Molecular Docking Suggests the Targets of Anti-Mycobacterial Natural Products
Abstract
1. Introduction
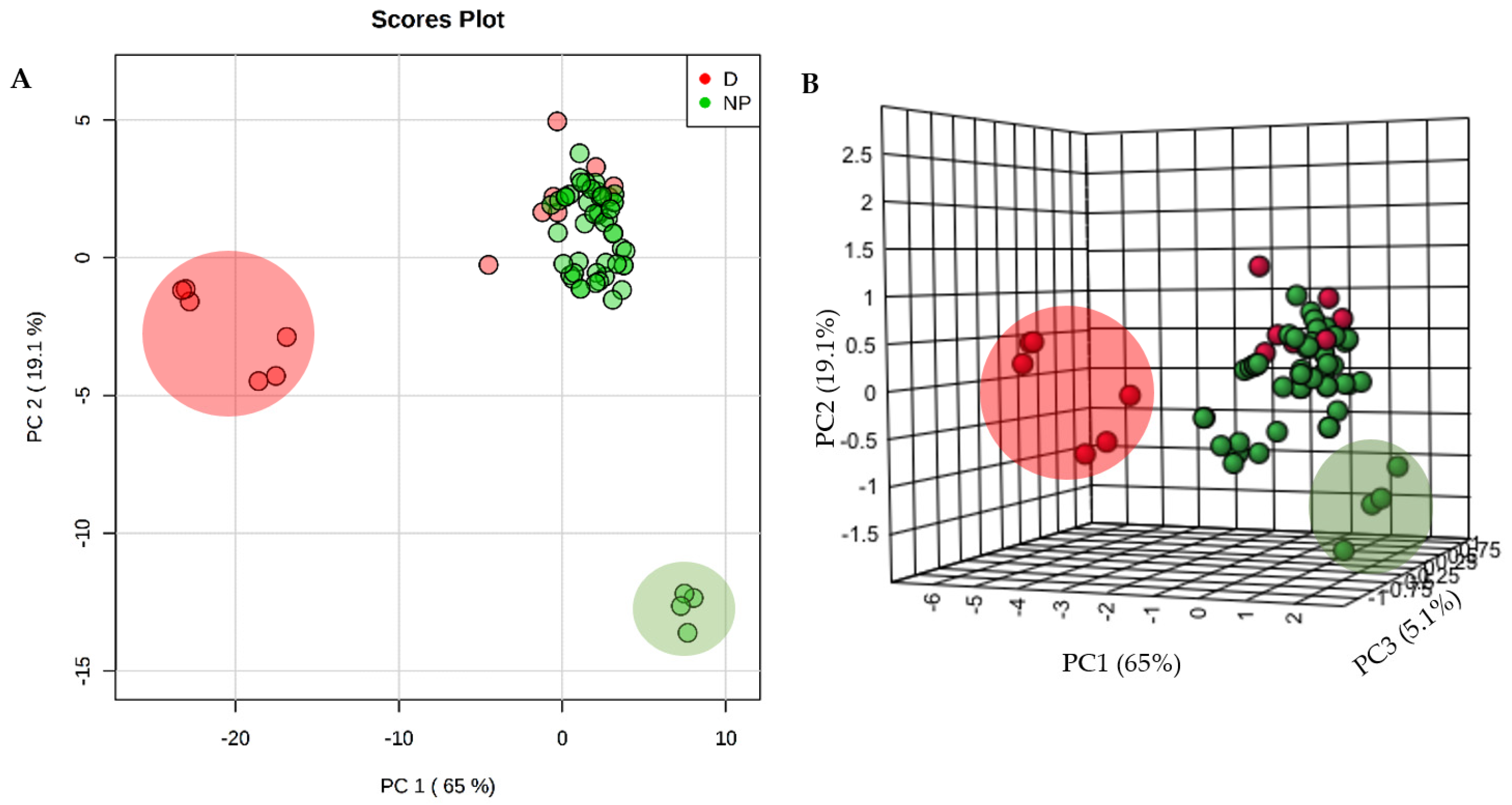
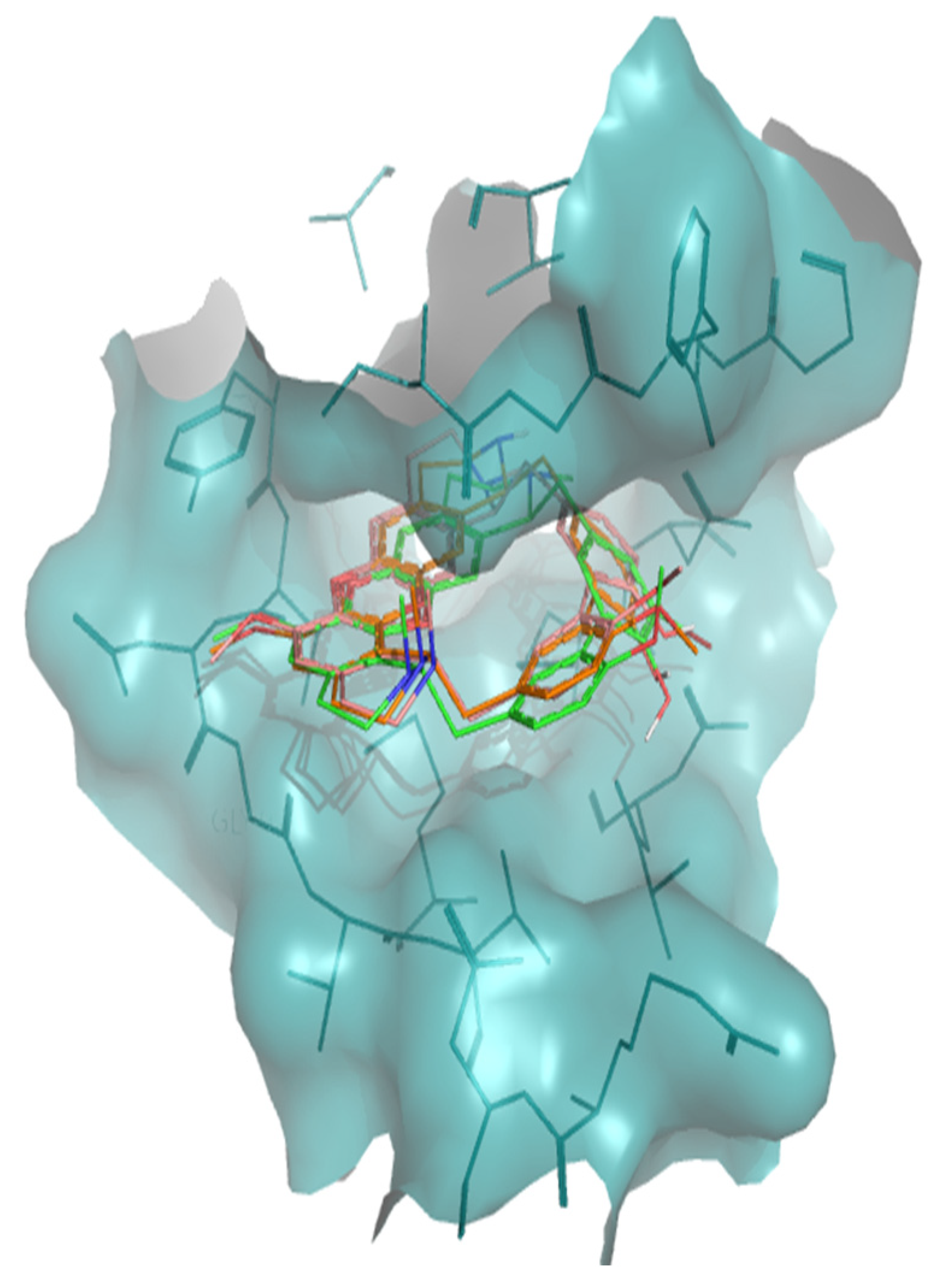
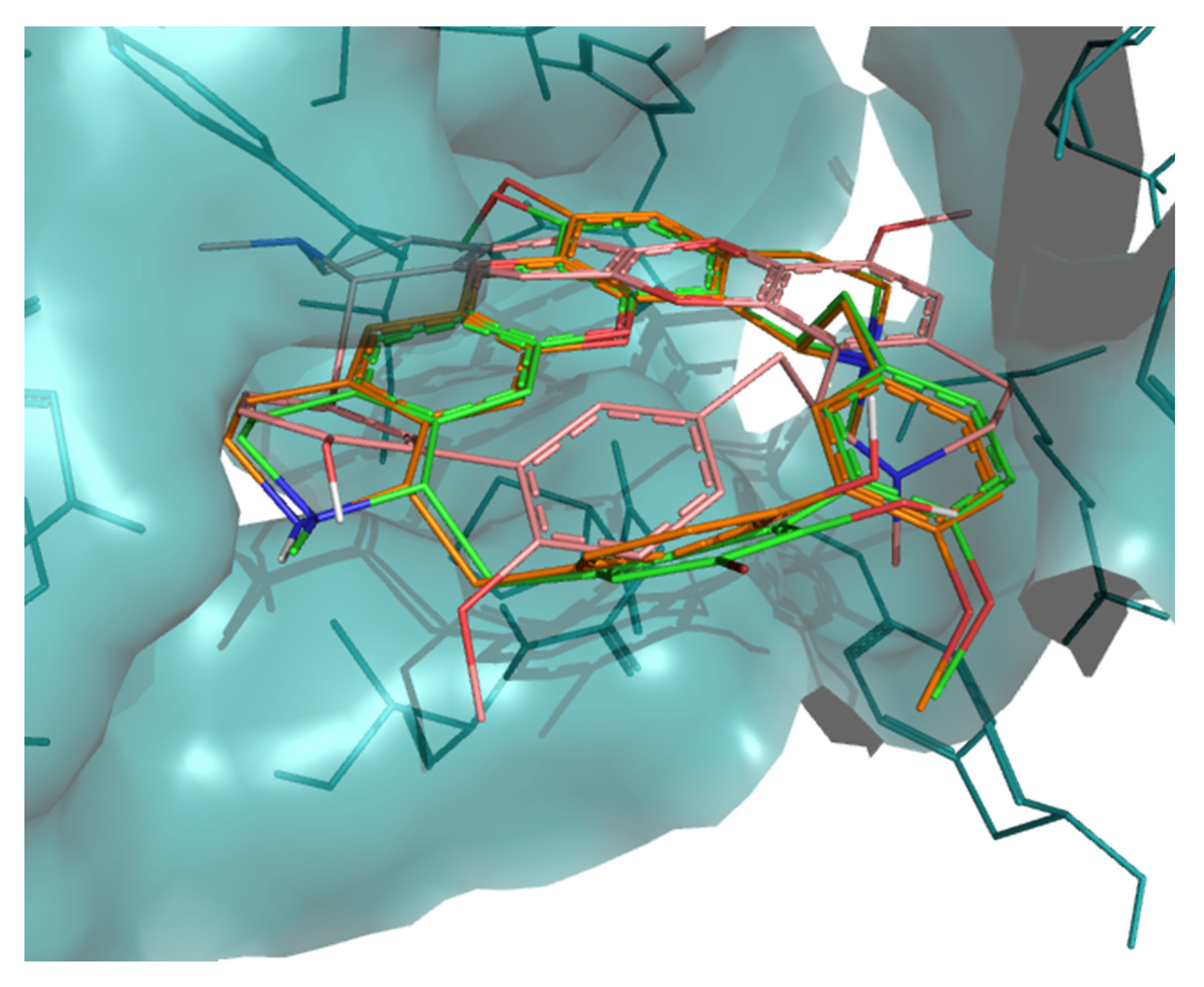
2. Results
3. Discussion
4. Materials and Methods
4.1. Selected Anti-Tubercular Natural Products
4.2. Ligand and Protein Selection
4.3. Physicochemical and Structural Properties
4.4. Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- WHO. Global Tuberculosis Report 2018; WHO Press: Geneva, Switzerland, 2018; ISBN 9789241565646. [Google Scholar]
- Baptista, R.; Bhowmick, S.; Nash, R.J.; Baillie, L.; Mur, L.A. Target discovery focused approaches to overcome bottlenecks in the exploitation of antimycobacterial natural products. Future Med. Chem. 2018, 10, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Kumar, M.M.; Bisht, D.; Kaushik, A. Plants in our combating strategies against Mycobacterium tuberculosis: Progress made and obstacles met. Pharm. Biol. 2017, 55, 1536–1544. [Google Scholar] [CrossRef] [PubMed]
- McConkey, B.J.; Sobolev, V.; Edelman, M. The performance of current methods in ligand-protein docking. Curr. Sci. 2002. [Google Scholar] [CrossRef]
- Hassan, A.H.E.; Yoo, S.Y.; Lee, K.W.; Yoon, Y.M.; Ryu, H.W.; Jeong, Y.; Shin, J.S.; Kang, S.Y.; Kim, S.Y.; Lee, H.H.; et al. Repurposing mosloflavone/5,6,7-trimethoxyflavone-resveratrol hybrids: Discovery of novel p38-α MAPK inhibitors as potent interceptors of macrophage-dependent production of proinflammatory mediators. Eur. J. Med. Chem. 2019, 180, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, Y.; Xu, J.; Qiu, L.; Xu, W.; Li, J.; Song, Y.; Lu, B.; Hu, Z.; Zhang, J. Esculentoside A suppresses lipopolysaccharide-induced pro-inflammatory molecule production partially by casein kinase 2. J. Ethnopharmacol. 2017, 198, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Kim, J.Y.; Anderson, J.R.; Akopian, T.; Hong, S.; Jin, Y.Y.; Kandror, O.; Kim, J.W.; Lee, I.A.; Lee, S.Y.; et al. The cyclic peptide ecumicin targeting CLpC1 is active against Mycobacterium tuberculosis in vivo. Antimicrob. Agents Chemother. 2015, 59, 880–889. [Google Scholar] [CrossRef]
- Schmitz, K.R.; Carney, D.W.; Sello, J.K.; Sauer, R.T. Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery. Proc. Natl. Acad. Sci. USA 2014, 111, E4587–E4595. [Google Scholar] [CrossRef] [PubMed]
- Chawla, Y.; Upadhyay, S.; Khan, S.; Nagarajan, S.N.; Forti, F.; Nandicoori, V.K. Protein kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J. Biol. Chem. 2014. [Google Scholar] [CrossRef]
