Modified Nucleosides, Nucleotides and Nucleic Acids via Click Azide-Alkyne Cycloaddition for Pharmacological Applications
Abstract
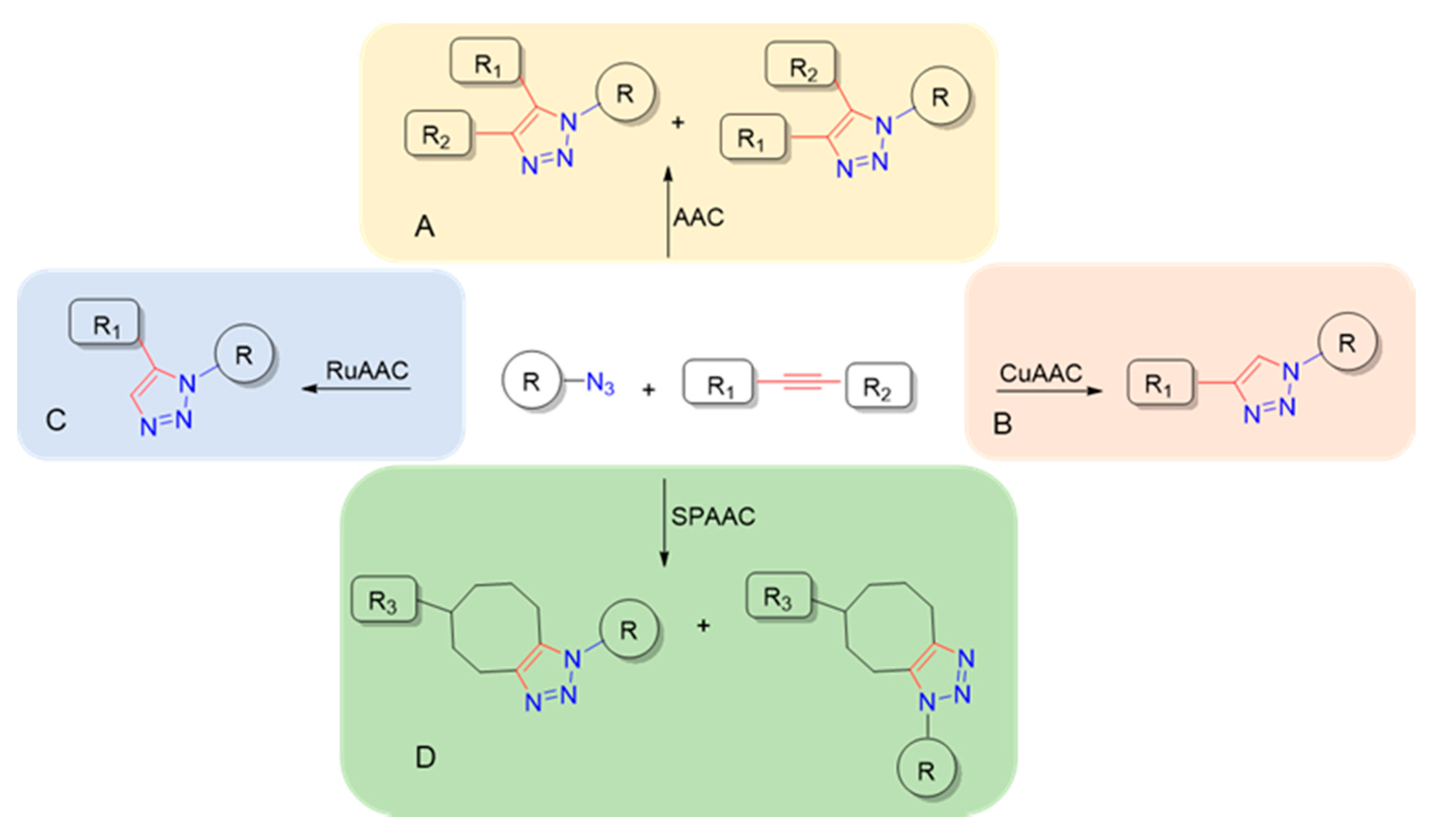
1. Introduction
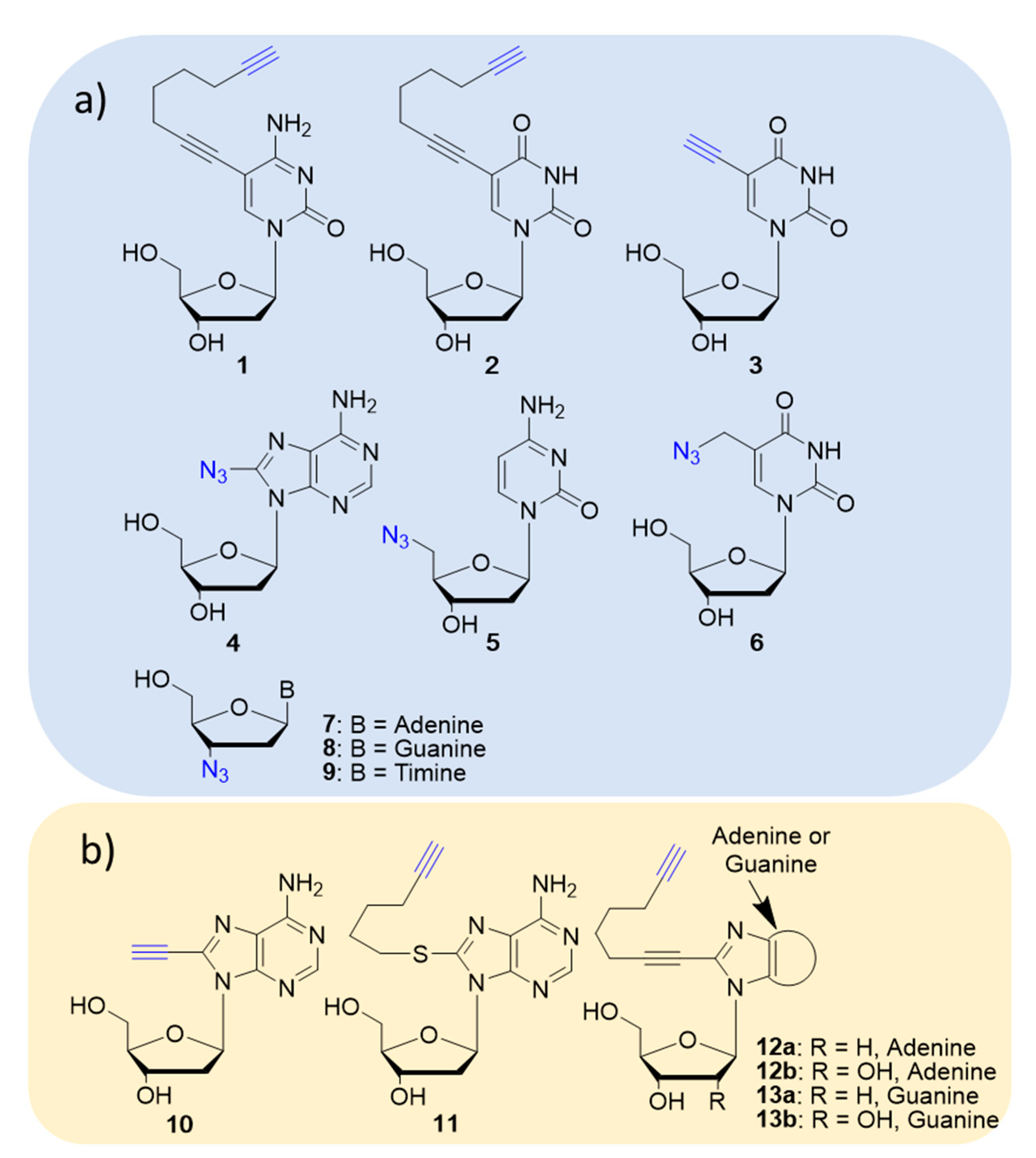
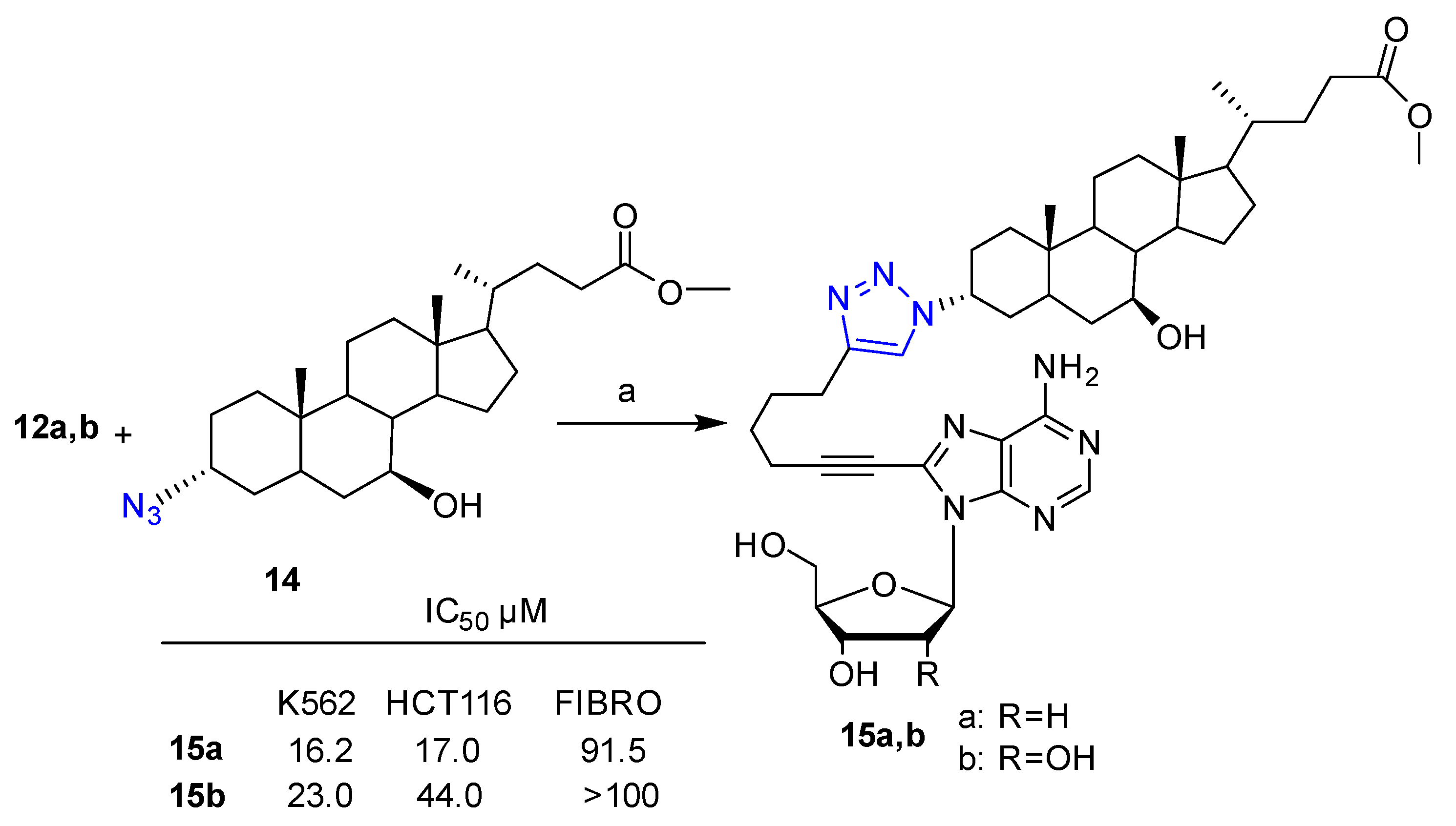
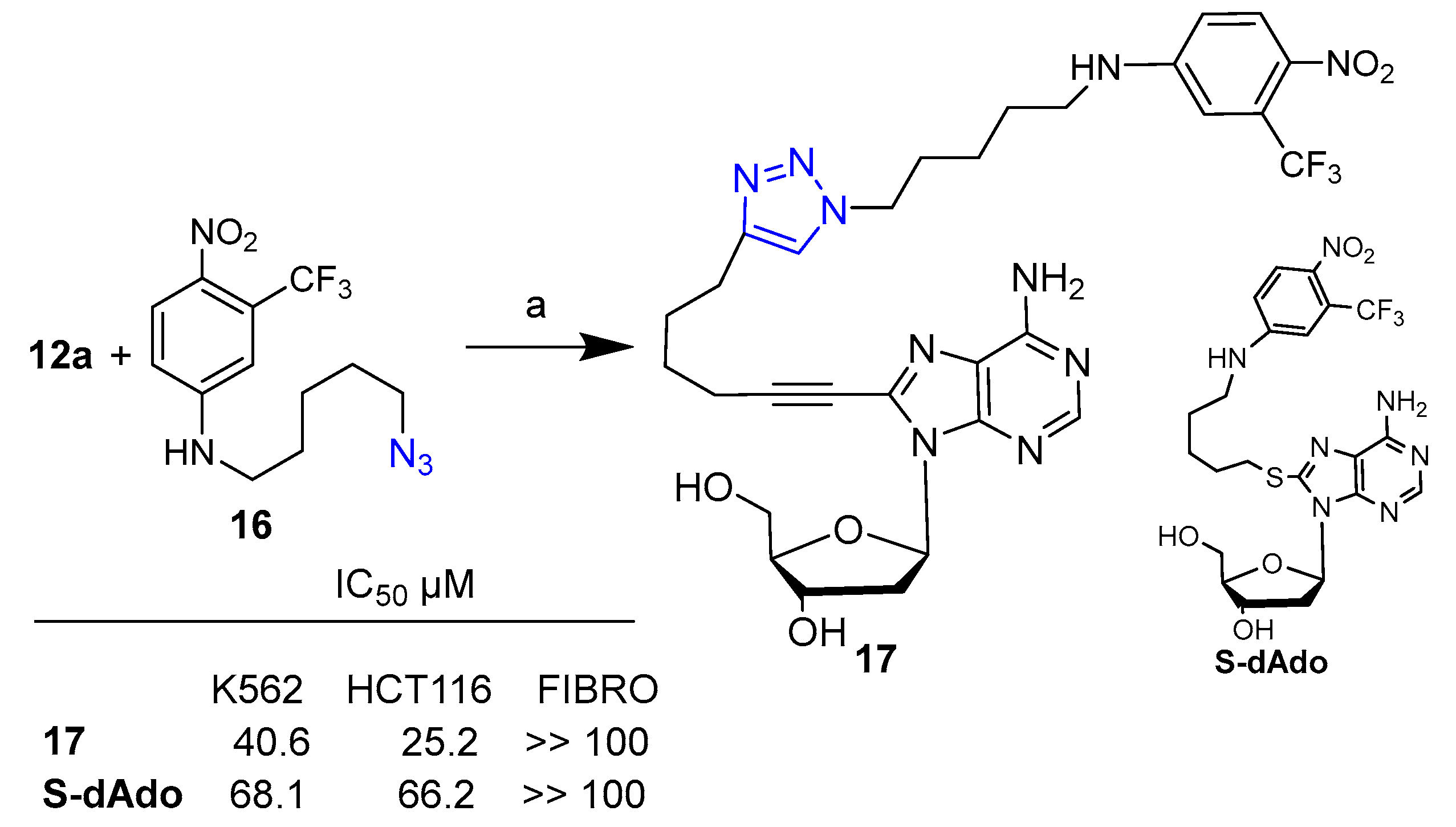
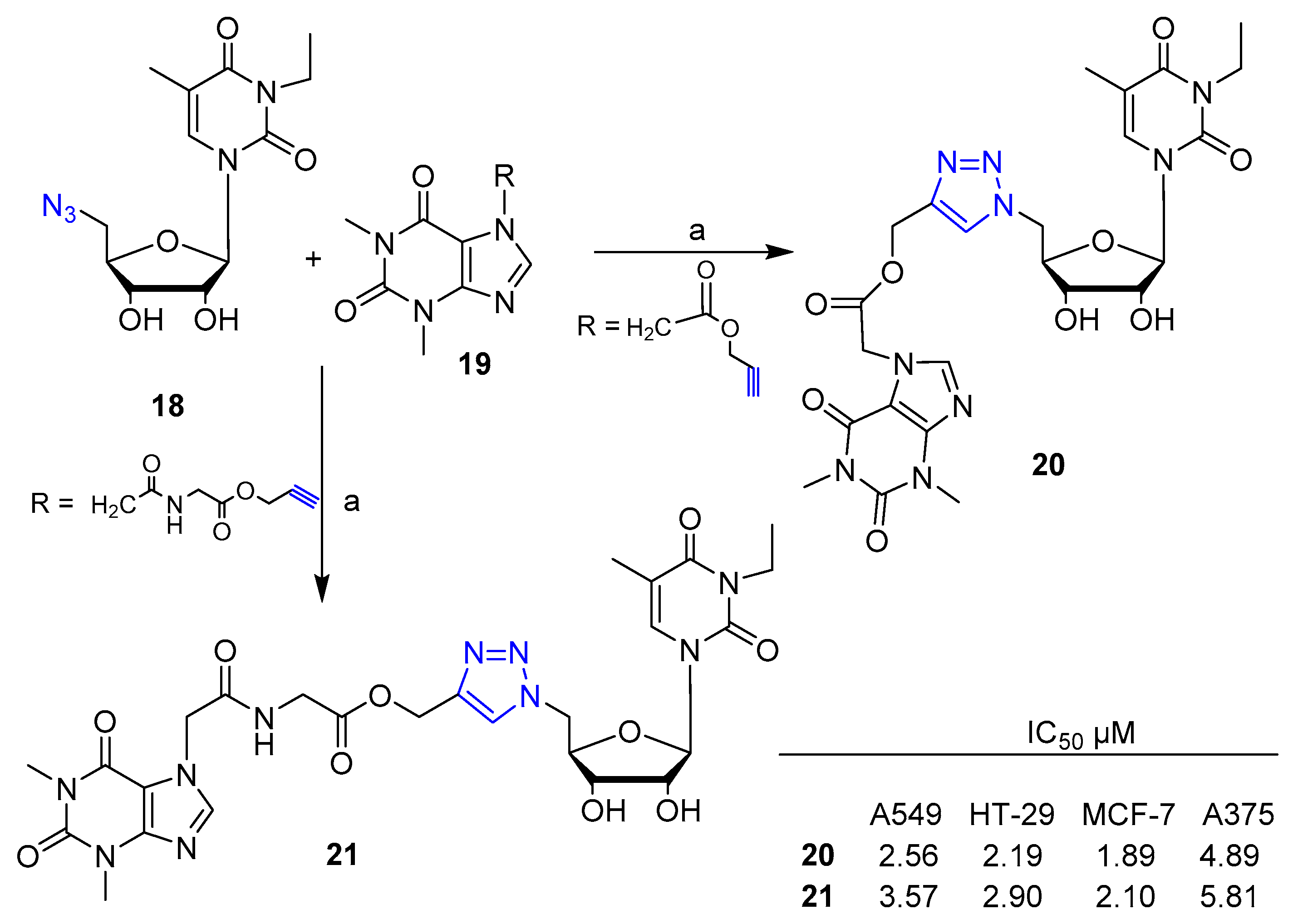
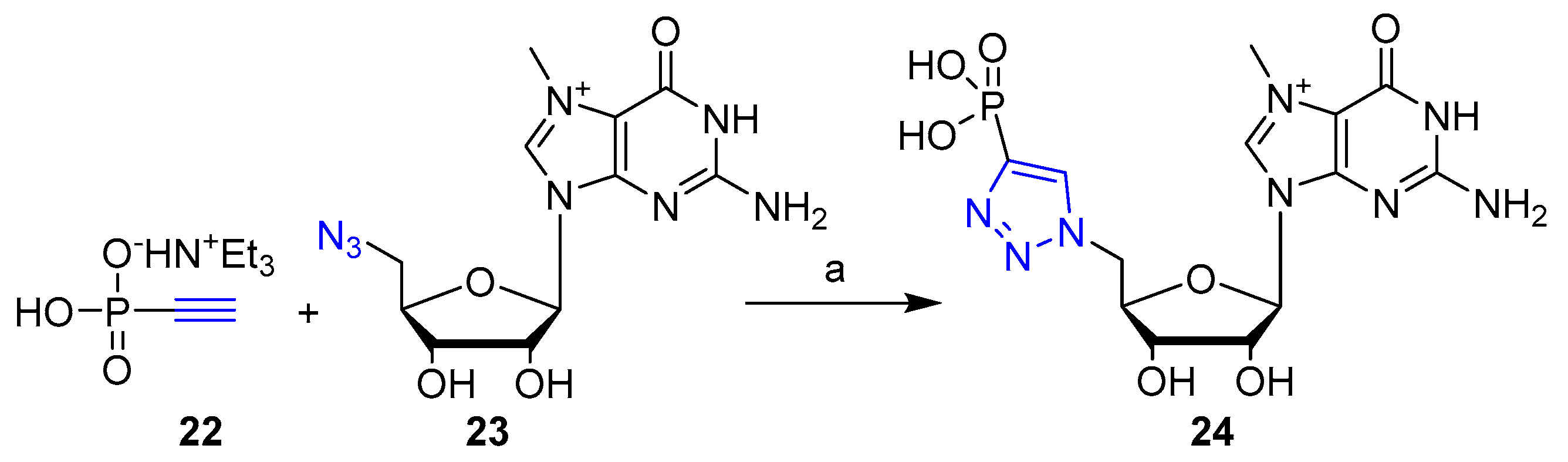
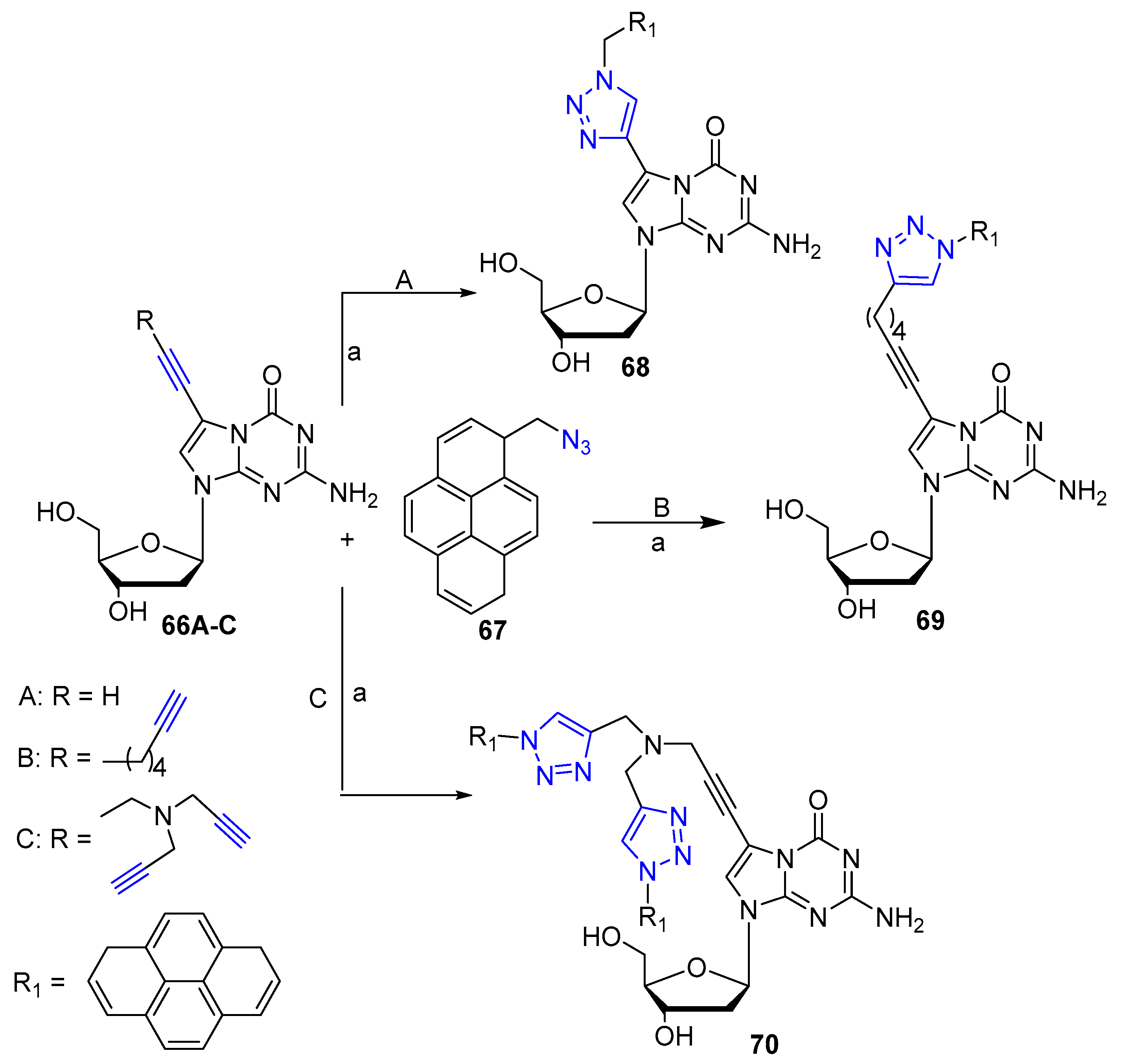
2. Clicked Nucleosides and Nucleotides
3. Clicked Nucleic Acids
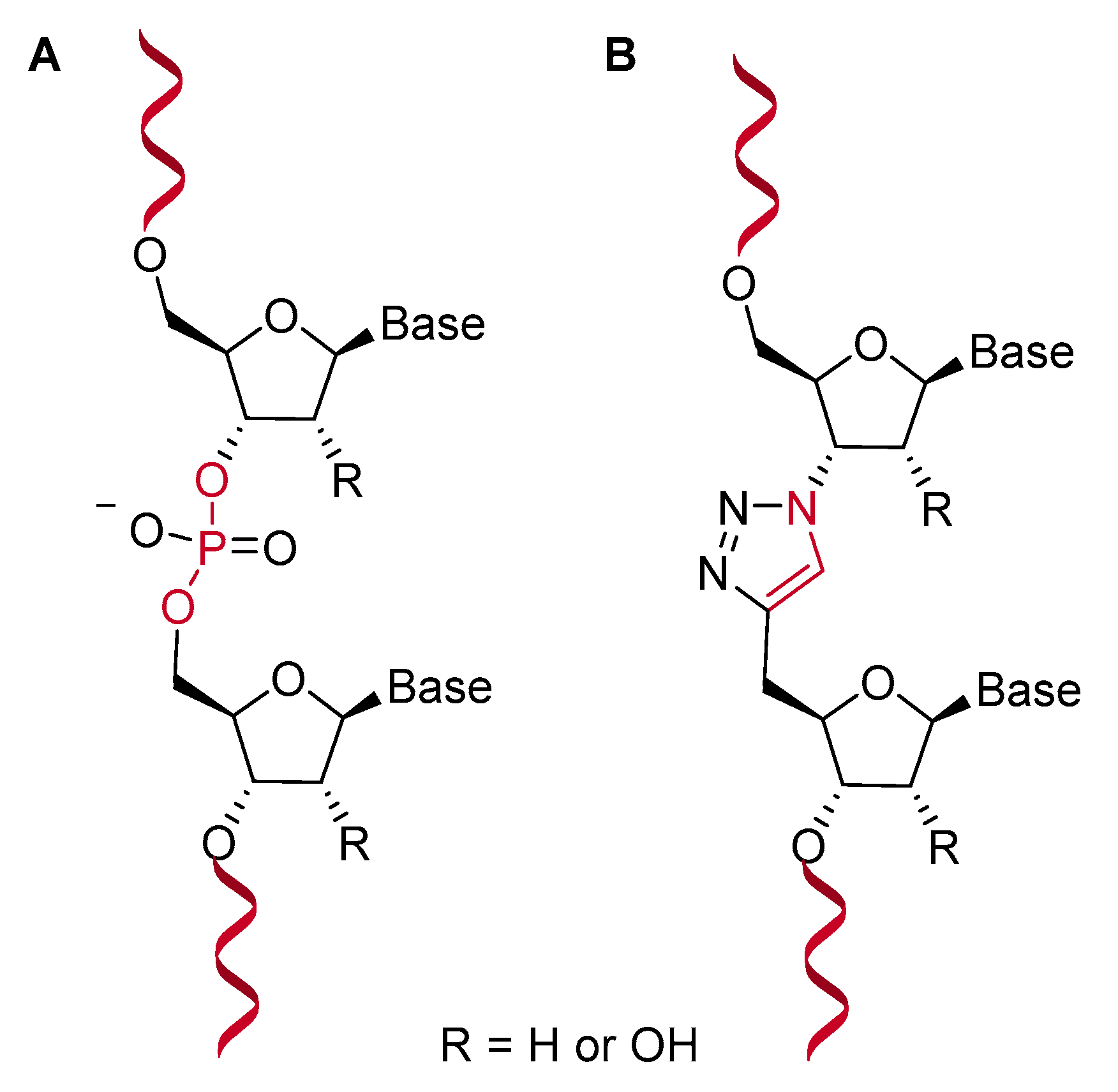
3.1. Triazole-Llinked Nucleic Acid Analogues
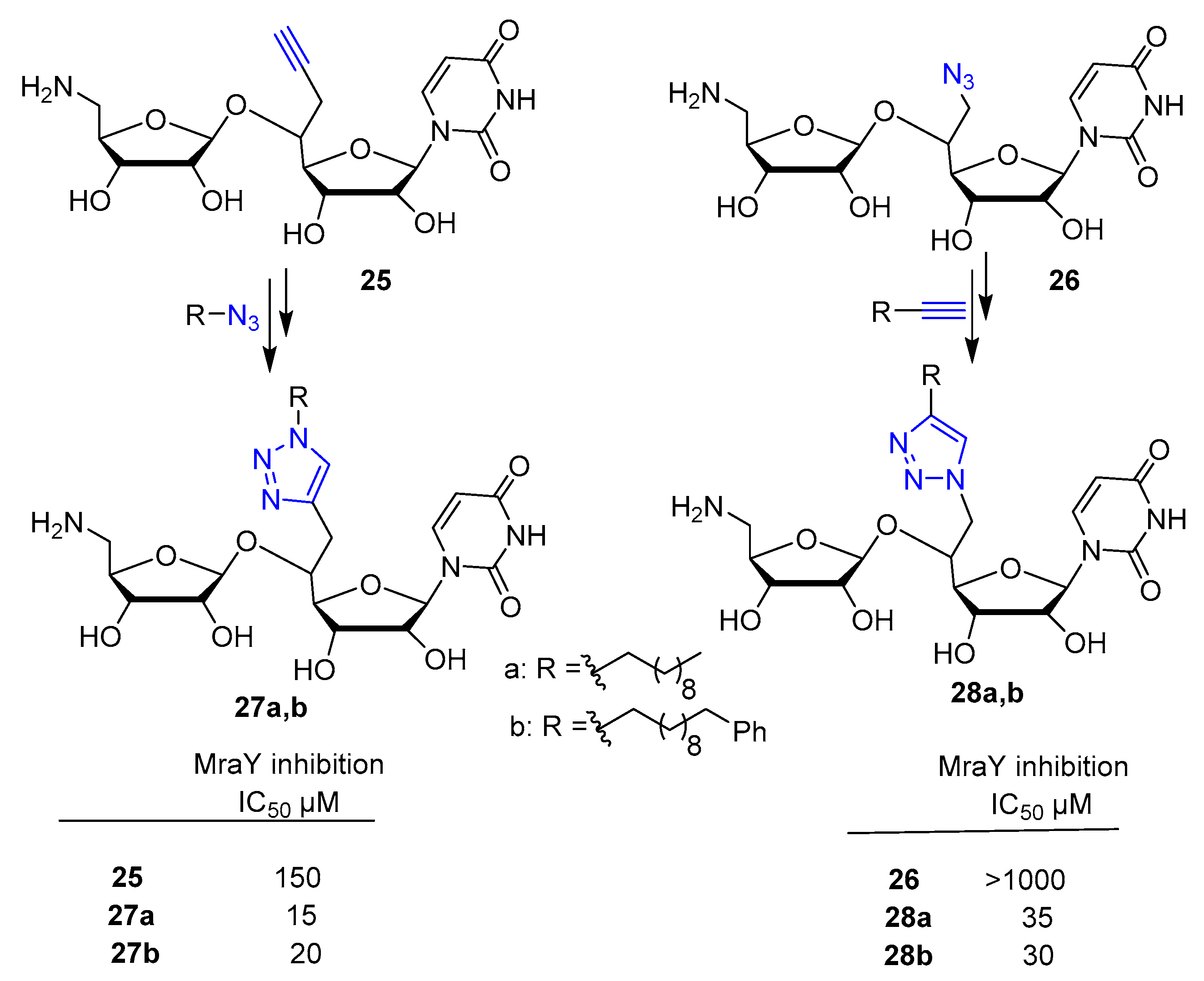
3.2. Nucleic Acid Conjugates via 1,2,3-Triazole Linkers
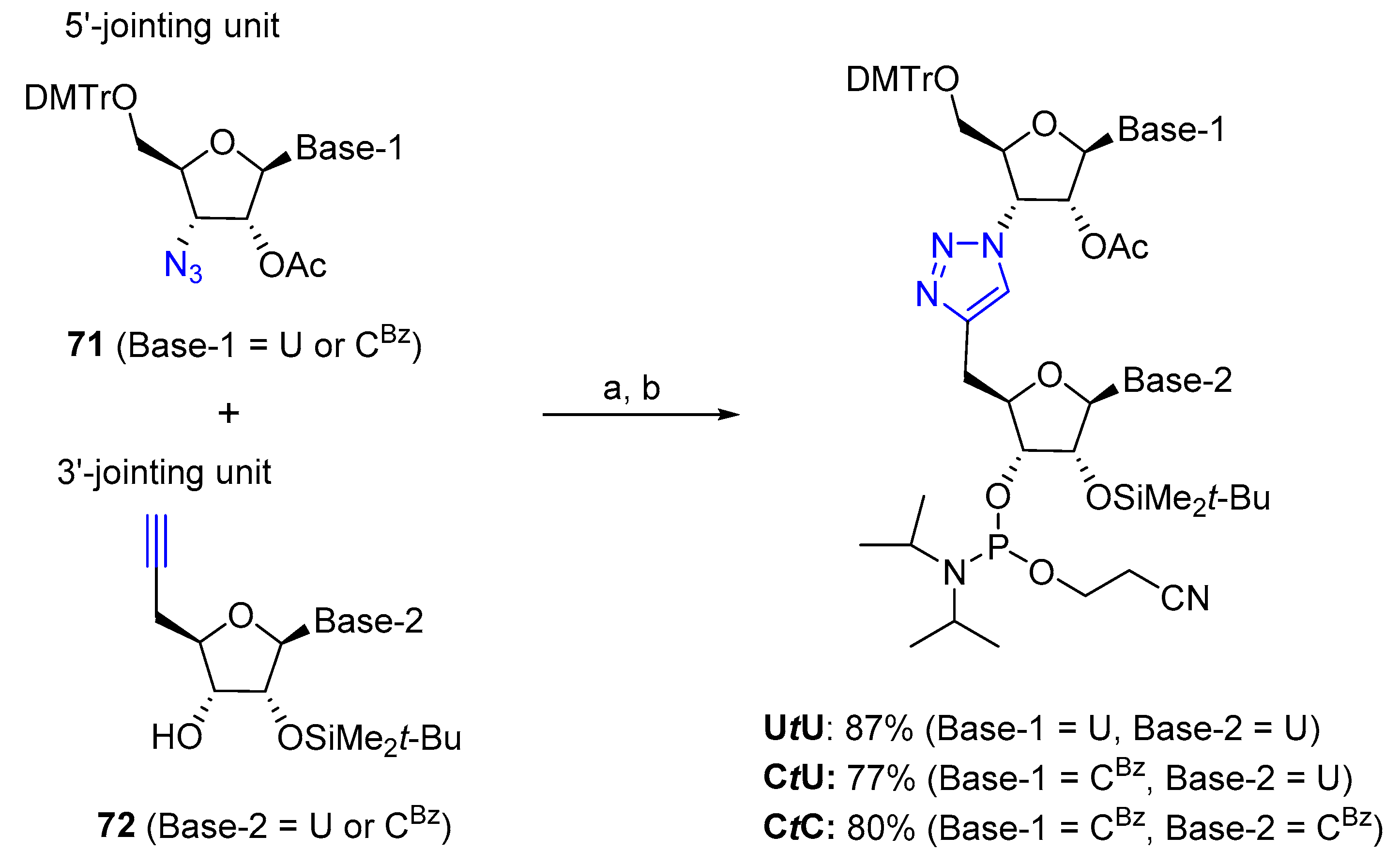
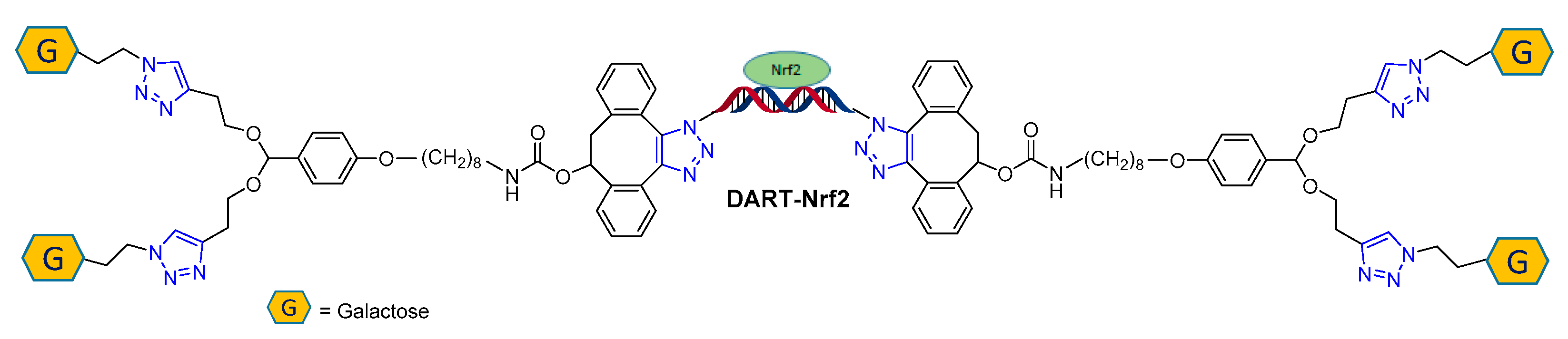
3.2.1. Solid Phase Supported Click Conjugation
3.2.2. Post-Synthetic-Click Conjugation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Tornøe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1,2,3]-Triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef] [PubMed]
- Huisgen, R. 1,3-Dipolar Cycloadditions. Past and Future. Angew. Chem. Int. Ed. Engl. 1963, 2, 565–598. [Google Scholar] [CrossRef]
- Huisgen, R. 1,3-Dipolar cycloaddition chemistry. Volumes 1 and 2. Edited by Albert Padwa. John Wiley and Sons. New York, 1984. Volume 1: XIII + 817 pages. Volume 2: XIII + 704 pages. ISBN 0-471-08364-X (set). $295.00 for the two-volume set. J. Heterocycl. Chem. 1986, 23, 1899. [Google Scholar] [CrossRef]
- Trost, B.M. Atom Economy—A Challenge for Organic Synthesis: Homogeneous Catalysis Leads the Way. Angew. Chem. Int. Ed. Engl. 1995, 34, 259–281. [Google Scholar] [CrossRef]
- Neumann, S.; Biewend, M.; Rana, S.; Binder, W.H. The CuAAC: Principles, Homogeneous and Heterogeneous Catalysts, and Novel Developments and Applications. Macromol. Rapid Commun. 2020, 41, 1900359. [Google Scholar] [CrossRef]
- Meldal, M.; Tornøe, C.W. Cu-Catalyzed Azide−Alkyne Cycloaddition. Chem. Rev. 2008, 108, 2952–3015. [Google Scholar] [CrossRef]
- Sreedhar, B.; Reddy, P.S. Sonochemical synthesis of 1,4-disubstituted 1,2,3-triazoles in aqueous medium. Synth. Commun. 2007, 37, 805–812. [Google Scholar] [CrossRef]
- Pickens, C.J.; Johnson, S.N.; Pressnall, M.M.; Leon, M.A.; Berkland, C.J. Practical Considerations, Challenges, and Limitations of Bioconjugation via Azide–Alkyne Cycloaddition. Bioconjug. Chem. 2018, 29, 686–701. [Google Scholar] [CrossRef]
- Hong, V.; Presolski, S.I.; Ma, C.; Finn, M.G. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew. Chem. Int. Ed. 2009, 48, 9879–9883. [Google Scholar] [CrossRef] [PubMed]
