Enzymatic Methods for Salivary Biomarkers Detection: Overview and Current Challenges
Abstract
:1. Biomarkers as a Diagnostic Tool
2. Advantages of Saliva Samples as a Biomarker Source
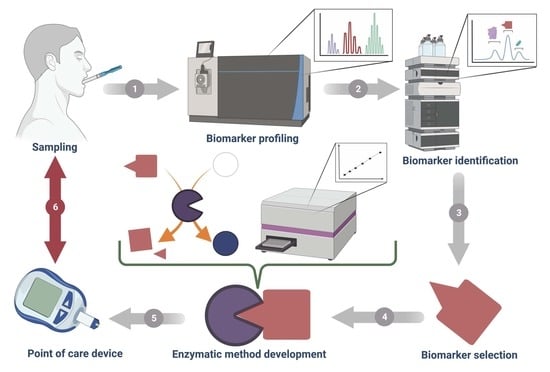
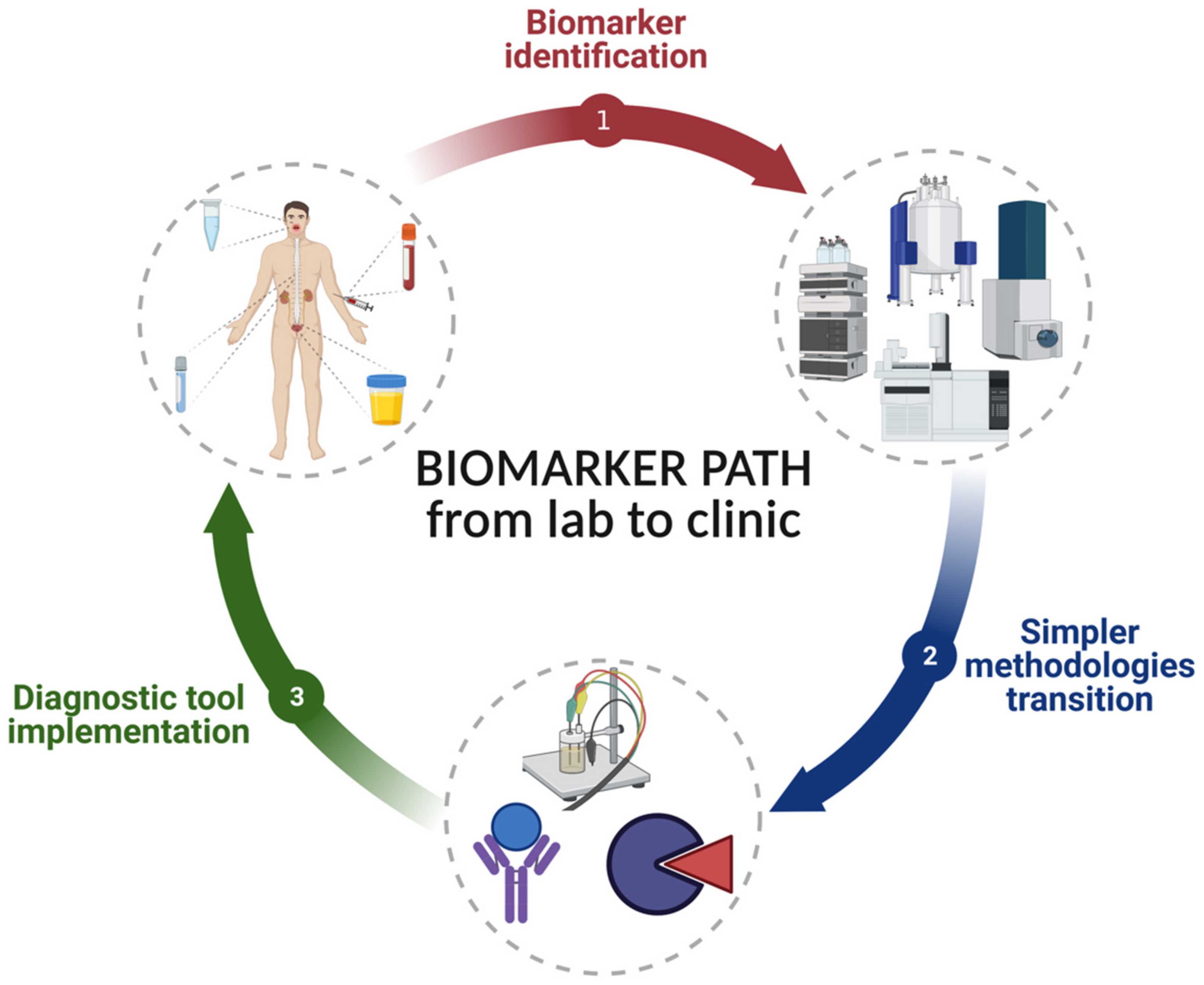
3. Current Methods for Salivary Biomarker Identification
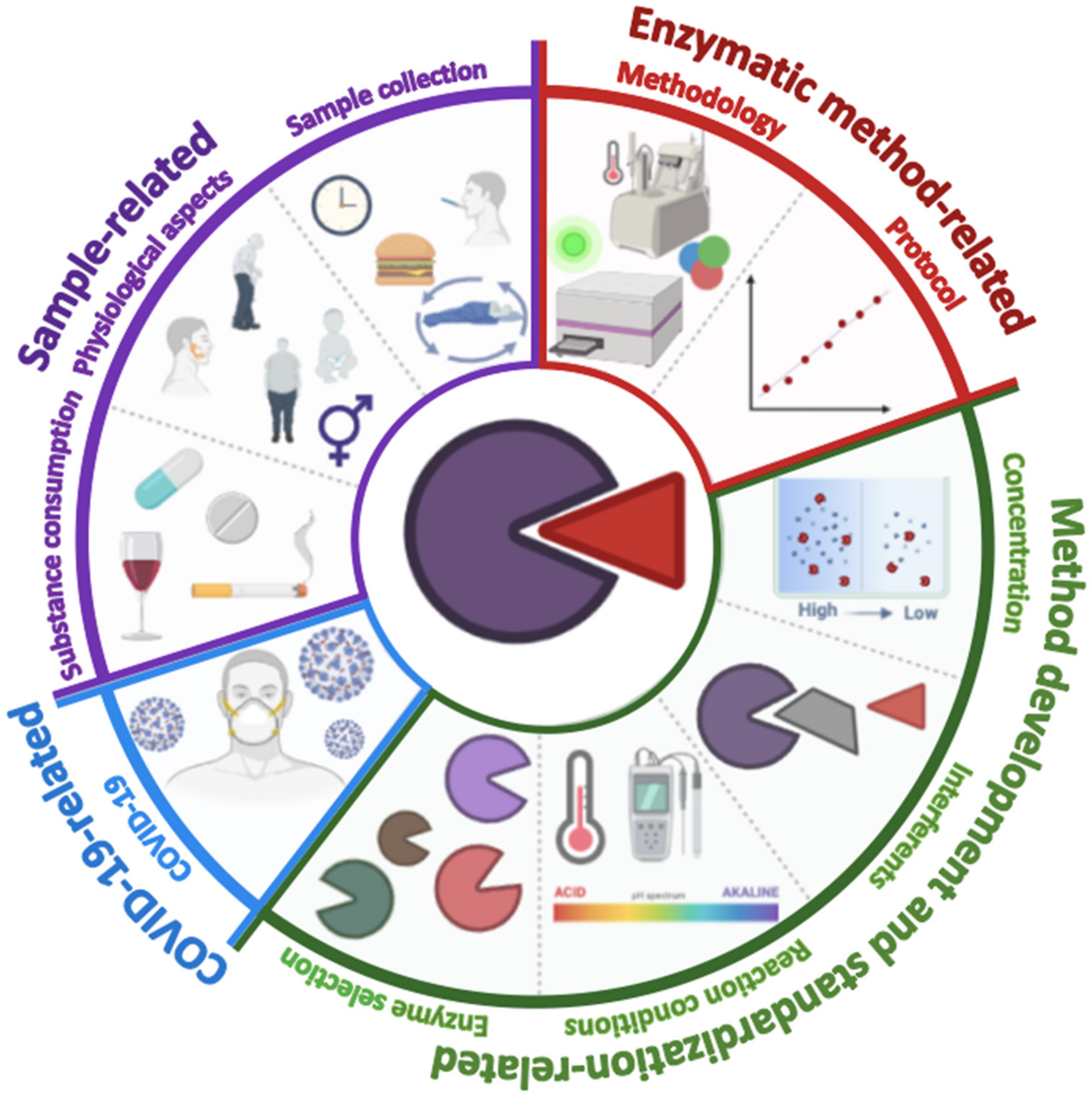
4. Challenges in Enzymatic Methods for Salivary Biomarkers Detection
4.1. Sample-Related Challenges
4.2. Enzymatic Method-Related Challenges
4.3. Method Development and Standardization-Related Challenges
4.4. Challenges in the COVID-19 Era
5. Current Trends and Future Perspectives in Enzymatic Methods for Salivary Biomarkers Detection
Author Contributions
Funding
Conflicts of Interest
References
- Adom, D.; Rowan, C.; Adeniyan, T.; Yang, J.; Paczesny, S. Biomarkers for Allogeneic HCT Outcomes. Front. Immunol. 2020, 11, 673. [Google Scholar] [CrossRef] [Green Version]
- Biomarkers Definitions Working Group. Biomarkers and Surrogate Endpoints: Preferred Definitions and Conceptual Framework. Clin. Pharm. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- Strimbu, K.; Tavel, J.A. What Are Biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef]
- Tolstikov, V.; Moser, A.J.; Sarangarajan, R.; Narain, N.R.; Kiebish, M.A. Current Status of Metabolomic Biomarker Discovery: Impact of Study Design and Demographic Characteristics. Metabolites 2020, 10, 224. [Google Scholar] [CrossRef] [PubMed]
- Ilyin, S.E.; Belkowski, S.M.; Plata-Salamán, C.R. Biomarker Discovery and Validation: Technologies and Integrative Approaches. Trends Biotechnol. 2004, 22, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for Biomarker Discovery: Moving to the Clinic. BioMed Res. Int. 2015, 2015, e354671. [Google Scholar] [CrossRef] [PubMed]
- Van Gool, A.; Corrales, F.; Čolović, M.; Krstić, D.; Oliver-Martos, B.; Martínez-Cáceres, E.; Jakasa, I.; Gajski, G.; Brun, V.; Kyriacou, K.; et al. Analytical Techniques for Multiplex Analysis of Protein Biomarkers. Expert Rev. Proteom. 2020, 17, 257–273. [Google Scholar] [CrossRef]
- Pirmohamed, M. Acceptance of Biomarker-Based Tests for Application in Clinical Practice: Criteria and Obstacles. Clin. Pharmacol. Ther. 2010, 88, 862–866. [Google Scholar] [CrossRef] [PubMed]
- O’Bryant, S.E.; Mielke, M.M.; Rissman, R.A.; Lista, S.; Vanderstichele, H.; Zetterberg, H.; Lewczuk, P.; Posner, H.; Hall, J.; Johnson, L.; et al. Blood-Based Biomarkers in Alzheimer Disease: Current State of the Science and a Novel Collaborative Paradigm for Advancing from Discovery to Clinic. Alzheimers Dement. 2017, 13, 45–58. [Google Scholar] [CrossRef] [Green Version]
- Basu, D.; Kulkarni, R. Overview of Blood Components and Their Preparation. Indian J. Anaesth. 2014, 58, 529–537. [Google Scholar] [CrossRef]
- Ngamchuea, K.; Chaisiwamongkhol, K.; Batchelor-McAuley, C.; Compton, R.G. Chemical Analysis in Saliva and the Search for Salivary Biomarkers—A Tutorial Review. Analyst 2017, 143, 81–99. [Google Scholar] [CrossRef]
- Dame, Z.T.; Aziat, F.; Mandal, R.; Krishnamurthy, R.; Bouatra, S.; Borzouie, S.; Guo, A.C.; Sajed, T.; Deng, L.; Lin, H.; et al. The Human Saliva Metabolome. Metabolomics 2015, 11, 1864–1883. [Google Scholar] [CrossRef]
- Chiappin, S.; Antonelli, G.; Gatti, R.; De Palo, E.F. Saliva Specimen: A New Laboratory Tool for Diagnostic and Basic Investigation. Clin. Chim. Acta 2007, 383, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Z.; Cheng, X.-Q.; Li, J.-Y.; Zhang, P.; Yi, P.; Xu, X.; Zhou, X.-D. Saliva in the Diagnosis of Diseases. Int. J. Oral Sci. 2016, 8, 133–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Leon, S.G.; Sarabia Sainz, J.A.; Ramos-Clamont Montfort, G.; Huerta-Ocampo, J.Á.; Ballesteros, M.N.; Guzman-Partida, A.M.; Robles-Burgueño, M.d.R.; Vazquez-Moreno, L. Nanoproteomic Approach for Isolation and Identification of Potential Biomarkers in Human Urine from Adults with Normal Weight, Overweight and Obesity. Molecules 2021, 26, 1803. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.E.; Maranian, M.J.; Spiteri, I.; Russell, R.; Ingle, S.; Luccarini, C.; Earl, H.M.; Pharoah, P.P.; Dunning, A.M.; Caldas, C. Saliva Samples Are a Viable Alternative to Blood Samples as a Source of DNA for High Throughput Genotyping. BMC Med. Genom. 2012, 5, 19. [Google Scholar] [CrossRef] [Green Version]