- Aggarwal, A.; Parai, M.K.; Shetty, N.; Wallis, D.; Woolhiser, L.; Hastings, C.; Dutta, N.K.; Galaviz, S.; Dhakal, R.C.; Shrestha, R.; et al. Development of a Novel Lead that Targets M. tuberculosis Polyketide Synthase 13. Cell 2017, 170, 249–259.e25. [Google Scholar] [CrossRef]
- Crellin, P.K.; Brammananth, R.; Coppel, R.L. Decaprenylphosphoryl-β-D-Ribose 2′-epimerase, the target of benzothiazinones and dinitrobenzamides, is an essential enzyme in Mycobacterium smegmatis. PLoS ONE 2011. [Google Scholar] [CrossRef]
- Brecik, M.; Centárová, I.; Mukherjee, R.; Kolly, G.S.; Huszár, S.; Bobovská, A.; Kilacsková, E.; Mokošová, V.; Svetlíková, Z.; Šarkan, M.; et al. DprE1 Is a Vulnerable Tuberculosis Drug Target Due to Its Cell Wall Localization. ACS Chem. Biol. 2015, 10, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Stanley, S.; Grant, S.; Kawate, T.; Iwase, N.; Shimizu, M.; Wivagg, C.; Silvis, M.; Kazyanskaya, E.; Aquadro, J.; Golas, A.; et al. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. Am. Chem. Soc. Chem. Biol. 2013, 7, 1377–1384. [Google Scholar]
- Dessen, A.; Quemard, A.; Blanchard, J.; Jacobs, W.; Sacchettini, J. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 1995, 267, 1638–1641. [Google Scholar] [CrossRef] [PubMed]
- Kapilashrami, K.; Bommineni, G.R.; MacHutta, C.A.; Kim, P.; Lai, C.T.; Simmerling, C.; Picart, F.; Tonge, P.J. Thiolactomycin-based β-ketoacyl-AcpM synthase a (KasA) inhibitors: Fragment-based inhibitor discovery using transient one-dimensional nuclear overhauser effect NMR spectroscopy. J. Biol. Chem. 2013, 288, 6045–6052. [Google Scholar] [CrossRef] [PubMed]
- Luckner, S.R.; Machutta, C.A.; Tonge, P.J.; Kisker, C. Crystal structures of Mycobacterium tuberculosis KasA show mode of action within cell wall biosynthesis and its inhibition by thiolactomycin. Structure 2009, 17, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Kumar, P.; Bhor, V.; Surolia, A.; Vijayan, M. Invariance and variability in bacterial PanK: A study based on the crystal structure of Mycobacterium tuberculosis PanK. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006. [Google Scholar] [CrossRef]
- Wehenkel, A.; Fernandez, P.; Bellinzoni, M.; Catherinot, V.; Barilone, N.; Labesse, G.; Jackson, M.; Alzari, P.M. The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria. FEBS Lett. 2006, 580, 3018–3022. [Google Scholar] [CrossRef]
- Arifullah, M.; Namsa, N.D.; Mandal, M.; Chiruvella, K.K.; Vikrama, P.; Gopal, G.R. Evaluation of anti-bacterial and anti-oxidant potential of andrographolide and echiodinin isolated from callus culture of Andrographis paniculata Nees. Asian Pac. J. Trop. Biomed. 2013. [Google Scholar] [CrossRef]
- Prabu, A.; Hassan, S.; Prabuseenivasan; Shainaba, A.S.; Hanna, L.E.; Kumar, V. Andrographolide: A potent antituberculosis compound that targets Aminoglycoside 2′-N-acetyltransferase in Mycobacterium tuberculosis. J. Mol. Graph. Model. 2015. [Google Scholar] [CrossRef]
