A Metabolomic Approach to Beer Characterization
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental
2.1.1. Sample Collection
2.1.2. Sample Preparation
- 10 min thawing in a water bath at room temperature;
- 20 min degassing in an ultrasonic bath in water at room temperature.
2.1.3. 1H-NMR Data Acquisition
2.2. Data Preprocessing and Data Analysis Methods
2.2.1. 1H-NMR Data Preparation
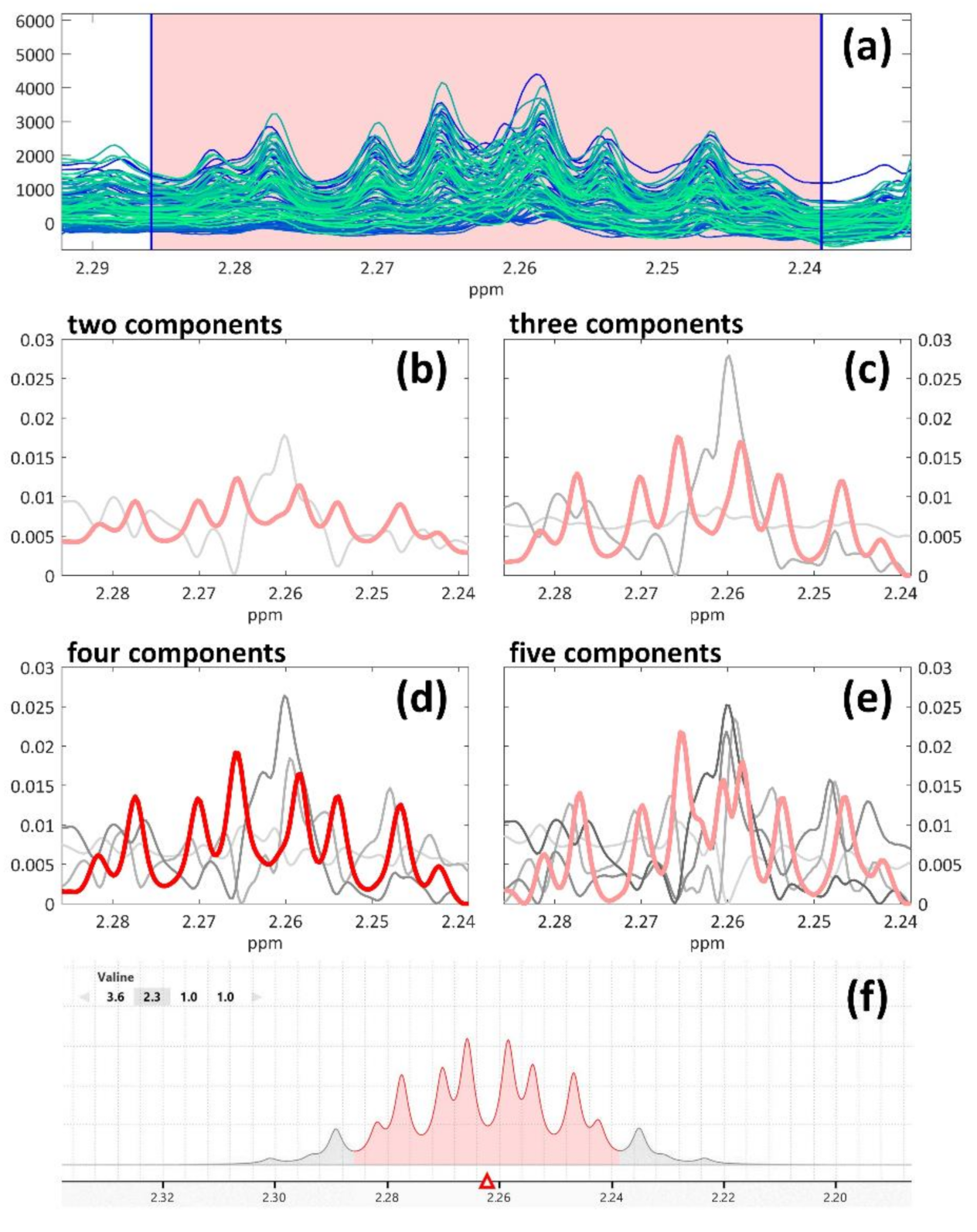
2.2.2. 1H-NMR Spectra Peaks’ Resolution by MCR
2.2.3. 1H-NMR Peak Identification and Assignment
2.2.4. Constitution of the Features Dataset
2.2.5. Multivariate Data Analysis Methods and Dataset Preprocessing
2.3. Software
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Thomé, K.; Soares, A.P.; Moura, J.V. Social Interaction and Beer Consumption. J. Food Prod. Mark. 2017, 23, 186–208. [Google Scholar] [CrossRef]
- Almeida, C.; Duarte, I.F.; Barros, A.; Rodrigues, J.; Spraul, A.M.; Gil, A.M. Composition of Beer by 1H NMR Spectroscopy: Effects of Brewing Site and Date of Production. J. Agric. Food Chem. 2006, 54, 700–706. [Google Scholar] [CrossRef]
- Donadini, G.; Fumi, M.D.; Lambri, M. A preliminary study investigating consumer preference for cheese and beer pairings. Food Qual. Prefer. 2013, 30, 217–228. [Google Scholar] [CrossRef]
- Aquilani, B.; Laureti, T.; Poponi, S.; Secondi, L. Beer choice and consumption determinants when craft beers are tasted: An exploratory study of consumer preferences. Food Qual. Prefer. 2015, 41, 214–224. [Google Scholar] [CrossRef]
- Marongiu, A.; Zara, G.; Legras, J.-L.; Del Caro, A.; Mascia, I.; Fadda, C.; Budroni, M. Novel starters for old processes: Use of Saccharomyces cerevisiae strains isolated from artisanal sourdough for craft beer production at a brewery scale. J. Ind. Microbiol. Biotechnol. 2015, 42, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Murray, D.W.; O’Neill, M.A. Craft beer: Penetrating a niche market. Br. Food J. 2012, 114, 899–909. [Google Scholar] [CrossRef]
- Elzinga, K.G.; Tremblay, C.H.; Tremblay, V.J. Craft Beer in the United States: History, Numbers, and Geography. J. Wine Econ. 2015, 10, 242–274. [Google Scholar] [CrossRef]
- Bevilacqua, M.; Bro, R.; Marini, F.; Rinnan, Å.; Rasmussen, M.A.; Skov, T. Recent chemometrics advances for foodomics. TrAC Trends Anal. Chem. 2017, 96, 42–51. [Google Scholar] [CrossRef]
- Hughey, C.A.; McMINN, C.M.; Phung, J. Beeromics: From quality control to identification of differentially expressed compounds in beer. Metabolomics 2016, 12, 11. [Google Scholar] [CrossRef]
- Donadini, G.; Fumi, M.; Kordialik-Bogacka, E.; Maggi, L.; Lambri, M.; Sckokai, P. Consumer interest in specialty beers in three European markets. Food Res. Int. 2016, 85, 301–314. [Google Scholar] [CrossRef]
