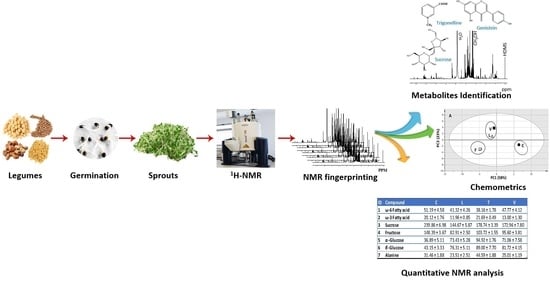
Nuclear Magnetic Resonance Metabolomics Approach for the Analysis of Major Legume Sprouts Coupled to Chemometrics
Abstract
1. Introduction
2. Results and Discussion
2.1. NMR Fingerprinting of Legume Sprouts
2.2. Quantification of Major Metabolites Detected Via 1H-NMR
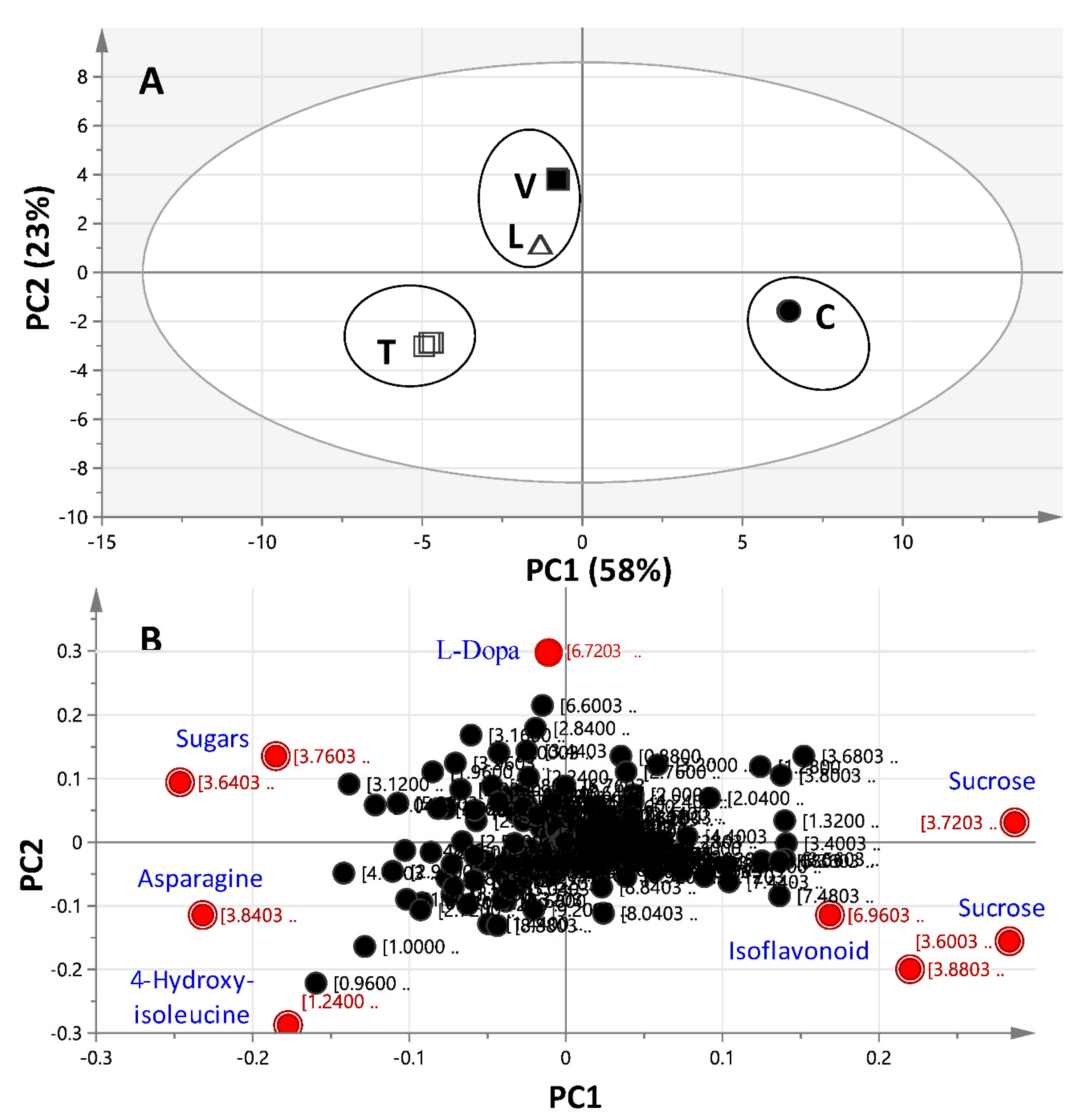
2.3. 1H-NMR Data Multivariate Data Analyses
2.3.1. Unsupervised Multivariate PCA of Full-Range 1H-NMR Data
2.3.2. Unsupervised Multivariate PCA of the Aromatic 1H-NMR Region Data
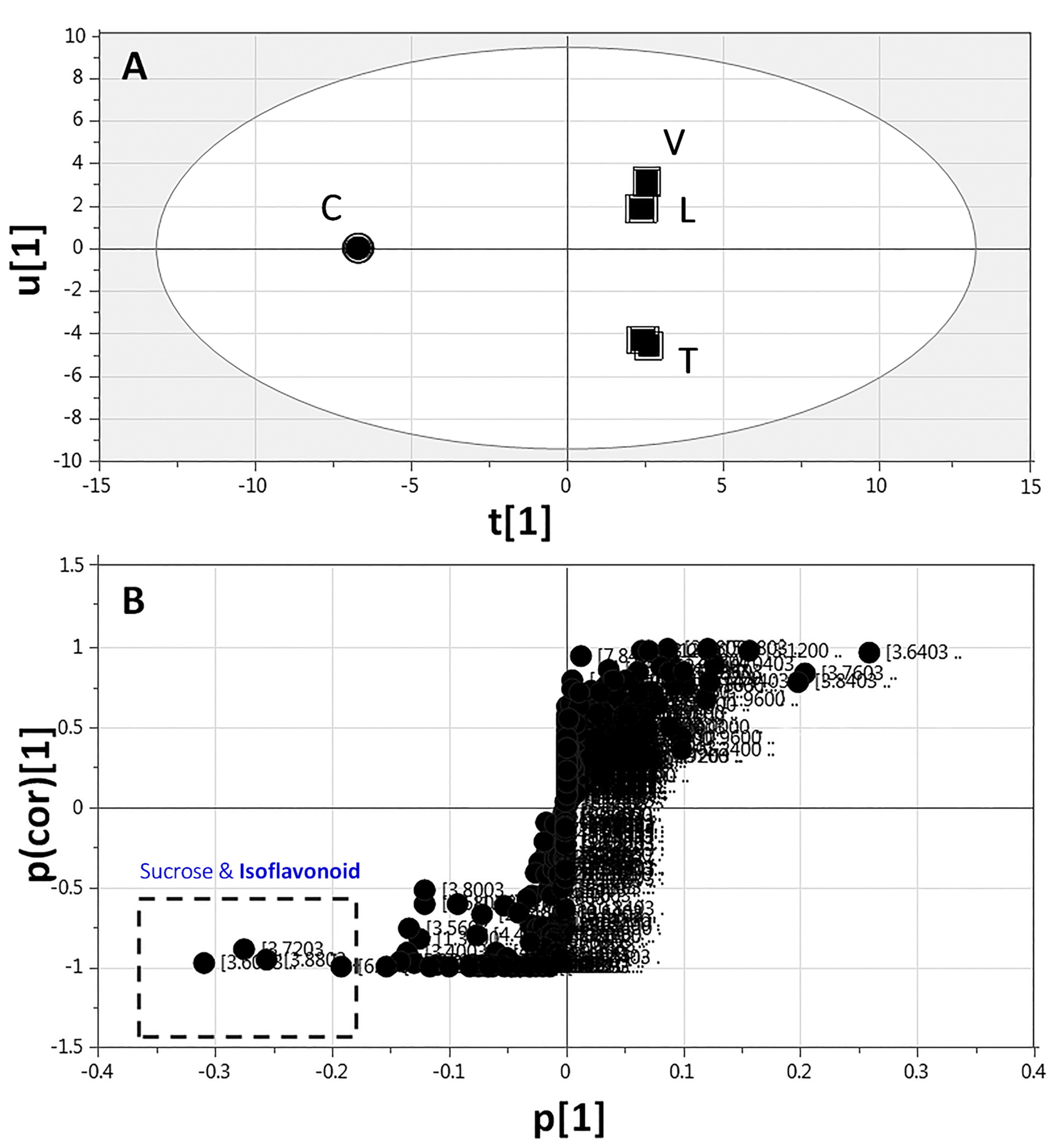
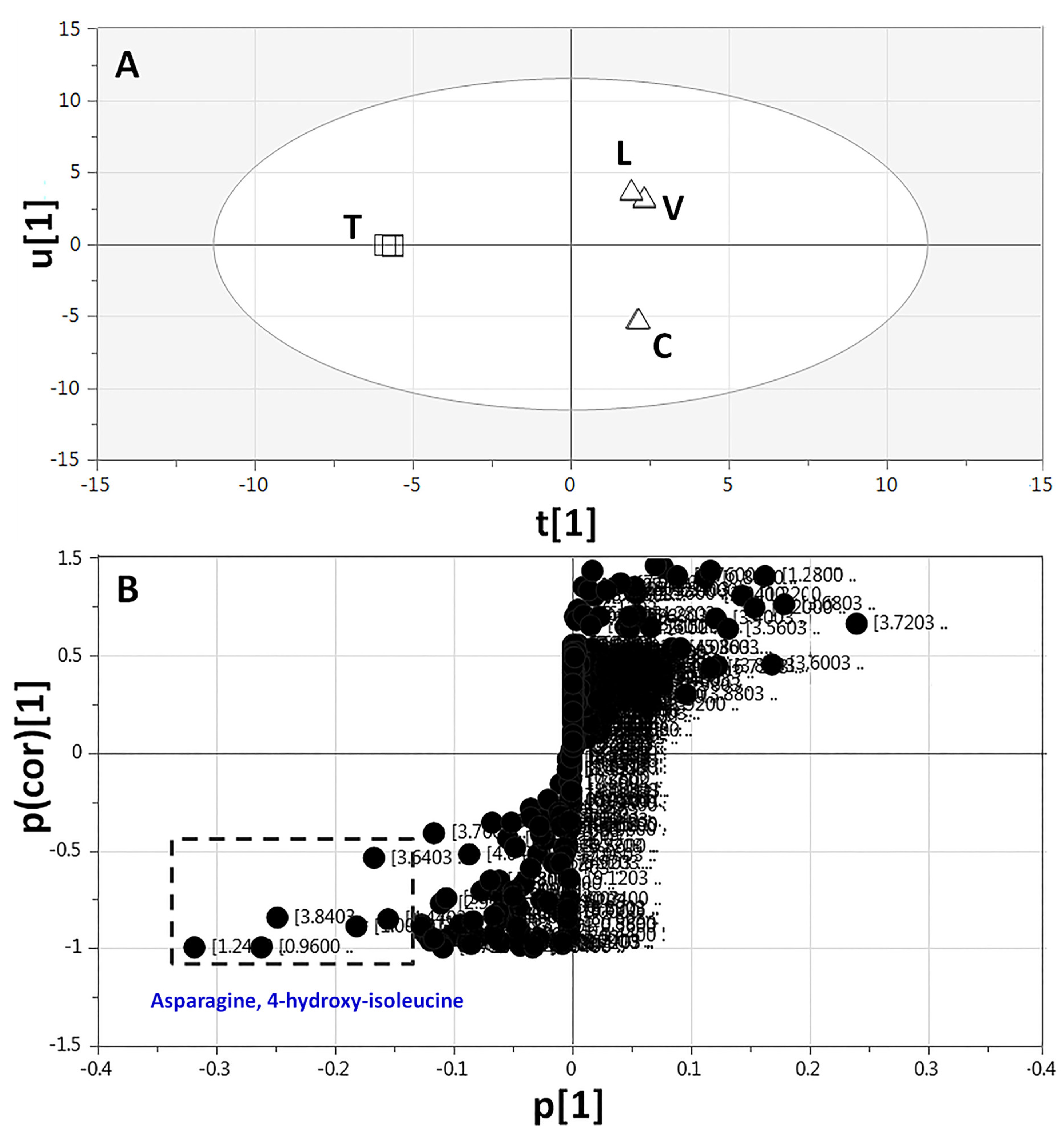
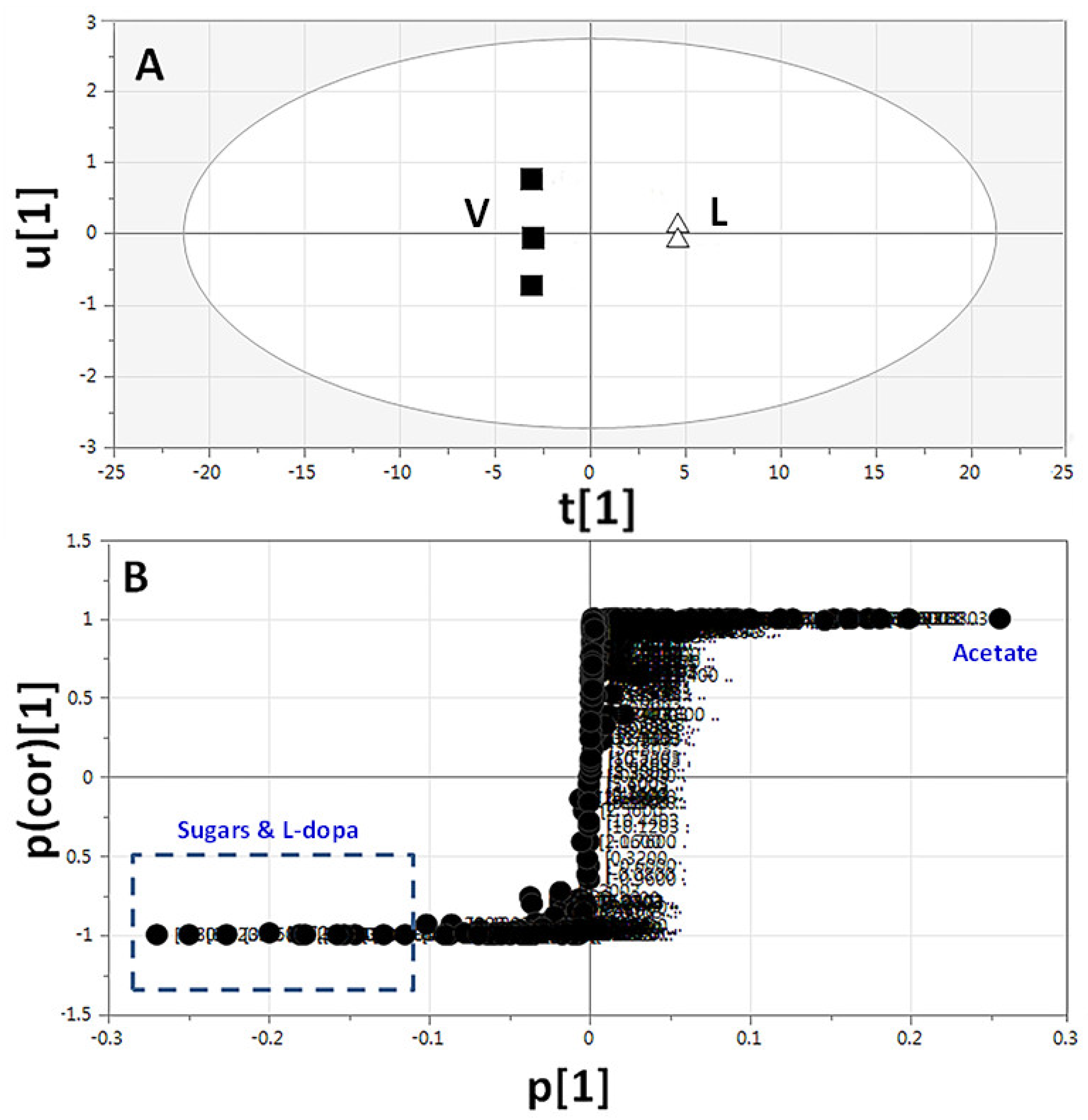
2.3.3. Supervised Multivariate OPLS-DA of 1H-NMR Data
3. Materials and Methods
3.1. Plant Material
3.2. Sprouting Procedures
3.3. Chemicals and Reagents
3.4. Extraction Procedure and Sample Preparation for NMR Analysis
3.5. NMR Analysis
3.6. NMR Quantification
3.7. NMR Data Processing and Multivariate Data Analysis
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Doyle, J.J.; Luckow, M.A. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 2003, 131, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Graham, P.H.; Allan, D.L. Biological nitrogen fixation: Phosphorus-a critical future need? In Nitrogen Fixation: From Molecules to Crop Productivity; Springer: Dordrecht, The Netherlands, 2000; pp. 509–514. [Google Scholar]
- Geil, P.B.; Anderson, J.W. Nutrition and health implications of dry beans: A review. J. Am. Coll. Nutr. 1994, 13, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Duranti, M. Grain legume proteins and nutraceutical properties. Fitoterapia 2006, 77, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.H.; Mansour, E.H. The effect of cooking, autoclaving and germination on the nutritional quality of faba beans. Food Chem. 1995, 54, 177–182. [Google Scholar] [CrossRef]
- Chevion, M.; Navok, T.; Glaser, G.; Mager, J. The chemistry of favism-inducing compounds: The properties of isouramil and divicine and their reaction with glutathione. Eur. J. Biochem. 1982, 127, 405–409. [Google Scholar] [CrossRef]
- Shakuntala, S.; Pura Naik, J.; Jeyarani, T.; Madhava Naidu, M.; Srinivas, P. Characterisation of germinated fenugreek (Trigonella foenum-graecum L.) seed fractions. Int. J. Food Sci. Technol. 2011, 46, 2337–2343. [Google Scholar] [CrossRef]
- El-Mahdy, A.R.; El-Sebaiy, L.A. Changes in carbohydrates of germinating fenugreek seeds (Trigonella foenum graecum L.). J. Sci. Food Agric. 1983, 34, 951–956. [Google Scholar] [CrossRef]
- Abd EI-Azim, M.A.; Abo EI-Azam, N.A.l.; Serage, A.O.; AbdaIIah, M.M.F. Sprouting using saline water on chemical composition, antinutritional compounds and amino acid profile of chickpea and lentil seeds. Arab Univ. J. Agric. Sci. 2018, 26, 2239–2251. [Google Scholar] [CrossRef]
- Cheng, Z.; Ayiguli, A.; Ma, Q.; Gao, Y.; Aisa, H. Determination of isoflavones in extracts of Cicer arietinum L and bean sprout by ultrabiolet spectrophotometry. Lishizhen Med. Mater. Med. Res. 2008, 19, 2612–2613. [Google Scholar]
- Abdelsalam, A.; Mahran, E.; Chowdhury, K.; Boroujerdi, A.; El-Bakry, A. NMR-based metabolomic analysis of wild, greenhouse, and in vitro regenerated shoots of Cymbopogon schoenanthus subsp. proximus with GC–MS assessment of proximadiol. Physiol. Mol. Biol. Plants. 2017, 23, 369–383. [Google Scholar] [CrossRef]
- Mbithi, S.; Van Camp, J.; Rodriguez, R.; Huyghebaert, A. Effects of sprouting on nutrient and antinutrient composition of kidney beans (Phaseolus vulgaris var. Rose coco). Eur. Food Res. Technol. 2001, 212, 188–191. [Google Scholar] [CrossRef]
