Biological Activity of Newly Synthesized Benzimidazole and Benzothizole 2,5-Disubstituted Furane Derivatives
Abstract
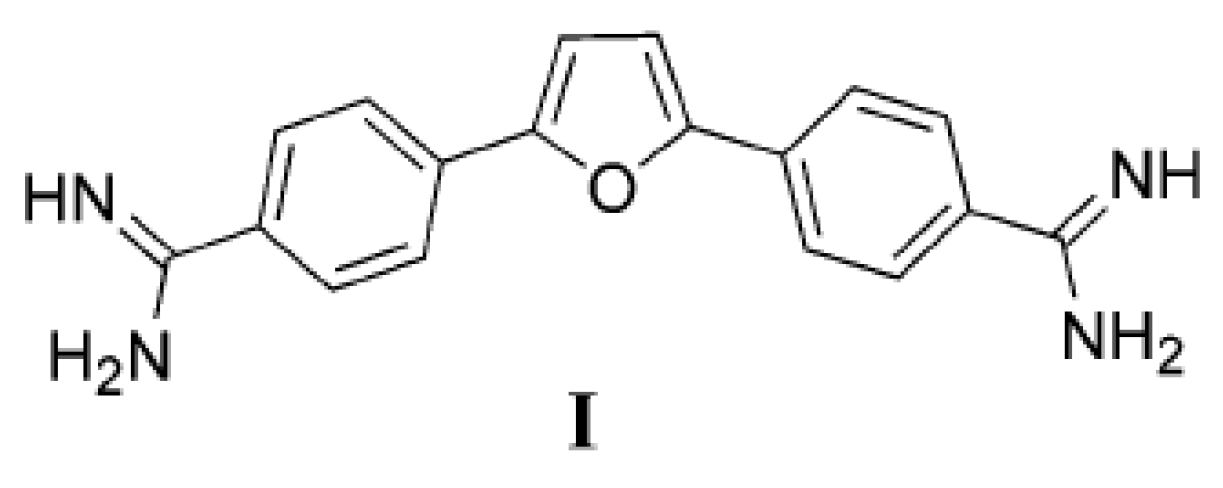
1. Introduction
2. Results and Discussion
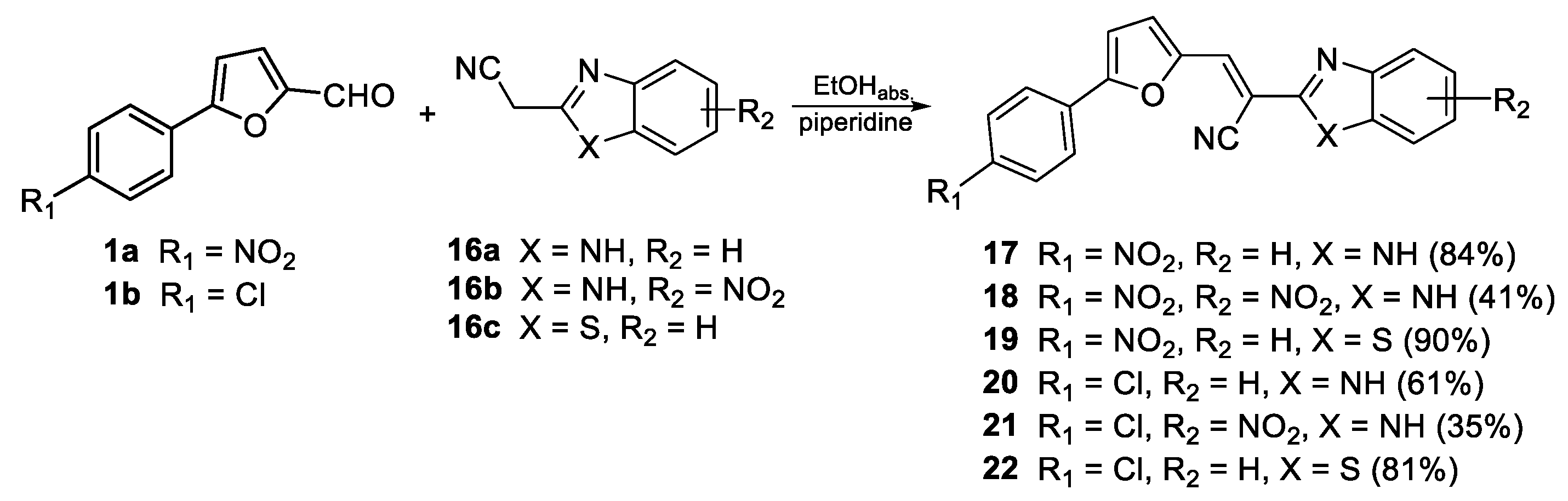
2.1. Chemistry
2.2. Biology
2.2.1. Antiproliferative Activity and Cytotoxicity Assays in 2D and 3D Cell Culture Assays In Vitro
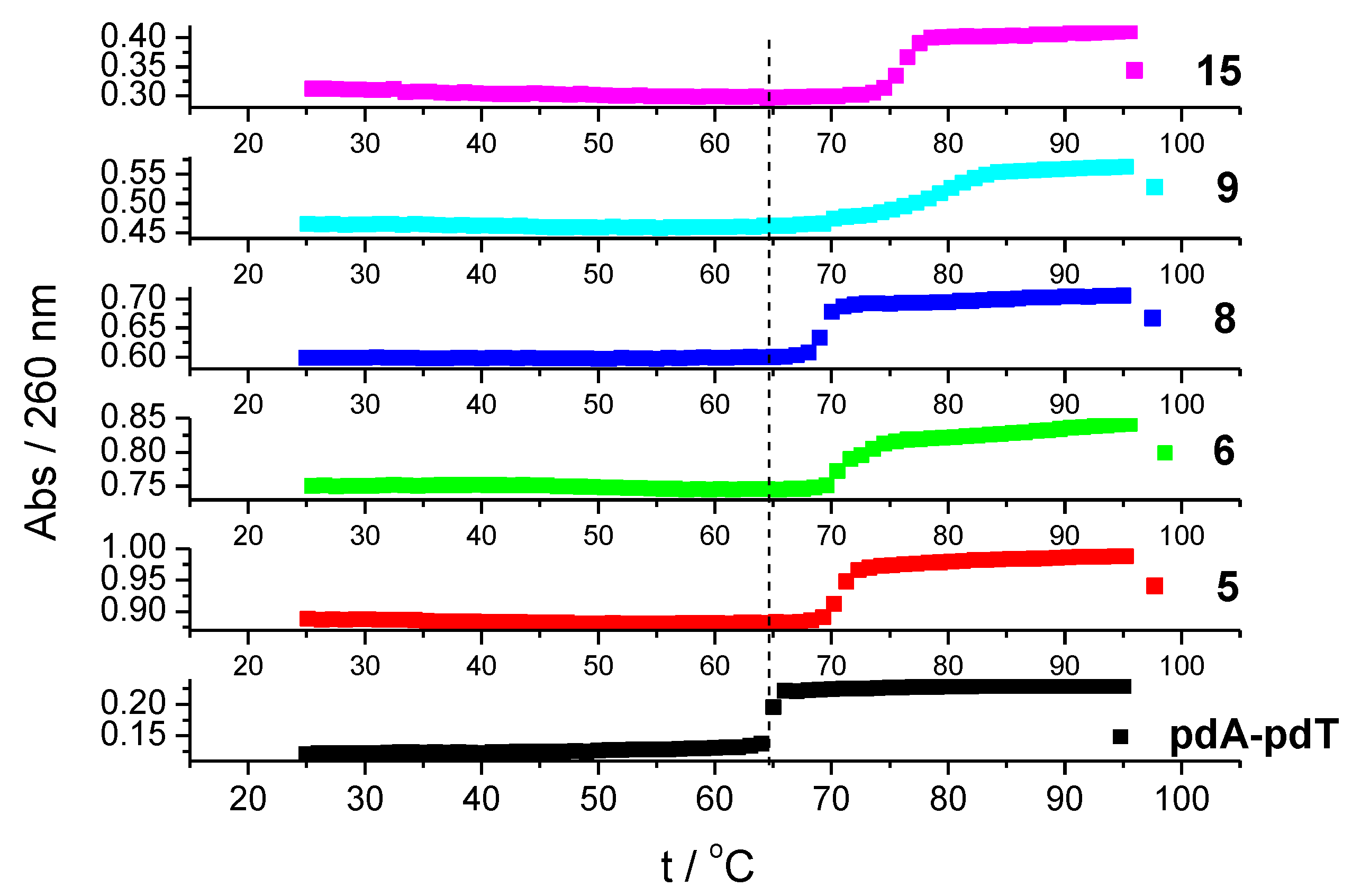
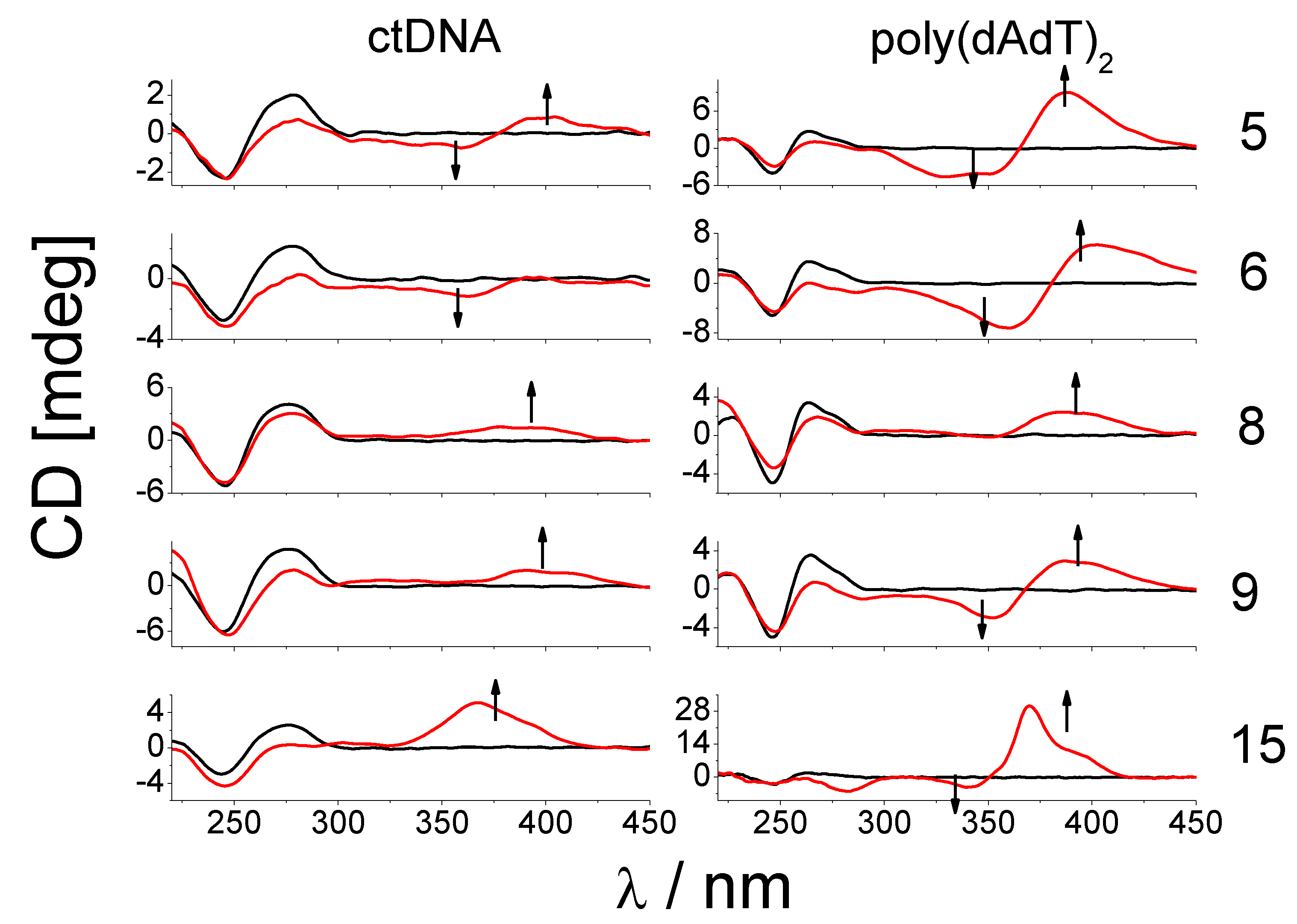
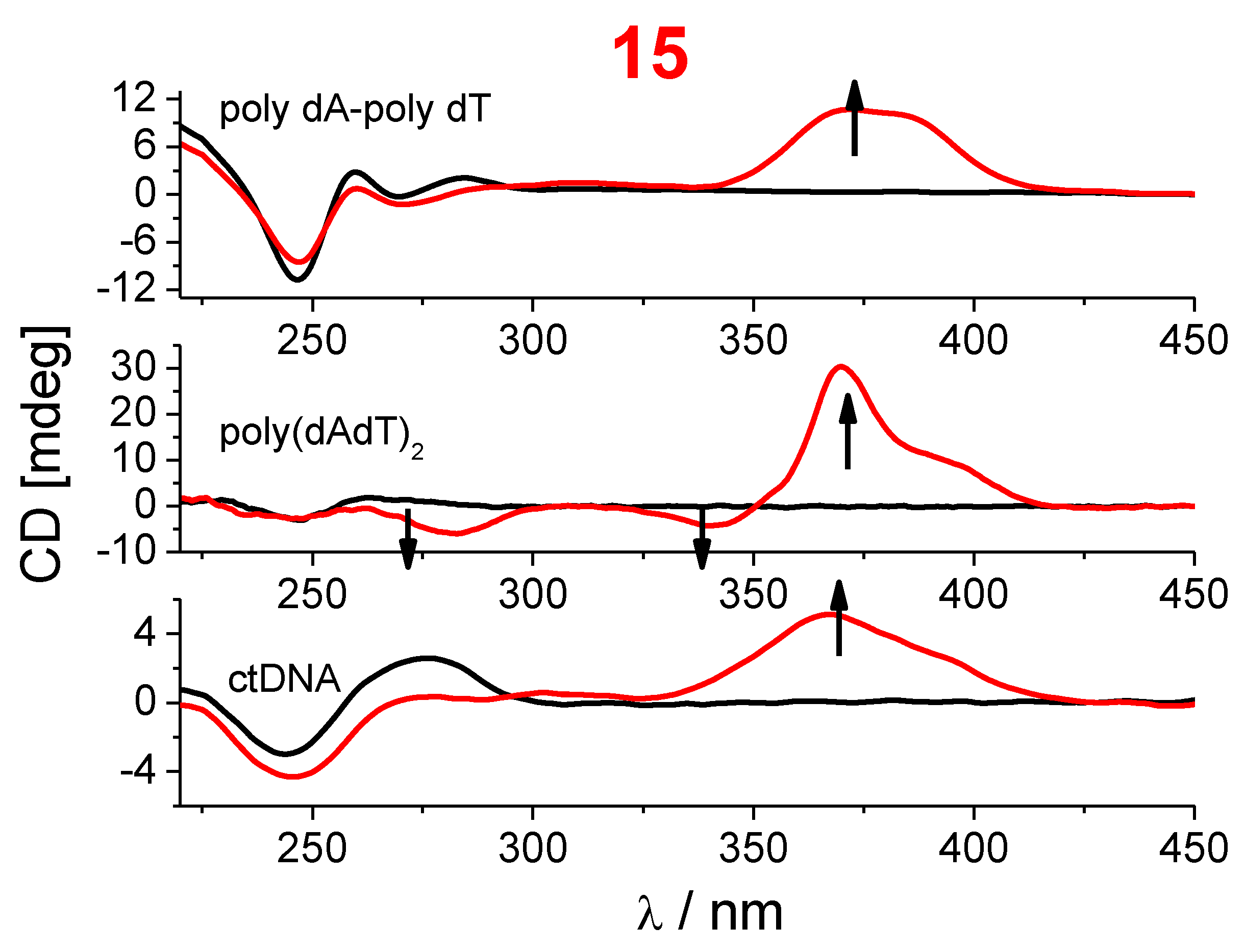
2.2.2. DNA Binding Study
3. Materials and Methods
3.1. Chemistry
3.1.1. General
3.1.2. General Method for Preparation of Compounds 4 and 7
3.1.3. General Method for Preparation of Compounds 5–6 and 8–9
3.1.4. General Method for Preparation of Compounds 10–15
3.1.5. General Method for Preparation of Compounds 17–22
3.2. Pharmacological/Biological Assays
3.2.1. Antitumor Activity in 2D and 3D Cell Culture Assays In Vitro
3.2.2. In Vitro Antimicrobial Activity
3.2.3. DNA Binding Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Sample Availability
References
- Silverman, R.B. The Organic Chemistry of Drug Design and Drug Action; Elsevier Academic Press: Amsterdam, The Netherlands, 2004. [Google Scholar]
- Sharma, P.C.; Sinhmar, A.; Sharma, A.; Rajak, H.; Pal Pathak, D. Medicinal significance of benzothiazole scaffold: An insight view. J. Enzyme Inhib. Med. Chem. 2013, 28, 240–266. [Google Scholar] [CrossRef] [PubMed]
- Bansal, Y.; Silakari, O. The therapeutic journey of benzimidazoles: A review. Bioorg. Med. Chem. 2012, 20, 6208–6236. [Google Scholar] [CrossRef] [PubMed]
- Yadav, G.; Ganguly, S. Structure activity relationship (SAR) study of benzimidazole scaffold for different biological activities: A mini-review. Eur. J. Med. Chem. 2015, 97, 419–443. [Google Scholar] [CrossRef] [PubMed]
- Irfan, A.; Batool, F.; Naqvi, S.A.Z.; Islam, A.; Osman, S.M.; Nocentini, A.; Alissa, S.A.; Supuran, C.T. Benzothiazole derivatives as anticancer agents. J. Enzyme Inhib. Med. Chem. 2019, 35, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Chhabra, S.; Shrivastava, S.K.; Mishra, P. Benzimidazole: A promising pharmacophore. Med. Chem. Res. 2013, 22, 5077–5104. [Google Scholar] [CrossRef]
- Rescifina, A.; Zagni, C.; Varrica, M.G.; Pistarà, V.; Corsaro, A. Recent advances in small organic molecules as DNA intercalating agents: Synthesis, activity, and modeling. Eur. J. Med. Chem. 2014, 74, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Rupinder Gill, K.; Rawal, R.K.; Bariwal, J. Recent Advances in the Chemistry and Biology of Benzothiazoles. Arch. Pharm. Chem. Life Sci. 2015, 348, 155–178. [Google Scholar] [CrossRef]
