Demonstration of Allium sativum Extract Inhibitory Effect on Biodeteriogenic Microbial Strain Growth, Biofilm Development, and Enzymatic and Organic Acid Production
Abstract
1. Introduction
2. Results
2.1. Chemical Composition and Antioxidant Activity
2.2. Qualitative Screening of the Antimicrobial Activity
2.2.1. Antifungal Activity
2.2.2. Antibacterial Activity
2.3. Quantitative Evaluation of the Antimicrobial Activity
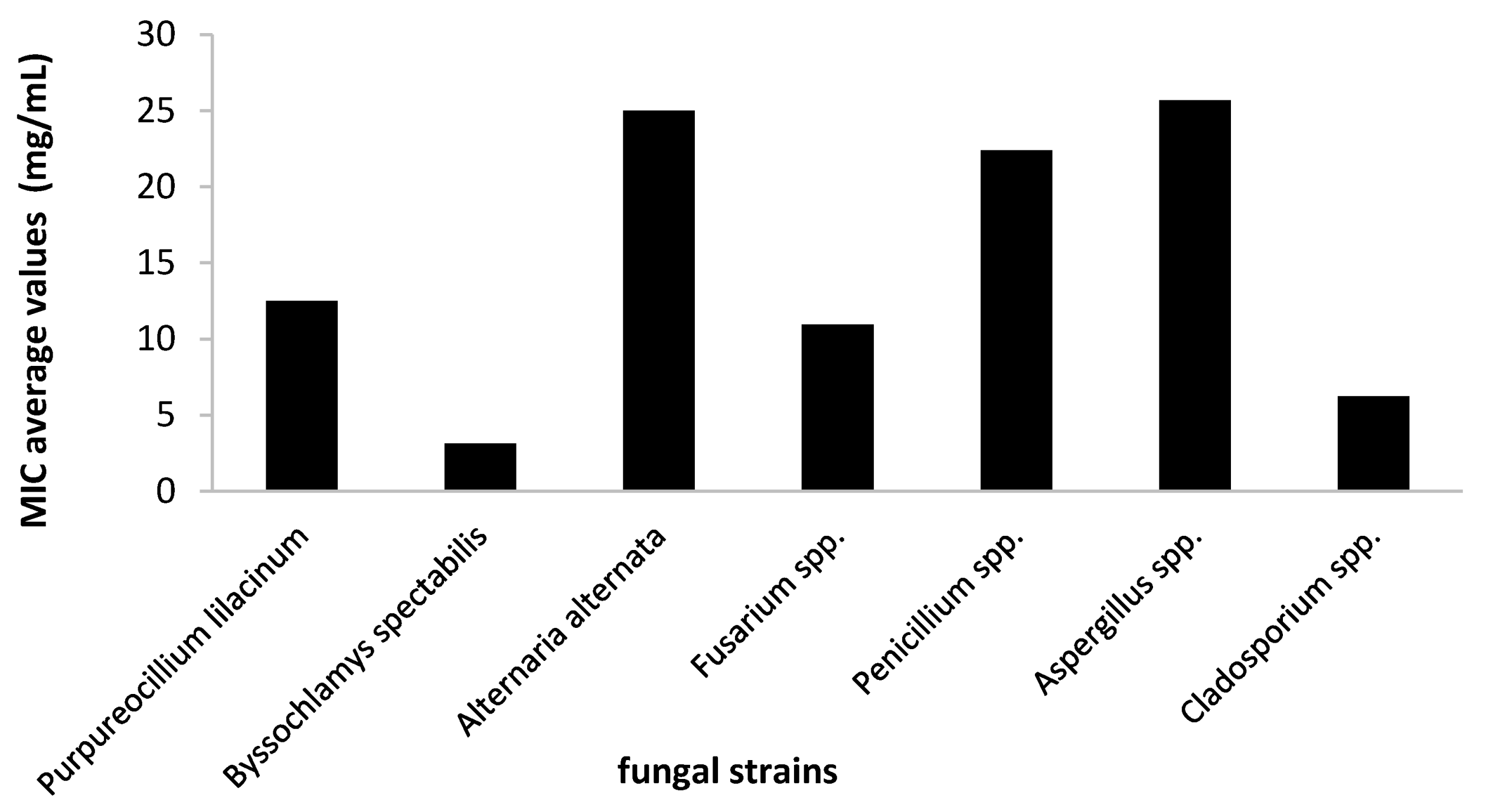
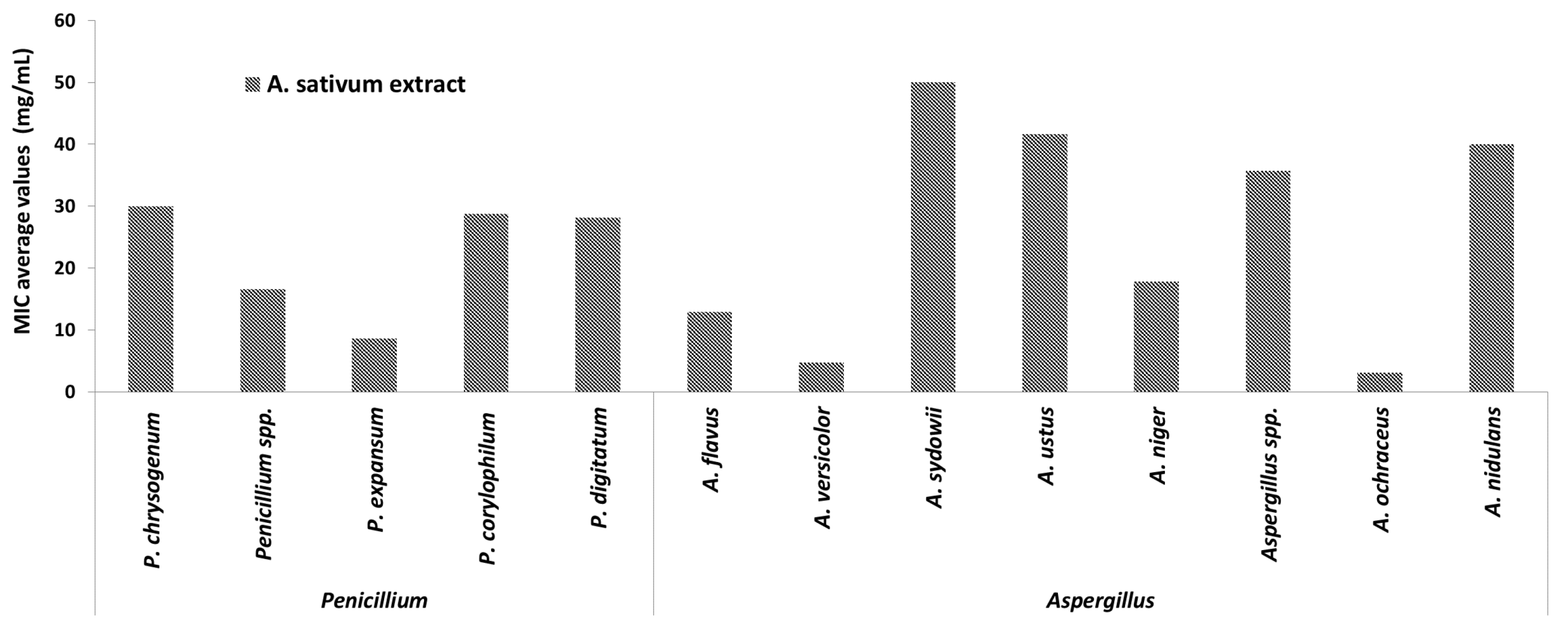
2.3.1. Antifungal Activity
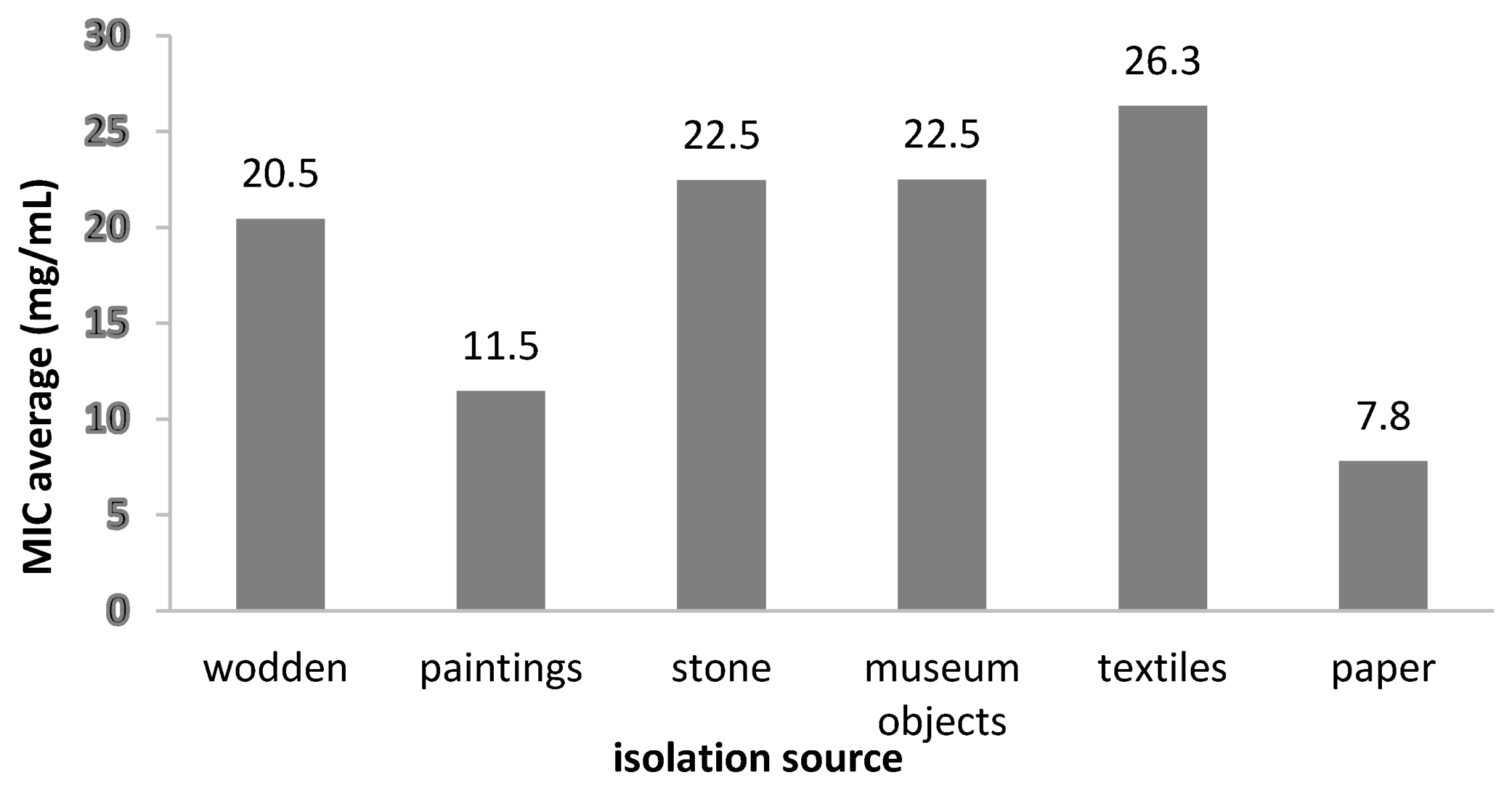
2.3.2. Antibacterial Activity
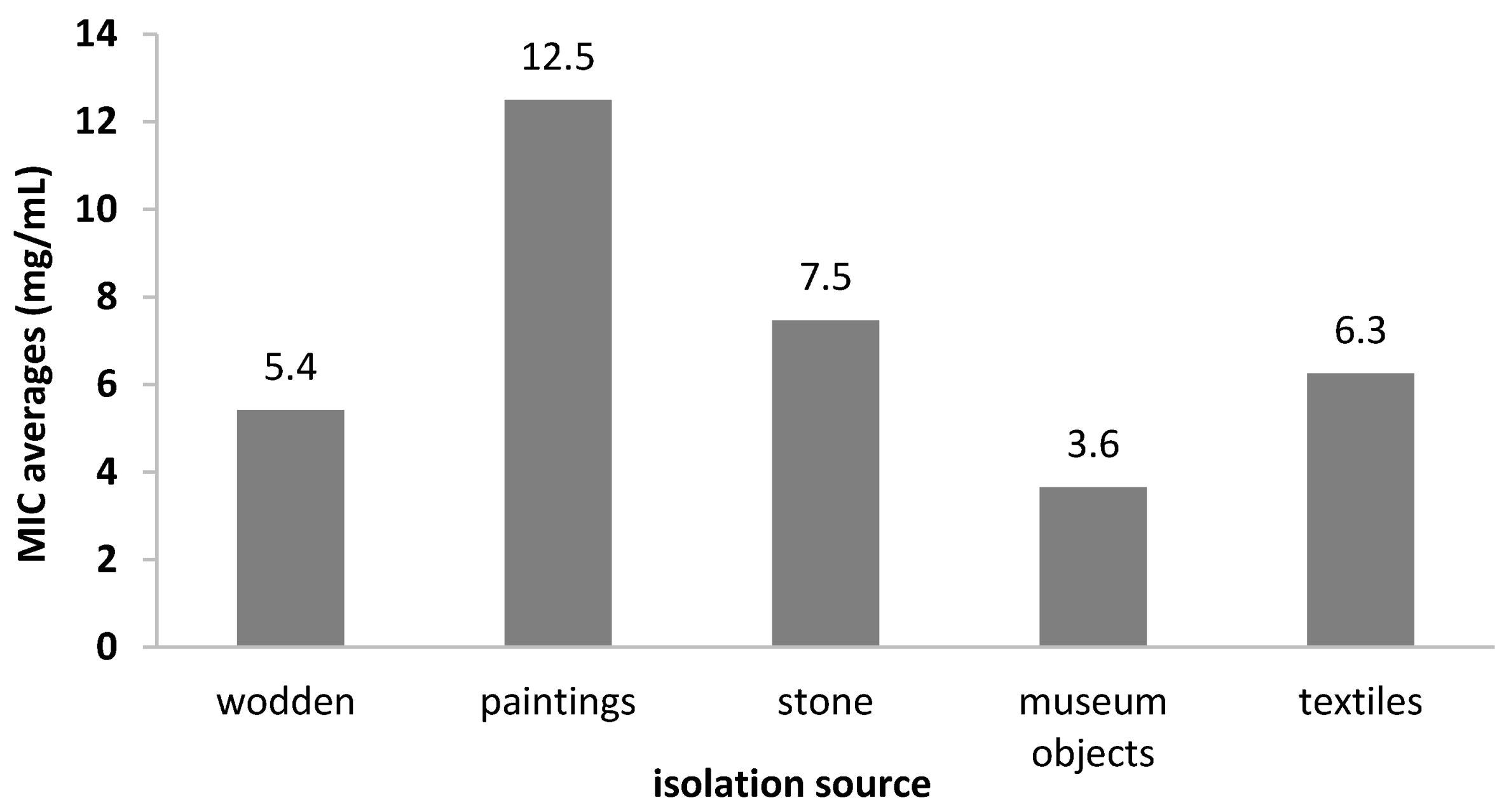
2.4. Antibiofilm Activity
2.5. Effects of A. sativum Extract on Microbial Soluble Factor Production
3. Discussion
4. Materials and Methods
4.1. Preparation of Plant Extract
4.2. The Total Phenolic Content (TPC) Assay
4.3. The Total Flavonoid Content Assay
4.4. Total Thiosulfinate Content Assay
4.5. Characterization of the Extract by UHPLC–MS/MS
4.6. Antioxidant Activity Assay
4.6.1. DPPH Assay
4.6.2. CUPRAC Method
4.6.3. FRAP Assay
4.7. Qualitative Screening of the Antimicrobial Activity of the Tested Plant Extract
4.8. Quantitative Evaluation of the Antimicrobial Activity of the Plant Extract
4.9. The Influence of A. sativum Extract on the Microbial Adherence Capacity to the Inert Surface
4.10. The Influence of A. sativum Hydro-Alcoholic Extract on the Production of Microbial Enzymes and Organic Acids Involved in Biodeterioration
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Sample Availability
References
- Rădulescu, H.C.; Perdum, E.; Mitran, C.E.; Dincă, L.C.; Lazăr, V. Biodeterioration capacity of a microfungal species isolated from textile cultural heritage items on contemporary wool materials. Publ. House Rom. Acad. Ser. B 2018, 20, 167–171. [Google Scholar]
- Sterflinger, K. Fungi: Their role in deterioration of cultural heritage. Fungal Biol. Rev. 2010, 24, 47–55. [Google Scholar] [CrossRef]
- Savković, Ž.; Stupar, M.; Unković, N.; Knežević, A.; Vukojević, J.; Grbić, M.L. Fungal Deterioration of Cultural Heritage Objects. In Biodegradation; IntechOpen: London, UK, 2021. [Google Scholar]
- Moza, M.I.; Mironescu, M.; Georgescu, C.; Florea, A.; Bucşa, L. Isolation and characterisation of moulds degrading mural paintings. Ann. Rscb 2012, 17, 136–142. [Google Scholar]
- Lupan, I.; Ianc, M.; Kelemen, B.; Carpa, R.; Rosca-Casian, O.; Chiriac, M.; Popescu, O. New and old microbial communities colonizing a seventeenth-century wooden church. Folia Microbiol. 2014, 59, 45–51. [Google Scholar] [CrossRef]
- IIies, D.; Onet, A.; Wendt, J.A.; Ilieş, M.; Timar, A.; Ilies, A.; Baias, Ş.; Herman, G. Study on microbial and fungal contamination of air and wooden surfaces inside of a historical Church from Romania. J. Environ. Biol. 2018, 39, 980–984. [Google Scholar]
- Rădulescu, H.C.; Gheorghe, I.; Gradisteanu, G.; Ispas, A.; Popescu, C.; Roşu, G.; Chifiriuc, M.C.; Lazăr, V. Molecular characterization based on Internal Transcribed Spacer (ITS) marker sequence of fungal strains isolated from heritage ethnographic textiles. Rom. Biotechnol. Lett. 2019, 24, 906–912. [Google Scholar] [CrossRef]
- Ilieș, D.C.; Oneț, A.; Grigore, H.; Liliana, I.; Alexandru, I.; Ligia, B.; Ovidiu, G.; Florin, M.; Ștefan, B.; Tudor, C. Exploring the indoor environment of heritage buildings and its role in the conservation of valuable objects. Environ. Eng. Manag. J. EEMJ 2019, 18, 2579–2586. [Google Scholar] [CrossRef]