- Marton, M.; Mandoki, Z.; Csapo-Kiss, Z.; Csapo, J. The role of sprouts in human nutrition. A review. Acta Univ. Sapientiae 2010, 3, 81–117. [Google Scholar]
- Peñas, E.; Gómez, R.; Frías, J.; Vidal-Valverde, C. Application of high-pressure treatment on alfalfa (Medicago sativa) and mung bean (Vigna radiata) seeds to enhance the microbiological safety of their sprouts. Food Control 2008, 19, 698–705. [Google Scholar] [CrossRef]
- Linnemann, A.R.; Benner, M.; Verkerk, R.; van Boekel, M.A. Consumer-driven food product development. Trends Food Sci. Technol. 2006, 17, 184–190. [Google Scholar] [CrossRef]
- Hertog, M.G.; Feskens, E.J.; Kromhout, D.; Hollman, P.; Katan, M. Dietary antioxidant flavonoids and risk of coronary heart disease: The Zutphen Elderly Study. Lancet 1993, 342, 1007–1011. [Google Scholar] [CrossRef]
- Diaz, M.N.; Frei, B.; Vita, J.A.; Keaney, J.F., Jr. Antioxidants and atherosclerotic heart disease. N. Engl. J. Med. 1997, 337, 408–416. [Google Scholar] [CrossRef]
- Ness, A.R.; Powles, J.W. Fruit and vegetables, and cardiovascular disease: A review. Int. J. Epidemiol. 1997, 26, 1–13. [Google Scholar] [CrossRef]
- Ito, N.; Hirose, M. Antioxidants-carcinogenic and chemopreventive properties. Adv. Cancer Res. 1989, 53, 247–302. [Google Scholar]
- Singhal, P.; Kaushik, G.; Mathur, P. Antidiabetic potential of commonly consumed legumes: A review. Crit. Rev. Food Sci. Nutr. 2014, 54, 655–672. [Google Scholar] [CrossRef]
- Vijayakumar, M.V.; Bhat, M.K. Hypoglycemic effect of a novel dialysed fenugreek seeds extract is sustainable and is mediated, in part, by the activation of hepatic enzymes. Phytother. Res. 2008, 22, 500–505. [Google Scholar] [CrossRef]
- Randhir, R.; Lin, Y.-T.; Shetty, K. Phenolics, their antioxidant and antimicrobial activity in dark germinated fenugreek sprouts in response to peptide and phytochemical elicitors. Asia Pac. J. Clin. Nutr. 2004, 13, 295–307. [Google Scholar] [PubMed]
- Farag, M.A.; Porzel, A.; Mahrous, E.A.; Mo’men, M.; Wessjohann, L.A. Integrated comparative metabolite profiling via MS and NMR techniques for Senna drug quality control analysis. Anal. Bioanal. Chem. 2015, 407, 1937–1949. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Gad, H.A.; Heiss, A.G.; Wessjohann, L.A. Metabolomics driven analysis of six Nigella species seeds via UPLC-qTOF-MS and GC–MS coupled to chemometrics. Food Chem. 2014, 151, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Labib, R.M.; Noleto, C.; Porzel, A.; Wessjohann, L.A. NMR approach for the authentication of 10 cinnamon spice accessions analyzed via chemometric tools. LWT 2018, 90, 491–498. [Google Scholar] [CrossRef]
- Otify, A.M.; El-Sayed, A.M.; Michel, C.G.; Farag, M.A. Metabolites profiling of date palm (Phoenix dactylifera L.) commercial by-products (pits and pollen) in relation to its antioxidant effect: A multiplex approach of MS and NMR metabolomics. Metabolomics 2019, 15, 119. [Google Scholar] [CrossRef]
- Krishnan, P.; Kruger, N.J.; Ratcliffe, R.G. Metabolite fingerprinting and profiling in plants using NMR. J. Exp. Bot. 2005, 56, 255–265. [Google Scholar] [CrossRef]
- Farag, M.A.; Sharaf El-Din, M.G.; Selim, M.A.-F.; Owis, A.I.; Abouzid, S.F. Mass spectrometry-based metabolites profiling of nutrients and anti-nutrients in major legume sprouts. Food Biosci. 2020, 100800. [Google Scholar]
- Zeisel, S.H.; da Costa, K.-A. Choline: An essential nutrient for public health. Nutr. Rev. 2009, 67, 615–623. [Google Scholar] [CrossRef]
