Exploring New Scaffolds for the Dual Inhibition of HIV-1 RT Polymerase and Ribonuclease Associated Functions
Abstract
1. Introduction
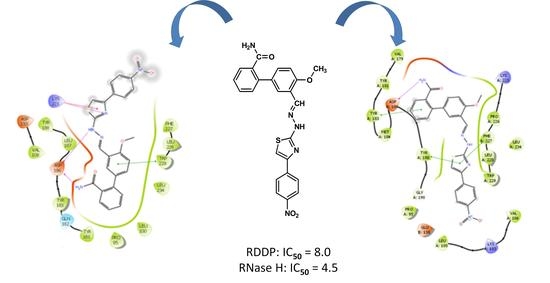
2. Results and Discussion
3. Materials and Method
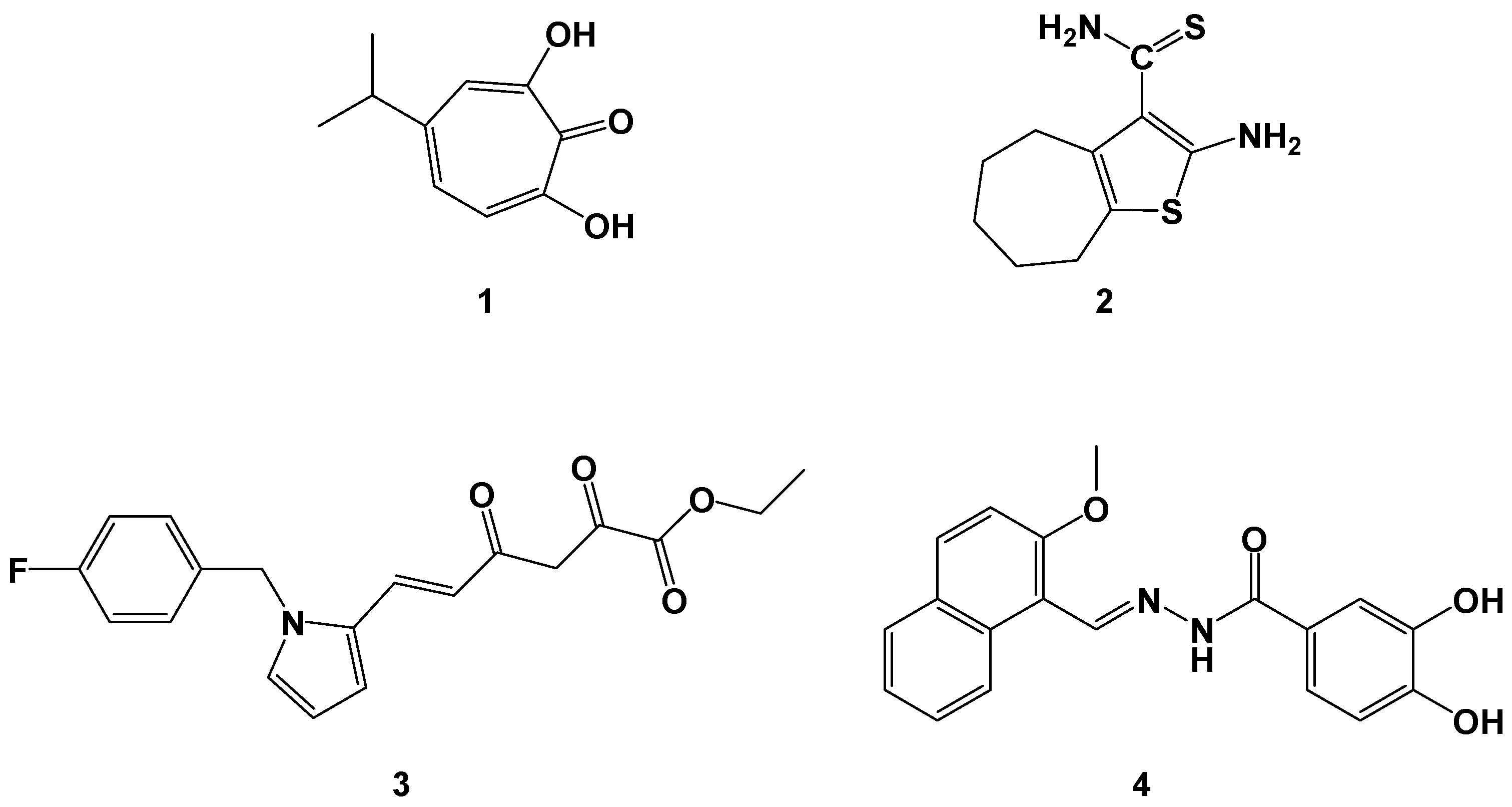
3.1. Chemistry
3.1.1. Synthetic Procedures
Preparation of 4-Methoxy-[1,1′-Biphenyl]-3-Carbaldehyde (IIIa)
Preparation of 2-[2-[(2-Methoxy-5-Phenylphenyl)methylidene]hydrazin-1-yl]-4-(4-Methoxyphenyl)-1,3-Thiazole Bromide (EMAC2056)
3.2. Biological Evaluation
3.2.1. HIV-1 RT-Associated DNA Polymerase-Independent RNase H Activity Determination
3.2.2. HIV-1 RT-Associated RNA Dependent DNA Polymerase Activity Determination
3.3. Molecular Medelling
3.3.1. Ligand Preparation
3.3.2. Protein Preparation
3.3.3. Docking Experiments
3.3.4. Figures
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Gallo, R.C.; Montagnier, L. The discovery of HIV as the cause of AIDS. N. Engl. J. Med. 2003, 349, 2283–2285. [Google Scholar] [CrossRef]
- Shaw, G.M.; Broder, S.; Essex, M.; Gallo, R.C. Human T-cell leukemia virus: Its discovery and role in leukemogenesis and immunosuppression. Adv. Intern. Med. 1984, 30, 1–27. [Google Scholar]
- Shridhar, T.D. Antiretroviral therapy—The future of HIV treatment. World J. Pharm. Res. 2020, 9, 669–673. [Google Scholar]
- Saag, M.S.; Gandhi, R.T.; Hoy, J.F.; Landovitz, R.J.; Thompson, M.A.; Sax, P.E.; Smith, D.M.; Benson, C.A.; Buchbinder, S.P.; del Rio, C.; et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2020 recommendations of the international antiviral society—USA panel. JAMA 2020, 324, 1651–1669. [Google Scholar] [CrossRef] [PubMed]
- Lal, H.; Kumar, T.; Dutta, S.; Ram, K. A concise review of existing therapies and recent advances in the management of HIV infection. Int. J. Pharm. Sci. Rev. Res. 2020, 64, 153–158. [Google Scholar] [CrossRef]
- Tseng, A.; Seet, J.; Phillips, E.J. The evolution of three decades of antiretroviral therapy: Challenges, triumphs and the promise of the future. Br. J. Clin. Pharmacol. 2015, 79, 182–194. [Google Scholar] [CrossRef]
- Fischl, M.A.; Richman, D.D.; Grieco, M.H.; Gottlieb, M.S.; Volberding, P.A.; Laskin, O.L.; Leedom, J.M.; Groopman, J.E.; Mildvan, D.; Schooley, R.T.; et al. The efficacy of Azidothymidine (AZT) in the treatment of patients with AIDS and AIDS-related complex. N. Engl. J. Med. 1987, 317, 185–191. [Google Scholar] [CrossRef]
- de Béthune, M.-P. Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: A review of the last 20 years (1989–2009). Antivir. Res. 2010, 85, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Flexner, C. HIV-protease inhibitors. N. Engl. J. Med. 1998, 338, 1281–1293. [Google Scholar] [CrossRef]