- Lima, D.P.; Diniz, D.G.; Moimaz, S.A.S.; Sumida, D.H.; Okamoto, A.C. Saliva: Reflection of the Body. Int. J. Infect. Dis. 2010, 14, e184–e188. [Google Scholar] [CrossRef] [Green Version]
- Bellagambi, F.G.; Lomonaco, T.; Salvo, P.; Vivaldi, F.; Hangouët, M.; Ghimenti, S.; Biagini, D.; Di Francesco, F.; Fuoco, R.; Errachid, A. Saliva Sampling: Methods and Devices. An Overview. TrAC Trends Anal. Chem. 2020, 124, 115781. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Trivedi, H.M.; Lima, P.O.; Camargo, P.M.; Giannobile, W.V.; Grogan, T.R.; Gleber-Netto, F.O.; Whiteman, Y.; Li, F.; Lee, H.J.; et al. Salivary ExRNA Biomarkers to Detect Gingivitis and Monitor Disease Regression. J. Clin. Periodontol. 2018, 45, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, G.; Radcenco, A.L.; Evaristo, J.; Monnerat, G. Novel Strategies for Clinical Investigation and Biomarker Discovery: A Guide to Applied Metabolomics. Horm. Mol. Biol. Clin. Investig. 2019, 38. [Google Scholar] [CrossRef]
- Crutchfield, C.A.; Thomas, S.N.; Sokoll, L.J.; Chan, D.W. Advances in Mass Spectrometry-Based Clinical Biomarker Discovery. Clin. Proteom. 2016, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Smolinska, A.; Blanchet, L.; Buydens, L.M.C.; Wijmenga, S.S. NMR and Pattern Recognition Methods in Metabolomics: From Data Acquisition to Biomarker Discovery: A Review. Anal. Chim. Acta 2012, 750, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Issaq, H.J.; Van, Q.N.; Waybright, T.J.; Muschik, G.M.; Veenstra, T.D. Analytical and Statistical Approaches to Metabolomics Research. J. Sep. Sci. 2009, 32, 2183–2199. [Google Scholar] [CrossRef]
- Zhang, W.; Segers, K.; Mangelings, D.; Eeckhaut, A.V.; Hankemeier, T.; Heyden, Y.V.; Ramautar, R. Assessing the Suitability of Capillary Electrophoresis-Mass Spectrometry for Biomarker Discovery in Plasma-Based Metabolomics. Electrophoresis 2019, 40, 2309–2320. [Google Scholar] [CrossRef]
- Kolch, W.; Neusüß, C.; Pelzing, M.; Mischak, H. Capillary Electrophoresis–Mass Spectrometry as a Powerful Tool in Clinical Diagnosis and Biomarker Discovery. Mass Spectrom. Rev. 2005, 24, 959–977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ramautar, R. CE-MS for Metabolomics: Developments and Applications in the Period 2018–2020. Electrophoresis 2021, 42, 381–401. [Google Scholar] [CrossRef]
- Daniel, R.M.; Dunn, R.V.; Finney, J.L.; Smith, J.C. The Role of Dynamics in Enzyme Activity. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 69–92. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, B. Words of Advice: Teaching Enzyme Kinetics. FEBS J. 2021, 288, 2068–2083. [Google Scholar] [CrossRef]
- Brown, T.M.; Prabhu, S.D. The Evolution of the Enzymatic Diagnosis of Myocardial Infarction. Am. J. Med. Sci. 2020, 359, 67–69. [Google Scholar] [CrossRef]
- Gaffney, E.M.; Lim, K.; Minteer, S.D. Breath Biosensing: Using Electrochemical Enzymatic Sensors for Detection of Biomarkers in Human Breath. Curr. Opin. Electrochem. 2020, 23, 26–30. [Google Scholar] [CrossRef]
- Moussa, S.; Horn, M.R.V.; Shah, A.; Pollegioni, L.; Thibodeaux, C.J.; Ruthazer, E.S.; Mauzeroll, J. Editors’ Choice—A Miniaturized Enzymatic Biosensor for Detection of Sensory-Evoked D-Serine Release in the Brain. J. Electrochem. Soc. 2021, 168, 025502. [Google Scholar] [CrossRef]
