrossRef]
- Macpherson, L.J.; Geierstanger, B.H.; Viswanath, V.; Bandell, M.; Eid, S.R.; Hwang, S.-W.; Patapoutian, A. The Pungency of Garlic: Activation of TRPA1 and TRPV1 in Response to Allicin. Curr. Biol. 2005, 15, 929–934. [Google Scholar] [CrossRef]
- Paulsen, C.; Armache, J.; Gao, Y.; Cheng, Y.; Julius, D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 2015, 520, 511–517. [Google Scholar] [CrossRef]
- Mihara, S.; Shibamoto, T. The role of flavor and fragrance chemicals in TRPA1 (transient receptor potential cation channel, member A1) activity associated with allergies. All. Asth. Clin. Immun. 2015, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, L.; Lee, C.Y.; Matsuo, T.; Wu, K.; Asher, G.; Tang, L.; Saitoh, T.; Russell, J.; Klewe-Nebenius, D.; et al. Large-scale forward genetics screening identifies Trpa1 as a chemosensor for predator odor-evoked innate fear behaviors. Nat. Commun. 2018, 9, 2041. [Google Scholar] [CrossRef]
- Ninkovic, V.; Rensing, M.; Dahlin, I.; Markovic, D. Who is my neighbor? Volatile cues in plant interactions. Plant. Signaling Behav. 2019, 14, 1634993. [Google Scholar] [CrossRef] [PubMed]
- Borlinghaus, J.; Albrecht, F.; Gruhlke, M.C.H.; Nwachukwu, I.D.; Slusarenko, A.J. Allicin: Chemistry and biological properties. Molecules 2014, 19, 12591–12618. [Google Scholar] [CrossRef]
- Kong, C.H.; Xuan, T.D.; Khanh, T.D.; Tran, H.D.; Trung, N.T. Allelochemicals and Signaling Chemicals in Plants. Molecules 2019, 24, 2737. [Google Scholar] [CrossRef]
- Block, E. Garlic and the other Alliums. The Lore and the Science, 1st ed.; RSC Publishing: Cambridge, UK, 2010. [Google Scholar]
- Anwar, A.; Groom, M.; Arbach, M.; Hamilton, C.J. How to Turn the Chemistry of Garlic into a ‘Botanical’ Pesticide. In Recent Advances in Redox Active Plant and Microbial Products: From Basic Chemistry to Widespread Applications in Medicine and Agriculture; Jacob, C., Slusarenko, A.J., Kirsch, G., Winyard, P., Eds.; Springer: Berlin, Germany, 2014; pp. 323–341. [Google Scholar]
- Miron, T.; Rabinkov, A.; Mirelman, D.; Wilchek, M.; Weiner, L. The mode of action of allicin: Its ready permeability through phospholipid membranes may contribute to its biological activity. Biochim. Biophys. Acta BBA Biomembranes 2000, 1463, 20–30. [Google Scholar] [CrossRef]
- Gruhlke, M.C.H.; Hemmis, B.; Noll, U.; Wagner, R.; Lühring, H.; Slusarenko, A.J. The defense substance allicin from garlic permeabilizes membranes of Beta vulgaris, Rhoeo discolor, Chara corallina and artificial lipid bilayers. Biochim. Biophys. Acta BBA 2015, 1850, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Gurtovenko, A.A.; Anwar, J. Modulating the Structure and Properties of Cell Membranes: The Molecular Mechanism of Action of Dimethyl Sulfoxide. J. Phys. Chem. B. 2007, 111, 10453–10460. [Google Scholar] [CrossRef]
- Rabinkov, A.; Miron, T.; Konstantinovski, L.; Wilchek, M.; Mirelman, D.; Weiner, L. The mode of action of allicin: Trapping of radicals and interaction with thiol containing proteins. Biochim. Biophys. Acta BBA 1998, 1379, 233–234. [Google Scholar] [CrossRef]
