In Vitro Confirmation of Siramesine as a Novel Antifungal Agent with In Silico Lead Proposals of Structurally Related Antifungals
Abstract
1. Introduction
2. Results and Discussion
2.1. Antifungal Activity of Siramesine on Planktonic and Biofilm-Formed Cells
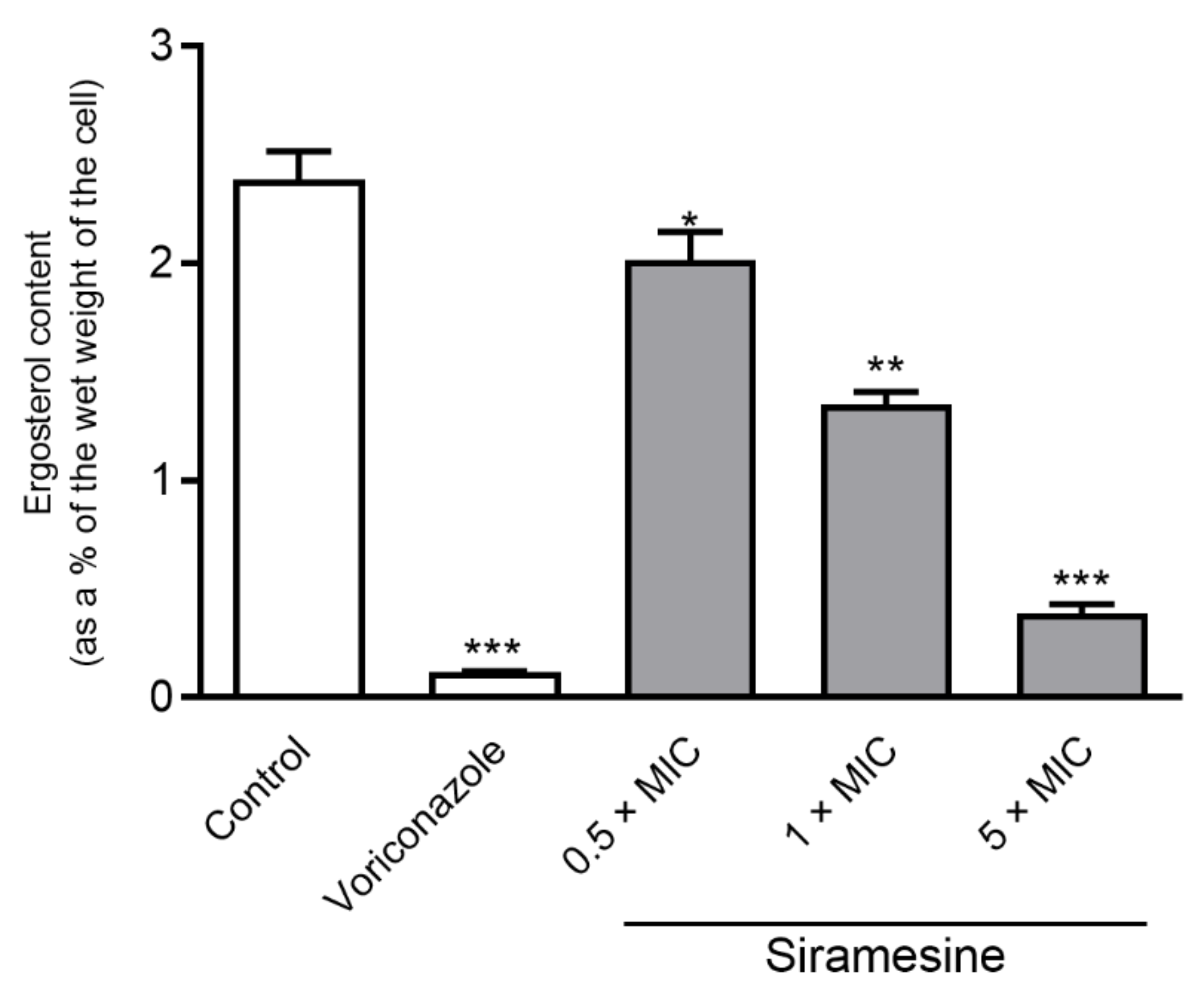
2.2. Ergosterol Content
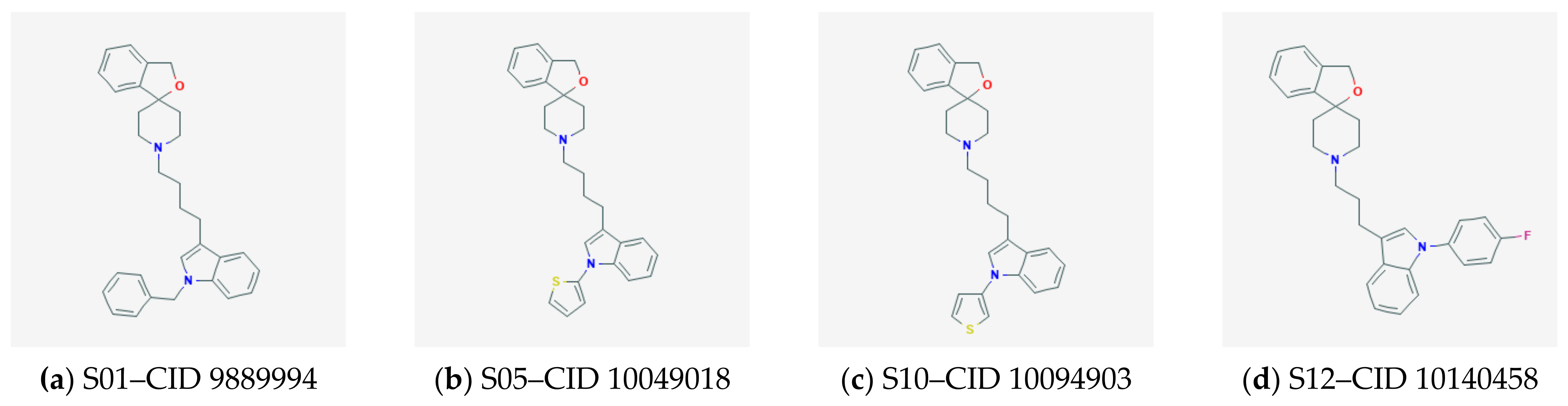
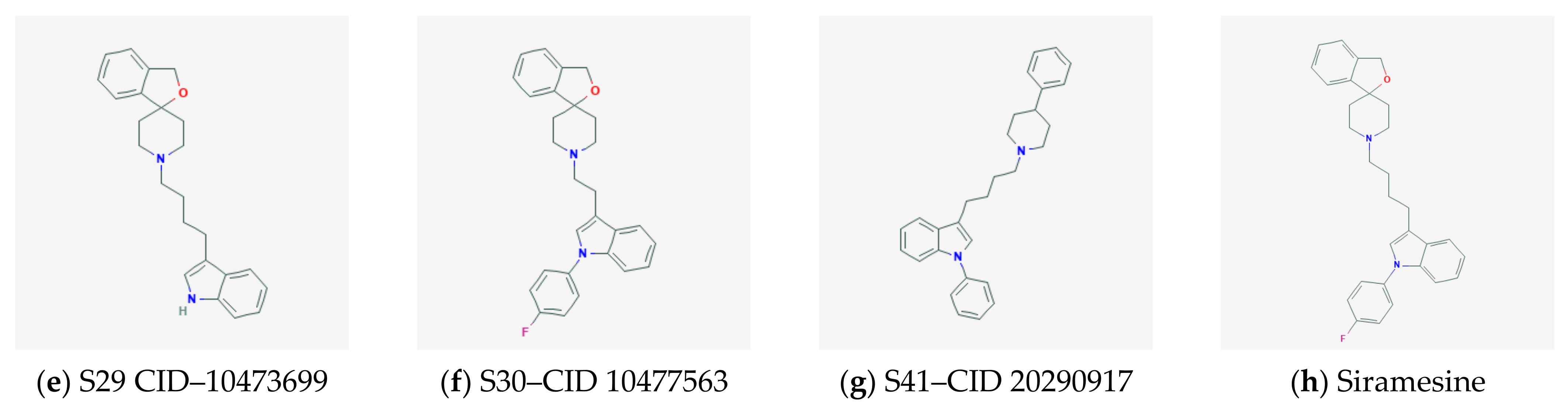
2.3. Docking and QSAR Modeling of Siramesine-Like Compounds
2.3.1. Docking to Sigma-1 Receptor
2.3.2. Docking to Erg2 Target
3. Materials and Methods
3.1. In Vitro Experiments
3.1.1. Determination of Antifungal Susceptibility
3.1.2. Minimum Biofilm Eradication Assay
3.1.3. Modulation of Ergosterol Biosynthesis
3.1.4. Statistical Analysis
3.2. In Silico Study
3.2.1. Homology Modeling
3.2.2. Molecular Docking and Machine Learning: Datasets
3.2.3. Molecular Docking
Sigma-1 Receptor Docking in Autodock4
Sigma-1 Receptor Docking in Gold
Parallel but Independent Molecular Docking Analyses
Sigma-1 Receptor QSAR
Erg2 Receptor Docking in Autodock4
Erg2 Receptor Docking in Gold
Erg2 QSAR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Richardson, M.; Lass-Flörl, C. Changing epidemiology of systemic fungal infections. Clin. Microbiol. Infect. 2008, 14, 5–24. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R. The antifungal pipeline: A reality check. Nat. Rev. Drug. Discov. 2017, 16, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Ademe, M.; Girma, F. Candida auris: From Multidrug Resistance to Pan-Resistant Strains. Infect. Drug Resist. 2020, 13, 1287–1294. [Google Scholar] [CrossRef]