- Bhide, A.; Cheeran, S.; Muthukumar, S.; Prasad, S. Enzymatic Low Volume Passive Sweat Based Assays for Multi-Biomarker Detection. Biosensors 2019, 9, 13. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, M. Science behind Human Saliva. J. Nat. Sci. Biol. Med. 2011, 2, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pushpass, R.-A.G.; Daly, B.; Kelly, C.; Proctor, G.; Carpenter, G.H. Altered Salivary Flow, Protein Composition, and Rheology Following Taste and TRP Stimulation in Older Adults. Front. Physiol. 2019, 10, 652. [Google Scholar] [CrossRef] [PubMed]
- Price, D.A.; Close, G.C.; Fielding, B.A. Age of Appearance of Circadian Rhythm in Salivary Cortisol Values in Infancy. Arch. Dis. Child. 1983, 58, 454–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, Å.M.; Garde, A.H.; Persson, R. Sources of Biological and Methodological Variation in Salivary Cortisol and Their Impact on Measurement among Healthy Adults: A Review. Scand. J. Clin. Lab. Investig. 2008, 68, 448–458. [Google Scholar] [CrossRef]
- Gozansky, W.S.; Lynn, J.S.; Laudenslager, M.L.; Kohrt, W.M. Salivary Cortisol Determined by Enzyme Immunoassay Is Preferable to Serum Total Cortisol for Assessment of Dynamic Hypothalamic–Pituitary–Adrenal Axis Activity. Clin. Endocrinol. 2005, 63, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Baliga, S.; Muglikar, S.; Kale, R. Salivary PH: A Diagnostic Biomarker. J. Indian Soc. Periodontol. 2013, 17, 461–465. [Google Scholar] [CrossRef]
- Batista, T.B.D.; Chaiben, C.L.; Penteado, C.A.S.; Nascimento, J.M.C.; Ventura, T.M.O.; Dionizio, A.; Rosa, E.A.R.; Buzalaf, M.A.R.; Azevedo-Alanis, L.R. Salivary Proteome Characterization of Alcohol and Tobacco Dependents. Drug Alcohol Depend. 2019, 204, 107510. [Google Scholar] [CrossRef]
- Nagler, R.M.; Hershkovich, O. Age-Related Changes in Unstimulated Salivary Function and Composition and Its Relations to Medications and Oral Sensorial Complaints. Aging Clin. Exp. Res. 2005, 17, 358–366. [Google Scholar] [CrossRef]
- Maier, H.; Bom, I.A.; Veith, S.; Adler, D.; Seitz, H.K. The Effect of Chronic Ethanol Consumption on Salivary Gland Morphology and Function in the Rat. Alcohol. Clin. Exp. Res. 1986, 10, 425–427. [Google Scholar] [CrossRef]
- Rodríguez-Rabassa, M.; López, P.; Rodríguez-Santiago, R.E.; Cases, A.; Felici, M.; Sánchez, R.; Yamamura, Y.; Rivera-Amill, V. Cigarette Smoking Modulation of Saliva Microbial Composition and Cytokine Levels. Int. J. Environ. Res. Public Health 2018, 15, 2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dukić, W.; Dobrijević, T.T.; Katunarić, M.; Lesić, S. Caries Prevalence in Chronic Alcoholics and the Relationship to Salivary Flow Rate and PH. Cent. Eur. J. Public Health 2013, 21, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Waszkiewicz, N.; Jelski, W.; Zalewska, A.; Szulc, A.; Szmitkowski, M.; Zwierz, K.; Szajda, S.D. Salivary Alcohol Dehydrogenase in Non-Smoking and Smoking Alcohol-Dependent Persons. Alcohol 2014, 48, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Enberg, N.; Alho, H.; Loimaranta, V.; Lenander-Lumikari, M. Saliva Flow Rate, Amylase Activity, and Protein and Electrolyte Concentrations in Saliva after Acute Alcohol Consumption. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2001, 92, 292–298. [Google Scholar] [CrossRef]
- Petrušić, N.; Posavac, M.; Sabol, I.; Mravak-Stipetić, M. The Effect of Tobacco Smoking on Salivation. Acta Stomatol. Croat 2015, 49, 309–315. [Google Scholar] [CrossRef]
- Kusumaningrum, D.M.; Tjahajawati, S.; Rizali, E. Differences of Young Adult Smokers and Non-Smokers Saliva Reviewed by Salivary PH, Viscosity, and Volume. Padjadjaran J. Dent. 2019, 31, 142–146. [Google Scholar] [CrossRef] [Green Version]
- Seethalakshmi, C.; Reddy, R.J.; Asifa, N.; Prabhu, S. Correlation of Salivary pH, Incidence of Dental Caries and Periodontal Status in Diabetes Mellitus Patients: A Cross-sectional Study. J. Clin. Diagn. Res. 2016, 10, ZC12–ZC14. [Google Scholar] [CrossRef]