- Navarro-García, V.M.; Luna-Herrera, J.; Rojas-Bribiesca, M.G.; Álvarez-Fitz, P.; Ríos, M.Y. Antibacterial activity of aristolochia brevipes against multidrug-resistant Mycobacterium tuberculosis. Molecules 2011, 16, 7357–7364. [Google Scholar] [CrossRef]
- León-Díaz, R.; Meckes, M.; Said-Fernández, S.; Molina-Salinas, G.M.; Vargas-Villarreal, J.; Torres, J.; Luna-Herrera, J.; Jiménez-Arellanes, A. Antimycobacterial neolignans isolated from Aristolochia taliscana. Mem. Inst. Oswaldo Cruz 2010. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Arellanes, A.; León-Díaz, R.; Meckes, M.; Tapia, A.; Molina-Salinas, G.M.; Luna-Herrera, J.; Yépez-Mulia, L. Antiprotozoal and antimycobacterial activities of pure compounds from Aristolochia elegans rhizomes. Evid. Based Complement. Altern. Med. 2012. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Arellanes, A.; Luna-Herrera, J.; Cornejo-Garrido, J.; López-García, S.; Castro-Mussot, M.E.; Meckes-Fischer, M.; Mata-Espinosa, D.; Marquina, B.; Torres, J.; Hernández-Pando, R. Ursolic and oleanolic acids as antimicrobial and immunomodulatory compounds for tuberculosis treatment. BMC Complement. Altern. Med. 2013. [Google Scholar] [CrossRef] [PubMed]
- Jyoti, M.A.; Nam, K.W.; Jang, W.S.; Kim, Y.H.; Kim, S.K.; Lee, B.E.; Song, H.Y. Antimycobacterial activity of methanolic plant extract of Artemisia capillaris containing ursolic acid and hydroquinone against Mycobacterium tuberculosis. J. Infect. Chemother. 2016. [Google Scholar] [CrossRef] [PubMed]
- Molina-Salinas, G.M.; Bórquez, J.; Ardiles, A.; Said-Fernández, S.; Loyola, L.A.; Yam-Puc, A.; Becerril-Montes, P.; Escalante-Erosa, F.; San-Martin, A.; González-Collado, I.; et al. Bioactive metabolites from the Andean flora. Antituberculosis activity of natural and semisynthetic azorellane and mulinane diterpenoids. Phytochem. Rev. 2010. [Google Scholar] [CrossRef]
- Chen, J.J.; Chou, E.T.; Peng, C.F.; Chen, I.S.; Yang, S.Z.; Huang, H.Y. Novel epoxyfuranoid lignans and antitubercular constituents from the leaves of Beilschmiedia tsangii. Planta Med. 2007. [Google Scholar] [CrossRef]
- Aponte, J.C.; Estevez, Y.; Gilman, R.H.; Lewis, W.H.; Rojas, R.; Sauvain, M.; Vaisberg, A.J.; Hammond, G.B. Anti-infective and cytotoxic compounds present in Blepharodon nitidum. Planta Med. 2008. [Google Scholar] [CrossRef]
- Mehta, A.; Srivastva, G.; Kachhwaha, S.; Sharma, M.; Kothari, S.L. Antimycobacterial activity of Citrullus colocynthis (L.) Schrad. against drug sensitive and drug resistant Mycobacterium tuberculosis and MOTT clinical isolates. J. Ethnopharmacol. 2013. [Google Scholar] [CrossRef]