- Craig, S.A. Betaine in human nutrition. Am. J. Clin. Nutr. 2004, 80, 539–549. [Google Scholar] [CrossRef]
- Gao, X.; Wang, Y.; Sun, G. High dietary choline and betaine intake is associated with low insulin resistance in the Newfoundland population. Nutrition 2017, 33, 28–34. [Google Scholar] [CrossRef]
- Goyoaga, C.; Burbano, C.; Cuadrado, C.; Varela, A.; Guillamón, E.; Pedrosa, M.M.; Muzquiz, M. Content and distribution of vicine, convicine and L-DOPA during germination and seedling growth of two Vicia faba L. varieties. Eur. Food Res. Technol. 2008, 227, 1537–1542. [Google Scholar] [CrossRef]
- Jenkins, T.A.; Nguyen, J.C.D.; Polglaze, K.E.; Bertrand, P.P. Influence of Tryptophan and Serotonin on Mood and Cognition with a Possible Role of the Gut-Brain Axis. Nutrients 2016, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Nakamura, T.; Shibakusa, T.; Sugita, M.; Naganuma, F.; Iida, T.; Miura, Y.; Mohsen, A.; Harada, R.; Yanai, K. Insufficient Intake of L-Histidine Reduces Brain Histamine and Causes Anxiety-Like Behaviors in Male Mice. J. Nutr. 2014, 144, 1637–1641. [Google Scholar] [CrossRef] [PubMed]
- Blank, I.; Lin, J.; Fumeaux, R.; Welti, D.H.; Fay, L.B. Formation of 3-hydroxy-4, 5-dimethyl-2 (5 H)-furanone (sotolone) from 4-hydroxy-l-isoleucine and 3-amino-4, 5-dimethyl-3, 4-dihydro-2 (5 H)-furanone. Agric. Food Chem. 1996, 44, 1851–1856. [Google Scholar] [CrossRef]
- Narender, T.; Puri, A.; Shweta; Khaliq, T.; Saxena, R.; Bhatia, G.; Chandra, R. 4-Hydroxyisoleucine an unusual amino acid as antidyslipidemic and antihyperglycemic agent. Bioorg. Med. Chem. Lett. 2006, 16, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Yoshinari, O.; Sato, H.; Igarashi, K. Anti-diabetic effects of pumpkin and its components, trigonelline and nicotinic acid, on Goto-kakizaki rats. Biosci. Biotechnol. Biochem. 2009, 73, 1033–1041. [Google Scholar] [CrossRef]
- Tramontano, W.A.; McGinley, P.A.; Ciancaglini, E.F.; Evans, L.S. A survey of trigonelline concentrations in dry seeds of the dicotyledoneae. Environ. Exp. Bot. 1986, 26, 197–205. [Google Scholar] [CrossRef]
- Ashihara, H. De novo Biosynthesis of Trigonelline in Fenugreek (Trigonella foenum-graecum) seedlings. Nat. Prod. Commun. 2016, 11, 1093–1096. [Google Scholar] [CrossRef]
- Shimizu, M.M.; Mazzafera, P. A Role for Trigonelline During Imbibition and Germination of Coffee Seeds. Plant Biol. 2000, 2, 605–611. [Google Scholar] [CrossRef]
- Singh, A.K.; Rehal, J.; Kaur, A.; Jyot, G. Enhancement of attributes of cereals by germination and fermentation: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 1575–1589. [Google Scholar] [CrossRef]
- Guleria, A.; Bajpai, N.K.; Rawat, A.; Khetrapal, C.L.; Prasad, N.; Kumar, D. Metabolite characterisation in peritoneal dialysis effluent using high-resolution 1H and 1H–13C NMR spectroscopy. Magn. Reson. Chem. 2014, 52, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Jain, K.; Chitre, T.; Miniyar, P.; Kathiravan, M.; Bendre, V.; Veer, V.; Shahane, S.; Shishoo, C. Biological and medicinal significance of pyrimidines. Curr. Sci. 2006, 793–803. [Google Scholar]
- Velásquez-Valle, R.; Villa-Ruano, N.; Hidalgo-Martínez, D.; Zepeda-Vallejo, L.G.; Pérez-Hernández, N.; Reyes-López, C.A.; Reyes-Cervantes, E.; Medina-Melchor, D.L.; Becerra-Martínez, E. Revealing the 1H NMR metabolome of mirasol chili peppers (Capsicum annuum) infected by Candidatus Phytoplasma trifolii. Food Res. Int. 2020, 131, 108863. [Google Scholar] [CrossRef] [PubMed]