- Carter, N.J.; Keating, G.M. Maraviroc. Drugs 2007, 67, 2277–2288. [Google Scholar] [CrossRef]
- Beccari, M.V.; Mogle, B.T.; Sidman, E.F.; Mastro, K.A.; Asiago-Reddy, E.; Kufel, W.D. Ibalizumab, a novel monoclonal antibody for the management of multidrug-resistant HIV-1 infection. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- LaBonte, J.; Lebbos, J.; Kirkpatrick, P. Enfuvirtide. Nat. Rev. Drug Discov. 2003, 2, 345–346. [Google Scholar] [CrossRef]
- Rossetti, B.; Montagnani, F.; De Luca, A. Current and emerging two-drug approaches for HIV-1 therapy in ART-naïve and ART-experienced, virologically suppressed patients. Expert Opin. Pharmacother. 2018, 19, 713–738. [Google Scholar] [CrossRef] [PubMed]
- Margolis, A.M.; Heverling, H.; Pham, P.A.; Stolbach, A. A review of the toxicity of HIV medications. J. Med. Toxicol. 2013, 10, 26–39. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Popovic-Nikolicb, M.R.; Nikolic, K.; Uliassi, E.; Bolognesi, M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med. 2018, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- de Castro, S.; Camarasa, M.-J. Polypharmacology in HIV inhibition: Can a drug with simultaneous action against two relevant targets be an alternative to combination therapy? Eur. J. Med. Chem. 2018, 150, 206–227. [Google Scholar] [CrossRef]
- Morphy, R.; Rankovic, Z. Chapter 27—Multi-target drugs: Strategies and challenges for medicinal chemists. In The Practice of Medicinal Chemistry, 3rd ed.; Wermuth, C.G., Ed.; Academic Press: New York, NY, USA, 2008; pp. 549–571. [Google Scholar]
- Velázquez, S.; Alvarez, R.; San-Felix, A.; Jimeno, M.L.; De Clercq, E.; Balzarini, J.; Camarasa, M.J. Synthesis and anti-HIV activity of [AZT]-[TSAO-T] and [AZT]-[HEPT] dimers as potential multifunctional inhibitors of HIV-1 reverse transcriptase. J. Med. Chem. 1995, 38, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, S.; Tuñón, V.; Jimeno, M.L.; Chamorro, C.; De Clercq, E.; Balzarini, J.; Camarasa, M.J. Potential multifunctional inhibitors of HIV-1 reverse transcriptase. Novel [AZT]-[TSAO-T] and [d4T]-[TSAO-T] heterodimers modified in the linker and in the dideoxynucleoside region. J. Med. Chem. 1999, 42, 5188–5196. [Google Scholar] [CrossRef]
- Telesnitsky, A.; Goff, S.P. Reverse Transcriptase and the Generation of Retroviral DNA; Cold Spring Harbour Laboratory Press: Cold Spring Harbour, NY, USA, 1997. [Google Scholar]