- Buric, A.J.; Dickerhoff, J.; Yang, D. Novel DNA Bis-Intercalator XR5944 as a Potent Anticancer Drug—Design and Mechanism of Action. Molecules 2021, 26, 4132. [Google Scholar] [CrossRef]
- Khadieva, A.; Mostovaya, O.; Padnya, P.; Kalinin, V.; Grishaev, D.; Tumakov, D.; Stoikov, I. Arylamine Analogs of Methylene Blue: Substituent Effect on Aggregation Behavior and DNA Binding. Int. J. Mol. Sci. 2021, 22, 5847. [Google Scholar] [CrossRef]
- Wróbel, A.; Baradyn, M.; Ratkiewicz, A.; Droydowska, D. Synthesis, Biological Activity, and Molecular Dynamics Study of Novel Series of a Trimethoprim Analogs as Multi-Targeted Compounds: Dihydrofolate Reductase (DHFR) Inhibitors and DNA-Binding Agents. Int. J. Mol. Sci. 2021, 22, 3685. [Google Scholar] [CrossRef]
- Depauw, S.; Lambert, M.; Jambon, S.; Paul, A.; Peixoto, P.; Nhili, R.; Marongiu, L.; Figeac, M.; Dassi, C.; Paul-Constant, C.; et al. Heterocyclic Diamidine DNA Ligands as HOXA9 Transcription Factor Inhibitors: Design, Molecular Evaluation, and Cellular Consequences in a HOXA9-Dependant Leukemia Cell Model. J. Med. Chem. 2019, 62, 1306–1329. [Google Scholar] [CrossRef]
- Patel, A.; Smith, H.J.; Sturzebecher, J. Introduction to the Principles of Drug Design and Action; Smith, H.J., Ed.; CRC Taylor & Francis: Boca Raton, FL, USA, 2006. [Google Scholar]
- Tanious, F.A.; Hamelberg, D.; Bailly, C.; Czarny, A.; Boykin, D.W.; Wilson, W.D. DNA Sequence Dependent Monomer–Dimer Binding Modulation of Asymmetric Benzimidazole Derivatives. J. Am. Chem. Soc. 2004, 126, 143–153. [Google Scholar] [CrossRef]
- Racané, L.; Tralić-Kulenović, V.; Kraljević Pavelić, S.; Ratkaj, I.; Peixoto, P.; Nhili, R.; Depauw, S.; Hildebrand, M.P.; David-Cordonnier, M.H.; Pavelić, K.; et al. Novel Diamidino-Substituted Derivatives of Phenyl Benzothiazolyl and Dibenzothiazolyl Furans and Thiophenes: Synthesis, Antiproliferative and DNA Binding Properties. J. Med. Chem. 2010, 53, 2418–2432. [Google Scholar] [CrossRef]