- Kanan, M.W.; Rozenman, M.M.; Sakural, K.; Snyder, T.M.; Liu, D.R. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature 2004, 431, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Besanceney-Webler, C.; Jiang, H.; Zheng, T.; Feng, L.; Soriano Del Amo, D.; Wang, W.; Klivansky, L.M.; Marlow, F.L.; Liu, Y.; Wu, P. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: A comparative study. Angew. Chem. Int. Ed. 2011, 50, 8051–8056. [Google Scholar] [CrossRef]
- Chan, T.R.; Hilgraf, R.; Sharpless, K.B.; Fokin, V.V. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org. Lett. 2004, 6, 2853–2855. [Google Scholar] [CrossRef] [PubMed]
- Abel, G.R.; Calabrese, Z.A.; Ayco, J.; Hein, J.E.; Ye, T. Measuring and Suppressing the Oxidative Damage to DNA during Cu(I)-Catalyzed Azide-Alkyne Cycloaddition. Bioconjug. Chem. 2016, 27, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, X.; Xue, P.; Sun, H.H.Y.; Williams, I.D.; Sharpless, K.B.; Fokin, V.V.; Jia, G. Ruthenium-catalyzed cycloaddition of alkynes and organic azides. J. Am. Chem. Soc. 2005, 127, 15998–15999. [Google Scholar] [CrossRef] [PubMed]
- Johansson, J.R.; Beke-Somfai, T.; Said Stålsmeden, A.; Kann, N. Ruthenium-Catalyzed Azide Alkyne Cycloaddition Reaction: Scope, Mechanism, and Applications. Chem. Rev. 2016, 116, 14726–14768. [Google Scholar] [CrossRef] [PubMed]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef]
- Baskin, J.M.; Prescher, J.A.; Laughlin, S.T.; Agard, N.J.; Chang, P.V.; Miller, I.A.; Lo, A.; Codelli, J.A.; Bertozzi, C.R. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. USA 2007, 104, 16793–16797. [Google Scholar] [CrossRef]
- Bonandi, E.; Christodoulou, M.S.; Fumagalli, G.; Perdicchia, D.; Rastelli, G.; Passarella, D. The 1,2,3-triazole ring as a bioisostere in medicinal chemistry. Drug Discov. Today 2017, 22, 1572–1581. [Google Scholar] [CrossRef]
- Agalave, S.G.; Maujan, S.R.; Pore, V.S. Click Chemistry: 1,2,3-Triazoles as Pharmacophores. Chem. Asian J. 2011, 6, 2696–2718. [Google Scholar] [CrossRef]
- Kharb, R.; Sharma, P.C.; Yar, M.S. Pharmacological significance of triazole scaffold. J. Enzym. Inhib. Med. Chem. 2011, 26, 1–21. [Google Scholar] [CrossRef]
- Malik, M.S.; Ahmed, S.A.; Althagafi, I.I.; Ansari, M.A.; Kamal, A. Application of triazoles as bioisosteres and linkers in the development of microtubule targeting agents. RSC Med. Chem. 2020, 11, 327–348. [Google Scholar] [CrossRef]
- Bozorov, K.; Zhao, J.; Aisa, H.A. 1,2,3-Triazole-containing hybrids as leads in medicinal chemistry: A recent overview. Bioorg. Med. Chem. 2019, 27, 3511–3531. [Google Scholar] [CrossRef]