- Lomazzi, M.; Moore, M.; Johnson, A.; Balasegaram, M.; Borisch, B. Antimicrobial resistance—Moving forward? BMC Public Health 2019, 19, 858. [Google Scholar] [CrossRef]
- Farha, M.A.; Brown, E.D. Drug repurposing for antimicrobial discovery. Nat. Microbiol. 2019, 4, 565–577. [Google Scholar] [CrossRef]
- Jović, O.; Šmuc, T. Combined Machine Learning and Molecular Modelling Workflow for the Recognition of Potentially Novel Fungicides. Molecules 2020, 25, 2198. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in us hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef]
- Cuéllar-Cruz, M.; López-Romero, E.; Villagómez-Castro, J.; Ruiz-Baca, E. Candida species: New insights into biofilm formation. Future Microbiol. 2012, 7, 755–771. [Google Scholar] [CrossRef]
- Søby, K.K.; Mikkelsen, J.D.; Meier, E.; Thomsen, C. Lu 28-179 labels a σ(2)-site in rat and human brain. Neuropharmacology 2002, 43, 95–100. [Google Scholar] [CrossRef]
- Ostenfeld, M.S.; Høyer-Hansen, M.; Bastholm, L.; Fehrenbacher, N.; Dines Olsen, O.; Groth-Pedersen, L.; Puustinen, P.; Sørensen, T.K.; Nylandsted, J.; Farkas, T.; et al. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation. Autophagy 2008, 4, 487–499. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef]