- Racané, L.; Kraljević Pavelić, S.; Nhili, R.; Depauw, S.; Paul-Constant, C.; Ratkaj, I.; David-Cordonnier, M.H.; Pavelić, K.; Tralić-Kulenović, V.; Karminski-Zamola, G. New anticancer active and selective phenylene-bisbenzothiazoles: Synthesis, antiproliferative evaluation and DNA binding. Eur. J. Med. Chem. 2013, 63, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Hranjec, M.; Kralj, M.; Piantanida, I.; Sedić, M.; Šuman, L.; Pavelić, K.; Karminski-Zamola, G. Novel Cyano- and Amidino-Substituted Derivatives of Styryl-2-Benzimidazoles and Benzimidazo[1,2-a]quinolines. Synthesis, Photochemical Synthesis, DNA Binding, and Antitumor Evaluation, Part 3. J. Med. Chem. 2007, 50, 5696–5711. [Google Scholar] [CrossRef]
- Perin, N.; Nhili, R.; Ester, K.; Laine, W.; Karminski-Zamola, G.; Kralj, M.; David-Cordonnier, M.H.; Hranjec, M. Synthesis, antiproliferative activity and DNA binding properties of novel 5-Aminobenzimidazo[1,2-a]quinoline-6-carbonitriles. Eur. J. Med. Chem. 2014, 80, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Loncar, B.; Perin, N.; Mioc, M.; Bocek, I.; Grgic, L.; Kralj, M.; Tomic, S.; Radic Stojkovic, M.; Hranjec, M. Novel amino substituted tetracyclic imidazo[4,5-b]pyridine derivatives: Design, synthesis, antiproliferative activity and DNA/RNA binding study. Eur. J. Med. Chem. 2021, 217, 113342. [Google Scholar] [CrossRef] [PubMed]
- Racané, L.; Stojković, R.; Tralić-Kulenović, V.; Cerić, H.; Đaković, M.; Ester, K.; Mišir Krpan, A.; Radić Stojković, M. Interactions with polynucleotides and antitumor activity of amidino and imidazolinyl substituted 2-phenylbenzothiazole mesylates. Eur. J. Med. Chem. 2014, 86, 406–419. [Google Scholar] [CrossRef]
- Cindrić, M.; Jambon, S.; Harej, A.; Depauw, S.; David-Cordonnier, M.H.; Kraljević Pavelić, S.; Karminski-Zamola, G.; Hranjec, M. Novel amidino substituted benzimidazole and benzothiazole benzo[b]thieno-2-carboxamides exert strong antiproliferative and DNA binding properties. Eur. J. Med. Chem. 2017, 136, 468–479. [Google Scholar] [CrossRef]
- King, H.; Lourie, E.M.; Yorke, W. Studies in chemotherapy. XIX. Further report on new trypanocidal substances. Ann. Trop. Med. Parasitol. 1938, 32, 177–192. [Google Scholar] [CrossRef]
- Lourie, E.M.; Yorke, W. Studies in chemotherapy. XVI. The trypanocidal action of synthalin. Ann. Trop. Med. Parasitol. 1939, 33, 289–304. [Google Scholar] [CrossRef]
- Lombardy, R.L.; Tanious, F.A.; Ramachandran, K.; Tidwell, R.R.; Wilson, W.D. Synthesis and DNA Interactions of Benzimidazole Dications Which Have Activity against Opportunistic Infections. J. Med. Chem. 1996, 39, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Blagburn, B.L.; Sundermann, C.A.; Lindsay, D.S.; Hall, J.E.; Tidwell, R.R. Inhibition of Cryptosporidium parvum in neonatal Hsd:(ICR)BR Swiss miceby polyether ionophores and aromatic amidines. Antimicrob. Agents Chemother. 1991, 35, 1520–1523. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazur, S.; Tanious, F.; Ding, D.; Kumar, A.; Boykin, D.W.; Neidle, S.; Wilson, W.D. A thermodynamic and structural analysis of DNA minor-groove complex formation. J. Mol. Biol. 2000, 300, 321–337. [Google Scholar] [CrossRef]