- Anderson, H.E.; Santos, I.C.; Hildenbrand, Z.L.; Schug, K.A. A review of the analytical methods used for beer ingredient and finished product analysis and quality control. Anal. Chim. Acta 2019, 1085, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Mutz, Y.S.; Rosario, D.K.A.; Conte-Junior, C.A. Insights into chemical and sensorial aspects to understand and manage beer aging using chemometrics. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3774–3801. [Google Scholar] [CrossRef] [PubMed]
- Duarte, I.F.; Barros, A.S.; Belton, P.S.; Righelato, R.; Spraul, M.; Humpfer, E.; Gil, A.M. High-Resolution Nuclear Magnetic Resonance Spectroscopy and Multivariate Analysis for the Characterization of Beer. J. Agric. Food Chem. 2002, 50, 2475–2481. [Google Scholar] [CrossRef] [PubMed]
- Nord, L.I.; Vaag, A.P.; Duus, J.Ø. Quantification of Organic and Amino Acids in Beer by 1H NMR Spectroscopy. Anal. Chem. 2004, 76, 4790–4798. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.E.; Gil, A.M. NMR methods for beer characterization and quality control. Magn. Reson. Chem. 2011, 49, S37–S45. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.A.; Flumignan, D.L.; Tininis, A.G.; Pezza, H.R.; Pezza, L. Discrimination of Brazilian lager beer by 1H NMR spectroscopy combined with chemometrics. Food Chem. 2019, 272, 488–493. [Google Scholar] [CrossRef]
- Jeong, J.-H.; Cho, S.-J.; Kim, Y. High-Resolution NMR Spectroscopy for the Classification of Beer. Bull. Korean Chem. Soc. 2017, 38, 466–470. [Google Scholar] [CrossRef]
- Sánchez-Estébanez, C.; Ferrero, S.; Alvarez, C.M.; Villafañe, F.; Caballero, I.; Blanco, C.A. Nuclear Magnetic Resonance Methodology for the Analysis of Regular and Non-Alcoholic Lager Beers. Food Anal. Methods 2018, 11, 11–22. [Google Scholar] [CrossRef]
- Palmioli, A.; Alberici, D.; Ciaramelli, C.; Airoldi, C. Metabolomic profiling of beers: Combining 1H NMR spectroscopy and chemometric approaches to discriminate craft and industrial products. Food Chem. 2020, 327, 127025. [Google Scholar] [CrossRef]
- Tian, J. Determination of several flavours in beer with headspace sampling–gas chromatography. Food Chem. 2010, 123, 1318–1321. [Google Scholar] [CrossRef]
- Da Silva, G.A.; Augusto, F.; Poppi, R.J. Exploratory analysis of the volatile profile of beers by HS–SPME–GC. Food Chem. 2008, 111, 1057–1063. [Google Scholar] [CrossRef]
- Tian, J. Application of static headspace gas chromatography for determination of acetaldehyde in beer. J. Food Compos. Anal. 2010, 23, 475–479. [Google Scholar] [CrossRef]
- Vivian, A.F.; Aoyagui, C.T.; De Oliveira, D.N.; Catharino, R.R. Mass spectrometry for the characterization of brewing process. Food Res. Int. 2016, 89, 281–288. [Google Scholar] [CrossRef]
- Grassi, S.; Amigo, J.M.; Lyndgaard, C.B.; Foschino, R.; Casiraghi, E. Beer fermentation: Monitoring of process parameters by FT-NIR and multivariate data analysis. Food Chem. 2014, 155, 279–286. [Google Scholar] [CrossRef]