- Sirghi, A.C.; Gheorghe, I.; Sarbu, I.; Marutescu, L.; Stoian, G.; Zhiyong, Z.; Chifiriuc, M.C. Identification of fungal strains isolated from buildings of cultural importance in Romania and antagonistic relationships amongst them. Rom. Biotechnol. Lett. 2018, 24, 1008–1014. [Google Scholar] [CrossRef]
- Gheorghe, I.; Sârbu, I.; Pecete, I.; Blăjan, I.; Balotescu, I. Multi-level characterization of microbial consortia involved in the biodeterioration of wooden and stone romanian heritage churches. Conserv. Sci. Cult. Herit. 2020, 20, 289–308. [Google Scholar]
- Palla, F.; Bruno, M.; Mercurio, F.; Tantillo, A.; Rotolo, V. Essential oils as natural biocides in conservation of cultural heritage. Molecules 2020, 25, 730. [Google Scholar] [CrossRef]
- Rosado, T.; Silva, M.; Dias, L.; Candeias, A.; Gil, M.; Mirão, J.; Pestana, J.; Caldeira, A.T. Microorganisms and the integrated conservation-intervention process of the renaissance mural paintings from Casas Pintadas in Évora–Know to act, act to preserve. J. King Saud Univ. Sci. 2017, 29, 478–486. [Google Scholar] [CrossRef]
- Gheorghe, I.; Avram, I.; Matis Corbu, V.; Măruţescu, L.; Popa, M.; Balotescu, I.; Blăjan, I.; Mateescu, V.; Zaharia, D.; Dumbravă, A.Ş. In vitro evaluation of MgB2 powders as novel tools to fight fungal biodeterioration of heritage buildings and objects. Front. Mater. 2021, 7, 458. [Google Scholar] [CrossRef]
- Yetgin, A.; Canlı, K.; Altuner, E.M. Comparison of antimicrobial activity of Allium sativum cloves from China and Taşköprü, Turkey. Adv. Pharmacol. Sci. 2018, 2018, 9302840. [Google Scholar]
- Grumezescu, A.M.; Andronescu, E.; Holban, A.M.; Ficai, A.; Ficai, D.; Voicu, G.; Grumezescu, V.; Balaure, P.C.; Chifiriuc, C.M. Water dispersible cross-linked magnetic chitosan beads for increasing the antimicrobial efficiency of aminoglycoside antibiotics. Int. J. Pharm. 2013, 454, 233–240. [Google Scholar] [CrossRef]
- Fierascu, I.; Dima, R.; Fierascu, R.C. Natural Extracts for preventing Artefacts Biodeterioration. In Proceedings of the International Conference on Cultural Heritage and New Technologies Vienna, Vienna, Austria, 5–7 November 2012. [Google Scholar]
- Fierascu, I.; Ion, R.M.; Radu, M.; Bunghez, I.; Avramescu, S.; Fierascu, R. Comparative study of antifungal effect of natural extracts and essential oils of Ocimum basilicum on selected artefacts. Rev. Roum. Chim. 2014, 59, 207–211. [Google Scholar]
- Wu, X.; Santos, R.R.; Fink-Gremmels, J. Analyzing the antibacterial effects of food ingredients: Model experiments with allicin and garlic extracts on biofilm formation and viability of Staphylococcus epidermidis. Food Sci. Nutr. 2015, 3, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Woods-Panzaru, S.; Nelson, D.; McCollum, G.; Ballard, L.M.; Millar, B.C.; Maeda, Y.; Goldsmith, C.E.; Rooney, P.J.; Loughrey, A.; Rao, J.R. An examination of antibacterial and antifungal properties of constituents described in traditional Ulster cures and remedies. Ulst. Med. J. 2009, 78, 13. [Google Scholar]
- Khan, S.; Imran, M.; Imran, M.; Pindari, N. Antimicrobial activity of various ethanolic plant extracts against pathogenic multi drug resistant Candida spp. Bioinformation 2017, 13, 67. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.I.; Neiva Correia, M.J.; Mateus, M.M.; Saraiva, J.A.; Vicente, A.A.; Moldão, M. Fourier transform infrared (FT-IR) spectroscopy as a possible rapid tool to evaluate abiotic stress effects on pineapple by-products. Appl. Sci. 2019, 9, 4141. [Google Scholar] [CrossRef]