- Nikolenko, G.N.; Palmer, S.; Maldarelli, F.; Mellors, J.W.; Coffin, J.M.; Pathak, V.K. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: Balance between RNase H activity and nucleotide excision. Proc. Natl. Acad. Sci. USA 2005, 102, 2093–2098. [Google Scholar] [CrossRef] [PubMed]
- Tisdale, M.; Schulze, T.; Larder, B.A.; Moelling, K. Mutations within the RNase H domain of Human Immunodeficiency Virus Type 1 reverse transcriptase abolish virus infectivity. J. Gen. Virol. 1991, 72, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Schatz, O.; Cromme, F.V.; Grüninger-Leitch, F.; Le Grice, S.F. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989, 257, 311–314. [Google Scholar] [CrossRef]
- Boyer, P.L.; Smith, S.J.; Zhao, X.Z.; Das, K.; Gruber, K.; Arnold, E.; Burke, T.R.; Hughes, S.H. Developing and evaluating inhibitors against the RNase H active site of HIV-1 reverse transcriptase. J. Virol. 2018, 92, e02203-17. [Google Scholar] [CrossRef]
- Wang, X.; Gao, P.; Menendez-Arias, L.; Liu, X.; Zhan, P. Update on recent developments in small molecular HIV-1 RNase H inhibitors (2013–2016): Opportunities and challenges. Curr. Med. Chem. 2018, 25, 1682–1702. [Google Scholar] [CrossRef]
- Figiel, M.; Krepl, M.; Poznański, J.; Gołąb, A.; Sponer, J.; Nowotny, M. Coordination between the polymerase and RNase H activity of HIV-1 reverse transcriptase. Nucleic Acids Res. 2017, 45, 3341–3352. [Google Scholar] [CrossRef]
- Himmel, D.M.; Maegley, K.A.; Pauly, T.A.; Bauman, J.D.; Das, K.; Dharia, C.; Clark, A.D.; Ryan, K.; Hickey, M.J.; Love, R.A.; et al. Structure of HIV-1 reverse transcriptase with the inhibitor β-thujaplicinol bound at the RNase H active site. Structure 2009, 17, 1625–1635. [Google Scholar] [CrossRef]
- Wendeler, M.; Lee, H.-F.; Bermingham, A.; Miller, J.T.; Chertov, O.; Bona, M.K.; Baichoo, N.S.; Ehteshami, M.; Beutler, J.; O’Keefe, B.R.; et al. Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity. ACS Chem. Biol. 2008, 3, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, K.; Mirzadegan, T. Recent progress in the design of small molecule inhibitors of HIV RNase H. Curr. Pharm. Des. 2006, 12, 1909–1922. [Google Scholar] [CrossRef]
- Himmel, D.M.; Sarafianos, S.G.; Dharmasena, S.; Hossain, M.M.; McCoy-Simandle, K.; Ilina, T.; Clark, A.D., Jr.; Knight, J.L.; Julias, J.G.; Clark, P.K.; et al. HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem. Biol. 2006, 1, 702–712. [Google Scholar] [CrossRef]
- Tramontano, E.; Corona, A.; Menéndez-Arias, L. Ribonuclease H, an unexploited target for antiviral intervention against HIV and hepatitis B virus. Antivir. Res. 2019, 171, 104613. [Google Scholar] [CrossRef]