- Rabinkov, A.; Miron, T.; Mirelman, D.; Wilchek, M.; Glozman, S.; Yavin, E.; Weiner, L. S-allylmercaptoglutathione: The reaction product of allicin with glutathione possesses SH-modifying and antioxidant properties. Biochim. Biophys. Acta BBA Mol. Cell Res. 2000, 1499, 144–153. [Google Scholar] [CrossRef]
- Giles, G.I.; Jacob, C. Reactive Sulfur Species: An Emerging Concept in Oxidative Stress. Biol. Chem. 2002, 383, 375–388. [Google Scholar] [CrossRef]
- Giles, G.I.; Tasker, K.M.; Collins, C.; Giles, N.M.; O’Rourke, E.; Jacob, C. Reactive sulphur species: An in vitro investigation of the oxidation properties of disulphide S-oxides. Biochem. J. 2001, 364, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Gruhlke, M.C.H.; Slusarenko, A.J. The biology of reactive sulfur species (RSS). Plant. Physiol. Biochem. 2012, 59, 98–107. [Google Scholar] [CrossRef]
- Müller, A.; Eller, J.; Albrecht, F.; Prochnow, P.; Kuhlmann, K.; Bandow, J.E.; Slusarenko, A.J.; Leichert, L.I. Allicin Induces Thiol Stress in Bacteria through S-Allylmercapto Modification of Protein Cysteines. J. Biol. Chem. 2016, 291, 11477–11490. [Google Scholar] [CrossRef]
- Schafer, F.Q.; Buettner, G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001, 30, 1191–1212. [Google Scholar] [CrossRef]
- Poole, L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015, 80, 148–157. [Google Scholar] [CrossRef]
- Horn, T.; Bettray, W.; Slusarenko, A.J.; Gruhlke, M.C.H. S-allylmercaptoglutathione Is a Substrate for Glutathione Reductase (E.C. 1.8.1.7) from Yeast (Saccharomyces cerevisiae). Antioxidants 2018, 7, 86. [Google Scholar] [CrossRef]
- Ankri, S.; Mirelman, D. Antimicrobial properties of allicin from garlic. Microbes Infect. 1999, 2, 125–129. [Google Scholar] [CrossRef]
- Slusarenko, A.J.; Patel, A.; Portz, D. Control of plant diseases by natural products: Allicin from garlic as a case study. Eur. J. Plant. Pathol. 2008, 121, 313–322. [Google Scholar] [CrossRef]
- Vaidya, V.; Ingold, K.U.; Pratt, D.A. Garlic: Source of the Ultimate Antioxidants—Sulfenic Acids. Angew. Chem. Int. Ed. 2009, 48, 157–160. [Google Scholar] [CrossRef]
- Banerjee, S.K.; Mukherjee, P.K.; Maulik, S.K. Garlic as an antioxidant: The good, the bad and the ugly. Phytother. Res. 2003, 17, 97–106. [Google Scholar] [CrossRef]
- Gruhlke, M.C.H.; Slusarenko, A.J. The Cellular “Thiolstat” as an Emerging Target of Some Plant Secondary Metabolites. In Recent Advances in Redox Active Plant and Microbial Products: From Basic Chemistry to Widespread Applications in Medicine and Agriculture; Jacob, C., Slusarenko, A.J., Kirsch, G., Winyard, P., Eds.; Springer: Berlin, Germany, 2014; pp. 235–262. [Google Scholar]
- Melino, S.; Sabelli, R.; Paci, M. Allyl sulfur compounds and cellular detoxification system: Effects and perspectives in cancer therapy. Amino Acids 2011, 41, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Bat-Chen, W.; Tal, G.; Irena, P.; Zvi, L.; Betty, S. Allicin purified from fresh garlic cloves induces apoptosis in colon cancer cells via Nrf2. Nutr. Cancer 2009, 62, 947–957. [Google Scholar] [CrossRef]