- Engelhard, S.; Löhmannsröben, H.-G.; Schael, F. Quantifying Ethanol Content of Beer Using Interpretive Near-Infrared Spectroscopy. Appl. Spectrosc. 2004, 58, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Takahashi, Y.; Kumagai, M.; Fujiwara, K.; Kikuchi, R.; Yoshimura, N.; Amano, T.; Lin, J.; Ogawa, N. A Chemometrics Approach for Distinguishing between Beers Using near Infrared Spectroscopy. J. Near Infrared Spectrosc. 2009, 17, 69–76. [Google Scholar] [CrossRef]
- Giovenzana, V.; Beghi, R.; Guidetti, R. Rapid evaluation of craft beer quality during fermentation process by vis/NIR spectroscopy. J. Food Eng. 2014, 142, 80–86. [Google Scholar] [CrossRef]
- Klein, O.; Roth, A.; Dornuf, F.; Schöller, O.; Mäntele, W. The Good Vibrations of Beer. The Use of Infrared and UV/Vis Spectroscopy and Chemometry for the Quantitative Analysis of Beverages. Zeitschrift Naturforsch. B 2012, 67, 1005–1015. [Google Scholar] [CrossRef]
- Biancolillo, A.; Bucci, R.; Magrì, A.L.; Magrì, A.D.; Marini, F. Data-fusion for multiplatform characterization of an italian craft beer aimed at its authentication. Anal. Chim. Acta 2014, 820, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Haddi, Z.; Amari, A.; Bouchikhi, B.; Mimendia, A.; Cetó, X.; Del Valle, M. Hybrid electronic tongue based on multisensor data fusion for discrimination of beers. Sens. Actuators B Chem. 2013, 177, 989–996. [Google Scholar] [CrossRef]
- Vera, L.; Aceña, L.; Guasch, J.; Boqué, R.; Mestres, M.; Busto, O. Characterization and classification of the aroma of beer samples by means of an MS e-nose and chemometric tools. Anal. Bioanal. Chem. 2011, 399, 2073–2081. [Google Scholar] [CrossRef] [PubMed]
- Vera, L.; Aceña, L.; Guasch, J.; Boqué, R.; Mestres, M.; Busto, O. Discrimination and sensory description of beers through data fusion. Talanta 2011, 87, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Sikorska, E.; Górecki, T.; Khmelinskii, I.V.; Sikorski, M.; De Keukeleire, D. Fluorescence Spectroscopy for Characterization and Differentiation of Beers. J. Inst. Brew. 2004, 110, 267–275. [Google Scholar] [CrossRef]
- Gordon, R.; Cozzolino, D.; Chandra, S.; Power, A.; Roberts, J.J.; Chapman, J. Analysis of Australian Beers Using Fluorescence Spectroscopy. Beverages 2017, 3, 57. [Google Scholar] [CrossRef]
- Sikorska, E.; Khmelinskii, I.; Sikorski, M. Fluorescence methods for analysis of beer. In Beer in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2008; pp. 963–976. ISBN 9780123738912. [Google Scholar]
- Dramićanin, T.; Zeković, I.; Periša, J.; Dramićanin, M.D. The Parallel Factor Analysis of Beer Fluorescence. J. Fluoresc. 2019, 29, 1103–1111. [Google Scholar] [CrossRef] [PubMed]
- De Juan, A.; Jaumot, J.; Tauler, R. Multivariate Curve Resolution (MCR). Solving the mixture analysis problem. Anal. Methods 2014, 6, 4964–4976. [Google Scholar] [CrossRef]