- Rojas, R.; Caviedes, L.; Aponte, J.C.; Vaisberg, A.J.; Lewis, W.H.; Lamas, G.; Sarasara, C.; Gilman, R.H.; Hammond, G.B. Aegicerin, the first oleanane triterpene with wide-ranging antimycobacterial activity, isolated from Clavija procera. J. Nat. Prod. 2006. [Google Scholar] [CrossRef]
- Changtam, C.; Hongmanee, P.; Suksamrarn, A. Isoxazole analogs of curcuminoids with highly potent multidrug-resistant antimycobacterial activity. Eur. J. Med. Chem. 2010. [Google Scholar] [CrossRef]
- Uc-Cachón, A.H.; Borges-Argáez, R.; Said-Fernández, S.; Vargas-Villarreal, J.; González-Salazar, F.; Méndez-González, M.; Cáceres-Farfán, M.; Molina-Salinas, G.M. Naphthoquinones isolated from Diospyros anisandra exhibit potent activity against pan-resistant first-line drugs Mycobacterium tuberculosis strains. Pulm. Pharmacol. Ther. 2014. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Ray, R.; Hazra, B. Antitubercular and antibacterial activity of quinonoid natural products against multi-drug resistant clinical isolates. Phytother. Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Lall, N.; Meyer, J.J.M.; Wang, Y.; Bapela, N.B.; Van Rensburg, C.E.J.; Fourie, B.; Franzblau, S.G. Characterization of intracellular activity of antitubercular constituents from the roots of Euclea natalensis. Pharm. Biol. 2005. [Google Scholar] [CrossRef]
- van der Kooy, F.; Meyer, J.J.M.; Lall, N. Antimycobacterial activity and possible mode of action of newly isolated neodiospyrin and other naphthoquinones from Euclea natalensis. S. Afr. J. Bot. 2006. [Google Scholar] [CrossRef]
- Al-Yahya, M.A.; Muhammad, I.; Mirza, H.H.; El-Feraly, F.S. Antibacterial constituents from the rhizomes of Ferula communis. Phytother. Res. 1998. [Google Scholar] [CrossRef]
- Gordien, A.Y.; Gray, A.I.; Franzblau, S.G.; Seidel, V. Antimycobacterial terpenoids from Juniperus communis L. (Cuppressaceae). J. Ethnopharmacol. 2009. [Google Scholar] [CrossRef]
- Mossa, J.S.; El-Feraly, F.S.; Muhammad, I. Antimycobacterial constituents from Juniperus procera, Ferula communis and Plumbago zeylanica and their in vitro synergistic activity with isonicotinic acid hydrazide. Phytother. Res. 2004. [Google Scholar] [CrossRef]
- Lakshmanan, D.; Werngren, J.; Jose, L.; Suja, K.P.; Nair, M.S.; Varma, R.L.; Mundayoor, S.; Hoffner, S.; Kumar, R.A. Ethyl p-methoxycinnamate isolated from a traditional anti-tuberculosis medicinal herb inhibits drug resistant strains of Mycobacterium tuberculosis in vitro. Fitoterapia 2011. [Google Scholar] [CrossRef]
- Jiménez-Arellanes, A.; Meckes, M.; Torres, J.; Luna-Herrera, J. Antimycobacterial triterpenoids from Lantana hispida (Verbenaceae). J. Ethnopharmacol. 2007. [Google Scholar] [CrossRef]