- Das, B.P.; Boykin, D.W. Synthesis and antiprotozoal activity of 2,5-bis(4-guanylphenyl)thiophenes and –pyrroles. J. Med. Chem. 1977, 20, 1219–1221. [Google Scholar] [CrossRef] [PubMed]
- Tidwell, R.R.; Bell, C.A. Pentamidine and Related Compounds in Treatment of Pneumocystis Carinii Infection. Pneumocystis carinii; Marcel Decker: New York, NY, USA, 1993; p. 561. [Google Scholar]
- Hopkins, K.T.; Wilson, W.D.; Bender, B.C.; McCurdy, D.R.; Hall, J.E.; Tidwell, R.R.; Kumar, A.; Bajic, M.; Boykin, D.W. Extended aromatic furan amidino derivatives as anti-Pneumocystis carinii agents. J. Med. Chem. 1998, 41, 3872–3878. [Google Scholar] [CrossRef] [PubMed]
- Boykin, D.W.; Kumar, A.; Xiao, G.; Wilson, W.D.; Bender, B.C.; McCurdy, D.R.; Hall, J.E.; Tidwell, R.R. 2,5-Bis[4-(N-alkylamidino)phenyl]furans as Anti-Pneumocystis carinii Agents. J. Med. Chem. 1998, 41, 124–129. [Google Scholar] [CrossRef]
- Ismail, M.A.; Brun, R.; Easterbrook, J.D.; Tanious, F.A.; Wilson, W.D.; Boykin, D.W. Synthesis and Antiprotozoal Activity of Aza-Analogues of Furamidine. J. Med. Chem. 2003, 46, 4761–4769. [Google Scholar] [CrossRef] [PubMed]
- Brajša, K.; Trzun, M.; Zlatar, I.; Jelić, D. Three-dimensional cell cultures as a new tool in drug discovery. Period. Biliogorum 2016, 118, 59–65. [Google Scholar] [CrossRef]
- Zlatar, I.; Jelić, D.; Kelava, V.; Cindrić, M.; Jarak, I.; Koštrun, S.; Karminski-Zamola, G.; Gabelica Marković, V.; Hranjec, M.; Brajša, K. Comparison of Antitumor Activity of Some Benzothiophene and Thienothiophene Carboxanilides and Quinolones in 2D and 3D Cell Culture System. Croat. Chem. Act. 2017, 90, 413–424. [Google Scholar] [CrossRef]
- Perin, N.; Bobanović, K.; Zlatar, I.; Jelić, D.; Kelava, V.; Koštrun, S.; Gabelica Marković, V.; Brajša, K.; Hranjec, M. Antiproliferative activity of amino substituted benzo[b]thieno[2,3-b]pyrido[1,2-a]benzimidazoles explored by 2D and 3D cell culture system. Eur. J. Med. Chem. 2017, 125, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, S.H.; Momen, A.A. Anticancer Drugs’ Deoxyribonucleic Acid (DNA) Interactions. In Biophysical Chemistry-Advance Applications; Khalid, M., Ed.; IntechOpen: London, UK, 2019; Available online: https://www.intechopen.com/books/biophysical-chemistry-advance-applications/anticancer-drugs-deoxyribonucleic-acid-dna-interactions (accessed on 14 July 2021). [CrossRef]