- Mata, A.D.; Marques, D.; Rocha, S.; Francisco, H.; Santos, C.; Mesquita, M.F.; Singh, J. Effects of Diabetes Mellitus on Salivary Secretion and Its Composition in the Human. Mol. Cell Biochem. 2004, 261, 137–142. [Google Scholar] [CrossRef]
- Bokor-Bratic, M.; Cankovic, M.; Dragnic, N. Unstimulated Whole Salivary Flow Rate and Anxiolytics Intake Are Independently Associated with Oral Candida Infection in Patients with Oral Lichen Planus. Eur. J. Oral Sci. 2013, 121, 427–433. [Google Scholar] [CrossRef]
- Letawsky, V.H.; Schreiber, A.-M.; Skoretz, S.A. A Tutorial on Saliva’s Role in Swallowing with a Focus on Sjögren’s Syndrome. Am. J. Speech-Lang. Pathol. 2020, 29, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Anuradha, B.R.; Katta, S.; Kode, V.S.; Praveena, C.; Sathe, N.; Sandeep, N.; Penumarty, S. Oral and Salivary Changes in Patients with Chronic Kidney Disease: A Clinical and Biochemical Study. J. Indian Soc. Periodontol. 2015, 19, 297–301. [Google Scholar] [CrossRef]
- Shpitzer, T.; Bahar, G.; Feinmesser, R.; Nagler, R.M. A Comprehensive Salivary Analysis for Oral Cancer Diagnosis. J. Cancer Res. Clin. Oncol. 2007, 133, 613–617. [Google Scholar] [CrossRef]
- Proctor, G.B.; Shaalan, A.M. Disease-Induced Changes in Salivary Gland Function and the Composition of Saliva. J. Dent. Res. 2021, 00220345211004842. [Google Scholar] [CrossRef]
- Panchbhai, A.S.; Degwekar, S.S.; Bhowte, R.R. Estimation of Salivary Glucose, Salivary Amylase, Salivary Total Protein and Salivary Flow Rate in Diabetics in India. J. Oral Sci. 2010, 52, 359–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez Cuesta, S.; Rahman, S.A.; Furnham, N.; Thornton, J.M. The Classification and Evolution of Enzyme Function. Biophys. J. 2015, 109, 1082–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veskoukis, A.S.; Kyparos, A.; Paschalis, V.; Nikolaidis, M.G. Spectrophotometric Assays for Measuring Redox Biomarkers in Blood. Biomarkers 2016, 21, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Mäntele, W.; Deniz, E. UV–VIS Absorption Spectroscopy: Lambert-Beer Reloaded. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 173, 965–968. [Google Scholar] [CrossRef]
- Karpińska, J. Derivative Spectrophotometry—Recent Applications and Directions of Developments. Talanta 2004, 64, 801–822. [Google Scholar] [CrossRef]
- Fereja, T.H.; Hymete, A.; Gunasekaran, T. A Recent Review on Chemiluminescence Reaction, Principle and Application on Pharmaceutical Analysis. ISRN Spectrosc. 2013, 2013, e230858. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, L.; Zezell, D.M.; Ribeiro, A.d.C.; Gomes, L.; Ito, A.S. Fluorescence Spectroscopy of Biological Tissues—A Review. Appl. Spectrosc. Rev. 2006, 41, 575–590. [Google Scholar] [CrossRef]
- Lee, P.-C.; Li, N.-S.; Hsu, Y.-P.; Peng, C.; Yang, H.-W. Direct Glucose Detection in Whole Blood by Colorimetric Assay Based on Glucose Oxidase-Conjugated Graphene Oxide/MnO2 Nanozymes. Analyst 2019, 144, 3038–3044. [Google Scholar] [CrossRef]
- Mohammadnejad, P.; Asl, S.S.; Aminzadeh, S.; Haghbeen, K. A New Sensitive Spectrophotometric Method for Determination of Saliva and Blood Glucose. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 229, 117897. [Google Scholar] [CrossRef] [PubMed]
- Tu, E.; Pearlmutter, P.; Tiangco, M.; Derose, G.; Begdache, L.; Koh, A. Comparison of Colorimetric Analyses to Determine Cortisol in Human Sweat. ACS Omega 2020, 5, 8211–8218. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Yeh, Y.-C. Detection of Tyrosine and Monitoring Tyrosinase Activity Using an Enzyme Cascade-Triggered Colorimetric Reaction. RSC Adv. 2020, 10, 29745–29750. [Google Scholar] [CrossRef]
- Yu, J.; Cao, M.; Wang, H.; Li, Y. Novel Manganese(II)-Based Metal-Organic Gels: Synthesis, Characterization and Application to Chemiluminescent Sensing of Hydrogen Peroxide and Glucose. Microchim. Acta 2019, 186, 696. [Google Scholar] [CrossRef] [PubMed]