- Gonzalez, R.E.; Soto, V.C.; Sance, M.M.; Camargo, A.B.; Galmarini, C.R. Variability of solids, organosulfur compounds, pungency and health-enhancing traits in garlic (Allium sativum L.) cultivars belonging to different ecophysiological groups. J. Agric. Food Chem. 2009, 57, 10282–10288. [Google Scholar] [CrossRef] [PubMed]
- Martínez, G.; Regente, M.; Jacobi, S.; Del Rio, M.; Pinedo, M.; de la Canal, L. Chlorogenic acid is a fungicide active against phytopathogenic fungi. Pestic. Biochem. Physiol. 2017, 140, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Lei, M.; Andargie, M.; Zeng, J.; Li, J. Antifungal activity and mechanism of action of tannic acid against Penicillium digitatum. Physiol. Mol. Plant Pathol. 2019, 107, 46–50. [Google Scholar] [CrossRef]
- Miron, T.; Rabinkov, A.; Mirelman, D.; Wilchek, M.; Weiner, L. The mode of action of allicin: Its ready permeability through phospholipid membranes may contribute to its biological activity. Biochim. Biophys. BBA Biomembr. 2000, 1463, 20–30. [Google Scholar] [CrossRef]
- Gruhlke, M.C.; Hemmis, B.; Noll, U.; Wagner, R.; Lühring, H.; Slusarenko, A.J. The defense substance allicin from garlic permeabilizes membranes of Beta vulgaris, Rhoeo discolor, Chara corallina and artificial lipid bilayers. Biochim. Biophys. Acta BBA Gen. Subj. 2015, 1850, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Leontiev, R.; Hohaus, N.; Jacob, C.; Gruhlke, M.C.; Slusarenko, A.J. A comparison of the antibacterial and antifungal activities of thiosulfinate analogues of allicin. Sci. Rep. 2018, 8, 6763. [Google Scholar] [CrossRef]
- Negri, M.; Salci, T.P.; Shinobu-Mesquita, C.S.; Capoci, I.R.; Svidzinski, T.I.; Kioshima, E.S. Early state research on antifungal natural products. Molecules 2014, 19, 2925–2956. [Google Scholar] [CrossRef]
- Choo, S.; Chong, P.; Tay, S.; Wong, E.; Madhavan, P.; Yong, P. Inhibition of sessile and biofilm growth in various Aspergillus species by allicin associated with disruption to structural changes in cell wall. Int. J. Infect. Dis. 2020, 101, 71. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Kim, K.S.; Han, I.; Kim, M.-H.; Jung, M.H.; Park, H.-K. Quantitative and qualitative analysis of the antifungal activity of allicin alone and in combination with antifungal drugs. PLoS ONE 2012, 7, e38242. [Google Scholar] [CrossRef]
- Gruhlke, M.C.; Portz, D.; Stitz, M.; Anwar, A.; Schneider, T.; Jacob, C.; Schlaich, N.L.; Slusarenko, A.J. Allicin disrupts the cell’s electrochemical potential and induces apoptosis in yeast. Free Radic. Biol. Med. 2010, 49, 1916–1924. [Google Scholar] [CrossRef]
- Aala, F.; Yusuf, U.K.; Nulit, R.; Rezaie, S. Inhibitory effect of allicin and garlic extracts on growth of cultured hyphae. Iran. J. Basic Med. Sci. 2014, 17, 150. [Google Scholar]
- Salehi, B.; Zucca, P.; Orhan, I.E.; Azzini, E.; Adetunji, C.O.; Mohammed, S.A.; Banerjee, S.K.; Sharopov, F.; Rigano, D.; Sharifi-Rad, J. Allicin and health: A comprehensive review. Trends Food Sci. Technol. 2019, 86, 502–516. [Google Scholar] [CrossRef]
- Rahman, M.S. Allicin and other functional active components in garlic: Health benefits and bioavailability. Int. J. Food Prop. 2007, 10, 245–268. [Google Scholar] [CrossRef]
- Sarfraz, M.; Nasim, M.J.; Jacob, C.; Gruhlke, M.C. Efficacy of allicin against plant pathogenic fungi and unveiling the underlying mode of action employing yeast based chemogenetic profiling approach. Appl. Sci. 2020, 10, 2563. [Google Scholar] [CrossRef]