- Abe, F.; Hiraki, T. Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim. Biophys. Acta 2009, 1788, 743–752. [Google Scholar] [CrossRef]
- Lu, H.; Shrivastava, M.; Whiteway, M.; Jiang, Y. Candida albicans targets that potentially synerzie with fluconazole. Crit. Rev. Microbiol. 2021. [Google Scholar] [CrossRef]
- Daum, G.; Wagner, A.; Czabany, T.; Grillitsch, K.; Athenstaedt, K. Lipid storage and mobilization pathways in yeast. Novartis Found. Symp. 2007, 286, 142–151. [Google Scholar]
- McLean-Bowen, C.A.; Parks, L.W. Corresponding changes in kynurenine hydroxylase activity, membrane fluidity, and sterol composition in Saccharomyces cerevisiae mitochondria. J. Bacteriol. 1981, 145, 1325–1333. [Google Scholar] [CrossRef]
- Onyewu, C.; Blankenship, J.R.; Del Poeta, M.; Heitman, J. Ergosterol Biosynthesis Inhibitors Become Fungicidal when Combined with Calcineurin Inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob. Agents Chemother. 2003, 47, 956–964. [Google Scholar] [CrossRef]
- Moebius, F.F.; Bermoser, K.; Reiter, R.J.; Hanner, M.; Glossmann, H. Yeast sterol C8-C7 isomerase: Identification and characterization of a high-affinity binding site for enzyme inhibitors. Biochemistry 1996, 35, 16871–16878. [Google Scholar] [CrossRef]
- FRAC Code List 2019: Fungicides Sorted by Mode of Action. Available online: http://www.frac.info/ (accessed on 26 December 2019).