- Surh, Y.J.; Kundu, J.K.; Na, H.K. Nrf2 as a Master Redox Switch in Turning on the Cellular Signaling Involved in the Induction of Cytoprotective Genes by Some Chemopreventive Phytochemicals. Planta Med. 2008, 74, 1526–1539. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Hampton, M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008, 45, 549–561. [Google Scholar] [CrossRef]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Garin, J.; Labarre, J.; Toledano, M.B. Yap1 andSkn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar] [CrossRef]
- Carmel-Harel, O.; Storz, G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 2000, 51, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Kuge, S.; Jones, N.; Nomoto, A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997, 16, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, K.; Rovinsky, S.A.; Coleman, S.T.; Moye-Rowley, W.S. Oxidant-specific folding of Yap1p regulates both transcriptional activation and nuclear localization. J. Biol. Chem. 2005, 280, 40524–40533. [Google Scholar] [CrossRef]
- Gruhlke, M.C.H.; Schlembach, I.; Leontiev, R.; Uebachs, A.; Gollwitzer, P.; Weiss, A.; Delaunay, A.; Toledano, M.B.; Slusarenko, A.J. Yap1p, the central regulator of the S. cerevisiae oxidative stress response, is activated by allicin, a natural oxidant and defence substance of garlic. Free Radic. Biol. Med. 2017, 108, 793–802. [Google Scholar] [CrossRef]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.L. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef]
- Niture, S.K.; Khatri, R.; Jaiswal, A.K. Regulation ofNrf2—an update. Free Radic. Biol. Med. 2014, 66, 36–44. [Google Scholar] [CrossRef]
- Liebler, D.C.; Guengerich, F.P. Elucidating mechanisms of drug-induced toxicity. Nat. Rev. Drug Discov. 2005, 4, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Schlembach, I.; Uebachs, A.; Caspers, T.; Fragoulis, A.; Slusarenko, A.J.; Gruhlke, M.C.H. Effect of prooxidative natural products: Comparison of the OSI1 (YKL071w) promoter luciferase construct from yeast with an Nrf2/Keap reporter system. Appl. Sci. 2020, 10, 3520. [Google Scholar] [CrossRef]
- Yu, L.; Guo, N.; Meng, R.; Liu, B.; Tang, X.; Jin, J.; Cui, Y.; Deng, X. Allicin-induced global gene expression profile of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2010, 88, 219–229. [Google Scholar] [CrossRef]
- Imlay, J.A. The Molecular Mechanisms and Physiological Consequences of Oxidative Stress: Lessons from a Model Bacterium. Nature Rev. Microbiol. 2013, 11, 443. [Google Scholar] [CrossRef]
- Horsburgh, M.J.; Clements, M.O.; Crossley, H.; Ingham, E.; Foster, S.J. PerR Controls Oxidative Stress Resistance and Iron Storage Proteins and Is Required for Virulence in Staphylococcus aureus. Infect. Immun. 2001, 69, 3744. [Google Scholar] [CrossRef]