- Meldal, M.; Diness, F. Recent Fascinating Aspects of the CuAAC Click Reaction. Trends Chem. 2020, 2, 569–584. [Google Scholar] [CrossRef]
- Hou, J.; Liu, X.; Shen, J.; Zhao, G.; Wang, P.G. The impact of click chemistry in medicinal chemistry. Expert Opin. Drug Discov. 2012, 7, 489–501. [Google Scholar] [CrossRef]
- Presolski, S.I.; Hong, V.P.; Finn, M.G. Copper-Catalyzed Azide–Alkyne Click Chemistry for Bioconjugation. Curr. Protoc. Chem. Biol. 2011, 3, 153–162. [Google Scholar] [CrossRef]
- Navacchia, M.; Marchesi, E.; Mari, L.; Chinaglia, N.; Gallerani, E.; Gavioli, R.; Capobianco, M.; Perrone, D. Rational Design of Nucleoside–Bile Acid Conjugates Incorporating a Triazole Moiety for Anticancer Evaluation and SAR Exploration. Molecules 2017, 22, 1710. [Google Scholar] [CrossRef]
- Perrone, D.; Navacchia, M.L. unpublished results.
- Navacchia, M.L.; Fraix, A.; Chinaglia, N.; Gallerani, E.; Perrone, D.; Cardile, V.; Graziano, A.C.E.; Capobianco, M.L.; Sortino, S. NO Photoreleaser-Deoxyadenosine and -Bile Acid Derivative Bioconjugates as Novel Potential Photochemotherapeutics. ACS Med. Chem. Lett. 2016, 7, 939–943. [Google Scholar] [CrossRef]
- Ruddarraju, R.R.; Murugulla, A.C.; Kotla, R.; Chandra Babu Tirumalasetty, M.; Wudayagiri, R.; Donthabakthuni, S.; Maroju, R.; Baburao, K.; Parasa, L.S. Design, synthesis, anticancer, antimicrobial activities and molecular docking studies of theophylline containing acetylenes and theophylline containing 1,2,3-triazoles with variant nucleoside derivatives. Eur. J. Med. Chem. 2016, 123, 379–396. [Google Scholar] [CrossRef]
- Kozarski, M.; Kubacka, D.; Wojtczak, B.A.; Kasprzyk, R.; Baranowski, M.R.; Kowalska, J. 7-Methylguanosine monophosphate analogues with 5′-(1,2,3-triazoyl) moiety: Synthesis and evaluation as the inhibitors of cNIIIB nucleotidase. Bioorg. Med. Chem. 2018, 26, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Fer, M.J.; Bouhss, A.; Patrão, M.; Le Corre, L.; Pietrancosta, N.; Amoroso, A.; Joris, B.; Mengin-Lecreulx, D.; Calvet-Vitale, S.; Gravier-Pelletier, C. 5′-Methylene-triazole-substituted-aminoribosyl uridines as MraY inhibitors: Synthesis, biological evaluation and molecular modeling. Org. Biomol. Chem. 2015, 13, 7193–7222. [Google Scholar] [CrossRef] [PubMed]
- Bodnár, B.; Mernyák, E.; Wölfling, J.; Schneider, G.; Herman, B.; Szécsi, M.; Sinka, I.; Zupkó, I.; Kupihár, Z.; Kovács, L. Synthesis and Biological Evaluation of Triazolyl 13α-Estrone–Nucleoside Bioconjugates. Molecules 2016, 21, 1212. [Google Scholar] [CrossRef] [PubMed]