- Racané, L.; Ptiček, L.; Fajdetić, G.; Tralić-Kulenović, V.; Klobučar, M.; Kraljević Pavelić, S.; Perić, M.; Čipčić Paljetak, H.; Verbanac, D.; Starčević, K. Green synthesis and biological evaluation of 6-substituted-2-(2-hydroxy/methoxy phenyl)benzothiazole derivatives as potential antioxidant, antibacterial and antitumor agents. Bioorg. Chem. 2020, 95, 103537. [Google Scholar] [CrossRef] [PubMed]
- Racanè, L.; Tralić-Kulenović, V.; Mihalić, Z.; Pavlović, G.; Karminski-Zamola, G. Synthesis of new amidino-substituted 2-aminothiophenoles: Mild basic ring opening of benzothiazole. Tetrahedron 2008, 64, 11594–11602. [Google Scholar] [CrossRef]
- Hranjec, M.; Pavlović, G.; Marjanović, M.; Karminski-Zamola, G. Benzimidazole derivatives related to 2,3-acrylonitriles, benzimidazo[1,2-a]quinolines and fluorenes. Eur. J. Med. Chem. 2010, 45, 2405–2417. [Google Scholar] [CrossRef]
- Estman, A. Improving anticancer drug development begins with cell culture: Misinformation perpetrated by the misuse of cytotoxicity assays. Oncotarget 2017, 8, 8854–8866. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Parish, J. Principles of Nucleic Acid Structure; Saenger, W., Ed.; Springer: New York, NY, USA, 1984; p. 556, DM 79; ISBN 3-540-90761-0. [Google Scholar]
- Scatchard, G. The attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Demeunynck, M.; Bailly, C.; Wilson, W.D. DNA and RNA Binders: From Small Molecules to Drugs; Wiley-VCH: Weinheim, Germany, 2002; Volume 1, Chapter 5. [Google Scholar]
- Rodger, A.; Nordén, B. Circular Dichroism and Linear Dichroism; Oxford University Press: New York, NY, USA, 1997. [Google Scholar]
- Eriksson, M.; Nordén, B. Linear and circular dichroism of drug-nucleic acid complexes. Methods Enzymol. 2001, 340, 68–98. [Google Scholar]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids–tutorial. Beilstein J. Org. Chem. 2017, 14, 84–105. [Google Scholar] [CrossRef] [PubMed]
- Jezuita, A.; Ejsmont, K.; Szatylowicz, H. Substituent effects of nitro group in cyclic compounds. Struct. Chem. 2021, 32, 179–203. [Google Scholar] [CrossRef]
- Neidle, S. Oxford Handbook of Nucleic Acid Structure; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Ganushchak, N.I.; Lesyuk, A.I.; Fedorovich, I.S.; Obushak, N.D.; Andrushko, V.N. 5-Aryl-2-furaldehydes in the Synthesis of 2-Substituted 1,3-Benzazoles. Russ. J. Org. Chem. 2003, 39, 1295–1300. [Google Scholar] [CrossRef]