- Ji, C.-J.; Kim, J.-H.; Won, Y.-B.; Lee, Y.-E.; Choi, T.-W.; Ju, S.-Y.; Youn, H.; Helmann, J.D.; Lee, J.-W. Staphylococcus aureus PerR Is a Hypersensitive Hydrogen Peroxide Sensor Using Iron-Mediated Histidine Oxidation. J. Biol. Chem. 2015, 290, 20374–20386. [Google Scholar] [CrossRef]
- Zuber, P. Management of Oxidative Stress in Bacillus. Annu. Rev. Microbiol. 2009, 63, 575–597. [Google Scholar] [CrossRef] [PubMed]
- Loi, V.V.; Huyen, N.T.T.; Busche, T.; Tung, Q.N.; Gruhlke, M.C.H.; Kalinowski, J.; Bernhardt, J.; Slusarenko, A.J.; Antelmann, H. Staphylococcus aureus responds to allicin by global S-thioallylation - Role of the Brx/BSH/YpdA pathway and the disulfide reductase MerA to overcome allicin stress. Free Radic. Biol. Med. 2019, 139, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Chi, B.K.; Huyen, N.T.T.; Loi, V.V.; Gruhlke, M.C.H.; Schaer, M.; Mäder, U.; Maaß, S.; Becher, D.; Bernhardt, J.; Arbach, M.; et al. The Disulfide Stress Response and Protein S-thioallylation Caused by Allicin and Diallyl Polysulfanes in Bacillus subtilis as Revealed by Transcriptomics and Proteomics. Antioxidants 2019, 8, 605. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. ASCORBATE AND GLUTATHIONE: Keeping Active Oxygen Under Control. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant. Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Gruhlke, M.C.H.; Portz, D.; Stitz, M.; Anwar, A.; Schneider, T.; Jacob, C.; Schlaich, N.L.; Slusarenko, A.J. Allicin disrupts the cell’s electrochemical potential and induces apoptosis in yeast. Free Radic. Biol. Med. 2010, 49, 1916–1924. [Google Scholar] [CrossRef] [PubMed]
- Toledano, M.B.; Delaunay-Moisan, A.; Outten, C.E.; Igbaria, A. Functions and Cellular Compartmentation of the Thioredoxin and Glutathione Pathways in Yeast. Antioxid. Redox Signal. 2013, 18, 1699–1711. [Google Scholar] [CrossRef]
- Gruhlke, M.C. Reactive Sulfur Species—A new player in plant physiology? In Reactive Oxygen, Nitrogen and Sulfur Species in Plants: Production, Metabolism, Signaling; Hasanuzzaman, M., Fotopoulos, V., Nahar, K., Fujita, M., Eds.; John Wiley & Sons Ltd.: Hoboken, NY, USA, 2019; pp. 715–728. [Google Scholar]
- Considine, M.J.; Foyer, C.H. Redox regulation of plant development. Antioxid. Redox Signal. 2014, 21, 1305–1326. [Google Scholar] [CrossRef]
- Rouhier, N.; Cerveau, D.; Couturier, J.; Reichheld, J.P.; Rey, P. Involvement of thiol-based mechanisms in plant development. Biochim. Biophys Acta 2015, 1850, 1479–1496. [Google Scholar] [CrossRef] [PubMed]
- Koprivova, A.; Mugford, S.; Kopriva, S. Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant. Cell Rep. 2010, 29, 1157–1167. [Google Scholar] [CrossRef]
- Vernoux, T.; Wilson, R.C.; Seeley, K.A.; Reichheld, J.P.; Muroy, S.; Brown, S.; Maughan, S.C.; Cobbett, C.S.; van Montagu, M.; Inzé, D.; et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant. Cell 2000, 12, 97–110. [Google Scholar] [CrossRef]
- Sanchez-Fernandez, R.; Fricker, M.; Corben, L.B.; White, N.S.; Sheard, N.; Leaver, C.J.; van Montagu, M.; Inzé, D.; May, M.J. Cell proliferation and hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc. Natl. Acad. Sci. USA 1997, 94, 2745–2750. [Google Scholar] [CrossRef] [PubMed]