- Curtis, H.; Noll, U.; Störmann, J.; Slusarenko, A.J. Broad-spectrum activity of the volatile phytoanticipin allicin in extracts of garlic (Allium sativum L.) against plant pathogenic bacteria, fungi and Oomycetes. Physiol. Mol. Plant Pathol. 2004, 65, 79–89. [Google Scholar] [CrossRef]
- Getti, G.; Poole, P. Allicin causes fragmentation of the peptidoglycan coat in Staphylococcus aureus by effecting synthesis and aiding hydrolysis: A determination by MALDI-TOF mass spectrometry on whole cells. J. Med. Microbiol. 2019, 68, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Mattio, L.; Catinella, G.; Iriti, M.; Vallone, L. Inhibitory activity of stilbenes against filamentous fungi. Ital. J. Food Saf. 2021, 10, 8461. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, V.; Carraro, E.; Auler, M.E.; Khalil, N.M. Quercetin and rutin as potential agents antifungal against Cryptococcus spp. Braz. J. Biol. 2016, 76, 1029–1034. [Google Scholar] [CrossRef]
- Lee, W.; Woo, E.R.; Lee, D.G. Phytol has antibacterial property by inducing oxidative stress response in Pseudomonas aeruginosa. Free Radic. Res. 2016, 50, 1309–1318. [Google Scholar] [CrossRef]
- Pei, K.; Ou, J.; Huang, J.; Ou, S. p-Coumaric acid and its conjugates: Dietary sources, pharmacokinetic properties and biological activities. J. Sci. Food Agric. 2016, 96, 2952–2962. [Google Scholar] [CrossRef]
- Zabka, M.; Pavela, R. Antifungal efficacy of some natural phenolic compounds against significant pathogenic and toxinogenic filamentous fungi. Chemosphere 2013, 93, 1051–1056. [Google Scholar] [CrossRef]
- Palla, F.; Rotolo, V.; Giordano, A. Biotechnology a Source of Knowledge in Agreement with Green Strategies for the Conservation of Cultural Assets. Conserv. Sci. Cult. Herit. 2019, 19, 69–80. [Google Scholar]
- Kutawa, A.B.; Danladi, M.D.; Haruna, A. Regular article antifungal activity of garlic (Allium sativum) extract on some selected fungi. J. Med. Herbs Ethnomed. 2018, 4, 12–14. [Google Scholar]
- Rotolo, V.; Barresi, G.; Di Carlo, E.; Giordano, A.; Lombardo, G.; Crimi, E.; Costa, E.; Bruno, M.; Palla, F. Plant extracts as green potential strategies to control the biodeterioration of cultural heritage. Int. J. Conserv. Sci. 2016, 839–846. [Google Scholar]
- Dakal, T.C.; Cameotra, S.S. Microbially induced deterioration of architectural heritages: Routes and mechanisms involved. Environ. Sci. Eur. 2012, 24, 36. [Google Scholar] [CrossRef]
- Deepika, M.S.; Thangam, R.; Sakthidhasan, P.; Arun, S.; Sivasubramanian, S.; Thirumurugan, R. Combined effect of a natural flavonoid rutin from Citrus sinensis and conventional antibiotic gentamicin on Pseudomonas aeruginosa biofilm formation. Food Control 2018, 90, 282–294. [Google Scholar] [CrossRef]
- Shao, D.; Li, J.; Li, J.; Tang, R.; Liu, L.; Shi, J.; Huang, Q.; Yang, H. Inhibition of gallic acid on the growth and biofilm formation of Escherichia coli and Streptococcus mutans. J. Food Sci. 2015, 80, M1299–M1305. [Google Scholar] [CrossRef]
- Teodoro, G.R.; Gontijo, A.V.; Salvador, M.J.; Tanaka, M.H.; Brighenti, F.L.; Delbem, A.C.; Delbem, Á.C.; Koga-Ito, C.Y. Effects of acetone fraction from Buchenavia tomentosa aqueous extract and gallic acid on Candida albicans biofilms and virulence factors. Front. Microbiol. 2018, 9, 647. [Google Scholar] [CrossRef] [PubMed]
- Myszka, K.; Schmidt, M.T.; Białas, W.; Olkowicz, M.; Leja, K.; Czaczyk, K. Role of gallic and p-coumaric acids in the AHL-dependent expression of flgA gene and in the process of biofilm formation in food-associated Pseudomonas fluorescens KM120. J. Sci. Food Agric. 2016, 96, 4037–4047. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Villalón, G.; Pérez-Giraldo, C. Effect of allicin on the production of polysaccharide intercellular adhesin in Staphylococcus epidermidis. J. Appl. Microbiol. 2011, 110, 723–728. [Google Scholar] [CrossRef]