- Chaires, J.B.; Dattagupta, N.; Crothers, D.M. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: Equilibrium binding studies on the interaction of daunomycin with deoxyribonucleic acid. Biochemistry 1982, 21, 3933–3940. [Google Scholar] [CrossRef]
- Chalikian, T.V.; Volker, J.; Plum, G.E.; Breslauer, K.J.A. A more unified picture for the thermodynamics of nucleic acid duplex melting: A characterization by calorimetric and volumetric techniques. Proc. Natl. Acad. Sci. USA 1999, 96, 7853–7858. [Google Scholar] [CrossRef] [PubMed]
- Trinquet, E.; Mathis, G. Fluorescence technologies for the investigation of chemical libraries. Mol. Biosyst. 2006, 2, 380–387. [Google Scholar] [CrossRef]
- Kurutos, A.; Crnolatac, I.; Orehovec, I.; Gadjev, N.; Piantanida, I.; Deligeorgiev, T. Novel synthetic approach to asymmetric monocationic trimethine cyanine dyes derived from N-ethyl quinolinum moiety. Combined fluorescent and ICD probes for AT-DNA labelling. J. Lumin. 2016, 174, 70–76. [Google Scholar] [CrossRef]
- Tumir, L.M.; Crnolatac, I.; Deligeorgiev, T.; Vasilev, A.; Kaloyanova, S.; Grabar Branilović, M.; Tomić, S.; Piantanida, I. Kinetic Differentiation between Homo- and Alternating AT DNA by Sterically Restricted Phosphonium Dyes. Chem. Eur. J. 2012, 18, 3859–3864. [Google Scholar] [CrossRef]
- Stefl, R.; Wu, H.; Ravindranathan, S.; Sklenář, V.; Feigon, J. DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum. Proc. Natl. Acad. Sci. USA 2004, 101, 1177–1182. [Google Scholar] [CrossRef] [PubMed]

| Cytotoxicity Assay IC50 Values (µM) | |||||||
|---|---|---|---|---|---|---|---|
| Comp. | IC50 (µM) ± SD; n = 2 | ||||||
| A549 | HCC827 | NCI-H358 | MRC-5 | ||||
| 2D | 3D | 2D | 3D | 2D | 3D | 2D | |
| 4 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| 5 | 8.76 ± 1.69 | 26.44 ± 13.37 | 19.33 ± 1.72 | 43.59 ± 6.45 | 9.97 ± 0.59 | 14.80 ± 0.57 | 21.65 ± 0.51 |
| 6 | 2.12 ± 0.21 | 4.01 ± 0.95 | 5.13 ± 0.97 | 7.02 ± 3.25 | 0.85 ± 0.05 | 1.73 ± 0.01 | 3.11 ± 0.26 |
| 7 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| 8 | 6.74 ± 0.19 | 9.31 ± 0.78 | 6.26 ± 0.33 | 20.46 ± 8.63 | 6.48 ± 0.11 | 16.00 ± 9.38 | 9.88 ± 2.38 |
| 9 | 4.39 ± 0.79 | 16.81 ± 2.61 | 5.9 ± 0.58 | 19.99 ± 7.92 | 4.48 ± 0.30 | 15.49 ± 0.07 | 6.65 ± 0.18 |