- Favela-Hernández, J.M.J.; García, A.; Garza-González, E.; Rivas-Galindo, V.M.; Camacho-Corona, M.R. Antibacterial and antimycobacterial lignans and flavonoids from Larrea tridentata. Phytother. Res. 2012. [Google Scholar] [CrossRef]
- Rijo, P.; Simões, M.F.; Francisco, A.P.; Rojas, R.; Gilman, R.H.; Vaisberg, A.J.; Rodríguez, B.; Moiteiro, C. Antimycobacterial metabolites from Plectranthus: Royleanone derivatives against Mycobacterium tuberculosis strains. Chem. Biodivers. 2010. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Singh, A.; Sharma, U.; Singh, D.; Dobhal, M.P.; Singh, S. Anti-mycobacterial activity of plumericin and isoplumericin against MDR Mycobacterium tuberculosis. Pulm. Pharmacol. Ther. 2013. [Google Scholar] [CrossRef]
- Leitão, F.; Leitão, S.G.; De Almeida, M.Z.; Cantos, J.; Coelho, T.; Da Silva, P.E.A. Medicinal plants from open-air markets in the State of Rio de Janeiro, Brazil as a potential source of new antimycobacterial agents. J. Ethnopharmacol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Sureram, S.; Senadeera, S.P.D.; Hongmanee, P.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Antimycobacterial activity of bisbenzylisoquinoline alkaloids from Tiliacora triandra against multidrug-resistant isolates of Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett. 2012, 22, 2902–2905. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Dwivedi, G.R.; Darokar, M.P.; Srivastava, S.K. Antimycobacterial activity of fractions and isolated compounds from Vetiveria zizanioides. Med. Chem. Res. 2012. [Google Scholar] [CrossRef]
- Azam, S.S.; Abbasi, S.W. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor. Biol. Med. Model. 2013, 10, 63. [Google Scholar] [CrossRef]
- Subramani, R.; Narayanasamy, M.; Feussner, K.D. Plant-derived antimicrobials to fight against multi-drug-resistant human pathogens. 3 Biotech 2017, 7, 172. [Google Scholar] [CrossRef]
- Espinoza-Moraga, M.; Njuguna, N.M.; Mugumbate, G.; Caballero, J.; Chibale, K. In silico Comparison of Antimycobacterial Natural Products with Known Antituberculosis Drugs. J. Chem. Inf. Model. 2013, 53, 649–660. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010. [Google Scholar] [CrossRef]
- Brenk, R.; Schipani, A.; James, D.; Krasowski, A.; Gilbert, I.H.; Frearson, J.; Wyatt, P.G. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem 2008. [Google Scholar] [CrossRef] [PubMed]
- Seyedi, S.S.; Shukri, M.; Hassandarvish, P.; Oo, A.; Shankar, E.M.; Abubakar, S.; Zandi, K. Computational Approach Towards Exploring Potential Anti-Chikungunya Activity of Selected Flavonoids. Sci. Rep. 2016, 6, 24027. [Google Scholar] [CrossRef]