- Velázquez-Libera, J.L.; Rossino, G.; Navarro-Retamal, C.; Collina, S.; Caballero, J. Docking, Interaction Fingerprint, and Three-Dimensional Quantitative Structure–Activity Relationship (3D-QSAR) of Sigma1 Receptor Ligands, Analogs of the Neuroprotective Agent RC-33. Front. Chem. 2019, 7, 20. [Google Scholar] [CrossRef]
- Laggner, C.; Schieferer, C.; Fiechtner, B.; Poles, G.; Hoffmann, R.D.; Glossmann, H.; Langer, T.; Moebius, F.F. Discovery of High-Affinity Ligands of ó1 Receptor, Erg2, and Emopamil Binding Protein by Pharmacophore Modeling and Virtual Screening. J. Med. Chem. 2005, 48, 4754–4764. [Google Scholar] [CrossRef]
- Oyer, H.M.; Sanders, C.M.; Kim, F.J. Small-Molecule Modulators of Sigma1 and Sigma2/TMEM97 in the Context of Cancer: Foundational Concepts and Emerging Themes. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Chahal, K.K.; Oto, P.; Nothias, L.-F.; Debnath, A.; McKerrow, J.H.; Podust, L.M.; Abagyan, R. Identification of Four Amoebicidal Nontoxic Compounds by a Molecular Docking Screen of Naegleria fowleri Sterol Δ8−Δ7- Isomerase and Phenotypic Assays. ACS Infect. Dis. 2019, 5, 2029–2038. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Arthington-Skaggs, B.; Lee, W.; Pierson, C.A.; Lees, N.D.; Eckstein, J.; Barbuch, R.; Bard, M. Candida albicans Sterol C-14 Reductase, Encoded by the ERG24 Gene, as a Potential Antifungal Target Site. Antimicrob. Agents Chemother. 2002, 46, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Hamdy, R.; Fayed, B.; Hamoda, A.M.; Rawas-Qalaji, M.; Haider, M.; Soliman, S.S.M. Essential Oil-Based Design and Development of Novel Anti-Candida Azoles Formulation. Molecules 2020, 25, 1463. [Google Scholar] [CrossRef]
- Donadu, M.G.; Peralta-Ruiz, Y.; Usai, D.; Maggio, F.; Molina-Hernandez, J.B.; Rizzo, D.; Bussu, F.; Rubino, S.; Zanetti, S.; Paparella, A.; et al. Colombian Essential Oil of Ruta graveolens against Nosocomial Antifungal Resistant Candida Strains. J. Fungi 2021, 7, 383. [Google Scholar] [CrossRef]
- Müller-Sepúlveda, A.; Chevecich, C.C.; Jara, J.A.; Belmar, C.; Sandoval, P.; Meyer, R.S.; Quijada, R.; Moura, S.; López-Muñoz, R.; Díaz-Dosque, M.; et al. Chemical Characterization of Lavandula dentata Essential Oil Cultivated in Chile and Its Antibiofilm Effect against Candida albicans. Planta Med. 2020, 86, 1225–1234. [Google Scholar] [CrossRef]