- Winning, H.; Larsen, F.; Bro, R.; Engelsen, S. Quantitative analysis of NMR spectra with chemometrics. J. Magn. Reson. 2008, 190, 26–32. [Google Scholar] [CrossRef]
- Khakimov, B.; Mobaraki, N.; Trimigno, A.; Aru, V.; Engelsen, S.B. Signature Mapping (SigMa): An efficient approach for processing complex human urine 1H NMR metabolomics data. Anal. Chim. Acta 2020, 1108, 142–151. [Google Scholar] [CrossRef]
- Puig-Castellví, F.; Alfonso, I.; Tauler, R. Untargeted assignment and automatic integration of 1 H NMR metabolomic datasets using a multivariate curve resolution approach. Anal. Chim. Acta 2017, 964, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Bro, R.; Kamstrup-Nielsen, M.H.; Engelsen, S.B.; Savorani, F.; Rasmussen, M.A.; Hansen, L.; Olsen, A.; Tjønneland, A.; Dragsted, L.O. Forecasting individual breast cancer risk using plasma metabolomics and biocontours. Metabolomics 2015, 11, 1376–1380. [Google Scholar] [CrossRef]
- Carbone, K.; Macchioni, V.; Petrella, G.; Cicero, D.O. Exploring the potential of microwaves and ultrasounds in the green extraction of bioactive compounds from Humulus lupulus for the food and pharmaceutical industry. Ind. Crop. Prod. 2020, 156, 112888. [Google Scholar] [CrossRef]
- Spevacek, A.R.; Benson, K.H.; Bamforth, C.W.; Slupsky, C.M. Beer metabolomics: Molecular details of the brewing process and the differential effects of late and dry hopping on yeast purine metabolism. J. Inst. Brew. 2016, 122, 21–28. [Google Scholar] [CrossRef]
- Dengis, P.B.; Nélissen, L.R.; Rouxhet, P.G. Mechanisms of yeast flocculation: Comparison of top- and bottom-fermenting strains. Appl. Environ. Microbiol. 1995, 61, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Duarte, I.F.; Godejohann, M.; Braumann, U.; Spraul, A.M.; Gil, A.M. Application of NMR Spectroscopy and LC-NMR/MS to the Identification of Carbohydrates in Beer. J. Agric. Food Chem. 2003, 51, 4847–4852. [Google Scholar] [CrossRef]
- Rodrigues, J.A.; Barros, A.S.; Carvalho, B.; Brandão, T.; Gil, A.M. Probing beer aging chemistry by nuclear magnetic resonance and multivariate analysis. Anal. Chim. Acta 2011, 702, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Lachenmeier, D.W. Rapid quality control of spirit drinks and beer using multivariate data analysis of Fourier transform infrared spectra. Food Chem. 2007, 101, 825–832. [Google Scholar] [CrossRef]
- Rodrigues, J.A.; Erny, G.L.; Barros, A.S.; Esteves, V.I.; Brandão, T.; Ferreira, A.A.; Cabrita, E.; Gil, A.M. Quantification of organic acids in beer by nuclear magnetic resonance (NMR)-based methods. Anal. Chim. Acta 2010, 674, 166–175. [Google Scholar] [CrossRef]