- Corona, A.; Ballana, E.; Distinto, S.; Rogolino, D.; Del Vecchio, C.; Carcelli, M.; Badia, R.; Riveira-Muñoz, E.; Esposito, F.; Parolin, C.; et al. Targeting HIV-1 RNase H: N’-(2-hydroxy-benzylidene)-3,4,5-trihydroxybenzoylhydrazone as selective inhibitor active against NNRTIs-resistant variants. Viruses 2020, 12, 729. [Google Scholar] [CrossRef]
- Tramontano, E.; Esposito, F.; Badas, R.; Costi, R.; Di Santo, R.; Colla, P. RDS1643, a novel diketo acid which selectively inhibits the HIV-1 multiplication in cell-based assays and the ribonuclease H activity in enzyme assays. In Proceedings of the 17th International Conference on Antiviral Research, Tucson, AR, USA, 2–6 May 2004; p. A35. [Google Scholar]
- Distinto, S.; Esposito, F.; Kirchmair, J.; Cardia, M.C.; Gaspari, M.; Maccioni, E.; Alcaro, S.; Markt, P.; Wolber, G.; Zinzula, L.; et al. Identification of HIV-1 reverse transcriptase dual inhibitors by a combined shape, 2D-fingerprint and pharmacophore-based virtual screening approach. Eur. J. Med. Chem. 2012, 50, 216–229. [Google Scholar] [CrossRef]
- Schneider, A.; Corona, A.; Spöring, I.; Jordan, M.; Buchholz, B.; Maccioni, E.; Di Santo, R.; Bodem, J.; Tramontano, E.; Wöhrl, B.M. Biochemical characterization of a multi-drug resistant HIV-1 subtype AG reverse transcriptase: Antagonism of AZT discrimination and excision pathways and sensitivity to RNase H inhibitors. Nucleic Acids Res. 2016, 44, 2310–2322. [Google Scholar] [CrossRef] [PubMed]
- Sonar, V.P.; Corona, A.; Distinto, S.; Maccioni, E.; Meleddu, R.; Fois, B.; Floris, C.; Malpure, N.V.; Alcaro, S.; Tramontano, E.; et al. Natural product-inspired esters and amides of ferulic and caffeic acid as dual inhibitors of HIV-1 reverse transcriptase. Eur. J. Med. Chem. 2017, 130, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Meleddu, R.; Distinto, S.; Corona, A.; Tramontano, E.; Bianco, G.; Melis, C.; Cottiglia, F.; Maccioni, E. Isatin thiazoline hybrids as dual inhibitors of HIV-1 reverse transcriptase. J. Enzym. Inhib. Med. Chem. 2016, 32, 130–136. [Google Scholar] [CrossRef]
- Corona, A.; Meleddu, R.; Esposito, F.; Distinto, S.; Bianco, G.; Masaoka, T.; Maccioni, E.; Menéndez-Arias, L.; Alcaro, S.; Le Grice, S.F.J.; et al. Ribonuclease H/DNA polymerase HIV-1 reverse transcriptase dual inhibitor: Mechanistic studies on the allosteric mode of action of isatin-based compound RMNC6. PLoS ONE 2016, 11, e0147225. [Google Scholar] [CrossRef]
- Meleddu, R.; Distinto, S.; Corona, A.; Bianco, G.; Cannas, V.; Esposito, F.; Artese, A.; Alcaro, S.; Matyus, P.; Bogdan, D.; et al. (3Z)-3-(2-[4-(aryl)-1,3-thiazol-2-yl]hydrazin-1-ylidene)-2,3-dihydro-1H-indol-2-one derivatives as dual inhibitors of HIV-1 reverse transcriptase. Eur. J. Med. Chem. 2015, 93, 452–460. [Google Scholar] [CrossRef]