- Bashandy, T.; Guilleminot, J.; Vernoux, T.; Caparros-Ruiz, D.; Ljung, K.; Meyer, Y.; Reichheld, J.P. Interplay between the NADP-Linked Thioredoxin and Glutathione Systems in Arabidopsis Auxin Signaling. Plant. Cell 2010, 22, 376–391. [Google Scholar] [CrossRef]
- Leontiev, R.; Hohaus, N.; Jacob, C.; Gruhlke, M.C.H.; Slusarenko, A.J. A Comparison of the Antibacterial and Antifungal Activities of Thiosulfinate Analogues of Allicin. Sci. Rep. 2018, 8, 6763. [Google Scholar] [CrossRef]
- Horn, T.; Bettray, W.; Noll, U.; Krauskopf, F.; Huang, M.-R.; Bolm, C.; Slusarenko, A.J.; Gruhlke, M.C.H. The Sulfilimine Analogue of Allicin, S-Allyl-S-(S-allyl)-N-Cyanosulfilimine, Is Antimicrobial and Reacts with Glutathione. Antioxidants 2020, 9, 1086. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Matsuyama, T.; Hashimoto, T. Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 1999, 206, 201–206. [Google Scholar] [CrossRef]
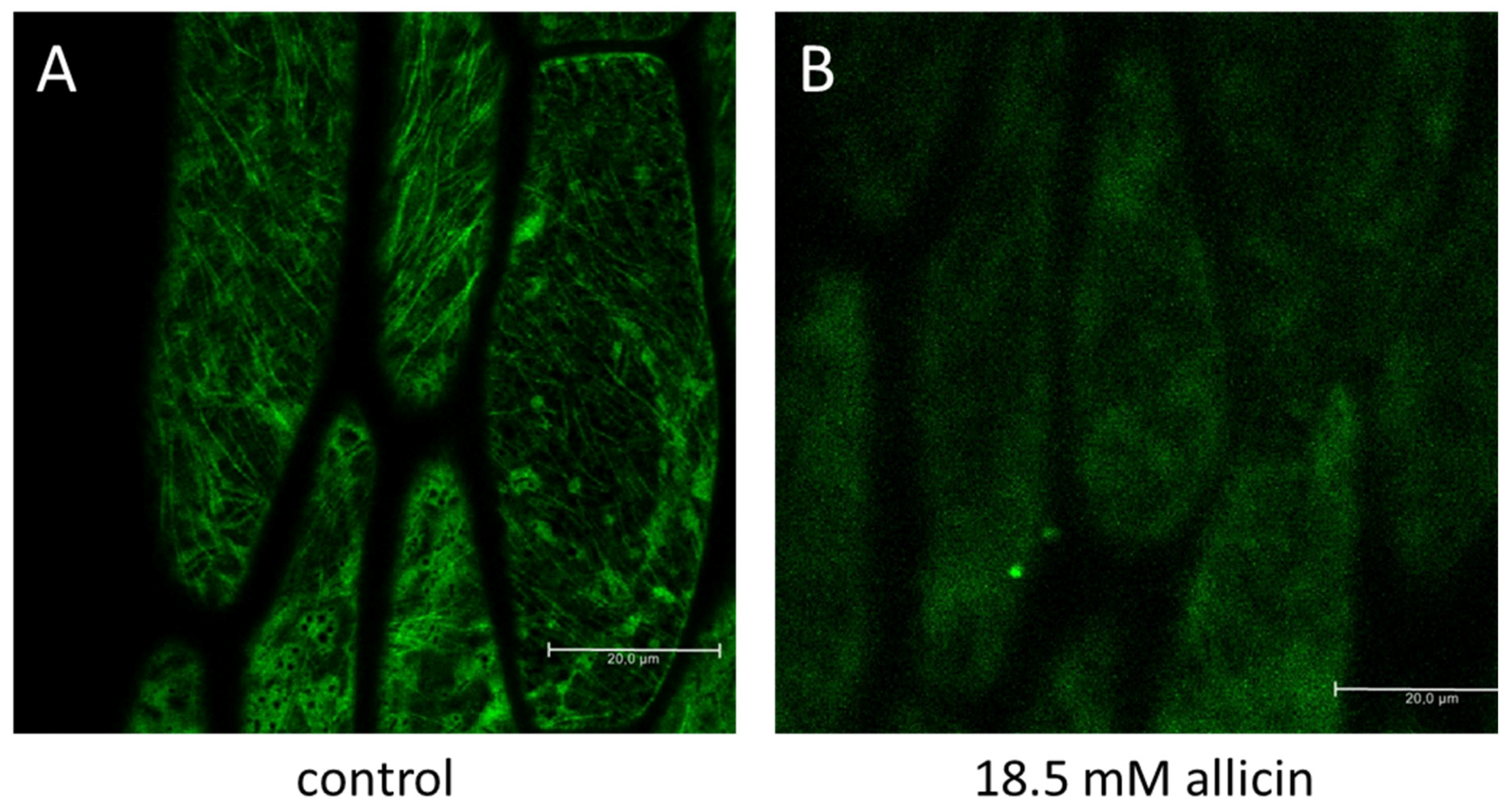
- Noll, U.; Schreiber, M.; Hermanns, M.; Mertes, C.A.; Slusarenko, A.J.; Gruhlke, M.C.H. Allicin from Garlic Disrupts the Cytoskeleton in Plants. bioRxiv 2021. [Google Scholar] [CrossRef]
- Leontiev, R.; Slusarenko, A.J. Finding the Starting Point for Mode-of-Action Studies of Novel Selenium Compounds: Yeast as a Genetic Toolkit. Curr. Org. Synth. 2017, 14, 1–7. [Google Scholar] [CrossRef]
- Borlinghaus, J.; Bolger, A.; Schier, C.; Vogel, A.; Usadel, B.; Gruhlke, M.C.H.; Slusarenko, A.J. Genetic and molecular characterization of multi-component resistance of Pseudomonas against allicin. Life Sci. Alliance 2020, 3, e202000670. [Google Scholar] [CrossRef] [PubMed]
- Wüllner, D.; Haupt, A.; Prochnow, P.; Leontiev, R.; Slusarenko, A.J.; Bandow, J.E. Interspecies Comparison of the Bacterial Response to Allicin Reveals Species-Specific Defense Strategies. Proteomics 2019, 19, 1900064. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.; Hübbers, A.M.; Albrecht, F.; Leichert, L.I.O.; Slusarenko, A.J. Allicin, a Natural Antimicrobial Defence Substance from Garlic, Inhibits DNA Gyrase Activity in Bacteria. Int. J. Med. Microbiol. 2020, 310, 151359. [Google Scholar] [CrossRef] [PubMed]
- Getti, G.T.M.; Poole, P.L. 2019 Allicin Causes Fragmentation of the Peptidoglycan Coat in Staphylococcus aureus by Effecting Synthesis and Aiding Hydrolysis: A Determination by MALDI-TOF Mass Spectrometry on Whole Cells. J. Med. Microbiol. 2019, 68, 667–677. [Google Scholar] [CrossRef]
- Slauch, J.M. How Does the Oxidative Burst of Macrophages Kill Bacteria? Still an Open Question. Mol. Microbiol. 2011, 80, 580–583. [Google Scholar] [CrossRef]
- Cutler, R.R.; Wilson, P. Antibacterial Activity of a New, Stable, Aqueous Extract of Allicin against Methicillin-Resistant Staphylococcus aureus. Brit. J. Biomed. Sci. 2004, 61, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Gardan, L.; Bella, P.; Meyer, J.M.; Christen, R.; Rott, P.; Achouak, W.; Samson, R. Pseudomonas salomonii sp. nov., pathogenic on garlic, and Pseudomonas palleroniana sp. nov., isolated from rice. Int. J. Syst. Evol. Microbiol. 2002, 52, 2065–2074. [Google Scholar] [PubMed]