| 10 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| 11 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| 12 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| 13 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| 14 | >50 | >100 | >50 | >100 | 31.07 ± 0.67 | >100 | >50 |
| 15 | 24.29 ± 0.40 | 55.84 ± 8.29 | 22.70 ± 0.04 | 65.42 ± 5.04 | 19.39 ± 0.57 | 81.62 ± 7.71 | >50 |
| 17 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| 18 | >50 | >100 | >50 | >100 | 3.50 ± 1.14 | >100 | >50 |
| 19 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| 20 | >50 | >100 | 43.53 ± 8.17 | 9.48 ± 1.15 | 31.08 ± 4.42 | >100 | >50 |
| 21 | >50 | >100 | >50 | >100 | 3.58 ± 0.95 | >100 | >50 |
| 22 | >50 | >100 | >50 | >100 | >50 | >100 | >50 |
| Doxorubicin | 1.31 ± 0.11 | 5.86 ± 1.10 | 0.32 ± 0.03 | 1.47 ± 0.12 | 0.15 ± 0.06 | 0.99 ± 0.15 | 0.60 ± 0.01 |
| Staurosporine | 0.047 ± 0.007 | 0.095 ± 0.01 | 0.033 ± 0.002 | 0.002 ± 0.0001 | 0.041 ± 0.007 | 0.002 ± 0.001 | 0.031 ± 0.011 |
| Vandetanib | >25 | >50 | 0.58 ± 0.03 | 0.16 ± 0.04 | 3.12 ± 35.99 | 0.38 ± 0.20 | 11.12 ± 0.00 |
| Proliferation Assay IC50 Values (µM) | ||||||
|---|---|---|---|---|---|---|
| Cpd. | IC50 (µM) ± SD; n = 2 | |||||
| A549 | HCC827 | NCI-H358 | ||||
| 2D | 3D | 2D | 3D | 2D | 3D | |
| 4 | >50 | >100 | >50 | >100 | >50 | >100 |
| 5 | 4.76 ± 1.03 | 65.93 ± 3.66 | 13.66 ± 3.13 | 14.69 ± 13.36 | 3.56 ± 0.18 | 14.29 ± 0.84 |
| 6 | 2.27 ± 0.13 | 5.52 ± 0.10 | 16.69 ± 9.39 | 7.63 ± 5.00 | 1.10 ± 0.001 | 2.36 ± 1.17 |
| 7 | >50 | >100 | >50 | >100 | >50 | >100 |
| 8 | 8.78 ± 3.62 | 19.94 ± 2.19 | 13.48 ± 0.81 | 17.75 ± 4.24 | 6.68 ± 0.15 | 11.27 ± 0.49 |
| 9 | 9.97 ± 5.37 | 17.01 ± 3.53 | 8.69 ± 2.79 | 20.28 ± 13.87 | 5.57 ± 0.81 | 6.39 ± 3.29 |
| 10 | >50 | >100 | >50 | >100 | >50 | >100 |
| 11 | >50 | >100 | >50 | >100 | >50 | >100 |
| 12 | >50 | >100 | >50 | 16.85 ± 8.33 | >50 | >100 |
| 13 | 22.73 ± 0.54 | >100 | >50 | >100 | 15.67 ± 0.86 | >100 |
| 14 | >50 | >100 | >50 | >100 | 20.20 ± 0.71 | >100 |
| 15 | >50 | >100 | >50 | >100 | 20.08 ± 2.82 | >100 |
| 17 | >50 | >100 | >50 | >100 | >50 | >100 |
| 18 | >50 | >100 | >50 | >100 | >50 | >100 |
| 19 | >50 | >100 | >50 | >100 | >50 | >100 |
| 20 | 43.86 ± 54.27 | >100 | >50 | 0.16 ± 0.11 | 14.36 ± 6.46 | >100 |
| 21 | >50 | >100 | >50 | >100 | >50 | >100 |
| 22 | >50 | >100 | >50 | >100 | >50 | >100 |
| Doxorubicin | 0.13 ± 0.01 | 0.48 ± 0.03 | 0.25 ± 0.28 | 1.89 ± 2.30 | 0.07 ± 0.04 | 2.30 ± 1.91 |
| Staurosporine | 0.035 ± 0.017 | 0.11 ± 0.001 | 0.004 ± 0.002 | 0.003 ± 0.002 | 0.0005 ± 0.00004 | 0.02 ± 0.01 |