- Qiu, J.-X.; Zhou, Z.-W.; He, Z.-X.; Zhang, X.; Zhou, S.-F.; Zhu, S. Estimation of the binding modes with important human cytochrome P450 enzymes, drug interaction potential, pharmacokinetics, and hepatotoxicity of ginger components using molecular docking, computational, and pharmacokinetic modeling studies. Drug Des. Dev. Ther. 2015, 9, 841–866. [Google Scholar] [CrossRef]
- Natarajan, A.; Sugumar, S.; Bitragunta, S.; Balasubramanyan, N. Molecular docking studies of (4Z, 12Z)-cyclopentadeca-4, 12-dienone from Grewia hirsuta with some targets related to type 2 diabetes. BMC Complement. Altern. Med. 2015, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.T.; Blicharska, N.; Shilpi, J.A.; Seidel, V. Investigation of the anti-TB potential of selected propolis constituents using a molecular docking approach. Sci. Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.M.; Unnikrishnan, M.; Rubin, D.H.F.; Krishnamoorthy, V.; Kandror, O.; Akopian, T.N.; Goldberg, A.L.; Rubin, E.J. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 2012. [Google Scholar] [CrossRef]
- Wolucka, B.A. Biosynthesis of D-arabinose in mycobacteria—A novel bacterial pathway with implications for antimycobacterial therapy. FEBS J. 2008, 275, 2691–2711. [Google Scholar] [CrossRef]
- Schaeffer, M.L.; Agnihotri, G.; Volker, C.; Kallender, H.; Brennan, P.J.; Lonsdale, J.T. Purification and Biochemical Characterization of the Mycobacterium tuberculosis Β-Ketoacyl-acyl Carrier Protein Synthases KasA and KasB. J. Biol. Chem. 2001. [Google Scholar] [CrossRef] [PubMed]
- Portevin, D.; de Sousa-D’Auria, C.; Houssin, C.; Grimaldi, C.; Chami, M.; Daffe, M.; Guilhot, C. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. USA 2004. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, I.; Loharch, S.; Gupta, P.; Madathil, R.; Parkesh, R. Structure, dynamics, and interaction of Mycobacterium tuberculosis (Mtb) DprE1 and DprE2 examined by molecular modeling, simulation, and electrostatic studies. PLoS ONE 2015, 10, e0119771. [Google Scholar] [CrossRef]
- De Pascoli, I.C.; Nascimento, I.R.; Lopes, L.M.X. Configurational analysis of cubebins and bicubebin from Aristolochia lagesiana and Aristolochia pubescens. Phytochemistry 2006. [Google Scholar] [CrossRef]
- Nascimento, I.R.; Murata, A.T.; Bortoli, S.A.; Lopes, L.M.X. Insecticidal activity of chemical constituents from Aristolochia pubescens against Anticarsia gemmatalis larvae. Pest Manag. Sci. 2004. [Google Scholar] [CrossRef]
- Harmatha, J.; Dinan, L. Biological activities of lignans and stilbenoids associated with plant-insect chemical interactions. Phytochem. Rev. 2003, 2, 321–330. [Google Scholar] [CrossRef]