- Ghirardello, M.; Perrone, D.; Chinaglia, N.; Sádaba, D.; Delso, I.; Tejero, T.; Marchesi, E.; Fogagnolo, M.; Rafie, K.; van Aalten, D.M.F.; et al. UDP-GlcNAc Analogues as Inhibitors of O -GlcNAc Transferase (OGT): Spectroscopic, Computational, and Biological Studies. Chem. Eur. J. 2018, 24, 7264–7272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Y.; Zheng, L.; Huang, X.; Wang, Y.; Chen, C.H.; Cheng, Y.Y.; Morris-Natschke, S.L.; Lee, K.H. Novel Betulinic Acid-Nucleoside Hybrids with Potent Anti-HIV Activity. ACS Med. Chem. Lett. 2020, 11, 2290–2293. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Peng, Y.; Lu, J.; Wang, J.; Ma, H.; Song, C.; Liu, B.; Qiao, Y.; Yu, W.; Wu, J.; et al. Design, synthesis, and biological evaluation of new 1,2,3-triazolo-2′-deoxy-2′-fluoro- 4′-azido nucleoside derivatives as potent anti-HBV agents. Eur. J. Med. Chem. 2018, 143, 137–149. [Google Scholar] [CrossRef]
- Sabat, N.; Migianu-Griffoni, E.; Tudela, T.; Lecouvey, M.; Kellouche, S.; Carreiras, F.; Gallier, F.; Uziel, J.; Lubin-Germain, N. Synthesis and antitumor activities investigation of a C-nucleoside analogue of ribavirin. Eur. J. Med. Chem. 2020, 188, 112009. [Google Scholar] [CrossRef]
- Cobb, A.J.A.; Dell’Isola, A.; Abdulsattar, B.O.; McLachlan, M.M.W.; Neuman, B.W.; Müller, C.; Shankland, K.; Al-Mulla, H.M.N.; Binks, A.W.D.; Elvidge, W. Synthesis and antiviral activity of novel spirocyclic nucleosides. New J. Chem. 2018, 42, 18363–18380. [Google Scholar] [CrossRef]
- Zayas, J.; Annoual, M.; Das, J.K.; Felty, Q.; Gonzalez, W.G.; Miksovska, J.; Sharifai, N.; Chiba, A.; Wnuk, S.F. Strain Promoted Click Chemistry of 2- or 8-Azidopurine and 5-Azidopyrimidine Nucleosides and 8-Azidoadenosine Triphosphate with Cyclooctynes. Application to Living Cell Fluorescent Imaging. Bioconjug. Chem. 2015, 26, 1519–1532. [Google Scholar] [CrossRef]
- Proskurin, G.V.; Orlov, A.A.; Brylev, V.A.; Kozlovskaya, L.I.; Chistov, A.A.; Karganova, G.G.; Palyulin, V.A.; Osolodkin, D.I.; Korshun, V.A.; Aralov, A.V. 3′-O-Substituted 5-(perylen-3-ylethynyl)-2′-deoxyuridines as tick-borne encephalitis virus reproduction inhibitors. Eur. J. Med. Chem. 2018, 155, 77–83. [Google Scholar] [CrossRef]
- Pogula, P.K.; De Chatterjee, A.; Chi, M.; VanKoten, H.W.; Das, S.; Patterson, S.E. Triazoxins: Novel nucleosides with anti-Giardia activity. Bioorg. Med. Chem. Lett. 2020, 30, 127175. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Tuttle, P.R.; Howlader, A.H.; Vasilyeva, A.; Gonzalez, L.; Tangar, A.; Lei, R.; Laverde, E.E.; Liu, Y.; Miksovska, J.; et al. Fluorescent 5-Pyrimidine and 8-Purine Nucleosides Modified with an N-Unsubstituted 1,2,3-Triazol-4-yl Moiety. J. Org. Chem. 2019, 84, 3624–3631. [Google Scholar] [CrossRef] [PubMed]