| Vandetanib | >25 | >50 | 0.20 ± 0.11 | 0.08 ± 0.02 | >25 | 0.38 ± 0.17 |
| Comp. | E. coli ATCC 25922 | S. aureus ATCC 29213 | S. cerevisiae ATCC 7752 |
|---|---|---|---|
| MIC (μM) | |||
| 4 | >100 | >100 | >100 |
| 5 | >100 | 50 | 12.5 |
| 6 | >100 | >100 | 1.6 |
| 7 | >100 | >100 | >100 |
| 8 | >100 | 12.5 | 6.25 |
| 9 | >100 | 25 | 3.13 |
| 10 | >100 | >100 | >100 |
| 11 | >100 | 100 | >100 |
| 12 | >100 | >100 | 100 |
| 13 | >100 | >100 | >100 |
| 14 | 50 | 12.5 | 50 |
| 15 | 25 | 6.25 | 6.25 |
| 17 | >100 | >100 | >100 |
| 18 | >100 | >100 | >100 |
| 19 | >100 | >100 | >100 |
| 20 | >100 | >100 | >100 |
| 21 | >100 | >100 | >100 |
| 22 | >100 | >100 | >100 |
| Ampicillin | 4 | 2 | >64 |
| Ceftazidime | <0.125 | 16 | >64 |
| Ciprofloxacin | <0.125 | 0.25 | >64 |
| Gentamycin | 0.5 | 0.25 | >64 |
| Voriconazole | >64 | >64 | <0.125 |
| br = 0.3 | 5 | 6 | 8 | 9 | 15 |
|---|---|---|---|---|---|
| ctDNA | 2.5 | 3.2 | 1.7 | 1.8 | 0.9 |
| poly (dAdT)2 | 4.8 | 3.6 | 3.6 | 4.6 | 6.2 |
| Poly dA-poly dT | 5.5 | 5.6 | 4.3 | 15.2 | 11.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Racané, L.; Zlatar, I.; Perin, N.; Cindrić, M.; Radovanović, V.; Banjanac, M.; Shanmugam, S.; Stojković, M.R.; Brajša, K.; Hranjec, M. Biological Activity of Newly Synthesized Benzimidazole and Benzothizole 2,5-Disubstituted Furane Derivatives. Molecules 2021, 26, 4935. https://doi.org/10.3390/molecules26164935
Racané L, Zlatar I, Perin N, Cindrić M, Radovanović V, Banjanac M, Shanmugam S, Stojković MR, Brajša K, Hranjec M. Biological Activity of Newly Synthesized Benzimidazole and Benzothizole 2,5-Disubstituted Furane Derivatives. Molecules. 2021; 26(16):4935. https://doi.org/10.3390/molecules26164935
Chicago/Turabian StyleRacané, Livio, Ivo Zlatar, Nataša Perin, Maja Cindrić, Vedrana Radovanović, Mihailo Banjanac, Suresh Shanmugam, Marijana Radić Stojković, Karmen Brajša, and Marijana Hranjec. 2021. "Biological Activity of Newly Synthesized Benzimidazole and Benzothizole 2,5-Disubstituted Furane Derivatives" Molecules 26, no. 16: 4935. https://doi.org/10.3390/molecules26164935
APA StyleRacané, L., Zlatar, I., Perin, N., Cindrić, M., Radovanović, V., Banjanac, M., Shanmugam, S., Stojković, M. R., Brajša, K., & Hranjec, M. (2021). Biological Activity of Newly Synthesized Benzimidazole and Benzothizole 2,5-Disubstituted Furane Derivatives. Molecules, 26(16), 4935. https://doi.org/10.3390/molecules26164935