- Lihua, L.; Jianhui, W.; Jialin, Y.; Yayin, L.; Guanxin, L. Effects of allicin on the formation of Pseudomonas aeruginosa biofilm and the production of quorum-sensing controlled virulence factors. Pol. J. Microbiol. 2013, 62, 243. [Google Scholar] [CrossRef] [PubMed]
- Girish, V.M.; Liang, H.; Aguilan, J.T.; Nosanchuk, J.D.; Friedman, J.M.; Nacharaju, P. Anti-biofilm activity of garlic extract loaded nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2019, 20, 102009. [Google Scholar] [CrossRef]
- Mazzoli, R.; Giuffrida, M.G.; Pessione, E. Back to the past: “Find the guilty bug—Microorganisms involved in the biodeterioration of archeological and historical artifacts”. Appl. Microbiol. Biotechnol. 2018, 102, 6393–6407. [Google Scholar] [CrossRef] [PubMed]
- Cappitelli, F.; Pasquariello, G.; Tarsitani, G.; Sorlini, C. Scripta manent? Assessing microbial risk to paper heritage. Trends Microbiol. 2010, 18, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P. A review of microbial decay types found in wooden objects of cultural heritage recovered from buried and waterlogged environments. J. Cult. Herit. 2012, 13, S16–S20. [Google Scholar] [CrossRef]
- El Hassni, M.; Laadouzaa, H.; El Hadrami, A.; Dihazi, A.; Rakibi, Y.; Lemjiber, N.; Naamani, K. An in vitro evaluation of the effect of hydroxycinnamic acids on the growth and hydrolytic enzyme production in Fusarium oxysporum f. sp. albedinis. Arch. Phytopathol. Plant Prot. 2021, 54, 1553–1567. [Google Scholar] [CrossRef]
- Zheng, W.; Zheng, Q.; Xue, Y.; Hu, J.; Gao, M.-T. Influence of rice straw polyphenols on cellulase production by Trichoderma reesei. J. Biosci. Bioeng. 2017, 123, 731–738. [Google Scholar] [CrossRef]
- Abu-Taleb, A.M.; El-Deeb, K.; Al-Otibi, F.O. Assessment of antifungal activity of Rumex vesicarius L. and Ziziphus spina-christi (L.) Willd. extracts against two phytopathogenic fungi. Afr. J. Microbiol. Res. 2011, 5, 1001–1011. [Google Scholar] [CrossRef]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar]
- Formagio, A.S.; Kassuya, C.A.; Neto, F.F.; Volobuff, C.R.; Iriguchi, E.K.; do C Vieira, M.; Foglio, M.A. The flavonoid content and antiproliferative, hypoglycaemic, anti-inflammatory and free radical scavenging activities of Annona dioica St. Hill. BMC Complement. Altern. Med. 2013, 13, 14. [Google Scholar] [CrossRef]
- Mansor, N.; Herng, H.J.; Samsudin, S.J.; Sufian, S.; Uemura, Y. Quantification and characterization of allicin in garlic extract. J. Med. Bioeng. 2016, 5, 24–27. [Google Scholar] [CrossRef]
- Marinas, I.C.; Oprea, E.; Geana, E.-I.; Tutunaru, O.; Pircalabioru, G.G.; Zgura, I.; Chifiriuc, M.C. Valorization of Gleditsia triacanthos Invasive Plant Cellulose Microfibers and Phenolic Compounds for Obtaining Multi-Functional Wound Dressings with Antimicrobial and Antioxidant Properties. Int. J. Mol. Sci. 2021, 22, 33. [Google Scholar] [CrossRef] [PubMed]
- Madhu, G.; Bose, V.C.; Aiswaryaraj, A.; Maniammal, K.; Biju, V. Defect dependent antioxidant activity of nanostructured nickel oxide synthesized through a novel chemical method. Colloids Surf. A Physicochem. Eng. Asp. 2013, 429, 44–50. [Google Scholar] [CrossRef]