- Distinto, S.; Maccioni, E.; Meleddu, R.; Corona, A.; Alcaro, S.; Tramontano, E. Molecular aspects of the RT/drug interactions. Perspective of dual inhibitors. Curr. Pharm. Des. 2013, 19, 1850–1859. [Google Scholar] [CrossRef]
- Corona, A.; Meleddu, R.; Esposito, F.; Distinto, S.; Bianco, G.; Maccioni, E.; Le Grice, S.S.; Tramontano, E. Site directed mutagenesis studies on HIV-1 reverse transcriptase (RT) shed light on the mechanism of action of a new ribonuclease H/ DNA polymerase RT dual inhibitor. Retrovirology 2013, 10, P19. [Google Scholar] [CrossRef][Green Version]
- Esposito, F.; Mandrone, M.; del Vecchio, C.; Carli, I.; Distinto, S.; Corona, A.; Lianza, M.; Piano, D.; Tacchini, M.; Maccioni, E.; et al. Multi-target activity of Hemidesmus indicus decoction against innovative HIV-1 drug targets and characterization of Lupeol mode of action. Pathog. Dis. 2017, 75, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Tramontano, E.; Esposito, F.; Badas, R.; Di Santo, R.; Costi, R.; La Colla, P. 6-[1-(4-Fluorophenyl)methyl-1H-pyrrol-2-yl)]-2,4-dioxo-5-hexenoic acid ethyl ester a novel diketo acid derivative which selectively inhibits the HIV-1 viral replication in cell culture and the ribonuclease H activity in vitro. Antivir. Res. 2005, 65, 117–124. [Google Scholar] [CrossRef]
- Sarafianos, S.G.; Das, K.; Hughes, S.H.; Arnold, E. Taking aim at a moving target: Designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr. Opin. Struct. Biol. 2004, 14, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Hah, J.-M.; Cho, A.E. Correlation between performance of QM/MM docking and simple classification of binding sites. J. Chem. Inf. Model. 2009, 49, 2382–2387. [Google Scholar] [CrossRef]
- Ellingson, S.R.; Miao, Y.; Baudry, J.; Smith, J.C. Multi-conformer ensemble docking to difficult protein targets. J. Phys. Chem. B 2014, 119, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Esnouf, R.; Garman, E.; Somers, D.; Ross, C.; Kirby, I.; Keeling, J.; Darby, G.; Jones, Y.; Stuart, D.; et al. High-resolution structures of HIV-1 RT from four RT–inhibitor complexes. Nat. Struct. Mol. Biol. 1995, 2, 293–302. [Google Scholar] [CrossRef]
- Das, K.; Bauman, J.D.; Clark, A.D.; Frenkel, Y.V.; Lewi, P.J.; Shatkin, A.J.; Hughes, S.H.; Arnold, E. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations. Proc. Natl. Acad. Sci. USA 2008, 105, 1466–1471. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Nichols, C.; Bird, L.E.; Fujiwara, T.; Sugimoto, H.; Stuart, D.; Stammers, D.K. Binding of the second-generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding. J. Biol. Chem. 2000, 275, 14316–14320. [Google Scholar] [CrossRef]