- Beigi, S.M.; Mesgari, F.; Hosseini, M.; Aghazadeh, M.; Ganjali, M.R. An Enhancement of Luminol Chemiluminescence by Cobalt Hydroxide Decorated Porous Graphene and Its Application in Glucose Analysis. Anal. Methods 2019, 11, 1346–1352. [Google Scholar] [CrossRef]
- Tang, X.; Shi, X.; Tang, Y.; Yue, Z.; He, Q. Flow-Injection Chemiluminescence Determination of Melamine in Urine and Plasma. Luminescence 2012, 27, 229–233. [Google Scholar] [CrossRef]
- Yuan, J.; Cen, Y.; Kong, X.-J.; Wu, S.; Liu, C.-L.; Yu, R.-Q.; Chu, X. MnO2-Nanosheet-Modified Upconversion Nanosystem for Sensitive Turn-On Fluorescence Detection of H2O2 and Glucose in Blood. ACS Appl. Mater. Interfaces 2015, 7, 10548–10555. [Google Scholar] [CrossRef]
- Li, F.; Feng, Y.; Huang, Y.; Yao, Q.; Huang, G.; Zhu, Y.; Chen, X. Colorimetric Sensing of Chloride in Sweat Based on Fluorescence Wavelength Shift via Halide Exchange of CsPbBr3 Perovskite Nanocrystals. Microchim. Acta 2021, 188, 2. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.; He, Y.; Ge, Y.; Song, G. A Novel Fluorescence and Naked Eye Sensor for Iodide in Urine Based on the Iodide Induced Oxidative Etching and Aggregation of Cu Nanoclusters. Sens. Actuators B Chem. 2015, 209, 147–153. [Google Scholar] [CrossRef]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular Interaction Studies Using Microscale Thermophoresis. ASSAY Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction Analysis and Beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef] [Green Version]
- Auriol, M.; Filali-Meknassi, Y.; Adams, C.D.; Tyagi, R.D. Natural and Synthetic Hormone Removal Using the Horseradish Peroxidase Enzyme: Temperature and PH Effects. Water Res. 2006, 40, 2847–2856. [Google Scholar] [CrossRef]
- Bisswanger, H. Enzyme Assays. Perspect. Sci. 2014, 1, 41–55. [Google Scholar] [CrossRef] [Green Version]
- Butré, C.I.; Wierenga, P.A.; Gruppen, H. Effects of Ionic Strength on the Enzymatic Hydrolysis of Diluted and Concentrated Whey Protein Isolate. J. Agric. Food Chem. 2012, 60, 5644–5651. [Google Scholar] [CrossRef] [PubMed]
- Herlet, J.; Kornberger, P.; Roessler, B.; Glanz, J.; Schwarz, W.H.; Liebl, W.; Zverlov, V.V. A New Method to Evaluate Temperature vs. PH Activity Profiles for Biotechnological Relevant Enzymes. Biotechnol. Biofuels 2017, 10, 234. [Google Scholar] [CrossRef] [Green Version]
- Synowiecki, J.; Grzybowska, B.; Zdziebło, A. Sources, Properties and Suitability of New Thermostable Enzymes in Food Processing. Crit. Rev. Food Sci. Nutr. 2006, 46, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Bollella, P.; Hibino, Y.; Kano, K.; Gorton, L.; Antiochia, R. The Influence of PH and Divalent/Monovalent Cations on the Internal Electron Transfer (IET), Enzymatic Activity, and Structure of Fructose Dehydrogenase. Anal. Bioanal. Chem. 2018, 410, 3253–3264. [Google Scholar] [CrossRef] [Green Version]
- Pavasovic, M.; Richardson, N.A.; Anderson, A.J.; Mann, D.; Mather, P.B. Effect of PH, Temperature and Diet on Digestive Enzyme Profiles in the Mud Crab, Scylla Serrata. Aquaculture 2004, 242, 641–654. [Google Scholar] [CrossRef]
- Blum, J.-M.; Su, Q.; Ma, Y.; Valverde-Pérez, B.; Domingo-Félez, C.; Jensen, M.M.; Smets, B.F. The PH Dependency of N-Converting Enzymatic Processes, Pathways and Microbes: Effect on Net N2O Production. Environ. Microbiol. 2018, 20, 1623–1640. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Wang, A.; Kan, J. Selective Uricase Biosensor Based on Polyaniline Synthesized in Ionic Liquid. Sens. Actuators B Chem. 2007, 124, 529–534. [Google Scholar] [CrossRef]
- Silvestre, M.P.C.; Carreira, R.L.; Silva, M.R.; Corgosinho, F.C.; Monteiro, M.R.P.; Morais, H.A. Effect of PH and Temperature on the Activity of Enzymatic Extracts from Pineapple Peel. Food Bioprocess Technol. 2012, 5, 1824–1831. [Google Scholar] [CrossRef]