- Meng, J.; Fang, Y.; Zhang, A.; Chen, S.; Xu, T.; Ren, Z.; Han, G.; Liu, J.; Li, H.; Zhang, Z. Phenolic content and antioxidant capacity of Chinese raisins produced in Xinjiang Province. Food Res. Int. 2011, 44, 2830–2836. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Najee, H.; Kamerzan, C.; Marutescu, L.; Gheorghe, I.; Popa, M.; Gradisteanu, G.; Lazar, V. Antifungal activity of some medicinal plant extracts against Candida albicans nosocomial isolates. Rom. Biotechnol. Lett. 2018, 23, 14073. [Google Scholar]
- Muhsin, T.M.; Al-Zubaidy, S.R.; Ali, E.T. Effect of garlic bulb extract on the growth and enzymatic activities of rhizosphere and rhizoplane fungi. Mycopathologia 2001, 152, 143–146. [Google Scholar] [CrossRef]
- Zajc, J.; Gostinčar, C.; Černoša, A.; Gunde-Cimerman, N. Stress-tolerant yeasts: Opportunistic pathogenicity versus biocontrol potential. Genes 2019, 10, 42. [Google Scholar] [CrossRef]
- Borrego, S.; Molina, A.; Santana, A. Fungi in archive repositories environments and the deterioration of the graphics documents. EC Microbiol. 2017, 11, 205–226. [Google Scholar]
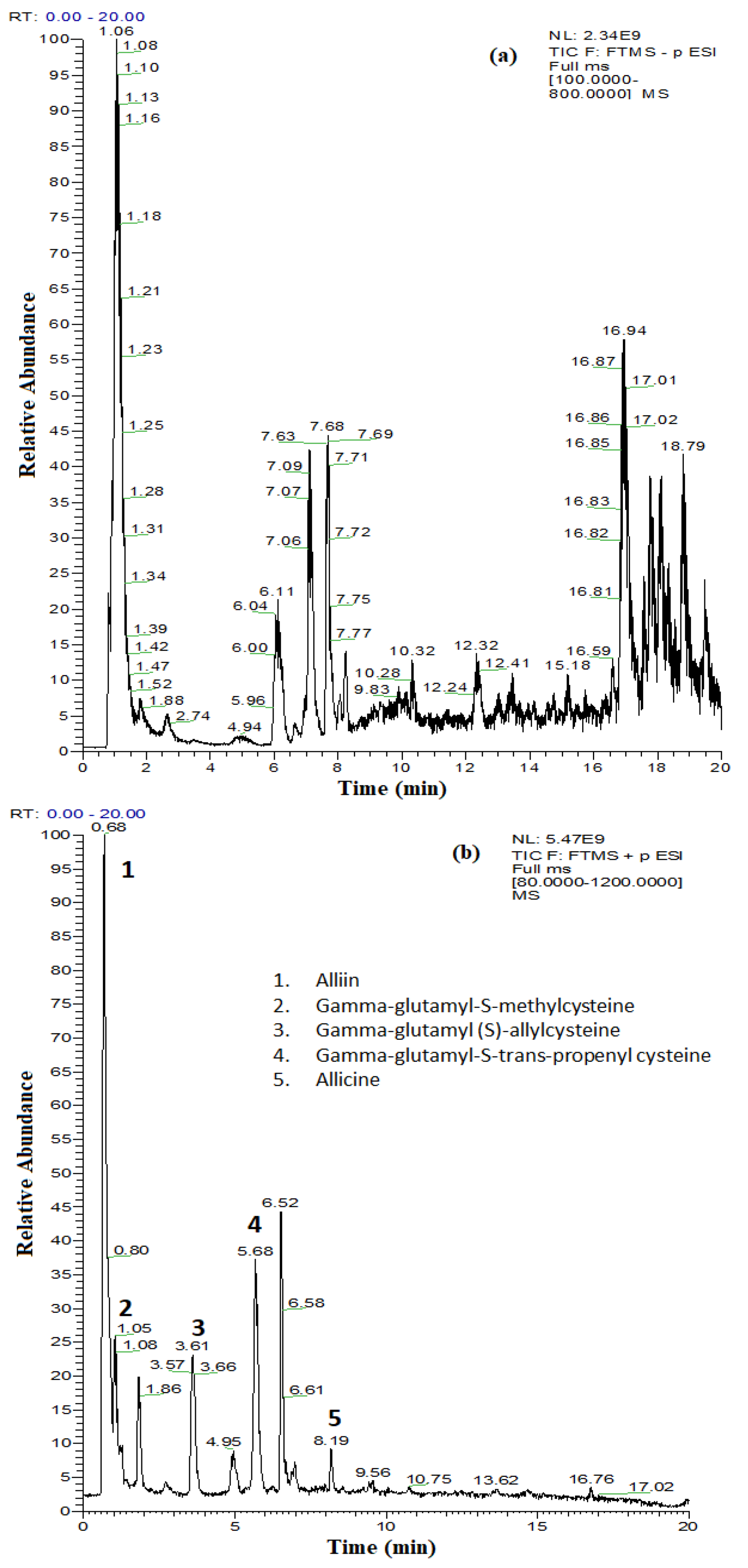

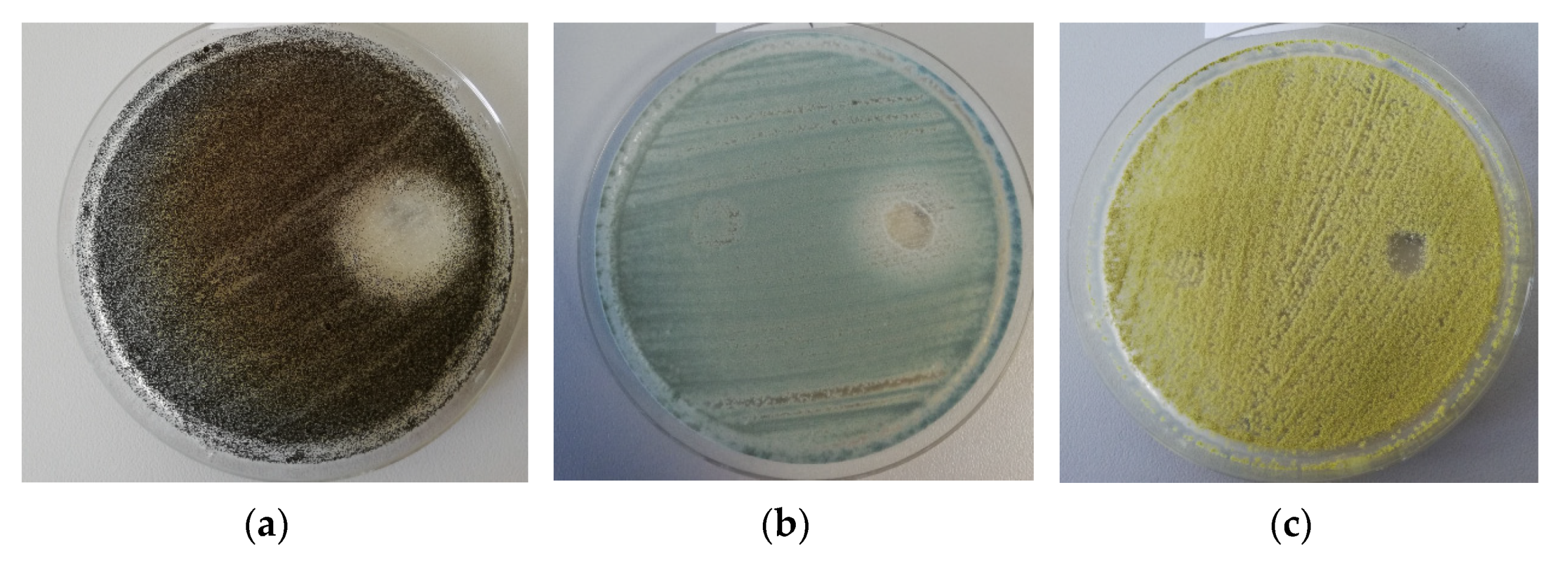
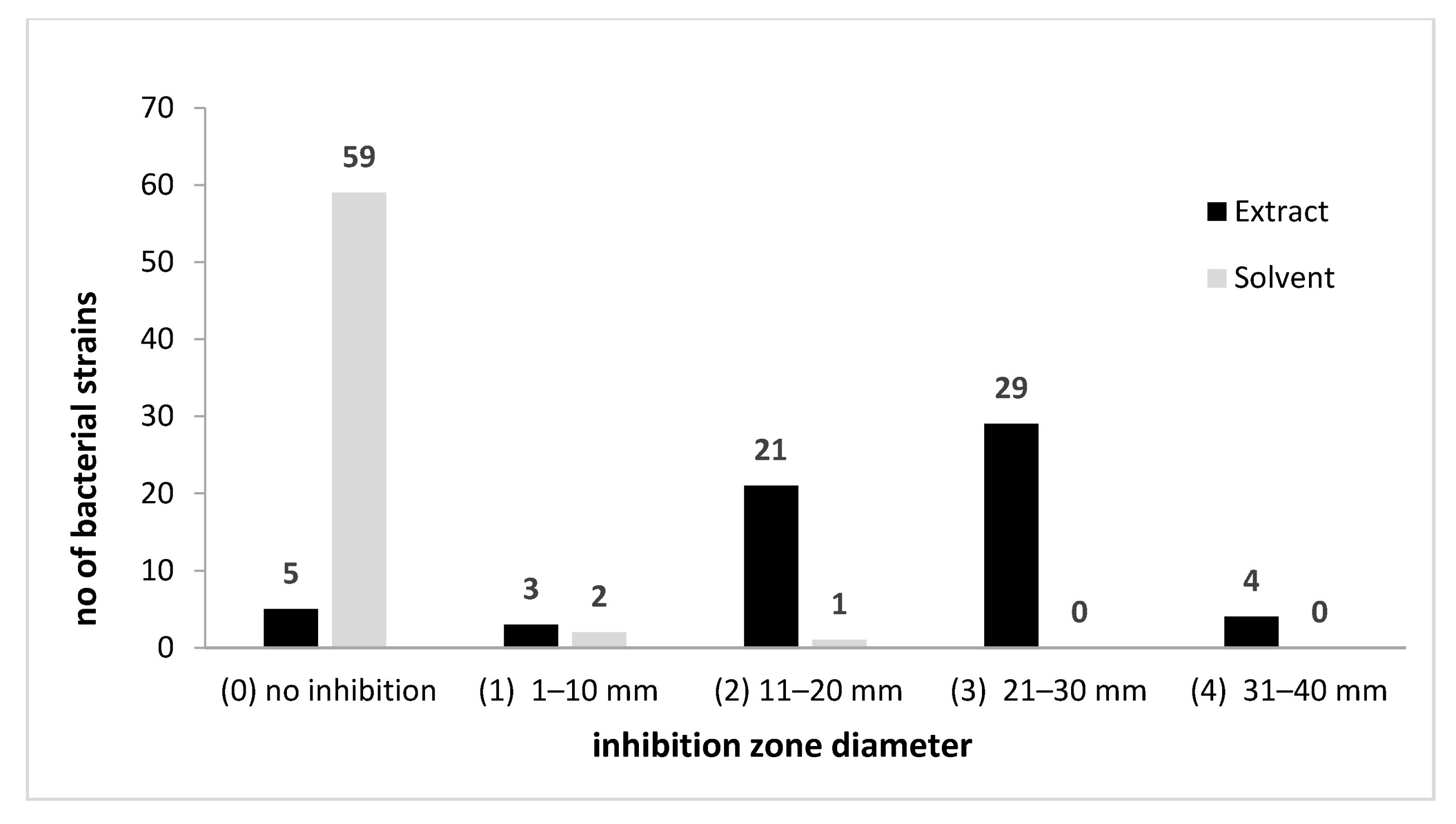
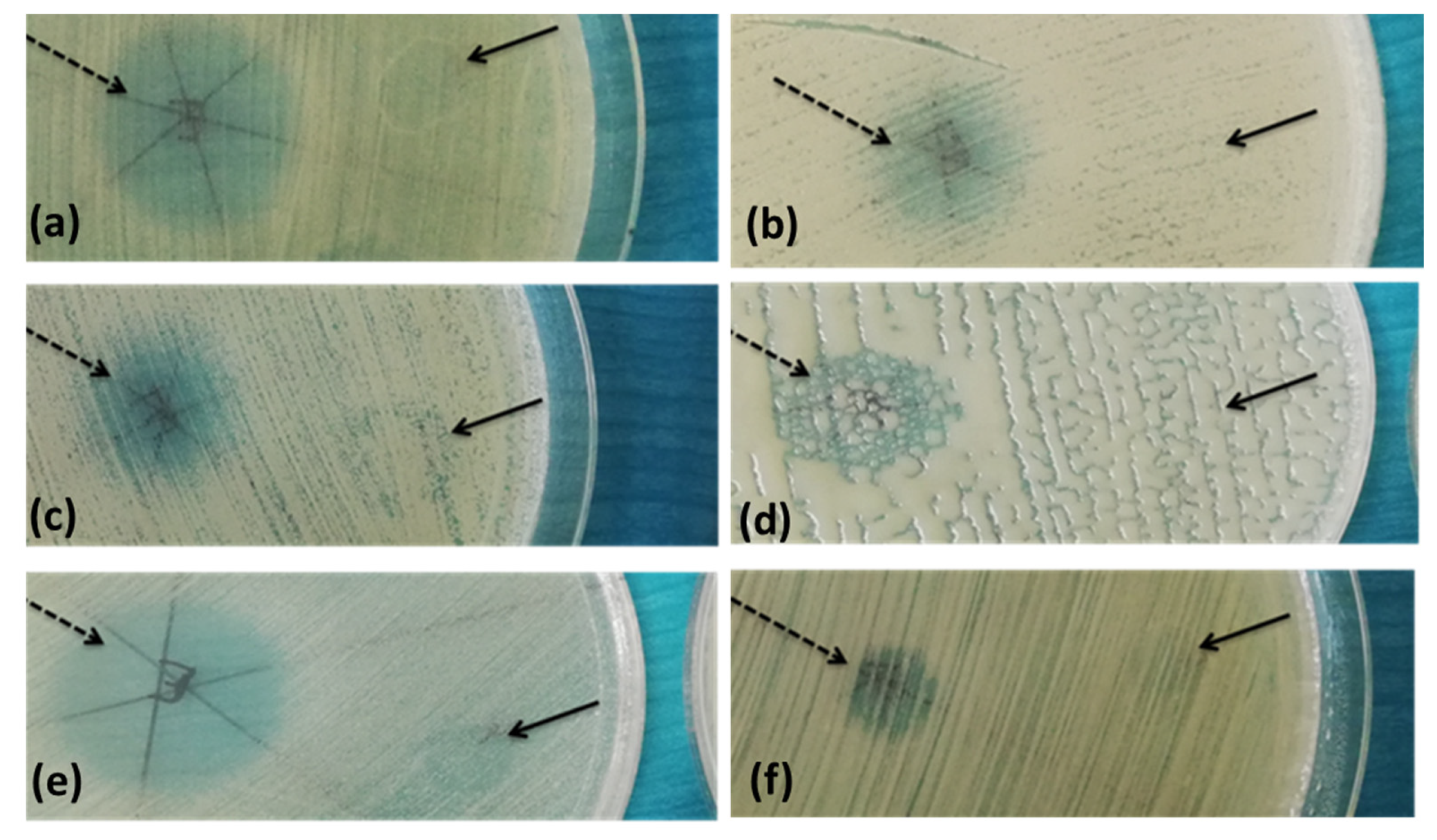

| Parameter | The Analyzed Extract in Relation to the Quantity Used (Mean ± SD) |
|---|---|
| TPC (mg GAE/g) | 0.95 ± 0.011 |
| Flavonoids (μg quercetin/g) | 64.33 ± 7.69 |
| Total thiosulfinate (µM/g) | 307.66 ± 0.043 |
| Antioxidant activity | Extract (0.1 g/mL) |
| DPPH (μM Trolox/mL) | 66.39 ± 5.50 |
| CUPRAC (μM Trolox/mL) | 52.25 ± 3.01 |
| FRAP (μM Trolox/mL) | 43.51 ± 1.49 |
| No. | Compound | Retention Time (min) | Accurate Mass [M − H]−/[M − H]+ | Mass Fragments | Concentration (µg/L) |
|---|---|---|---|---|---|
| Polyphenolic compounds by UHPLC-MS/MS in negative ionization mode | |||||
| 1 | Gallic acid | 0.68 | 169.0133 | 125.0231 | 0.4 |
| 2 | 3,4-Dihydroxybenzoic acid | 1.59 | 153.0183 | 109.0281 | 0.8 |
| 3 | 4-Hydroxybenzoic acid | 5.40 | 137.0232 | 93.0331 | 4 |
| 4 | Chlorogenic acid | 7.55 | 353.0879 | 191.0553 | 0.8 |
| 5 | Syringic acid | 8.03 | 197.0450 | 182.0212, 166.9976, 153.0547, 138.0311, 123.0075 | 7.2 |
| 6 | Caffeic acid | 8.08 | 179.0338 | 135.044 | 6.4 |
| 7 | Vanillic acid | 8.31 | 167.0343 | 152.0105, 124.0154, 111.0075, 139.0025, 95.0125 | 65.2 |
| 8 | p-Coumaric acid | 8.59 | 163.0392 | 119.0489 | 1.2 |
| 9 | t-Ferulic acid | 8.83 | 193.0500 | 178.0262, 134.0361 | 18.8 |
| 10 | Ellagic acid | 9.66 | 300.9990 | 300.9990 | 13.2 |
| 11 | Cinnamic acid | 10.45 | 147.0441 | 119.0489, 103.0387 | 7.6 |
| 12 | Abscisic acid | 10.04 | 263.1288 | 179.9803, 191.9454 | 2.4 |
| Ʃ phenolic acids | 128 | ||||
| 13 | Epi-catechin | 7.98 | 289.0719 | 109.0282, 125.0232, 137.0232, 151.0390, 203.0708, 245.0817 | 41.2 |
| 14 | Quercetin | 10.74 | 301.0356 | 151.0226, 178.9977, 121.0282, 107.0125 | 0.8 |
| 15 | Naringin | 9.25 | 579.1718 | 363.0721 | 3.6 |
| 16 | Hesperidin | 9.37 | 609.1824 | 377.0876 | 14 |
| 17 | Rutin | 9.43 | 609.1462 | 3345.0614 | 10 |
| 18 | Kaempferol | 11.62 | 285.0406 | 151.0389, 117.0180 | 2.8 |
| 19 | Isorhamnetin | 11.80 | 315.0512 | 300.0276 | 9.6 |
| 20 | Apigenin | 11.86 | 269.0457 | 117.0333, 151.0027, 107.0126 | 4.4 |
| 21 | Pinocembrin | 12.70 | 255.0663 | 213.0551, 151.0026, 107.0125 | 1.6 |
| 22 | Chrysin | 13.52 | 253.0506 | 143.0491, 145.0284, 107.0125, 209.0603, 63.0226, 65.0019 | 3.6 |
| 23 | Galangin | 13.77 | 269.0458 | 169.0650, 143.0491 | 2.8 |
| 24 | Pinostrobin | 14.84 | 269.081 | 179.0554 | 2.8 |
| Ʃ flavonoids | 97.2 | ||||
| 25 | t-Resveratrol | 9.55 | 227.0707 | 185.0813, 143.0337 | 0.8 |
| Thiosulfinate compounds by UHPLC-MS/MS in positive ionization mode | |||||