- Xin, X.F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A Model Pathogen for Probing Disease Susceptibility and Hormone Signaling in Plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar]
- Stice, S.P.; Stumpf, S.D.; Gitaitis, R.D.; Kvitko, B.H.; Dutta, B. Pantoea ananatis Genetic Diversity Analysis Reveals Limited Genomic Diversity as Well as Accessory Genes Correlated with Onion Pathogenicity. Front. Microbiol. 2018, 9, 184. [Google Scholar] [CrossRef]
- Stice, S.P.; Thao, K.K.; Khang, C.H.; Baltrus, D.A.; Dutta, B.; Kvitko, B.H. Thiosulfinate Tolerance Is a Virulence Strategy of an Atypical Bacterial Pathogen of Onion. Curr. Biol. 2020, 30, 3130–3140.e6. [Google Scholar] [CrossRef]
- Loi, V.V.; Busche, T.; Tedin, K.; Bernhardt, J.; Wollenhaupt, J.; Huyen, N.T.T.; Weise, C.; Kalinowski, J.; Wahl, M.C.; Fulde, M.; et al. Redox-Sensing Under Hypochlorite Stress and Infection Conditions by the Rrf2-Family Repressor HypR in Staphylococcus aureus. Antioxid. Redox Signal. 2018, 29, 615–636. [Google Scholar] [CrossRef]
- Reiter, J.; Levina, N.; van der Linden, M.; Gruhlke, M.C.H.; Slusarenko, A.J. Diallylthiosulfinate (Allicin), a volatile antimicrobial from garlic (Allium sativum), kills human lung pathogenic bacteria, including MDR strains, as a vapor. Molecules 2017, 22, 1711. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.R.; Weiss, A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol. 2004, 4, 301–308. [Google Scholar] [CrossRef]
- Gruhlke, M.C.H.; Antelmann, H.; Bernhardt, J.; Kloubert, V.; Rink, L.; Slusarenko, A.J. The human allicin-proteome: S-thioallylation of proteins by the garlic defence substance allicin and its biological effects. Free Radic. Biol. Med. 2019, 131, 144–153. [Google Scholar] [CrossRef]
- Gruhlke, M.C.; Nicco, C.; Batteux, F.; Slusarenko, A.J. The Effects of Allicin, a Reactive Sulfur Species from Garlic, on a Selection of Mammalian Cell Lines. Antioxidants 2016, 6, 1. [Google Scholar] [CrossRef]
- Haase, H.; Hieke, N.; Plum, L.M.; Gruhlke, M.C.H.; Slusarenko, A.J.; Rink, L. Impact of allicin on macrophage activity. Food Chem. 2012, 134, 141–148. [Google Scholar] [CrossRef]
- Lawson, L.D.; Hunsaker, S.M. Allicin Bioavailability and Bioequivalence from Garlic Supplements and Garlic Foods. Nutrients 2018, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Scheffler, L.; Sauermann, Y.; Heinlein, A.; Sharapa, C.; Buettner, A. Detection of Volatile Metabolites Derived from Garlic (Allium sativum) in Human Urine. Metabolites 2016, 6, 43. [Google Scholar] [CrossRef]
- Scheffler, L.; Sharapa, C.; Amar, T.; Buettner, A. Identification and Quantification of Volatile Ramson-Derived Metabolites in Humans. Front. Chem. 2018, 6, 410. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.J.; Block, E.; Uden, P.C.; Quimby, B.D.; Sullivan, J.J. Allium Chemistry: Identification of Natural Abundance Organoselenium Compounds in Human Breath after Ingestion of Garlic Using Gas Chromatography with Atomic Emission Detection. J. Agric. Food Chem. 1995, 43, 1751–1753. [Google Scholar] [CrossRef]