- Das, K.; Bauman, J.D.; Rim, A.S.; Dharia, C.; Clark, A.D., Jr.; Camarasa, M.J.; Balzarini, J.; Arnold, E. Crystal structure of tert-butyldimethylsilyl-spiroaminooxathioledioxide-thymine (TSAO-T) in complex with HIV-1 reverse transcriptase (RT) redefines the elastic limits of the non-nucleoside inhibitor-binding pocket. J. Med. Chem. 2011, 54, 2727–2737. [Google Scholar] [CrossRef]
- Pata, J.; Stirtan, W.G.; Goldstein, S.W.; Steitz, T.A. Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant reverse transcriptases resistant to other nonnucleoside inhibitors. Proc. Natl. Acad. Sci. USA 2004, 101, 10548–10553. [Google Scholar] [CrossRef]
- Su, H.-P.; Yan, Y.; Prasad, G.S.; Smith, R.F.; Daniels, C.L.; Abeywickrema, P.D.; Reid, J.C.; Loughran, H.M.; Kornienko, M.; Sharma, S.; et al. Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors. J. Virol. 2010, 84, 7625–7633. [Google Scholar] [CrossRef]
- Corona, A.; Di Leva, F.S.; Thierry, S.; Pescatori, L.; Crucitti, G.C.; Subra, F.; Delelis, O.; Esposito, F.; Rigogliuso, G.; Costi, R.; et al. Identification of highly conserved residues involved in inhibition of HIV-1 RNase H function by diketo acid derivatives. Antimicrob. Agents Chemother. 2014, 58, 6101–6110. [Google Scholar] [CrossRef]
- Suchaud, V.; Bailly, F.; Lion, C.; Tramontano, E.; Esposito, F.; Corona, A.; Christ, F.; Debyser, Z.; Cotelle, P. Development of a series of 3-hydroxyquinolin-2(1H)-ones as selective inhibitors of HIV-1 reverse transcriptase associated RNase H activity. Bioorganic Med. Chem. Lett. 2012, 22, 3988–3992. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.; Rocca, R.; Corona, A.; Grandi, N.; Moraca, F.; Romeo, I.; Talarico, C.; Gagliardi, M.G.; Ambrosio, F.A.; Ortuso, F.; et al. Novel natural non-nucleoside inhibitors of HIV-1 reverse transcriptase identified by shape- and structure-based virtual screening techniques. Eur. J. Med. Chem. 2019, 161, 1–10. [Google Scholar] [CrossRef]
- Mohamadi, F.; Richards, N.G.J.; Guida, W.C.; Liskamp, R.; Lipton, M.; Caufield, C.; Chang, G.; Hendrickson, T.; Still, W.C. Macromodel—An integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 1990, 11, 440–467. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J. Comput. Chem. 1996, 17, 520–552. [Google Scholar] [CrossRef]
- Still, W.C.; Tempczyk, A.; Hawley, R.C.; Hendrickson, T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990, 112, 6127–6129. [Google Scholar] [CrossRef]
- QM-Polarized Protocol; Schrodinger LLC: New York, NY, USA, 2021.