- Bhat, S.; McLaughlin, J.L.H.; Emslie, K.R. Effect of Sustained Elevated Temperature Prior to Amplification on Template Copy Number Estimation Using Digital Polymerase Chain Reaction. Analyst 2011, 136, 724–732. [Google Scholar] [CrossRef]
- Daniel, R.M.; Peterson, M.E.; Danson, M.J.; Price, N.C.; Kelly, S.M.; Monk, C.R.; Weinberg, C.S.; Oudshoorn, M.L.; Lee, C.K. The Molecular Basis of the Effect of Temperature on Enzyme Activity. Biochem. J. 2009, 425, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T. The Definition and Unit of Ionic Strength. J. Chem. Educ. 2001, 78, 1691. [Google Scholar] [CrossRef]
- Nordbö, H.; Darwish, S.; Bhatnagar, R.S. Salivary Viscosity and Lubrication: Influence of PH and Calcium. Eur. J. Oral Sci. 1984, 92, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Durdiaková, J.; Fábryová, H.; Koborová, I.; Ostatníková, D.; Celec, P. The Effects of Saliva Collection, Handling and Storage on Salivary Testosterone Measurement. Steroids 2013, 78, 1325–1331. [Google Scholar] [CrossRef]
- Marshall, T.; Williams, K.M. Electrophoresis Indicates Protein Loss on Centrifugation of Whole Saliva. Clin. Chem. 1987, 33, 1263–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapkota, D.; Søland, T.M.; Galtung, H.K.; Sand, L.P.; Giannecchini, S.; To, K.K.W.; Mendes-Correa, M.C.; Giglio, D.; Hasséus, B.; Braz-Silva, P.H. COVID-19 Salivary Signature: Diagnostic and Research Opportunities. J. Clin. Pathol. 2020, 74, 344–349. [Google Scholar] [CrossRef]
- Hamid, H.; Khurshid, Z.; Adanir, N.; Zafar, M.S.; Zohaib, S. COVID-19 Pandemic and Role of Human Saliva as a Testing Biofluid in Point-of-Care Technology. Eur. J. Dent. 2020, 14, S123–S129. [Google Scholar] [CrossRef]
- Das, A.K.; Islam, N.; Billah, M.; Sarker, A. COVID-19 Pandemic and Healthcare Solid Waste Management Strategy—A Mini-Review. Sci. Total Environ. 2021, 778, 146220. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, S.; Srivastava, R.R.; Kim, H. Disinfection Technology and Strategies for COVID-19 Hospital and Bio-Medical Waste Management. Sci. Total Environ. 2020, 749, 141652. [Google Scholar] [CrossRef]
- Joung, Y.-H. Development of Implantable Medical Devices: From an Engineering Perspective. Int. Neurourol. J. 2013, 17, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Eom, K.S.; Shin, K.-S.; Kang, J.Y.; Lee, S.H. Enzyme-Loaded Paper Combined Impedimetric Sensor for the Determination of the Low-Level of Cholesterol in Saliva. Sens. Actuators B Chem. 2018, 271, 73–81. [Google Scholar] [CrossRef]
- Mahosenaho, M.; Caprio, F.; Micheli, L.; Sesay, A.M.; Palleschi, G.; Virtanen, V. A Disposable Biosensor for the Determination of Alpha-Amylase in Human Saliva. Microchim. Acta 2010, 170, 243–249. [Google Scholar] [CrossRef]
- Tan, W.; Sabet, L.; Li, Y.; Yu, T.; Klokkevold, P.R.; Wong, D.T.; Ho, C.-M. Optical Protein Sensor for Detecting Cancer Markers in Saliva. Biosens. Bioelectron. 2008, 24, 266–271. [Google Scholar] [CrossRef] [Green Version]
- Vargas, E.; Teymourian, H.; Tehrani, F.; Eksin, E.; Sánchez-Tirado, E.; Warren, P.; Erdem, A.; Dassau, E.; Wang, J. Enzymatic/Immunoassay Dual-Biomarker Sensing Chip: Towards Decentralized Insulin/Glucose Detection. Angew. Chem. Int. Ed. 2019, 58, 6376–6379. [Google Scholar] [CrossRef]
- Pandey, C.M.; Augustine, S.; Kumar, S.; Kumar, S.; Nara, S.; Srivastava, S.; Malhotra, B.D. Microfluidics Based Point-of-Care Diagnostics. Biotechnol. J. 2018, 13, 1700047. [Google Scholar] [CrossRef]
- Luppa, P.B.; Müller, C.; Schlichtiger, A.; Schlebusch, H. Point-of-Care Testing (POCT): Current Techniques and Future Perspectives. TrAC Trends Anal. Chem. 2011, 30, 887–898. [Google Scholar] [CrossRef]
- Chan, H.N.; Tan, M.J.A.; Wu, H. Point-of-Care Testing: Applications of 3D Printing. Lab Chip 2017, 17, 2713–2739. [Google Scholar] [CrossRef]
- Campuzano, S.; Yánez-Sedeño, P.; Pingarrón, J.M. Electrochemical Bioaffinity Sensors for Salivary Biomarkers Detection. TrAC Trends Anal. Chem. 2017, 86, 14–24. [Google Scholar] [CrossRef]