- Schwachtje, J.; Fischer, A.; Erban, A.; Kopka, J. Primed primary metabolism in systemic leaves: A functional systems analysis. Sci. Rep. 2018, 8, 216. [Google Scholar] [CrossRef]
- Vaughan, J.G.; Geissler, C.; Nicholson, B. The New Oxford Book of Food Plants; Oxford University Press: Oxford, UK; New York, NY, USA, 1997; p. 239. [Google Scholar]
- Goad, L.J.; Akihisa, T. Analysis of Sterols, 1st ed.; Blackie Academic & Professional: London, UK, 1997; p. 437. [Google Scholar]
- Kumar, Y.; Dholakia, B.B.; Panigrahi, P.; Kadoo, N.Y.; Giri, A.P.; Gupta, V.S. Metabolic profiling of chickpea-Fusarium interaction identifies differential modulation of disease resistance pathways. Phytochemistry 2015, 116, 120–129. [Google Scholar] [CrossRef]
- Caligiani, A.; Palla, G.; Maietti, A.; Cirlini, M.; Brandolini, V. 1H NMR fingerprinting of soybean extracts, with emphasis on identification and quantification of isoflavones. Nutrients 2010, 2, 280–289. [Google Scholar] [CrossRef]
- Wu, Z.; Song, L.; Feng, S.; Liu, Y.; He, G.; Yioe, Y.; Liu, S.Q.; Huang, D. Germination dramatically increases isoflavonoid content and diversity in chickpea (Cicer arietinum L.) seeds. Agric. Food Chem. 2012, 60, 8606–8615. [Google Scholar] [CrossRef]
- Lv, Q.; Yang, Y.; Zhao, Y.; Gu, D.; He, D.; Yili, A.; Ma, Q.; Cheng, Z.; Gao, Y.; Aisa, H.A. Comparative study on separation and purification of isoflavones from the seeds and sprouts of chickpea by high-speed countercurrent chromatography. J. Liq. Chrom. Relat. Technol. 2009, 32, 2879–2892. [Google Scholar] [CrossRef]
- HaiRong, M.; HuaBo, W.; Zhen, C.; Yi, Y.; ZhengHua, W.; Madina, H.; Xu, C.; Akber, A.H. The estrogenic activity of isoflavones extracted from chickpea Cicer arietinum L sprouts in vitro. Phytother. Res. 2013, 27, 1237–1242. [Google Scholar] [CrossRef]
- Kunzru, R.; Sinha, S. “Cicerin” a new phytoalexin associated with blight of gram. In Proceedings of Plant Disease Problems; Raychaudhuri, S.P., Ed.; New Delhi, India, 1970; pp. 724–732. [Google Scholar]
- McCue, P.; Shetty, K. Health benefits of soy isoflavonoids and strategies for enhancement: A review. Crit. Rev. Food Sci. Nutr. 2004, 44, 361–367. [Google Scholar] [CrossRef]
- Masood, T.; Shah, H.; Zeb, A. Effect of sprouting time on proximate composition and ascorbic acid level of mung bean (Vigna radiate L.) and chickpea (Cicer arietinum L.) seeds. J. Anim. Plant Sci. 2014, 24, 850–859. [Google Scholar]
- Zafar, M.I.; Gao, F. 4-Hydroxyisoleucine: A Potential New Treatment for Type 2 Diabetes Mellitus. Biodrugs 2016, 30, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, S.; Zeng, S. Experimental diabetes treated with trigonelline: Effect on β cell and pancreatic oxidative parameters. Fundam. Clin. Pharmacol. 2013, 27, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Porzel, A.; Wessjohann, L.A. Comparative metabolite profiling and fingerprinting of medicinal licorice roots using a multiplex approach of GC-MS, LC-MS and 1D NMR techniques. Phytochemistry 2012, 76, 60–72. [Google Scholar] [CrossRef]
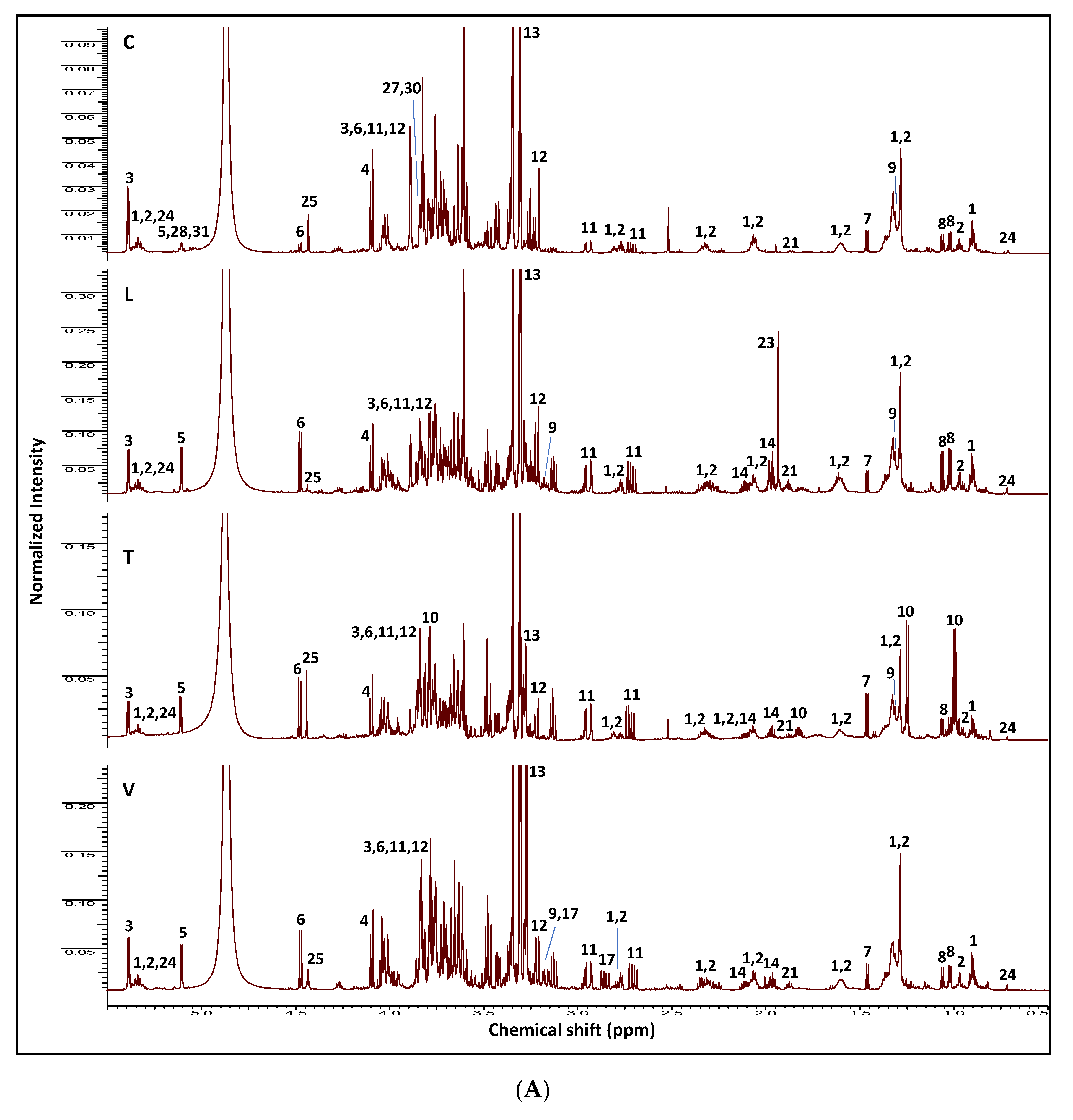

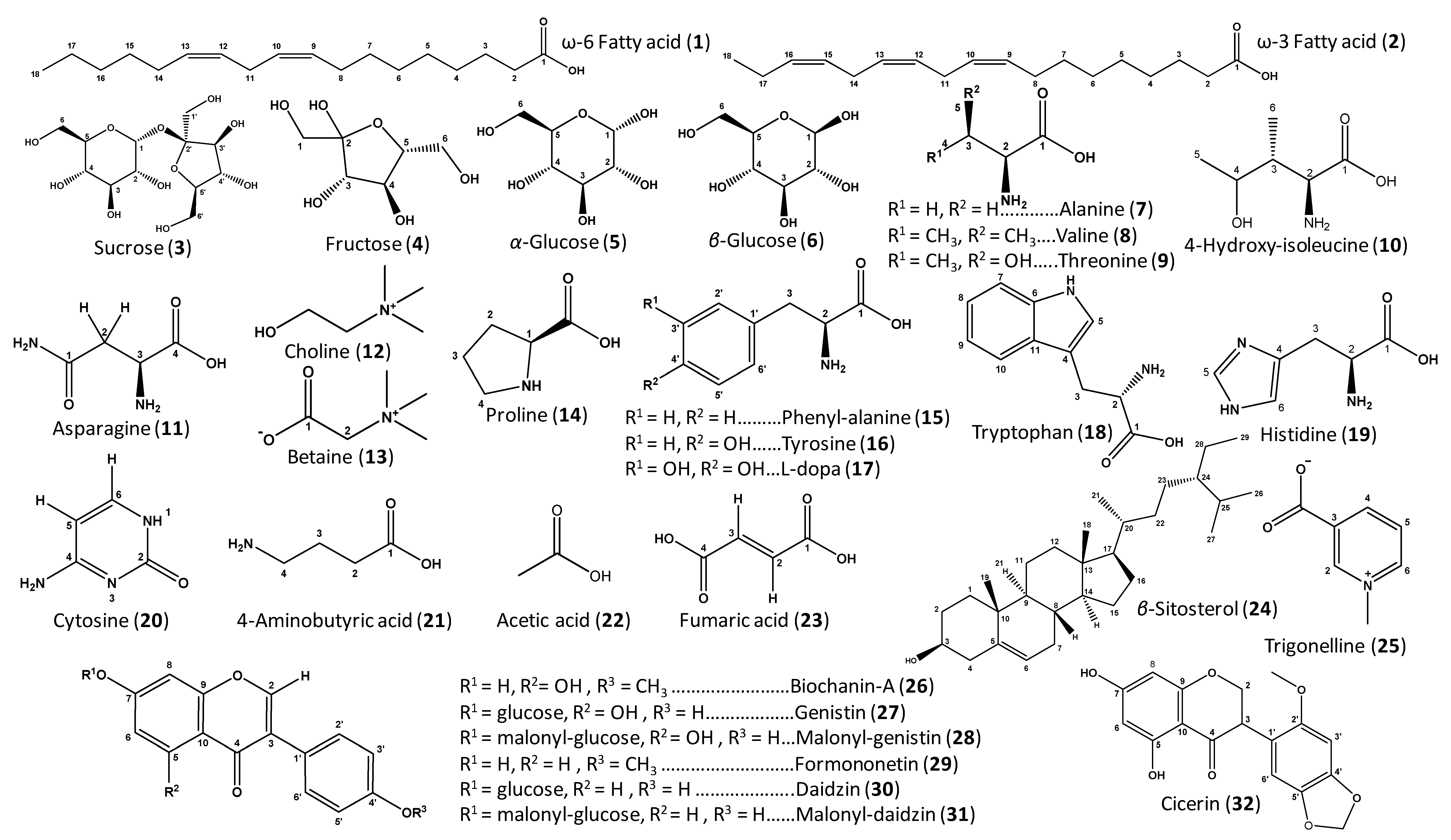
| ID | Metabolite | Assignment | δ1H (ppm) | δ13C in HSQC (ppm) | δ1H in COSY (ppm) | HMBC correlations δ13C (ppm) | C | L | T | V | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–2 | ω-6 and ω-3 Fatty acids | (CH2)n | 1.27–1.39 (br. s) | 30.8 | 0.91 (t-CH3), 1.61 (H-3), 2.07 (allylic CH2) | 30.8 (CH2)n., 131.9 (olefinic C) | + | + | + | + | |
| C-2 | 2.31 (m) | 35.5 | 1.61 (H-3) | 26.2 (C-3), 30.8 (CH2)n, 175.8 (C-1) | |||||||
| C-3 | 1.61 (m) | 26.2 | 2.31 (H-2), 1.33 (CH2)n | 30.8 (CH2)n, 35.5 (C-2), 175.8 (C-1) | |||||||