- Esperandim, V.R.; da Silva Ferreira, D.; Rezende, K.C.S.; Cunha, W.R.; Saraiva, J.; Bastos, J.K.; e Silva, M.L.A.; de Albuquerque, S. Evaluation of the in vivo therapeutic properties of (−)-cubebin and (−)-hinokinin against Trypanosoma cruzi. Exp. Parasitol. 2013, 133, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Mascarello, A.; Mori, M.; Chiaradia-Delatorre, L.D.; Menegatti, A.C.O.; Monache, F.D.; Ferrari, F.; Yunes, R.A.; Nunes, R.J.; Terenzi, H.; Botta, B.; et al. Discovery of Mycobacterium tuberculosis Protein Tyrosine Phosphatase B (PtpB) Inhibitors from Natural Products. PLoS ONE 2013, 8, e77081. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.; Rudolph, I.; Möllmann, U.; Voigt, K.; Chung, C.; Singh, O.M.P.; Rees, M.; Mendoza-Losana, A.; Bates, R.; Ballell, L.; et al. Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1. Sci. Rep. 2018, 8, 13473. [Google Scholar] [CrossRef] [PubMed]
- Bjorkelid, C.; Bergfors, T.; Raichurkar, A.K.V.; Mukherjee, K.; Malolanarasimhan, K.; Bandodkar, B.; Jones, T.A. Structural and biochemical characterization of compounds inhibiting Mycobacterium tuberculosis pantothenate kinase. J. Biol. Chem. 2013, 288, 18260–18270. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Umamaheswari, M.; Aji, C.S.; Asokkumar, K.; Sivashanmugam, T.; Subhadradevi, V.; Jagannath, P.; Madeswaran, A. In silico docking studies of aldose reductase inhibitory activity of selected flavonoids. Int. J. Drug Dev. Res. 2012. [Google Scholar] [CrossRef][Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminform. 2011. [Google Scholar] [CrossRef] [PubMed]



| Plant Names | Active Phytomolecules | MIC (μg/mL) | References |
|---|---|---|---|
| Andrographis paniculata | Andrographolide | - | [19,20] |
| Aristolochia brevipes Benth. | 6α-7-Dehydro-N-formyl-nornantenine | >50 a,b,d | [21] |
| N-Formylnornantenine | >50 a,b,d | ||
| Aristolactam I | 12.5–25 a,b,d | ||
| Aristolochia taliscana Hook and Arn. | Licarin A | 3.12–25 a,b,d | [22] |
| Licarin B | 12.5–5 a,b,d | ||
| Eupomatenoid-7 | 6.25–50 a,b,d | ||
| Aristolochia elegans Mast. | Fargesin | 12–50 a,b,d | [23] |
| (8R,8’R,9R)-Cubebin or α-Cubebin | 50–100 a,b,d | ||
| Artemisia capillaris Thunb. | Ursolic acid | 12.5–50 a,b,c,d | [24,25] |
| Hydroquinone | 12.5–25 a,b,c,d | [25] | |
| Azorella compacta Phil., A. madreporica Clos. | Azorellanol | 12.5 b,d | [26] |
| Mulin-11,13-dien-20-oic acid | 25–50 b,d | ||
| Mulinol | 12.5–25 b,d | ||
| Beilschmiedia tsangii Merr. | Beilschmin A | 2.5 d | [27] |
| Blepharodon nitidum (Vell.) J.F. Macbr. | 25-Hydroperoxycycloart-23-en-3β-ol | 25 b | [28] |
| Citrullus colocynthis (L.) Schrad. | Cucurbitacin-E-2-o-β-d-glucopyranoside | 25–62.5 a,b,c,d | [29] |