- Zorić, N.; Kosalec, I.; Tomić, S.; Bobnjarić, I.; Jug, M.; Vlainić, T.; Vlainić, J. Membrane of Candida albicans as a target of berberine. BMC Complement. Altern. Med. 2017, 17, 268. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Dierich, M.P.; Fuchs, D.; Semenitz, E.; Jenewein, I.; Ledochowski, M. Antifungal properties of selective serotonin reuptake inhibitors against Aspergillus species in vitro. J. Antimicrob. Chemother. 2001, 48, 775–779. [Google Scholar] [CrossRef]
- Oliveira, A.S.; Martinez-de-Oliveira, J.; Donders, G.G.G.; Palmeira-de-Oliveira, R.; Palmeira-de-Oliveira, A. Anti-Candida activity of antidepressants sertraline and fluoxetine: Effect upon pre-formed biofilms. Med. Microbiol. Immunol. 2018, 207, 195–200. [Google Scholar] [CrossRef]
- Levkovitz, Y.; Ben-Shushan, G.; Hershkovitz, A.; Isaac, R.; Gil-Ad, I.; Shvartsman, D.; Ronen, D.; Weizman, A.; Zick, Y. Antidepressants induce cellular insulin resistance by activation of IRS-1 kinases. Mol. Cell Neurosci. 2007, 36, 305–312. [Google Scholar] [CrossRef]
- Reddy, K.K.; Lefkove, B.; Chen, L.B.; Govindarajan, B.; Carracedo, A.; Velasco, G.; Carrillo, C.O.; Bhandarkar, S.S.; Owens, M.J.; Mechta-Grigoriou, F.; et al. The antidepressant sertraline downregulates Akt and has activity against melanoma cells. Pigment Cell Melanoma Res. 2008, 21, 451–456. [Google Scholar] [CrossRef]
- Kim, J.H.; Cheng, L.W.; Chan, K.L.; Tam, C.C.; Mahoney, N.; Friedman, M.; Shilman, M.M.; Land, K.M. Antifungal Drug Repurposing. Antibiotics 2020, 9, 812. [Google Scholar] [CrossRef]
- Rossi, D.; Rui, M.; Giacomo, M.D.; Schepmann, D.; Wuensch, B.; Monteleone, S.; Liedl, K.R.; Collina, S. Gaining in Pan-Affinity Towards Sigma 1 and Sigma 2 Receptors. SAR studies on arylalkylamines. Bioorg. Med. Chem. 2017, 25, 11–19. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing Determination of Method for the determination of broth dilution minimum inhibitory concentrations (MICs) of antibacterial antifungal agents by broth micro dilution for yeasts. EUCAST Definitive document E. DEF 7.3.2. 2020. EUCAST Discussion Document. Clin. Microbiol. Infect. 2013, 9, 1–10. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_7.3.2_Yeast_testing_definitive_revised_2020.pdf (accessed on 2 February 2021).