- Kondhare, D.; Zhang, A.; Leonard, P.; Seela, F. Alkynylated and Dendronized 5-Aza-7-deazaguanine Nucleosides: Cross-Coupling with Tripropargylamine and Linear Alkynes, Click Functionalization, and Fluorescence of Pyrene Adducts. J. Org. Chem. 2020, 85, 10525–10538. [Google Scholar] [CrossRef] [PubMed]
- Herkt, M.; Thum, T. Pharmacokinetics and Proceedings in Clinical Application of Nucleic Acid Therapeutics. Mol. Ther. 2021, 29, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Opalinska, J.B.; Gewirtz, A.M. Nucleic-acid therapeutics: Basic principles and recent applications. Nat. Rev. Drug Discov. 2002, 1, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Zanardi, T.A.; Kim, T.W.; Shen, L.; Serota, D.; Papagiannis, C.; Park, S.Y.; Kim, Y.; Henry, S.P. Chronic Toxicity Assessment of 2′-O-Methoxyethyl Antisense Oligonucleotides in Mice. Nucleic Acid Ther. 2018, 28, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Catani, M.; De Luca, C.; Medeiros Garcia Alcântara, J.; Manfredini, N.; Perrone, D.; Marchesi, E.; Weldon, R.; Müller-Späth, T.; Cavazzini, A.; Morbidelli, M.; et al. Oligonucleotides: Current Trends and Innovative Applications in the Synthesis, Characterization, and Purification. Biotechnol. J. 2020, 15, 1900226. [Google Scholar] [CrossRef] [PubMed]
- Seo, T.S.; Li, Z.; Ruparel, H.; Ju, J. Click chemistry to construct fluorescent oligonucleotides for DNA sequencing. J. Org. Chem. 2003, 68, 609–612. [Google Scholar] [CrossRef]
- Gierlich, J.; Burley, G.A.; Gramlich, P.M.E.; Hammond, D.M.; Carell, T. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org. Lett. 2006, 8, 3639–3642. [Google Scholar] [CrossRef]
- Gramlich, P.M.E.; Warncke, S.; Gierlich, J.; Carell, T. Click-click-click: Single to triple modification of DNA. Angew. Chem. Int. Ed. 2008, 47, 3442–3444. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Sanzone, A.P.; Gao, R.; Tavassoli, A.; Brown, T. Biocompatible artificial DNA linker that is read through by DNA polymerases and is functional in Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 11338–11343. [Google Scholar] [CrossRef] [PubMed]
- Fantoni, N.Z.; El-Sagheer, A.H.; Brown, T. A Hitchhiker’s Guide to Click-Chemistry with Nucleic Acids. Chem. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Dallmann, A.; El-Sagheer, A.H.; Dehmel, L.; Mügge, C.; Griesinger, C.; Ernsting, N.P.; Brown, T. Structure and Dynamics of Triazole-Linked DNA: Biocompatibility Explained. Chem. Eur. J. 2011, 17, 14714–14717. [Google Scholar] [CrossRef] [PubMed]