- Duarte, I.F.; Barros, A.S.; Almeida, C.; Spraul, M.; Gil, A.M. Multivariate Analysis of NMR and FTIR Data as a Potential Tool for the Quality Control of Beer. J. Agric. Food Chem. 2004, 52, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Gil, A.M.; Duarte, I.F.; Godejohann, M.; Braumann, U.; Maraschin, M.; Spraul, M. Characterization of the aromatic composition of some liquid foods by nuclear magnetic resonance spectrometry and liquid chromatography with nuclear magnetic resonance and mass spectrometric detection. Anal. Chim. Acta 2003, 488, 35–51. [Google Scholar] [CrossRef]
- Lachenmeier, D.W.; Frank, W.; Humpfer, E.; Schäfer, H.; Keller, S.; Mörtter, M.; Spraul, M. Quality control of beer using high-resolution nuclear magnetic resonance spectroscopy and multivariate analysis. Eur. Food Res. Technol. 2005, 220, 215–221. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Savorani, F.; Rasmussen, M.A.; Rinnan, Å.; Engelsen, S.B. Interval-Based Chemometric Methods in NMR Foodomics. In Data Handling in Science and Technology; Ruckebusch, C., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 449–486. ISBN 978-0-444-63638-6. [Google Scholar]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. icoshift: A versatile tool for the rapid alignment of 1D NMR spectra. J. Magn. Reson. 2010, 202, 190–202. [Google Scholar] [CrossRef]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. Alignment of 1D NMR Data using the iCoshift Tool: A Tutorial. In Magnetic Resonance in Food Science: Food for Thought; van Duynhoven, J., Belton, P.S., Webb., G.A., van As, H., Eds.; Royal Society of Chemistry: London, UK, 2013; pp. 14–24. ISBN 978-1-84973-634-3. [Google Scholar]
- Metrulas, L.K.; McNeil, C.; Slupsky, C.M.; Bamforth, C.W. The application of metabolomics to ascertain the significance of prolonged maturation in the production of lager-style beers. J. Inst. Brew. 2019, 125, 242–249. [Google Scholar] [CrossRef]
- Khatib, A.; Wilson, E.G.; Kim, H.K.; Lefeber, A.W.; Erkelens, C.; Choi, Y.H.; Verpoorte, R. Application of two-dimensional J-resolved nuclear magnetic resonance spectroscopy to differentiation of beer. Anal. Chim. Acta 2006, 559, 264–270. [Google Scholar] [CrossRef]
- Tian, J.; Yu, J.; Chen, X.; Zhang, W. Determination and quantitative analysis of acetoin in beer with headspace sampling-gas chromatography. Food Chem. 2009, 112, 1079–1083. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef]
- Viant, M.R.; Kurland, I.J.; Jones, M.R.; Dunn, W.B. How close are we to complete annotation of metabolomes? Curr. Opin. Chem. Biol. 2017, 36, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Chemical Analysis Working Group (CAWG). Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- De Juan, A.; Tauler, R. Multivariate Curve Resolution (MCR) from 2000: Progress in Concepts and Applications. Crit. Rev. Anal. Chem. 2006, 36, 163–176. [Google Scholar] [CrossRef]