- Tommaso, P.D.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.-M.; Taly, J.-F.; Notredame, C. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39, W13–W17. [Google Scholar] [CrossRef]
- Schmidt, H.R.; Zheng, S.; Gurpinar, E.; Koehl, A.; Manglik, A.; Kruse, A.C. Crystal structure of the human σ1 receptor. Nat. Lett. 2016, 532, 527–530. [Google Scholar] [CrossRef]
- Centner, V.; Massart, D.L.; De Noord, O.E.; De Jong, S.; Vandeginste, B.M.; Sterna, C. Elimination of Uninformative Variables for Multivariate Calibration. Anal. Chem. 1996, 68, 3851–3858. [Google Scholar] [CrossRef]
- Heading, C. Siramesine H Lundbeck. Curr. Opin. Investig. Drugs 2001, 2, 266–270. [Google Scholar]


| Fungal Species | Talarozole | Ozagrel | Siramesine | L-778123 | MBX2982 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC * | MBEC ** | MIC | MBEC | MIC | MBEC | MIC | MBEC | MIC | MBEC | |
| Candida albicans ATCC 10231 | 250 | 500 | 250 | 500 | 12.5 | 50 | 250 | 500 | 250 | 500 |
| Candida albicans ATCC 90028 | 250 | 500 | 250 | 500 | 12.5 | 50 | 250 | 500 | 250 | 500 |
| Candida tropicalis ATCC 750 | 250 | 500 | 250 | 500 | 12.5 | 62.5 | 250 | 500 | >250 | >500 |
| Candida kefyr ATCC 2512 | 250 | 500 | 250 | 500 | 12.5 | 62.5 | 250 | 500 | >250 | >500 |
| Candida parapsylosis ATCC 22019 | 250 | 500 | 250 | 500 | 12.5 | 50 | 250 | 500 | >250 | >500 |
| Candida krusei ATCC 14243 | 250 | 500 | 250 | 500 | 12.5 | 50 | 250 | 500 | 250 | 500 |
| Issatchenkia orientalis ATCC 6258 | >250 | >500 | >250 | >500 | >250 | >500 | >250 | >500 | >250 | >500 |
| Aspergillus brasiliensis ATCC 16404 | >250 | NP *** | >250 | NP | >250 | NP | >250 | NP | >250 | NP |
| Comp | Exp pKi | Gold pKi | Adt pKi |
|---|---|---|---|
| 3 | 7.41 | 7.39 | 7.59 |
| 8 | 6 | 5.83 | 6.24 |
| 13 | 7.72 | 7.15 | 7.29 |
| 18 | 8.1 | 8.38 | 8.31 |
| 23 | 8.27 | 8.91 | 8.76 |
| 28 | 9.07 | 8.24 | 8.32 |
| 33 | 8.89 | 8.67 | 8.68 |
| 38 | 8.11 | 7.91 | 7.93 |
| 43 | 6 | 6.63 | 6.30 |
| 48 | 6 | 6.58 | 6.74 |
| 53 | 7.2 | 6.78 | 7.35 |
| 58 | 8.16 | 8.65 | 8.89 |
| 63 | 7.12 | 7.25 | 7.01 |
| 68 | 6.37 | 6.75 | 6.89 |
| 73 | 8 | 7.89 | 7.89 |
| 78 | 7.21 | 7.18 | 7.47 |
| Pentazocine | 7.66 | 8.18 | 8.36 |
| Data | pKi Range | No. var | LV | RMSEC | R2 (Train) | RMSECV | RMSEP | R2 (val/CVtr) |
|---|---|---|---|---|---|---|---|---|
| σ1 Gold | 6.0–9.16 | 99 | 4 | 0.401 | 0.821 | 0.514 | 0.436 | 0.788 (val) |
| σ1 Adt | 6.0–9.16 | 140 | 4 | 0.397 | 0.824 | 0.526 | 0.436 | 0.813 (val) |
| Erg2 Gold | 4.0–10.3 | 191 | 8 | 0.380 | 0.961 | 0.832 | - * | 0.774 * (CVtr) |
| Erg2 Adt | 4.0–10.3 | 162 | 9 | 0.284 | 0.979 | 0.697 | - * | 0.841 * (CVtr) |