| Olefinic Cs | 5.30–5.38 (br. m) | 129–132 | 2.77, 2.81 (bis-allylic CH2), 2.07 (allylic CH2) | 26.8 (bis-allylic CH2), 130.2 (olefinic C) | |||||||
| allylic CH2 | 2.05–2.09 (m) | 28.2–28.8 | 1.37 (CH2)n, 5.30–5.38 (olefinic Hs) | 14.9 (t-CH3), 30.8 (CH2)n, 130.2, 131.9 (olefinic Cs) | |||||||
| 1 | ω-6 Fatty acid | bis-allylic CH2 | 2.77 (t, J = 6.6 Hz) | 26.8 | 5.30–5.38 (olefinic Hs) | 130.2, 131.9 (olefinic Cs) | |||||
| t-CH3 | 0.91 (t, J = 6.2 Hz) | 14.9 | 1.33 (CH2)n | 23.9 (ω-2 C) | |||||||
| 2 | ω-3 Fatty acid | bis-allylic CH2 | 2.81 (t, J = 6.9 Hz) | 26.8 | 5.30–5.38 (olefinic Hs) | 130.2, 131.9 (olefinic Cs) | |||||
| t-CH3 | 0.97 (t, J = 7.5 Hz) | 14.9 | 2.07 (allylic CH2) | 22.1 (ω-2 C), 133.7 (olefinic C) | |||||||
| 3 | Sucrose | C-1 | 5.39 (d, J = 3.8 Hz) | 94.5 | 3.42 (H-2) | 73.9 (C-2), 105.6 (C-2′) | + | + | + | + | |
| C-2 | 3.42 (dd, J = 9.8, 3.8 Hz) | 73.9 | 5.39 (H-1), 3.71 (H-3) | 75.3 (C-3) | |||||||
| C-1′ | 3.61 (s) | 64.7 | - | 79.9 (C-3′), 105.6 (C-2′) | |||||||
| 4 | Fructose | C-3 | 4.10 | 78.6 | 4.03 (H-3) | 62.0 (C-1), 74.6 (C-4) | + | + | + | + | |
| C-4 | 4.03 * | 74.6 | 4.10 (H-4) | - | |||||||
| 5 | α-Glucose | C-1 | 5.11 (d, J = 3.7 Hz) | 94.5 | 3.35 (H-2) | 73.2 (C-5), 74.7 (C-2) | + | + | + | + | |
| C-2 | 3.35 (br. s) | 74.7 | 5.11 (H-1), 3.67 (H-3) | - | |||||||
| 6 | β-Glucose | C-1 | 4.48 (d, J = 7.8 Hz) | 97.9 | 3.13 (H-2) | 75.2 (C-2), 77.3 (C-3) | + | + | + | + | |
| C-2 | 3.13 (dd, J = 7.8, 9.0 Hz) | 75.2 | 3.35 (H-3) | 77.3 (C-3) | |||||||
| 7 | Alanine | C-2 | 3.59 * | 51.9 | 1.46 | 17.6 (C-3), 176.6 (C-1) | + | + | + | + | |
| C-3 | 1.46 (d, J = 7.2 Hz) | 17.6 | 3.59 | 51.9 (C-2), 176.6 (C-1) | |||||||
| 8 | Valine | C-2 | 3.42 * | 61.9 | 2.25 (H-3) | 19.6 (C-5), 30.7 (C-3) | + | + | + | + | |
| C-3 | 2.25 (m) | 30.7 | 1.02 (H-4), 1.06 (H-5), 3.42 (H-2) | 180.2 (C-1) | |||||||
| C-4 | 1.02 (d, J = 7.0 Hz) | 18.2 | 2.25 (H-3) | 19.6 (C-5), 30.7 (C-3), 61.9 (C-2) | |||||||
| C-5 | 1.06 (d, J = 7.0 Hz) | 19.6 | 2.25 (H-3) | 18.2 (C-4), 30.7 (C-3), 61.9 (C-2) | |||||||
| 9 | Threonine | C-2 | 3.18 * | nd | 4.12 (H-3) | - | + | + | + | + | |
| C-3 | 4.12 * | nd | 1.31 (H-4), 3.18 (H-2) | 174.0 (C-1) | |||||||
| C-4 | 1.31 * | 21.6 | 4.12 (H-3) | 62.6 (C-2), 67.7 (C-3) | |||||||
| 10 | 4-Hydroxy-isoleucine | C-2 | 3.81 (d, J = 5.5 Hz) | 58.7 | 1.82 (H-3) | 13.6 (C-6), 43.4 (C-3), 175.1 (C-1) | - | - | + | - | |
| C-3 | 1.82 (m) | 43.4 | 3.81 (H-2), 3.79 (H-4) | 13.6 (C-6), 22.9 (C-5), 58.7 (C-2), 72.0 (C-4), 175.1 (C-1) | |||||||
| C-4 | 3.79 * | 72.0 | 1.82 (H-3), 1.24 (H-5) | 58.7 (C-2), 72.0 (C-4), | |||||||
| C-5 | 1.24 (d, J = 6.3 Hz) | 22.9 | 3.79 (H-4) | 43.4 (C-3), 72.0 (C-4) | |||||||
| C-6 | 0.99 (d, J = 7.2 Hz) | 13.6 | 3.79 (H-4) | 43.4 (C-3), 58.7 (C-2), 72.0 (C-4) | |||||||
| 11 | Asparagine | C-2a | 2.72 (dd, J = 9.3, 17.0 Hz) | 36.2 | 3.84 (H-3) | 53.4 (C-3), 174.7, 176.4 (C-1 and C-4) | + | + | + | + | |
| C-2b | 2.94 (dd, J = 3.6, 17.0 Hz) | 36.2 | 3.84 (H-3) | 53.4 (C-3), 174.7, 176.4 (C-1 and C-4) | |||||||
| C-3 | 3.84 * | 53.4 | 2.72 (H-2a), 2.94 (H-2b) | 36.2 (C-2), 174.7, 176.4 (C-1 and C-4) | |||||||
| 12 | Choline | N-(CH3)3 | 3.21 (s) | 55.4 | - | 55.4 (N-CH3), 69.3 (N-CH2) | + | + | + | + | |
| N-CH2 | 3.47 * | 69.3 | 4.00 (O-CH2) | 55.4 (N-CH3), 69.3 (N-CH2), 57.3 (O-CH3) | |||||||
| O-CH2 | 4.00 * | 57.3 | 3.47 (N-CH2) | - | |||||||
| 13 | Betaine | N-(CH3)3 | 3.27 (s) | 52.7 | - | 52.7 (N-CH3), 66.7 (C-2), 168.3 (CO) | + | + | + | + | |
| N-CH32 | 3.83 (s) | 66.7 | - | 53.7 (N-CH3), 168.3 (CO) | |||||||