| 1 | Alliin | 0.68 | 178.0530 | 175.1188, 116.0709 | - |
| 2 | Gamma-glutamyl-S-methylcysteine | 1.05 | 265.0850 | 182.0811, 132.1020 | - |
| 3 | Gamma-glutamyl (S)-allylcysteine | 3.61 | 291.1005 | 162.0582, 139.0502 | - |
| 4 | Gamma-glutamyl-S-trans-propenyl cysteine | 5.68 | 291.1006 | 162.0582 | - |
| 5 | Allicin | 8.19 | 163.0245 | 120.9779 | - |
| Species | Isolation Source | MIC (mg/mL) | MBEC 1 (mg/mL) | IIBG 2 (%) | p Value (Relative to Solvent Control) |
|---|---|---|---|---|---|
| A. niger | textiles | 25 | 25 | 95.08 ± 0.91 | <0.0001 |
| P. chrysogenum | textiles | 25 | 6.25 | 89.11 ± 0.91 | <0.01 |
| Penicilium spp. | stone | 25 | 12.5 | 90.71 ± 0.70 | <0.001 |
| P. corylophilum | textiles | 25 | 6.25 | 80.47 ± 2.73 | <0.05 |
| A. flavus | textiles | 25 | 12.5 | 88.57 ± 1.30 | <0.01 |
| Aspergillus spp. | stone | 25 | 6.25 | 90.26 ± 1.01 | <0.000001 |
| A. sydowii | stone | 50 | 12.5 | 93.54 ± 1.09 | <0.01 |
| A. niger | wood | 6.25 | 12.5 | 88.74 ± 1.31 | <0.001 |
| A. flavus | textiles | 12.5 | 6.25 | 88.19 ± 2.40 | <0.01 |
| P. lilacium | stone | 12.5 | NA | ||
| P. lilacium | stone | 12.5 | NA | ||
| Aspergillus spp. | stone | 25 | 25 | 84.28 ± 1.86 | <0.01 |
| P. chrysogenum | wood | 50 | 12.5 | 93.87 ± 0.03 | 0.072 |
| P. corylophilum | wood | 12.5 | 6.25 | 92.50 ± 0.00 | <0.00001 |
| P. corylophilum | wood | 6.25 | 12.5 | 92.30 ± 1.92 | <0.0001 |
| P. corylophilum | wood | 25 | 12.5 | 93.42 ± 1.38 | <0.001 |
| A. niger | textiles | 25 | 12.5 | 91.96 ± 0.71 | <0.001 |
| B. megaterium | wood | 6.25 | NA | ||
| B. subtilis | wood | 6.25 | NA | ||
| B. cereus | ceramic | 1.56 | NA | ||
| B. pumilus | wood | 0.78 | NA | ||
| P. koreensis | stone | 12.5 | NA | ||
| B. arthopahaeus | stone | 6.25 | 12.5 | 44.62 ± 7.30 | 0.342 |
| B. megaterium | wood | 3.125 | NA | ||
| B. thuringiensis | stone | 3.125 | 12.5 | 87.56 ± 4.60 | <0.01 |
| B. pumilus | stone | 6.25 | NA | ||
| A. aurascens | wood | 12.5 | NA | ||
| B. cereus | wood | 3.125 | NA | ||
| B. megaterium | wood | 3.125 | NA | ||
| B. megaterium | wood | 3.125 | NA | ||
| B. pumilus | painting | 12.5 | NA | ||
| R. erythrophilus | wood | 3.125 | NA | ||
| B. pumilus | wood | 3.125 | NA | ||
| B. megaterium | textiles | 6.25 | NA | ||
| A. globiformis | wood | 6.25 | NA | ||
| B. cereus | wood | 6.25 | 12.5 | 57.72 ± 5.11 | <0.05 |
| Fungal Strains | ||||||||||
| Species | Isolation Sources | Cellulase | Esterase | Acid Production | ||||||
| A. sativum Extract | Ethanol 50% | p-Value | A. sativum Extract | Ethanol 50% | p-Value | A. Sativum Extract | Ethanol 50% | p-Value | ||
| A. niger | textile | 105.00 ± 8.66 | 140.00 ± 17.32 | 0.0535 | 66.67 ± 4.44 | 58.97 ± 4.44 | 0.1012 | 79.09 ± 3.86 | 92.73 ± 0.00 | <0.05 |
| P. chrysogenum | textile | 111.77 ± 5.09 | 108.82 ± 5.09 | 0.5185 | - | - | - | 70.37 ± 8.49 | 98.15 ± 3.21 | <0.05 |
| Penicilium spp. | stone | 272.73 ± 0.01 | 327.27 ± 0.05 | <0.0001 | 78.26 ± 13.04 | 86.96 ± 19.92 | 0.5666 | 116.67 ± 5.77 | 96.67 ± 11.55 | 0.0765 |
| P. corylophilum | textile | 105.56 ± 7.86 | 107.41 ± 6.42 | 0.7683 | - | - | - | - | - | - |
| A. flavus | textile | - | - | - | 108.00 ± 0.00 | 88.00 ± 6.93 | <0.05 | 83.91 ± 7.18 | 81.61 ± 3.98 | 0.6596 |
| Aspergillus spp. | stone | 108.16 ± 3.53 | 106.16 ± 12.75 | 0.8113 | - | - | - | 72.00 ± 12.00 | 104 ± 18.33 | 0.0746 |
| A. sydowii | stone | 143.75 ± 8.84 | 150.00 ± 21.65 | 0.1918 | - | - | - | 68.18 ± 6.43 | 60.61 ± 5.24 | 0.6788 |
| A. niger | wood | - | - | - | - | - | - | 92.00 ± 13.86 | 136.00 ± 6.93 | <0.05 |