| Comp | Gold pKi | Adt pKi | Comp | Gold pKi | Adt pKi | Comp | Gold pKi | Adt pKi |
|---|---|---|---|---|---|---|---|---|
| Siramesine | 7.24 | 7.22 | S21 | 7.40 | 7.23 | S45 * (G&A) | 8.48 | 8.21 |
| S01 * (G&A) | 8.19 | 8.21 | S22 * (A) | 7.51 | 7.72 | S46 | 7.46 | 7.26 |
| S02 * (G&A) | 8.28 | 8.14 | S23 | 6.68 | 7.06 | S47 * (G&A) | 7.73 | 7.83 |
| S03 | 6.32 | 6.89 | S24 * (G&A) | 7.88 | 7.90 | S48 * (G&A) | 7.70 | 7.99 |
| S04 | 6.95 | 7.05 | S25 * (G&A) | 7.80 | 7.75 | S49 | 7.14 | 7.32 |
| S05 * (G&A) | 8.08 | 7.88 | S26 | 7.08 | 7.15 | S54 | 6.97 | 7.35 |
| S06 | 7.25 | 7.44 | S27 | 7.07 | 7.17 | S55 | 7.25 | 7.24 |
| S07 | 7.34 | 7.66 | S28 * (G&A) | 7.76 | 7.87 | S56 | 7.19 | 7.26 |
| S08 * (G&A) | 8.22 | 8.14 | S29 * (G&A) | 8.25 | 7.85 | S57 | 7.14 | 7.27 |
| S09 | 7.31 | 7.40 | S30 * (G&A) | 7.81 | 7.93 | S58 | 7.04 | 6.98 |
| S10 * (G&A) | 8.20 | 8.45 | S31 | 6.50 | 7.30 | S59 | 6.77 | 6.88 |
| S11 | 7.27 | 7.19 | S33 | 7.15 | 7.39 | S62 | 6.62 | 7.22 |
| S12 * (G&A) | 8.05 | 8.08 | S34 | - | 7.32 | S63 | 6.61 | 7.28 |
| S13 | 7.08 | 7.41 | S36 * (G) | 7.70 | 7.57 | S64 | 6.65 | 6.64 |
| S14 * (G&A) | 7.77 | 8.06 | S37 | 7.15 | 7.22 | S65 | 7.48 | 7.46 |
| S15 | 7.33 | 7.52 | S38 * (G&A) | 7.71 | 7.75 | S71 | 7.67 | 7.19 |
| S16 | 6.82 | 7.20 | S39 * (G&A) | 8.17 | 7.95 | S72 | 7.66 | 7.42 |
| S17 * (G) | 7.77 | 7.62 | S40 | 7.01 | 7.17 | S80 * (G&A) | 8.58 | 8.59 |
| S18 | 7.35 | 7.22 | S41 * (G&A) | 8.17 | 8.17 | S84 * (A) | 7.19 | 7.78 |
| S19 | 6.81 | 6.80 | S43 | 7.54 | 7.54 | |||
| S20 * (G&A) | 8.49 | 8.63 | S44 | 7.05 | 7.00 | Stand. err. | 0.44 | 0.44 |
| Compound | Exp pKi | Gold pKi | Adt pKi | Compound | Exp pKi | Gold pKi | Adt pKi |
|---|---|---|---|---|---|---|---|
| 1,3-di-o-tolylguanidine | 5.7 | 6.82 | 5.73 | MDL-28815 | 9.4 | 8.68 | 9.37 |
| carbisocaine | 5.5 | 6.84 | 5.79 | Pentazocine | 6 | 5.33 | 6.36 |
| AY-9944 | 7.19 | 7.84 | 7.89 | jervine | 6.55 | 5.94 | 5.90 |
| buflumedil | 5.15 | 2.85 | 3.67 | progesterone | 5.35 | 5.29 | 5.62 |
| BM-15766 | 4.78 | 6.38 | 6.97 | opipramol | 7.77 | 6.22 | 6.12 |
| azidopamil * | 5 * | 5.73 | 4.99 | SR-31747 | 8.05 | 8.54 | 8.20 |
| amiodarone | 7.21 | 6.85 | 6.75 | ronipamil | 7.89 | 7.84 | 7.48 |
| 3-PPP | 5.92 | 6.20 | 6.43 | terbinafine * | 4.3 * | 5.95 | 5.17 |
| enclomiphene | 6.8 | 7.76 | 8.13 | tamoxifene | 5.82 | 6.20 | 5.85 |
| CP-74932-4 | 4.5 | 4.89 | 3.61 | raloxifene | 7.18 | 7.23 | 7.38 |
| Haloperidol | 9.3 | 9.25 | 8.96 | solanidine * | 4 * | 3.78 | 3.62 |
| Emopamil | 7.19 | 7.52 | 7.14 | solasodine | 5.93 | 6.05 | 5.87 |
| fenpropimorph | 10.3 | 9.71 | 9.14 | testosterone | 5.11 | 5.18 | 4.42 |