| 14 | Proline | CO | - | 173.6 | - | - | + | + | + | + | |
| C-1 | 3.98 * | 62.0 | 2.11 (H-2) | 24.2 (C-3), 29.6 (C-2), 173.6 (CO) | |||||||
| C-2 | 2.11, 2.30 (m) | 29.6 | 3.98 (H-1) | 46.2 (C-4), 62.0 (C-1) | |||||||
| C-3 | 1.96 (m) | 24.2 | 3.25, 3.37 (H-4) | 46.2 (C-4), 62.0 (C-1) | |||||||
| C-4 | 3.25, 3.37 * | 46.1 | 1.96 (H-3) | 24.2 (C-3), 29.6 (C-2) | |||||||
| 15 | Phenylalanine | C-3 | 3.33 * | 38.80 | - | 55.9, (C-2), 131.5 (C-3′/C-5′), 138.5 (C-1′) | - | + | + | + | |
| C-4′ | 7.33 * | 129.0 | - | 131.5 (C-3′/C-5′) | |||||||
| C-3′/C-5′ | 7.33 * | 131.0 | 7.28 (H-2′/H-6′) | 138.5 (C-1′) | |||||||
| C-2′/C-6′ | 7.28 (d, J = 6.7 Hz) | 129.0 | 7.33 (H-3′/H-5′) | 38.8 (C-3), 131.5 (C-3′/C-5′) | |||||||
| 16 | Tyrosine | C-3′/C-5′ | 6.76 * | 112.5 | 7.12 (H-2′/H-6′) | - | - | + | + | + | |
| C-2′/C-6′ | 7.12 (d, J = 8.5 Hz) | 131.0 | 6.76 (H-3′/H-5′) | 36.6 (C-3), 157.7 (C-4′) | |||||||
| 17 | L-Dopa | C-2 | 3.70 * | 56.6 | 2.86, 3.17 (H-3) | 120.9 (C-2′, C-6′), 173.6 (C-1) | - | - | - | + | |
| C-3a | 2.86 (dd, J = 14.7, 9.0 Hz) | 36.6 | 3.70 (H-2) | 56.6 (C-2), 116.5 (C-5′), 120.9 (C-2′,C-6′), 127.9 (C-1′), 173.6 (C-1) | |||||||
| C-3b | 3.17 (dd, J = 14.7, 4.2 Hz) | 36.6 | 3.70 (H-2) | 56.6 (C-2), 116.5 (C-5′), 120.9 (C-2′,C-6′), 127.9 (C-1′), 173.6 (C-1) | |||||||
| C-2′ | 6.73 (br. s) | 120.9 | 6.61 (H-6′) | 36.6 (C-3), 127.9 (C-1′), 145.5 (C-4′) | |||||||
| C-5′ | 6.75 * | 116.5 | 6.61 (H-6′) | 36.6 (C-3), 120.9 (C-2′,C-6′), 127.9 (C-1′), 145.5 (C-4′) | |||||||
| C-6′ | 6.61 (dd, J = 8.1, 2.1 Hz) | 120.9 | 6.73 (H-2′), 6.75 (H-5′) | 36.6 (C-3), 116.5 (C-5′), 145.5 (C-4′) | |||||||
| 18 | Tryptophan | C-5 | 7.19 (s) | 125.7 | - | 109.9 (C-4), 129.2 (C-11), 138.9 (C-6) | + | + | + | + | |
| C-7 | 7.33 * | 113.1 | 7.10 (H-8) | 121.1 (C-9), 129.2 (C-11) | |||||||
| C-8 | 7.10 (t, J = 7.8 Hz) | 123.0 | 7.33 (H-7) | 119.7 (C-10), 138.9 (C-6) | |||||||
| C-9 | 7.05 * | 121.1 | 7.63 (H-10) | 113.1 (C-7), 129.2 (C-11) | |||||||
| C-10 | 7.63 (d, J = 7.8 Hz) | 119.7 | 7.05 (H-9) | 123.0 (C-8), 138.9 (C-6) | |||||||
| 19 | Histidine | C-5 | 7.75 (s) | 136.5 | - | 134.2 (H-4), 117.0 (H-6) | + | + | + | + | |
| C-6 | 7.01 (s) | 117.0 | - | 134.2 (H-4), 136.5 (H-5) | |||||||
| 20 | Cytosine | C-5 | 5.70 (d, J = 8.7 Hz) | 97.1 | 8.01 | - | + | + | + | + | |
| C-6 | 8.01 (d, J = 8.7 Hz) | 143.6 | 5.70 | 156.4 (C-2), 167.0 (C-4) | |||||||
| 21 | 4-Aminobutyric acid | C-2 | 2.36 (t, J = 6.9 Hz) | 35.5 | 1.88 (H-3) | 24.9 (C-3), 41.4 (C-4), 180.2 (C-1) | + | + | + | + | |
| C-3 | 1.88 (m) | 24.9 | 2.36 (H-2), 2.96 (H-4) | 35.2 (C-2), 41.4 (C-4), 180.2 (C-1) | |||||||
| C-4 | 2.96 * | 41.4 | 1.88 (H-3) | 24.9 (C-3), 35.2 (C-2) | |||||||
| 22 | Acetic acid | CH3 | 1.92 (s) | 22.8 | - | 174.0 (CO) | - | + | - | - | |
| 23 | Fumaric acid | C-2/C-3 | 6.67 (s) | 136.9 | - | 173.2 (C-1, C-4) | + | + | + | + | |
| 24 | β-Sitosterol | C-6 | 5.34 * | 123.0 | - | - | + | + | + | + | |
| C-18 | 0.72 (s) | 14.9 | - | 41.4 (C-12), 44.0 (C-13), 58.9 (C-14) | + | + | + | + | |||
| C-19 | 1.02* | 20.2 | - | 143.3 (C-5) | |||||||
| C-26/C-27 | 0.83 * | 19.6 | - | 47.5 (C-24) | |||||||
| 25 | Trigonelline | C-2 | 9.23 (s) | 148.2 | - | 49.4 (N-CH3), 141.0 (C-3), 146.9 (C-4), 168.4 (CO) | + | + | + | + | |
| C-4 | 8.91 (d, J = 8.1 Hz) | 146.9 | 8.07 (H-5) | 147.6 (C-6), 168.4 (CO) | |||||||
| C-5 | 8.07 (dd, J = 8.1, 6.2 Hz) | 129.0 | 8.91 (H-4), 8.88 (H-6) | 141.0 (C-3), 146.9 (C-4) | |||||||
| C-6 | 8.88 d (J = 6.2 Hz) | 147.6 | 8.07 (H-5) | 49.4 (N-CH3), 129.0 (C-5), 148.2 (C-2) | |||||||