- Rutan, S.C.; de Juan, A.; Tauler, R. Introduction to Multivariate Curve Resolution. In Comprehensive Chemometrics; Elsevier: Amsterdam, The Netherlands, 2009; pp. 249–259. ISBN 9780444527011. [Google Scholar]
- Euceda, L.R.; Giskeødegård, G.F.; Bathen, T.F. Preprocessing of NMR metabolomics data. Scand. J. Clin. Lab. Investig. 2015, 75, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Bro, R.; Papalexakis, E.E.; Acar, E.; Sidiropoulos, N.D. Coclustering-a useful tool for chemometrics. J. Chemom. 2012, 26, 256–263. [Google Scholar] [CrossRef]
- Witten, D.M.; Tibshirani, R.; Hastie, T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics 2009, 10, 515–534. [Google Scholar] [CrossRef] [PubMed]
- Quifer-Rada, P.; Vallverdú-Queralt, A.; Martínez-Huélamo, M.; Chiva-Blanch, G.; Jáuregui, O.; Estruch, R.; Lamuela-Raventós, R. A comprehensive characterisation of beer polyphenols by high resolution mass spectrometry (LC-ESI-LTQ-Orbitrap-MS). Food Chem. 2015, 169, 336–343. [Google Scholar] [CrossRef]
- Steenackers, B.; De Cooman, L.; De Vos, D. Chemical transformations of characteristic hop secondary metabolites in relation to beer properties and the brewing process: A review. Food Chem. 2015, 172, 742–756. [Google Scholar] [CrossRef]


| Fermentation Style (Yeast Strain) | % ABV Range | Beer Styles | ||
|---|---|---|---|---|
| Top, “ales” (S. cerevisiae) | 40 | 5.7 ± 1.3% | Ale | 18 |
| India pale ale (IPA) | 19 | |||
| Imperial India pale ale (IIPA) | 3 | |||
| Bottom, “lagers” (S. carlsbergensis) | 57 | 4.8 ± 1.0% | Lager | 30 |
| Lager (pale) | 27 | |||
| Unclassified 1 | 3 | 4.8–5.2–6.0% | Organic ginger brew–Oktoberfest–Kristallweizen |
| Tentative Compound Name | Chemical Shift (δ, ppm) | Multiplicity and Assignment | References c | |
|---|---|---|---|---|
| 1 | 2′-Deoxyguanosine b | 7.98 | s | [56] b; Chenomx; HMDB0000085 |
| 2 | 2′-Deoxyuridine a | 7.83 | d | [56] b; Chenomx; HMDB0000012 |
| 6.28 | t | |||
| 3 | Acetaldehyde a | 9.66 | q, CHO | [13]; HMDB0000990 |
| 2.23 | d, CH3 | |||
| 4 | Acetoin b | 2.21 | s, COCH3 | [20,43,58] b; Chenomx; HMDB0003243 |
| 1.35 | d, CH3 | |||
| 5 | Adenine | 8.21 | s | [57] (not 8.21 but 8.32 ppm); Chenomx |
| 8.18 | s | |||
| 6 | Alanine | 1.47 | d, β-CH3 | [13,14,17,19]; Chenomx |
| 7 | Butanone a | 2.19 | s | Chenomx |
| 8 | Choline | 3.18 | s, N-CH3 | [57]; Chenomx |
| 9 | Oligosaccharides I | 5.35 | C1H glyc. bond | also called “dextrins” in [13] |
| 10 | Oligosaccharides II a | 3.54 | also called “dextrins” in [17] | |
| 5.01 | d | |||
| 11 | Oligosaccharides III a | 5.08 | d | also called “dextrins” in [45] or generally “carbohydrates” in [14,53] |
| 5.08 | d | |||
| 4.57 | m | |||
| 12 | Ethanol | 3.64 | q, CH2 | [2,13,14,17,45,46,50] |
| 1.17 | t, CH3 | |||
| 13 | Gallic acid | 7.04 | s, C2H, C6H | [13,19,46]; Chenomx |
| 14 | Glucose a | 3.22 | dd | Chenomx; HMDB0000122 |