| L-690404 | 8.3 | 8.72 | 8.03 | triparanol | 8.7 | 8.47 | 8.31 |
| ifenprodil | 9 | 7.73 | 8.48 | testosterone propionate | 5.72 | 5.23 | 5.69 |
| N-Allylnormetazocine | 5.33 | 5.62 | 5.45 | tridemorph | 10.05 | 10.00 | 10.39 |
| cyclopamine | 6.3 | 6.95 | 7.12 | trifluoperazine | 6.3 | 5.85 | 6.00 |
| naftifine | 6.51 | 4.72 | 5.65 | trifluperidol | 9.82 | 10.04 | 9.62 |
| corticosterone * | 4 * | 4.97 | 5.21 | U-18666A | 9.7 | 9.23 | 10.20 |
| R-59494 | 5.55 | 5.02 | 5.71 | VUF-8410 | 6.77 | 7.38 | 7.17 |
| MDL-5332 | 9.15 | 8.72 | 8.66 | zuclomiphene | 8.7 | 7.69 | 8.57 |
| nafoxidine | 6.63 | 6.95 | 6.54 | tomatidine | 6.41 | 7.30 | 6.50 |
| Comp | Gold pKi | Adt pKi | Comp | Gold pKi | Adt pKi | Comp | Gold pKi | Adt pKi |
|---|---|---|---|---|---|---|---|---|
| Siramesine | 6.63 | 7.13 | S20 | 7.47 * | 7.64 | S39 | 5.89 | 6.51 |
| S01 ** | 7.97 * | 8.43 * | S22 | - | 7.75 | S41 ** | 7.92 * | 8.21 * |
| S02 | 7.15 | 7.10 | S24 | 7.32 | 7.02 | S45 | 6.75 | 6.79 |
| S05 ** | 7.59 * | 7.96 * | S25 | 7.57* | 7.77 | S47 | 7.21 | 5.73 |
| S08 | 7.36 | 6.98 | S28 | 5.97 | 7.04 | S48 | 6.09 | 6.42 |
| S10 ** | 7.55 * | 9.07 * | S29 ** | 7.61 * | 7.93 * | S80 | 7.80 * | 7.83 |
| S12 ** | 7.69 * | 8.05 * | S30 ** | 7.51 * | 8.11 * | S84 | - | 6.72 |
| S14 | 7.06 | 8.32 * | S36 | 7.21 | - | |||
| S17 | 6.29 | - | S38 | 6.63 | 6.91 | Stand. err. | 0.83 | 0.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlainić, J.; Jović, O.; Kosalec, I.; Vugrek, O.; Čož-Rakovac, R.; Šmuc, T. In Vitro Confirmation of Siramesine as a Novel Antifungal Agent with In Silico Lead Proposals of Structurally Related Antifungals. Molecules 2021, 26, 3504. https://doi.org/10.3390/molecules26123504
Vlainić J, Jović O, Kosalec I, Vugrek O, Čož-Rakovac R, Šmuc T. In Vitro Confirmation of Siramesine as a Novel Antifungal Agent with In Silico Lead Proposals of Structurally Related Antifungals. Molecules. 2021; 26(12):3504. https://doi.org/10.3390/molecules26123504
Chicago/Turabian StyleVlainić, Josipa, Ozren Jović, Ivan Kosalec, Oliver Vugrek, Rozelindra Čož-Rakovac, and Tomislav Šmuc. 2021. "In Vitro Confirmation of Siramesine as a Novel Antifungal Agent with In Silico Lead Proposals of Structurally Related Antifungals" Molecules 26, no. 12: 3504. https://doi.org/10.3390/molecules26123504
APA StyleVlainić, J., Jović, O., Kosalec, I., Vugrek, O., Čož-Rakovac, R., & Šmuc, T. (2021). In Vitro Confirmation of Siramesine as a Novel Antifungal Agent with In Silico Lead Proposals of Structurally Related Antifungals. Molecules, 26(12), 3504. https://doi.org/10.3390/molecules26123504