| N-CH3 | 4.44 (s) | 49.4 | - | 147.6 (C-6) | |||||||
| 26–31 | Isoflavone derivatives | C-3′/C-5′ | 6.99 * | 115.7 | 7.49 (H-3′/H-5′) | 124–128 (C-3, C-1′), 131.8 (C-2′/C-6′), 161.7 (C-4′) | + | - | - | - | |
| C-2′/C-6′ | 7.49 * | 132.3 | 6.99 (H-2′/H-6′) | 115.7 (C-3′/C-5′), 124–128 (C-3, C-1′), 161.7 (C-4′) | |||||||
| 26–28 | Genistein derivatives | C-2 | 8.08 (s) | 155.5 | - | 124–128 (C-3, C-1′), 160.9 (C-9), 182.9 (C-4) | + | - | - | - | |
| 8.17 (s) | 156.2 | - | 124–128 (C-3, C-1′), 159.9 (C-9), 183.7 (C-4) | ||||||||
| 8.20 (s) | 156.6 | - | 124–128 (C-3, C-1′), 159.9 (C-9), 183.7 (C-4) | ||||||||
| 29–31 | Daidzein derivatives | C-2 | 8.15 (s) | 155.6 | - | 124–128 (C-3, C-1′), 159.9 (C-9), 179.3 (C-4) | + | - | - | - | |
| 8.23 (s) | 156.2 | - | 124–128 (C-3, C-1′), 179.3 (C-4) | ||||||||
| 8.27 (s) | 156.2 | - | 124–128 (C-3, C-1′), 159.9 (C-9), 179.3 (C-4) | ||||||||
| 26 | Biochanin-A | C-6 | 6.23 (d, J = 2.1 Hz) | 101.2 | 6.35 (H-8) | 107.2 (C-10), 167.0 (C-7) | + | - | - | - | |
| C-8 | 6.35 (d, J = 2.1 Hz) | 95.1 | 6.23 (H-6) | 101.2 (C-6), 160.9 (C-9), 167.0 (C-7) | |||||||
| O-CH3 | 3.83 (s) | 56.0 | - | 161.7 (C-4′) | |||||||
| 27 | Genistin | C-6 | 6.52 (d, J = 2.1 Hz) | 101.8 | 6.71 (H-8) | 96.5 (C-8), 109.0 (C-10), 164.6 (C-5) | + | - | - | - | |
| C-8 | 6.71 (d, J = 2.1 Hz) | 96.5 | 6.52 (H-6) | 101.8 (C-6), 109.0 (C-10), 159.9 (C-9), 165.3 (C-7) | |||||||
| C-1″ | 5.06 (d, J = 7.8 Hz) | 102.5 | 3.50 (H-2″) | 78.3 (C-2″), 165.3 (C-7) | |||||||
| 28 | Malonyl-genistin | C-6 | 6.50 (d, J = 2.3 Hz) | 101.8 | 6.72 (H-8) | 96.5 (C-8), 109.0 (C-10), 164.6 (C-5) | + | - | - | - | |
| C-8 | 6.72 (d, J = 2.3 Hz) | 96.5 | 6.50 (H-6) | 101.8 (C-6), 109.0 (C-10), 159.9 (C-9), 165.3 (C-7) | |||||||
| Malonyl CH2 | 3.17 (s) | 42.1 | - | - | |||||||
| 29 | Formononetin | C-5 | 8.05 (d, J = 9.0 Hz) | 129.0 | 6.94 (H-6) | 160.9 (C-9), 165.3 (C-7), 179.3 (C-4) | + | - | - | - | |
| C-6 | 6.94 (dd, J = 9.0, 2.3 Hz) | 117.0 | 6.86 (H-8), 8.05 (H-5) | 103.8 (C-8), 118.7 (C-10) | |||||||
| C-8 | 6.86 (d, J = 2.3 Hz) | 103.8 | 6.94 (H-6) | 117.0 (C-6), 118.7 (C-10), 160.9 (C-9), 165.3 (C-7) | |||||||
| O-CH3 | 3.83 (s) | 56.0 | - | 161.7 (C-4′) | |||||||
| 30 | Daidzin | C-5 | 8.14 (d, J = 8.7 Hz) | 129.0 | 7.25 (H-8) | 105.8 (C-8), 159.9 (C-9), 164.6 (C-7) | + | - | - | - | |
| C-6 | 7.19 (dd, J = 8.7, 2.2 Hz) | 117.7 | 8.14 (H-5) | 105.8 (C-8), 164.4 (C-7) | |||||||
| C-8 | 7.25 (d, J = 2.2 Hz) | 105.8 | 7.19 (H-6), 8.14 (H-5) | 117.7 (C-6), 159.9 (C-9), 164.4 (C-7) | |||||||
| C-1″ | 5.10 * | 102.5 | 3.52 (H-2″) | 78.3 (C-3″), 164.6 (C-7) | |||||||
| 31 | Malonyl-daidzin | C-6 | 7.22 (dd, J = 8.7, 2.2 Hz) | 117.7 | 8.14 (H-5) | 105.8 (C-8), 164.4 (C-7) | + | - | - | - | |
| C-8 | 7.27 (d, J = 2.4 Hz) | 105.8 | 7.22 (H-6), 8.14 (H-5) | 117.7 (C-6), 159.9 (C-9), 164.4 (C-7) | |||||||
| Malonyl CH2 | 3.17 (s) | 42.1 | - | - | |||||||
| 32 | Cicerin | OCH2O | 5.98 * | 103.2 | - | 150.3 (C-4′) | + | - | - | - | |
| C-6 | 5.98 * | 94.5 | - | - | |||||||
| C-8 | 5.96 (br. s) | 91.9 | - | - | |||||||
| C-3′ | 6.37 * | 98.5 | - | 150.3 (C-4′), 156.5 (C-2′) | |||||||
| C-6′ | 6.80 * | 106.4 | - | 150.3 (C-4′), 156.5 (C-2′) |
| ID | Compound | Amount µg/mg Dry Matter | |||
|---|---|---|---|---|---|
| C | L | T | V | ||
| 1 | ω-6 Fatty acid | 51.19 ± 4.58 a | 41.32 ± 4.26 bc | 38.16 ± 1.78 c | 47.77 ± 4.12 ab |
| 2 | ω-3 Fatty acid | 20.12 ± 1.76 a | 11.96 ± 0.85 b | 21.69 ± 0.49 a | 13.00 ± 1.30 b |
| 3 | Sucrose | 239.82 ± 6.98 a | 144.67 ± 5.87 c | 178.74 ± 3.39 b | 172.96 ± 7.80 b |