- Minchin, W.D. A Study in Tubercule Virus Polymorphism, and the Treatment of Tuberculosis and Lupus with Oleum allii; Bailliere, Tindall and Cox: London, UK, 1927. [Google Scholar]
- Reiter, J.; Borlinghaus, J.; Dörner, P.; Schröder, W.; Gruhlke, M.C.; Klaas, M.; Slusarenko, A.J. Investigation of the deposition behaviour and antibacterial effectivity of allicin aerosols and vapour using a lung model. Exp. Ther. Med. 2020, 19, 1541–1549. [Google Scholar] [CrossRef]
- Dörner, P.; Müller, P.M.; Reiter, J.; Gruhlke, M.C.; Slusarenko, A.J.; Schröder, W.; Klaas, M. Feasibility study of a surface-coated lung model to quantify active agent deposition for preclinical studies. Clin. Biomech. 2020, 76, 10529. [Google Scholar] [CrossRef] [PubMed]
- Shekh, S.; Reddy, K.K.A.; Gowd, K.H. In silico allicin induced S-thioallylation of SARS-CoV-2 main protease. J. Sulf. Chem. 2021, 42, 109–120. [Google Scholar] [CrossRef]
- Weßels, I.; Rolles, B.; Slusarenko, A.J.; Rink, L. Zinc deficiency as a possible risk factor for increased susceptibility and severe progression of COVID-19. Br. J. Nutr. 2021, 1–42. [Google Scholar] [CrossRef]
- Cha, J.H.; Choi, Y.J.; Cha, S.H.; Choi, C.H.; Cho, W.H. Allicin inhibits cell growth and induces apoptosis in U87MG human glioblastoma cells through an ERK-dependent pathway. Oncol. Rep. 2012, 28, 41–48. [Google Scholar] [CrossRef]
- Chen, H.; Zhu, B.; Zhao, L.; Liu, Y.; Zhao, F.; Feng, J.; Jin, Y.; Sun, J.; Geng, R.; Wei, Y. Allicin Inhibits Proliferation and Invasion in Vitro and in Vivo via SHP-1-Mediated STAT3 Signaling in Cholangiocarcinoma. Cell. Physiol. Biochem. 2018, 47, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ni, J.; Tang, Y.; Wang, X.; Tang, H.; Li, H.; Zhang, S.; Shen, X. Allicin inhibits mouse colorectal tumorigenesis through suppressing the activation of STAT3 signaling pathway. Nat. Prod. Res. 2019, 33, 2722–2725. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Li, Y.; Chi, Y. Role of p38 MAPK activation and mitochondrial cytochrome-c release in allicin-induced apoptosis in SK-N-SH cells. Anticancer Drugs 2016, 27, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Liang, J.; Sun, J.; Hu, X.; Lei, L.; Wu, D.; Liu, L. Allicin sensitizes hepatocellular cancer cells to anti-tumor activity of 5-fluorouracil through ROS-mediated mitochondrial pathway. J. Pharmacol. Sci. 2016, 131, 233–240. [Google Scholar] [CrossRef]
- Gao, X.Y.; Geng, X.J.; Zhai, W.L.; Zhang, X.W.; Wei, Y.; Hou, G.J. Effect of combined treatment with cyclophosphamidum and allicin on neuroblastoma-bearing mice. Asian Pac. J. Trop. Med. 2015, 8, 137–141. [Google Scholar] [CrossRef]
- Jiang, W.; Huang, Y.; Wang, J.P.; Yu, X.Y.; Zhang, L.Y. The synergistic anticancer effect of artesunate combined with allicin in osteosarcoma cell line in vitro and in vivo. Asian Pac. J. Cancer Prev. 2013, 14, 4615–4619. [Google Scholar] [CrossRef] [PubMed]