- Isobe, H.; Fujino, T. Triazole-Linked Analogues of DNA and RNA (TL DNA and TL RNA): Synthesis and Functions. Chem. Rec. 2014, 14, 41–51. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.H.; Brown, T. A triazole linkage that mimics the DNA phosphodiester group in living systems. Q. Rev. Biophys. 2015, 48, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Nuzzi, A.; Massi, A.; Dondoni, A. Model Studies Toward the Synthesis of Thymidine Oligonucleotides with Triazole Internucleosidic LinkagesVia Iterative Cu(I)-Promoted Azide–Alkyne Ligation Chemistry. QSAR Comb. Sci. 2007, 26, 1191–1199. [Google Scholar] [CrossRef]
- Isobe, H.; Fujino, T.; Yamazaki, N.; Guillot-Nieckowski, M.; Nakamura, E. Triazole-linked analogue of deoxyribonucleic acid (TLDNA): Design, synthesis, and double-strand formation with natural DNA. Org. Lett. 2008, 10, 3729–3732. [Google Scholar] [CrossRef]
- Fujino, T.; Kogashi, K.; Okada, K.; Mattarella, M.; Suzuki, T.; Yasumoto, K.; Sogawa, K.; Isobe, H. Chimeric RNA Oligonucleotides with Triazole and Phosphate Linkages: Synthesis and RNA Interference. Chem. Asian J. 2015, 10, 2683–2688. [Google Scholar] [CrossRef]
- Palframan, M.J.; Alharthy, R.D.; Powalowska, P.K.; Hayes, C.J. Synthesis of triazole-linked morpholino oligonucleotides via CuI catalysed cycloaddition. Org. Biomol. Chem. 2016, 14, 3112–3119. [Google Scholar] [CrossRef]
- Kaur, H.; Babu, B.R.; Maiti, S. Perspectives on Chemistry and Therapeutic Applications of Locked Nucleic Acid (LNA). Chem. Rev. 2007, 107, 4672–4697. [Google Scholar] [CrossRef]
- Watts, J.K. Locked nucleic acid: Tighter is different. Chem. Commun. 2013, 49, 5618–5620. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Singh, S.K.; Krishnamurthy, P.M.; Alterman, J.F.; Haraszti, R.A.; Khvorova, A.; Prasad, A.K.; Watts, J.K. Synthesis and biological properties of triazole-linked locked nucleic acid. Chem. Commun. 2017, 53, 8906–8909. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; El-Sagheer, A.H.; Truong, L.; Brown, T. Locked nucleic acid (LNA) enhances binding affinity of triazole-linked DNA towards RNA. Chem. Commun. 2017, 53, 8910–8913. [Google Scholar] [CrossRef]
- El-Sagheer, A.H.; Brown, T. New strategy for the synthesis of chemically modified RNA constructs exemplified by hairpin and hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 2010, 107, 15329–15334. [Google Scholar] [CrossRef] [PubMed]
- Strzyz, P. CRISPR-Cas9 wins Nobel. Nat. Rev. Mol. Cell Biol. 2020, 21, 714. [Google Scholar] [CrossRef] [PubMed]
- Taemaitree, L.; Shivalingam, A.; El-Sagheer, A.H.; Brown, T. An artificial triazole backbone linkage provides a split-and-click strategy to bioactive chemically modified CRISPR sgRNA. Nat. Commun. 2019, 10, 1610. [Google Scholar] [CrossRef]