| 15 | Guanosine b | 8.00 | s | [56] b, [19]; Chenomx; HMDB0000133 |
| 16 | 5-Hydroxymethylfurfural a | 9.44 | s | [46]; HMDB0034355 |
| 17 | Inosine | 8.22 | s, C4′H | [13,19]; Chenomx |
| 18 | Isobutanol/Isopentanol | 0.88 | d, CH3 | [13,17] |
| 19 | Isoleucine | 0.99 | d, ε-CH3 | [14]; Chenomx |
| 0.95 | t, δ-CH3 | |||
| 20 | Isopentanol | 1.42 | CH | [13,48] |
| 21 | Leucine | 0.96 | t, δ-CH3 | [14]; Chenomx; HMDB0000687 |
| 22 | Maltose | 5.22 | d, α-C1H | [13]; Chenomx |
| 4.64 | d, β-C1H | |||
| 3.42 | dd, α/β-C4H | |||
| 3.26 | dd, β-C2H | |||
| 23 | Methionine a | 2.12 | m, β-CH2 | [14]; Chenomx |
| 24 | N-acetyltyrosine a | 6.84 | m | Chenomx; HMDB0000866 |
| 25 | Phenylalanine | 7.43 | m, H2/H6 | [14,19,50]; Chenomx |
| 7.37 | m, H4 | |||
| 7.33 | m, H3/H5 | |||
| 26 | Phosphocholine | 3.22 | s, N-CH3 | [57]; Chenomx |
| 27 | Polyphenols a | 7.74 | [2] | |
| 7.75 | ||||
| 7.77 | ||||
| 28 | Proline | 2.36 | m, β-CH2 | [13,14,17,48,57]; Chenomx |
| 2.34 | m, β-CH2 | |||
| 1.98 | m, γ-CH2 | |||
| 29 | Propanol | 1.53 | m, CH2 | [13]; HMDB0000820 |
| 0.88 | t, CH3 | [13,46] | ||
| 30 | Pyroglutamate a | 2.39 | m, CH2 | [43,56] b; Chenomx; HMDB0000267 |
| 31 | Pyruvate | 2.36 | s, CH3 | [2,13,14,17,46,48]; Chenomx |
| 32 | Pyruvate hydrate a | 1.58 | s, CH3 | [14] |
| 33 | Trehalose | 5.18 | d | [2]; Chenomx |
| 34 | Trigonelline b | 9.11 | s | [19,43,56] b; Chenomx |
| 8.82 | m | |||
| 35 | Tryptophan | 7.72 | bd, C4H | [13,14,19,50]; Chenomx |
| 36 | Tyrosine | 7.18 | d, C2H, C6H | [13,14,17,19,50,57]; Chenomx |
| 6.89 | d, C3H, C5H | + [2] | ||
| 37 | Unknown 1 | 10.2 | s | |
| 38 | Unknown 2 d | 6.35 | s | [46] d |
| 39 | Unknown 3 | 2.22 | s | |
| 40 | Unknown 4 | 5.79 | m | |
| 41 | Uracilb | 5.79 | d | [56] b, [19]; Chenomx; HMDB0000300 |
| 42 | Uridine | 7.86 | d, C6H | [2,13,17,19,50]; Chenomx |
| 43 | Uridine/Guanosine b | 5.89 | m, C1′H | [13,17,50,57], [56] b; Chenomx; HMDB0000296 (Uri), HMDB0000133 (Guan) |
| 44 | Valine | 2.26 | m, β-CH | [14,19]; Chenomx |
| 1.03 | d, γ-CH3 | |||
| 0.97 | d, γ-CH3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavallini, N.; Savorani, F.; Bro, R.; Cocchi, M. A Metabolomic Approach to Beer Characterization. Molecules 2021, 26, 1472. https://doi.org/10.3390/molecules26051472
Cavallini N, Savorani F, Bro R, Cocchi M. A Metabolomic Approach to Beer Characterization. Molecules. 2021; 26(5):1472. https://doi.org/10.3390/molecules26051472
Chicago/Turabian StyleCavallini, Nicola, Francesco Savorani, Rasmus Bro, and Marina Cocchi. 2021. "A Metabolomic Approach to Beer Characterization" Molecules 26, no. 5: 1472. https://doi.org/10.3390/molecules26051472
APA StyleCavallini, N., Savorani, F., Bro, R., & Cocchi, M. (2021). A Metabolomic Approach to Beer Characterization. Molecules, 26(5), 1472. https://doi.org/10.3390/molecules26051472