- Jobani, B.M.; Najafzadeh, N.; Mazani, M.; Arzanlou, M.; Vardin, M.M. Molecular mechanism and cytotoxicity of allicin and all-trans retinoic acid against CD44(+) versus CD117(+) melanoma cells. Phytomedicine 2018, 48, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Schultz, C.R.; Gruhlke, M.C.H.; Slusarenko, A.J.; Bachmann, A.S. Allicin, a Potent New Ornithine Decarboxylase Inhibitor in Neuroblastoma Cells. J. Nat. Prod. 2020, 83, 2518–2527. [Google Scholar] [CrossRef]
- Estrela, J.M.; Ortega, A.; Obrador, E. Glutathione in cancer biology and therapy. Crit. Rev. Clin. Lab. Sci. 2006, 43, 143–181. [Google Scholar] [CrossRef]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of Glutathione in Cancer Progression and Chemoresistance. Oxid. Med. Cell. Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, P.J.; Hamilton, T.C.; LaCreta, F.P.; Gallo, J.M.; Kilpatrick, D.; Halbherr, T.; Brennan, J.; Bookman, M.A.; Hoffman, J.; Young, R.C.; et al. Phase I trial of buthionine sulfoximine in combination with melphalan in patients with cancer. J. Clin. Oncol. 1996, 14, 249–256. [Google Scholar] [CrossRef]
- Bailey, H.H.; Ripple, G.; Tutsch, K.D.; Arzoomanian, R.Z.; Alberti, D.; Feierabend, C.; Mahvi, D.; Schink, J.; Pomplun, M.; Mulcahy, R.T.; et al. Phase I study of continuous-infusion L-S,R-buthionine sulfoximine with intravenous melphalan. J. Nat. Cancer Inst. 1997, 89, 1789–1796. [Google Scholar] [CrossRef]
- Hernandez-Breijo, B.; Monserrat, J.; Ramirez-Rubio, S.; Cuevas, E.P.; Vara, D.; Díaz-Laviada, I.; Fernández-Moreno, M.D.; Román, I.D.; Gisbert, J.P.; Guijarro, L.G. Preclinical evaluation of azathioprine plus buthionine sulfoximine in the treatment of human hepatocarcinoma and colon carcinoma. World, J. Gastroenterol. 2011, 17, 3899–3911. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borlinghaus, J.; Foerster, J.; Kappler, U.; Antelmann, H.; Noll, U.; Gruhlke, M.C.H.; Slusarenko, A.J. Allicin, the Odor of Freshly Crushed Garlic: A Review of Recent Progress in Understanding Allicin’s Effects on Cells. Molecules 2021, 26, 1505. https://doi.org/10.3390/molecules26061505
Borlinghaus J, Foerster J, Kappler U, Antelmann H, Noll U, Gruhlke MCH, Slusarenko AJ. Allicin, the Odor of Freshly Crushed Garlic: A Review of Recent Progress in Understanding Allicin’s Effects on Cells. Molecules. 2021; 26(6):1505. https://doi.org/10.3390/molecules26061505
Chicago/Turabian StyleBorlinghaus, Jan, Jana Foerster (née Reiter), Ulrike Kappler, Haike Antelmann, Ulrike Noll, Martin C. H. Gruhlke, and Alan J. Slusarenko. 2021. "Allicin, the Odor of Freshly Crushed Garlic: A Review of Recent Progress in Understanding Allicin’s Effects on Cells" Molecules 26, no. 6: 1505. https://doi.org/10.3390/molecules26061505
APA StyleBorlinghaus, J., Foerster, J., Kappler, U., Antelmann, H., Noll, U., Gruhlke, M. C. H., & Slusarenko, A. J. (2021). Allicin, the Odor of Freshly Crushed Garlic: A Review of Recent Progress in Understanding Allicin’s Effects on Cells. Molecules, 26(6), 1505. https://doi.org/10.3390/molecules26061505