- Honcharenko, M.; Honcharenko, D.; Strömberg, R. Efficient Conjugation to Phosphorothioate Oligonucleotides by Cu-Catalyzed Huisgen 1,3-Dipolar Cycloaddition. Bioconjug. Chem. 2019, 30, 1622–1628. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Vickers, T.A.; Liang, X.H. Phosphorothioate modified oligonucleotide-protein interactions. Nucleic Acids Res. 2020, 48, 5235–5253. [Google Scholar] [CrossRef]
- Ora, M.; Peltomäki, M.; Oivanen, M.; Lönnberg, H. Metal-Ion-Promoted Cleavage, Isomerization, and Desulfurization of the Diastereomeric Phosphoromonothioate Analogues of Uridylyl(3′,5′)uridine. J. Org. Chem. 1998, 63, 2939–2947. [Google Scholar] [CrossRef]
- Khan, R.S.; Yu, C.; Kastin, A.J.; He, Y.; Ehrensing, R.H.; Hsuchou, H.; Stone, K.P.; Pan, W. Brain Activation by Peptide Pro-Leu-Gly-NH2 (MIF-1). Int. J. Pept. 2010, 2010, 537639. [Google Scholar] [CrossRef][Green Version]
- Farzan, V.M.; Ulashchik, E.A.; Martynenko-Makaev, Y.V.; Kvach, M.V.; Aparin, I.O.; Brylev, V.A.; Prikazchikova, T.A.; Maklakova, S.Y.; Majouga, A.G.; Ustinov, A.V.; et al. Automated Solid-Phase Click Synthesis of Oligonucleotide Conjugates: From Small Molecules to Diverse N-Acetylgalactosamine Clusters. Bioconjug. Chem. 2017, 28, 2599–2607. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Patisiran: First Global Approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Givosiran: First Approval. Drugs 2020, 80, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Massaad-Massade, L.; Boutary, S.; Caillaud, M.; Gracia, C.; Parola, B.; Gnaouiya, S.B.; Stella, B.; Arpicco, S.; Buchy, E.; Desmaële, D.; et al. New Formulation for the Delivery of Oligonucleotides Using “clickable” siRNA-Polyisoprenoid-Conjugated Nanoparticles: Application to Cancers Harboring Fusion Oncogenes. Bioconjug. Chem. 2018, 29, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Zewge, D.; Butora, G.; Sherer, E.C.; Tellers, D.M.; Sidler, D.R.; Gouker, J.; Copeland, G.; Jadhav, V.; Li, Z.; Armstrong, J.; et al. Post-Synthetic Modification of Oligonucleotides via Orthogonal Amidation and Copper Catalyzed Cycloaddition Reactions. Bioconjug. Chem. 2018, 29, 1859–1865. [Google Scholar] [CrossRef]
- Good, L.; Sandberg, R.; Larsson, O.; Nielsen, P.E.; Wahlestedt, C. Antisense PNA effects in Escherichia coli are limited by the outer-membrane LPS layer. Microbiology 2000, 146, 2665–2670. [Google Scholar] [CrossRef]
- Giedyk, M.; Jackowska, A.; Równicki, M.; Kolanowska, M.; Trylska, J.; Gryko, D. Vitamin B12 transports modified RNA into E. coli and S. Typhimurium cells. Chem. Commun. 2019, 55, 763–766. [Google Scholar] [CrossRef]