- Singh, N.K.; Ray, P.; Carlin, A.F.; Magallanes, C.; Morgan, S.C.; Laurent, L.C.; Aronoff-Spencer, E.S.; Hall, D.A. Hitting the Diagnostic Sweet Spot: Point-of-Care SARS-CoV-2 Salivary Antigen Testing with an off-the-Shelf Glucometer. Biosens. Bioelectron. 2021, 180, 113111. [Google Scholar] [CrossRef]
- Tripoliti, E.E.; Ioannidou, P.; Toumpaniaris, P.; Rammos, A.; Pacitto, D.; Lourme, J.-C.; Goletsis, Y.; Naka, K.K.; Errachid, A.; Fotiadis, D.I. Point-of-Care Testing Devices for Heart Failure Analyzing Blood and Saliva Samples. IEEE Rev. Biomed. Eng. 2020, 13, 17–31. [Google Scholar] [CrossRef]
- Bermejo-Pareja, F.; Antequera, D.; Vargas, T.; Molina, J.A.; Carro, E. Saliva Levels of Abeta1-42 as Potential Biomarker of Alzheimer’s Disease: A Pilot Study. BMC Neurol. 2010, 10, 108. [Google Scholar] [CrossRef] [Green Version]
- Viswanath, B.; Choi, C.S.; Lee, K.; Kim, S. Recent Trends in the Development of Diagnostic Tools for Diabetes Mellitus Using Patient Saliva. TrAC Trends Anal. Chem. 2017, 89, 60–67. [Google Scholar] [CrossRef]
- Boras, V.V.; Lukač, J.; Brailo, V.; Picek, P.; Kordić, D.; Žilić, I.A. Salivary Interleukin-6 and Tumor Necrosis Factor-α in Patients with Recurrent Aphthous Ulceration. J. Oral Pathol. Med. 2006, 35, 241–243. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Deutsch, O.; Zaks, B.; Krief, G.; Chaushu, S.; Palmon, A. Identification of Salivary Protein Biomarkers for Orthodontically Induced Inflammatory Root Resorption. Proteom. Clin. Appl. 2017, 11, 1600119. [Google Scholar] [CrossRef]
- Li, F.; Kaczor-Urbanowicz, K.E.; Sun, J.; Majem, B.; Lo, H.-C.; Kim, Y.; Koyano, K.; Rao, S.L.; Kang, S.Y.; Kim, S.M.; et al. Characterization of Human Salivary Extracellular RNA by Next-Generation Sequencing. Clin. Chem. 2018, 64, 1085–1095. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, S.; Tiwari, S.; Srivastava, S.; Srivastava, M.; Yadav, B.K.; Kumar, S.; Tran, T.T.; Dewan, A.K.; Mulchandani, A.; et al. Biofunctionalized Nanostructured Zirconia for Biomedical Application: A Smart Approach for Oral Cancer Detection. Adv. Sci. 2015, 2, 1500048. [Google Scholar] [CrossRef]
- Ahmed, A.; Rushworth, J.V.; Wright, J.D.; Millner, P.A. Novel Impedimetric Immunosensor for Detection of Pathogenic Bacteria Streptococcus Pyogenes in Human Saliva. Anal. Chem. 2013, 85, 12118–12125. [Google Scholar] [CrossRef]
- Chohan, B.H.; Lavreys, L.; Mandaliya, K.N.; Kreiss, J.K.; Bwayo, J.J.; Ndinya-Achola, J.O.; Martin, H.L. Validation of a Modified Commercial Enzyme-Linked Immunoassay for Detection of Human Immunodeficiency Virus Type 1 Immunoglobulin G Antibodies in Saliva. Clin. Diagn. Lab. Immunol. 2001, 8, 346–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.J.; Jaedicke, K.M.; van de Merwe, R.C.; Bissett, S.M.; Landsdowne, N.; Whall, K.M.; Pickering, K.; Thornton, V.; Lawson, V.; Yatsuda, H.; et al. A Prototype Antibody-Based Biosensor for Measurement of Salivary MMP-8 in Periodontitis Using Surface Acoustic Wave Technology. Sci. Rep. 2019, 9, 11034. [Google Scholar] [CrossRef] [Green Version]
- Dalirirad, S.; Han, D.; Steckl, A.J. Aptamer-Based Lateral Flow Biosensor for Rapid Detection of Salivary Cortisol. ACS Omega 2020, 5, 32890–32898. [Google Scholar] [CrossRef] [PubMed]
- Shetty, V.; Yamaguchi, M. Salivary Biosensors for Screening Trauma-Related Psychopathology. Oral Maxillofac. Surg. Clin. N. Am. 2010, 22, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Imani, S.; de Araujo, W.R.; Warchall, J.; Valdés-Ramírez, G.; Paixão, T.R.L.C.; Mercier, P.P.; Wang, J. Wearable Salivary Uric Acid Mouthguard Biosensor with Integrated Wireless Electronics. Biosens. Bioelectron. 2015, 74, 1061–1068. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, M.; Le Goff, A.; Cosnier, S. Nanomaterials for Biosensing Applications: A Review. Front. Chem. 2014, 2, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahcen, A.A.; Rauf, S.; Beduk, T.; Durmus, C.; Aljedaibi, A.; Timur, S.; Alshareef, H.N.; Amine, A.; Wolfbeis, O.S.; Salama, K.N. Electrochemical Sensors and Biosensors Using Laser-Derived Graphene: A Comprehensive Review. Biosens. Bioelectron. 2020, 168, 112565. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Creran, B.; Rotello, V.M. Gold Nanoparticles: Preparation, Properties, and Applications in Bionanotechnology. Nanoscale 2012, 4, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Martynenko, I.V.; Litvin, A.P.; Purcell-Milton, F.; Baranov, A.V.; Fedorov, A.V.; Gun’ko, Y.K. Application of Semiconductor Quantum Dots in Bioimaging and Biosensing. J. Mater. Chem. B 2017, 5, 6701–6727. [Google Scholar] [CrossRef]
- Adhikari, C. Polymer Nanoparticles-Preparations, Applications and Future Insights: A Concise Review. Polym.-Plast. Technol. Mater. 2021, 1–29. [Google Scholar] [CrossRef]
- Sireesha, M.; Babu, V.J.; Kiran, A.S.K.; Ramakrishna, S. A Review on Carbon Nanotubes in Biosensor Devices and Their Applications in Medicine. Nanocomposites 2018, 4, 36–57. [Google Scholar] [CrossRef]
- Turcheniuk, K.; Mochalin, V.N. Biomedical Applications of Nanodiamond (Review). Nanotechnology 2017, 28, 252001. [Google Scholar] [CrossRef] [PubMed]
- Rievaj, M.; Culková, E.; Šandorová, D.; Lukáčová-Chomisteková, Z.; Bellová, R.; Durdiak, J.; Tomčík, P. Electroanalytical Techniques for the Detection of Selenium as a Biologically and Environmentally Significant Analyte—A Short Review. Molecules 2021, 26, 1768. [Google Scholar] [CrossRef]
- Kou, X.; Tong, L.; Shen, Y.; Zhu, W.; Yin, L.; Huang, S.; Zhu, F.; Chen, G.; Ouyang, G. Smartphone-Assisted Robust Enzymes@MOFs-Based Paper Biosensor for Point-of-Care Detection. Biosens. Bioelectron. 2020, 156, 112095. [Google Scholar] [CrossRef] [PubMed]
- Lardo, A.; Mancini, D.; Paoloni, N.; Russo, G. The Perspective of Capability Providers in Creating a Sustainable I4.0 Environment. Manag. Decis. 2020, 58, 1759–1777. [Google Scholar] [CrossRef]
| Method | Principle | Detection Range | Biofluid | Biomarker | Detection Limit | Reference |
|---|---|---|---|---|---|---|
| Colorimetric | Absorption of radiation in the visible area by colored substances | M–nM | Blood | Glucose | 31 µg mL−1 | [62] |
| Saliva | Glucose | 0.36 µg mL−1 | [63] | |||
| Sweat | Cortisol | 97 ng mL−1 | [64] | |||
| Urine | Tyrosine | 2.54 µM | [65] | |||
| Luminescent | Light emitted by a molecule when receiving radiant energy | mM–nM | Blood | Glucose | 80 nM | [66] |
| Saliva | Glucose | 0.63 nM | [67] | |||
| Urine | Melamine | 3.5 ng mL−1 | [68] | |||
| Fluorescent | Light emitted by a molecule when receiving radiant energy | mM–nM | Blood | Glucose | 3.7 µM | [69] |
| Sweat | Chloride | 3 mM | [70] | |||
| Urine | Iodide | 100 nM | [71] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ornelas-González, A.; Ortiz-Martínez, M.; González-González, M.; Rito-Palomares, M. Enzymatic Methods for Salivary Biomarkers Detection: Overview and Current Challenges. Molecules 2021, 26, 7026. https://doi.org/10.3390/molecules26227026
Ornelas-González A, Ortiz-Martínez M, González-González M, Rito-Palomares M. Enzymatic Methods for Salivary Biomarkers Detection: Overview and Current Challenges. Molecules. 2021; 26(22):7026. https://doi.org/10.3390/molecules26227026
Chicago/Turabian StyleOrnelas-González, Alonso, Margarita Ortiz-Martínez, Mirna González-González, and Marco Rito-Palomares. 2021. "Enzymatic Methods for Salivary Biomarkers Detection: Overview and Current Challenges" Molecules 26, no. 22: 7026. https://doi.org/10.3390/molecules26227026
APA StyleOrnelas-González, A., Ortiz-Martínez, M., González-González, M., & Rito-Palomares, M. (2021). Enzymatic Methods for Salivary Biomarkers Detection: Overview and Current Challenges. Molecules, 26(22), 7026. https://doi.org/10.3390/molecules26227026