| A. flavus | textile | 87.50 ± 8.66 | 90.00 ± 12.99 | 0.7972 | 97.96 ± 6.12 | 79.59 ± 0.00 | <0.05 | 63.16 ± 0.00 | 68.42 ± 4.56 | 0.1835 |
| P. lilacium | stone | 76.92 ± 0.00 | 100.00 ± 10.88 | 0.0667 | 94.44 ± 9.62 | 111.11 ± 9.62 | 0.1012 | 110.71 ± 6.19 | 103.57 ± 6.19 | 0.2302 |
| P. lilacium | stone | 118.52 ± 16.97 | 133.33 ± 0.00 | 0.4226 | 93.75 ± 0.00 | 87.50 ± 10.83 | 0.2697 | 137.5 ± 5.41 | 118.75 ± 5.41 | <0.05 |
| Aspergillus spp. | stone | 97.50 ± 13.00 | 60.00 ± 7.50 | <0.05 | 94.29 ± 0.00 | 62.86 ± 4.95 | <0.01 | 76.67 ± 5.77 | 130 ± 17.32 | <0.05 |
| P. chrysogenum | wood | 93.94 ± 6.31 | 93.94 ± 9.26 | >1 | - | - | - | 82.14 ± 6.19 | 60.71 ± 6.19 | <0.05 |
| P. corylophilum | wood | 126.67 ± 11.55 | 70.00 ± 14.14 | <0.01 | - | - | - | 144.44 ± 19.25 | 120.83 ± 5.89 | 0.1589 |
| P. corylophilum | wood | 71.80 ± 11.75 | 97.44 ± 8.88 | <0.05 | - | - | - | - | - | - |
| A. niger | textile | - | - | - | - | - | - | 138.46 ± 0.01 | 115.39 ± 0.11 | <0.0001 |
| Bacterial Strains | ||||||||||
| Species | Isolation Sources | Cellulase | Esterase | Acid Production | ||||||
| A. sativum Extract | Ethanol 50% | p-Value | A. sativum Extract | Ethanol 50% | p-Value | A. sativum Extract | Ethanol 50% | p-Value | ||
| B. megaterium | wood | - | - | - | 120.00 ± 0.00 | 90.00 ± 42.43 | 0.3454 | 100.00 ± 11.47 | 105.00 ± 7.50 | 0.5667 |
| B. subtilis | wood | - | - | - | 100.00 ± 21.65 | 0 | <0.05 | - | - | - |
| P. koreensis | stone | - | - | - | - | - | - | 89.80 ± 3.53 | 95.92 ± 3.53 | 0.1013 |
| B. arthopahaeuss | stone | - | - | - | 71.43 ± 12.37 | 78.57 ± 12.37 | 0.5186 | - | - | - |
| B. megaterium | wood | - | - | - | - | - | - | 183.33 ± 23.57 | 183.33 ± 23.57 | >1 |
| B. cereus | wood | - | - | - | 75.00 ± 9.64 | 68.19 ± 0.00 | 0.3457 | - | - | - |
| B. pumilus | painting | - | - | - | 116.67 ± 57.70 | 0 | 0.0728 | 93.333 ± 11.55 | 106.67 ± 11.55 | 0.2301 |
| B. megaterium | textile | - | - | - | - | - | - | 0 | 75.76 ± 13.89 | <0.05 |
| B. cereus | wooden | - | - | - | 87.50 ± 5.41 | 75.00 ± 0.00 | 0 | 76.67 ± 4.71 | <0.01 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corbu, V.M.; Gheorghe, I.; Marinaș, I.C.; Geană, E.I.; Moza, M.I.; Csutak, O.; Chifiriuc, M.C. Demonstration of Allium sativum Extract Inhibitory Effect on Biodeteriogenic Microbial Strain Growth, Biofilm Development, and Enzymatic and Organic Acid Production. Molecules 2021, 26, 7195. https://doi.org/10.3390/molecules26237195
Corbu VM, Gheorghe I, Marinaș IC, Geană EI, Moza MI, Csutak O, Chifiriuc MC. Demonstration of Allium sativum Extract Inhibitory Effect on Biodeteriogenic Microbial Strain Growth, Biofilm Development, and Enzymatic and Organic Acid Production. Molecules. 2021; 26(23):7195. https://doi.org/10.3390/molecules26237195
Chicago/Turabian StyleCorbu, Viorica Maria, Irina Gheorghe, Ioana Cristina Marinaș, Elisabeta Irina Geană, Maria Iasmina Moza, Ortansa Csutak, and Mariana Carmen Chifiriuc. 2021. "Demonstration of Allium sativum Extract Inhibitory Effect on Biodeteriogenic Microbial Strain Growth, Biofilm Development, and Enzymatic and Organic Acid Production" Molecules 26, no. 23: 7195. https://doi.org/10.3390/molecules26237195
APA StyleCorbu, V. M., Gheorghe, I., Marinaș, I. C., Geană, E. I., Moza, M. I., Csutak, O., & Chifiriuc, M. C. (2021). Demonstration of Allium sativum Extract Inhibitory Effect on Biodeteriogenic Microbial Strain Growth, Biofilm Development, and Enzymatic and Organic Acid Production. Molecules, 26(23), 7195. https://doi.org/10.3390/molecules26237195