| 4 | Fructose | 148.39 ± 3.67 a | 82.91 ± 2.50 d | 103.72 ± 1.55 b | 95.60 ± 3.81 c |
| 5 | α-Glucose | 36.89 ± 5.11 c | 73.43 ± 5.28 b | 94.92 ± 1.76 a | 71.06 ± 7.58 b |
| 6 | β-Glucose | 43.15 ± 3.33 c | 76.31 ± 5.11 b | 89.00 ± 7.70 a | 81.72 ± 4.15 ab |
| 7 | Alanine | 31.46 ± 1.88 b | 23.51 ± 2.51 c | 44.59 ± 1.88 a | 25.01 ± 1.19 c |
| 8 | Valine | 12.61 ± 0.63 b | 14.59 ± 0.95 a | 12.57 ± 0.75 b | 10.26 ± 0.83 c |
| 10 | 4-Hydroxyisoleucine | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 51.13 ± 3.53 a | 0.0 ± 0.0 b |
| 11 | Asparagine | 61.05 ± 4.51 b | 73.46 ± 8.09 b | 93.43 ± 4.29 a | 72.71 ± 9.68 b |
| 12 | Choline | 19.06 ± 0.54 a | 16.91 ± 1.09 b | 9.06 ± 0.20 c | 9.94 ± 0.95 c |
| 13 | Betaine | 12.98 ± 0.62 b | 10.06 ± 0.87 bc | 5.06 ± 1.10 c | 109.16 ± 5.49 a |
| 15 | Phenylalanine | 0.0 ± 0.0 b | 8.61 ± 0.62 a | 8.69 ± 0.94 a | 9.07 ± 1.45 a |
| 16 | Tyrosine | 0.0 ± 0.0 c | 8.59 ± 0.53 b | 8.93 ± 0.68 b | 15.57 ± 2.69 a |
| 17 | L-Dopa | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 112.40 ± 13.16 a |
| 18 | Tryptophan | 24.16 ± 5.02 a | 22.82 ± 3.70 a | 22.05 ± 2.32 a | 10.36 ± 2.99 b |
| 19 | Histidine | 4.23 ± 0.25 c | 11.22 ± 1.77 a | 7.43 ± 1.91 b | 11.07 ± 1.77 a |
| 20 | Cytosine | 9.39 ± 1.95 a | 6.16 ± 1.30 b | 5.53 ± 0.72 b | 7.30 ± 1.45 ab |
| 22 | Acetic acid | 0.0 ± 0.0 b | 10.51 ± 0.46 a | 0.0 ± 0.0 b | 0.0 ± 0.0 b |
| 23 | Fumaric acid | 2.18 ± 0.19 c | 2.51 ± 0.25 bc | 3.11 ± 0.16 a | 2.84 ± 0.34 ab |
| 24 | β-Sitosterol | 8.95 ± 0.67 b | 10.12 ± 0.78 ab | 8.56 ± 1.03 b | 10.77 ± 0.90 a |
| 25 | Trigonelline | 18.03 ± 0.97 b | 8.11 ± 1.02 d | 24.73 ± 1.02 a | 11.59 ± 1.34 c |
| 26 | Biochanin A | 32.04 ± 2.12 a | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 b |
| 27 | Genistin | 43.86 ± 4.87 a | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 b |
| 28 | Malonyl-genistin | 78.88 ± 1.46 a | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 b |
| 29 | Formononetin | 35.52 ± 2.00 a | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 b |
| 30 | Daidzin | 49.27 ± 3.10 a | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 b |
| 31 | Malonyl-daidzin | 80.22 ± 3.56 a | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 b |
| 32 | Cicerin | 33.19 ± 2.84 a | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farag, M.A.; Sharaf El-Din, M.G.; Selim, M.A.; Owis, A.I.; Abouzid, S.F.; Porzel, A.; Wessjohann, L.A.; Otify, A. Nuclear Magnetic Resonance Metabolomics Approach for the Analysis of Major Legume Sprouts Coupled to Chemometrics. Molecules 2021, 26, 761. https://doi.org/10.3390/molecules26030761
Farag MA, Sharaf El-Din MG, Selim MA, Owis AI, Abouzid SF, Porzel A, Wessjohann LA, Otify A. Nuclear Magnetic Resonance Metabolomics Approach for the Analysis of Major Legume Sprouts Coupled to Chemometrics. Molecules. 2021; 26(3):761. https://doi.org/10.3390/molecules26030761
Chicago/Turabian StyleFarag, Mohamed A., Mohamed G. Sharaf El-Din, Mohamed A. Selim, Asmaa I. Owis, Sameh F. Abouzid, Andrea Porzel, Ludger A. Wessjohann, and Asmaa Otify. 2021. "Nuclear Magnetic Resonance Metabolomics Approach for the Analysis of Major Legume Sprouts Coupled to Chemometrics" Molecules 26, no. 3: 761. https://doi.org/10.3390/molecules26030761
APA StyleFarag, M. A., Sharaf El-Din, M. G., Selim, M. A., Owis, A. I., Abouzid, S. F., Porzel, A., Wessjohann, L. A., & Otify, A. (2021). Nuclear Magnetic Resonance Metabolomics Approach for the Analysis of Major Legume Sprouts Coupled to Chemometrics. Molecules, 26(3), 761. https://doi.org/10.3390/molecules26030761