| Clavija procera B. Ståhl | Aegicerin | 1.6–3.12 a,b,d | [30] |
| Curcuma longa L. | Curcumin | 100 d | [31] |
| Demethoxycurcumin | 50 d | ||
| Bisdemethoxycurcumin | 25 d | ||
| Diospyros anisandra S.F. Blake | Plumbagin | 1.5–62.5 b,c,d | [32,33] |
| Maritinone or 8,8′-Biplumbagin | 3.12 b,d | [32] | |
| 3,3′-Biplumbagin | 3.12 b,d | ||
| Diospyros montana | Diospyrin | 8–250 b,c,d | [33,34] |
| Euclea natalensis A. DC. | 7-Methyljuglone | 0.5–1.25 a,b,d | [34] |
| Mamegakinone | 100 d | [35] | |
| Isodiospyrin | 10 d | ||
| Neodiospyrin | 10 d | ||
| Shinanolone | 100 d | ||
| Ferula communis Linn. | Ferulenol | 1.25 c | [36] |
| Feniculum vulgare Mill. | 5-Hydroxy-furanocoumarin or bergaptol | 100–200 b | [37] |
| Juniperus communis subsp. communis var. communis L. | Totarol | 2–25 a,c,d | [38] |
| Ferruginol | 5 c | [39] | |
| Sandaracopimeric acid | 30 c | ||
| 4-Epiabietol | 60 c | ||
| Justicia adhatoda L. or Adhatoda vesica | Vasicine | 200 d | |
| Kaempferia galangal L. | Ethyl-p-methoxycinnamate | 50–100 b,d | [40] |
| Lantana hispida Kunth | Oleanolic acid | 25–100 a,b,c,d | [24,41] |
| Larrea tridentata Coville | Dihydroguaiaretic acid | 12–50 b,d | [42] |
| 4-Epi-larreatricin | 25–50 b,d | ||
| Plectranthus grandidentatus Gurke | Abietane | 3.12–25 b,d | [43] |
| Plumeria bicolor Ruiz and Pav. | Plumericin | 1.5–2 b,d | [44] |
| Isoplumericin | 2–2.5 b,d | ||
| Struthanthus concinnus | Obtusifoliol | 50 d | [45] |
| Tabernemontana elegans Stapf. or Tiliacora triandra | Tiliacorinine | 3.12–6.25 b,d | [46] |
| 2′-Nortiliacorinine | 1.5–6.25 b,d | ||
| 13′-Bromotiliacorinine | 1.5–6.25 b,d | ||
| Ventilago madraspatana | Emodin | 4–128 b,c | [33] |
| Vetiveria zizanioides | α-Curcumene | 31.25–125 a,b,c | [47] |
| Valencene | 62.5–250 a,b,c | ||
| Selina-3,7(11)-diene |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baptista, R.; Bhowmick, S.; Shen, J.; Mur, L.A.J. Molecular Docking Suggests the Targets of Anti-Mycobacterial Natural Products. Molecules 2021, 26, 475. https://doi.org/10.3390/molecules26020475
Baptista R, Bhowmick S, Shen J, Mur LAJ. Molecular Docking Suggests the Targets of Anti-Mycobacterial Natural Products. Molecules. 2021; 26(2):475. https://doi.org/10.3390/molecules26020475
Chicago/Turabian StyleBaptista, Rafael, Sumana Bhowmick, Jianying Shen, and Luis A. J. Mur. 2021. "Molecular Docking Suggests the Targets of Anti-Mycobacterial Natural Products" Molecules 26, no. 2: 475. https://doi.org/10.3390/molecules26020475
APA StyleBaptista, R., Bhowmick, S., Shen, J., & Mur, L. A. J. (2021). Molecular Docking Suggests the Targets of Anti-Mycobacterial Natural Products. Molecules, 26(2), 475. https://doi.org/10.3390/molecules26020475