- Wolber, G.; Dornhofer, A.A.; Langer, T. Efficient overlay of small organic molecules using 3D pharmacophores. J. Comput. Mol. Des. 2006, 20, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Wolber, G.; Langer, T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2004, 45, 160–169. [Google Scholar] [CrossRef]


| Compound | R | R’ | RNase H a IC50 (µM) | RDDP b IC50 (µM) |
|---|---|---|---|---|
| EMAC2056 | H | 4-OCH3 | 19.5 ± 5.5 | 36.0 ± 5.0 |
| EMAC2057 | H | 3,4-Cl | 57.0 ± 5.0 | 88.0 ± 12.0 |
| EMAC2058 | H | 4-Cl | 46.0 ± 1.0 | 56.5 ± 9.5 |
| EMAC2059 | H | 4-NO2 | 12.8 ± 4.3 | 39.5 ± 11.5 |
| EMAC2060 | 2-CONH2 | 4-OCH3 | 21.5 ± 3.5 | 27.0 ± 1.0 |
| EMAC2061 | 2-CONH2 | 3,4-Cl | 6.5 ± 2.5 | 19.5 ± 0.5 |
| EMAC2062 | 2-CONH2 | 4-Cl | 8.0 ± 2.0 | 17.0 ± 2.0 |
| EMAC2063 | 2-CONH2 | 4-NO2 | 4.5 ± 0.5 | 8.0 ± 0.0 |
| EMAC2064 | 2-Br | 4-OCH3 | 13.0 ± 4.0 | 32.5 ± 2.5 |
| EMAC2065 | 2-Br | 3,4-Cl | 15.0 ± 1.0 | 53.5 ± 15.5 |
| EMAC2066 | 2-Br | 4-Cl | 47.0 ± 3.0 | 71.5 ± 6.5 |
| EMAC2067 | 2-Br | 4-NO2 | 27.0 ± 5.0 | 30.0 ± 3.0 |
| EMAC2068 | 4-F | 4-OCH3 | 29.5 ± 3.5 | 61.5 ± 8.5 |
| EMAC2069 | 4-F | 3,4-Cl | 52.5 ± 10.5 | 66.0 ± 2.0 |
| EMAC2070 | 4-F | 4-Cl | 55.5 ± 1.5 | 59.0 ± 6.0 |
| EMAC2071 | 4-F | 4-NO2 | 19.0 ± 1.0 | 47.5 ± 11.5 |
| Efavirenz | // | // | * | 0.023 ± 0.002 |
| β-Thujaplicinol | // | // | 0.19 ± 0.03 | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meleddu, R.; Corona, A.; Distinto, S.; Cottiglia, F.; Deplano, S.; Sequeira, L.; Secci, D.; Onali, A.; Sanna, E.; Esposito, F.; et al. Exploring New Scaffolds for the Dual Inhibition of HIV-1 RT Polymerase and Ribonuclease Associated Functions. Molecules 2021, 26, 3821. https://doi.org/10.3390/molecules26133821
Meleddu R, Corona A, Distinto S, Cottiglia F, Deplano S, Sequeira L, Secci D, Onali A, Sanna E, Esposito F, et al. Exploring New Scaffolds for the Dual Inhibition of HIV-1 RT Polymerase and Ribonuclease Associated Functions. Molecules. 2021; 26(13):3821. https://doi.org/10.3390/molecules26133821
Chicago/Turabian StyleMeleddu, Rita, Angela Corona, Simona Distinto, Filippo Cottiglia, Serenella Deplano, Lisa Sequeira, Daniela Secci, Alessia Onali, Erica Sanna, Francesca Esposito, and et al. 2021. "Exploring New Scaffolds for the Dual Inhibition of HIV-1 RT Polymerase and Ribonuclease Associated Functions" Molecules 26, no. 13: 3821. https://doi.org/10.3390/molecules26133821
APA StyleMeleddu, R., Corona, A., Distinto, S., Cottiglia, F., Deplano, S., Sequeira, L., Secci, D., Onali, A., Sanna, E., Esposito, F., Cirone, I., Ortuso, F., Alcaro, S., Tramontano, E., Mátyus, P., & Maccioni, E. (2021). Exploring New Scaffolds for the Dual Inhibition of HIV-1 RT Polymerase and Ribonuclease Associated Functions. Molecules, 26(13), 3821. https://doi.org/10.3390/molecules26133821