- Barth, R.F.; Coderre, J.A.; Graça, M.; Vicente, H.; Blue, T.E. Boron Neutron Capture Therapy of Cancer: Current Status and Future Prospects. Clin. Cancer Res. 2005, 11, 3987–4002. [Google Scholar] [CrossRef]
- Ebenryter-Olbińska, K.; Kaniowski, D.; Sobczak, M.; Wojtczak, B.A.; Janczak, S.; Wielgus, E.; Nawrot, B.; Leśnikowski, Z.J. Versatile Method for the Site-Specific Modification of DNA with Boron Clusters: Anti-Epidermal Growth Factor Receptor (EGFR) Antisense Oligonucleotide Case. Chem. Eur. J. 2017, 23, 16535–16546. [Google Scholar] [CrossRef]
- Moreno, P.M.D.; Pêgo, A.P. Therapeutic antisense oligonucleotides against cancer: Hurdling to the clinic. Front. Chem. 2014, 2, 87. [Google Scholar] [CrossRef]
- Kaniowski, D.; Ebenryter-Olbińska, K.; Sobczak, M.; Wojtczak, B.; Janczak, S.; Leśnikowski, Z.; Nawrot, B. High Boron-loaded DNA-Oligomers as Potential Boron Neutron Capture Therapy and Antisense Oligonucleotide Dual-Action Anticancer Agents. Molecules 2017, 22, 1393. [Google Scholar] [CrossRef] [PubMed]
- Shimo, T.; Maruyama, R.; Yokota, T. Designing Effective Antisense Oligonucleotides for Exon Skipping. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2018; Volume 1687, pp. 143–155. [Google Scholar]
- Smith, C.I.E.; Zain, R. Therapeutic Oligonucleotides: State of the Art. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 605–630. [Google Scholar] [CrossRef] [PubMed]
- Shabanpoor, F.; McClorey, G.; Saleh, A.F.; Jarver, P.; Wood, M.J.A.; Gait, M.J. Bi-specific splice-switching PMO oligonucleotides conjugated via a single peptide active in a mouse model of Duchenne muscular dystrophy. Nucleic Acids Res. 2015, 43, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Rafi, M.; Wang, X.; Aran, K.; Feng, X.; Lo Sterzo, C.; Tang, R.; Lingampalli, N.; Kim, H.J.; Murthy, N. In vivo delivery of transcription factors with multifunctional oligonucleotides. Nat. Mater. 2015, 14, 701–706. [Google Scholar] [CrossRef]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef]
- Tolle, F.; Brändle, G.M.; Matzner, D.; Mayer, G. A Versatile Approach Towards Nucleobase-Modified Aptamers. Angew. Chem. Int. Ed. 2015, 54, 10971–10974. [Google Scholar] [CrossRef]
- Romanelli, A.; Affinito, A.; Avitabile, C.; Catuogno, S.; Ceriotti, P.; Iaboni, M.; Modica, J.; Condorelli, G.; Catalucci, D. An anti-PDGFRβ aptamer for selective delivery of small therapeutic peptide to cardiac cells. PLoS ONE 2018, 13, e0193392. [Google Scholar] [CrossRef]
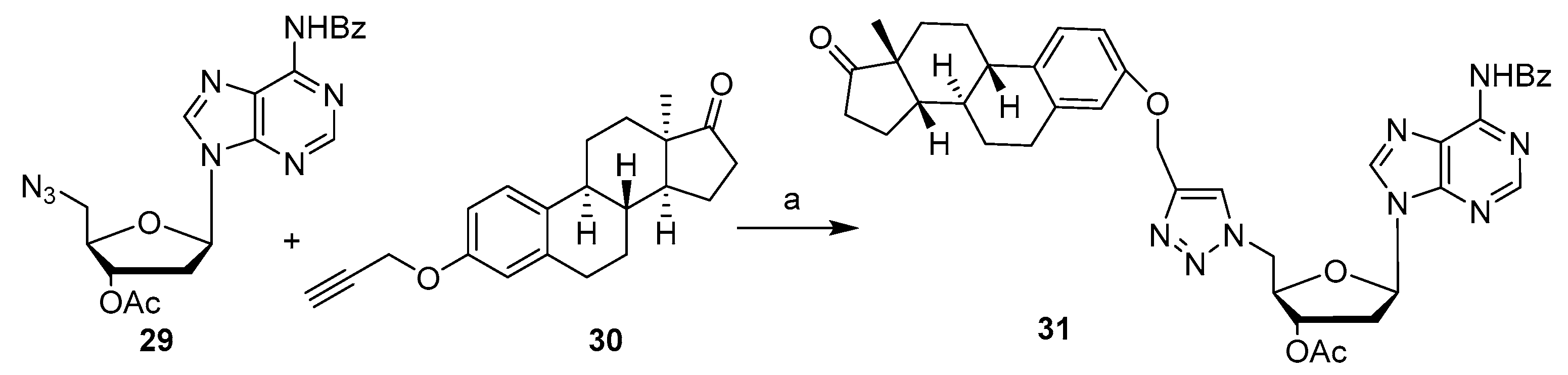
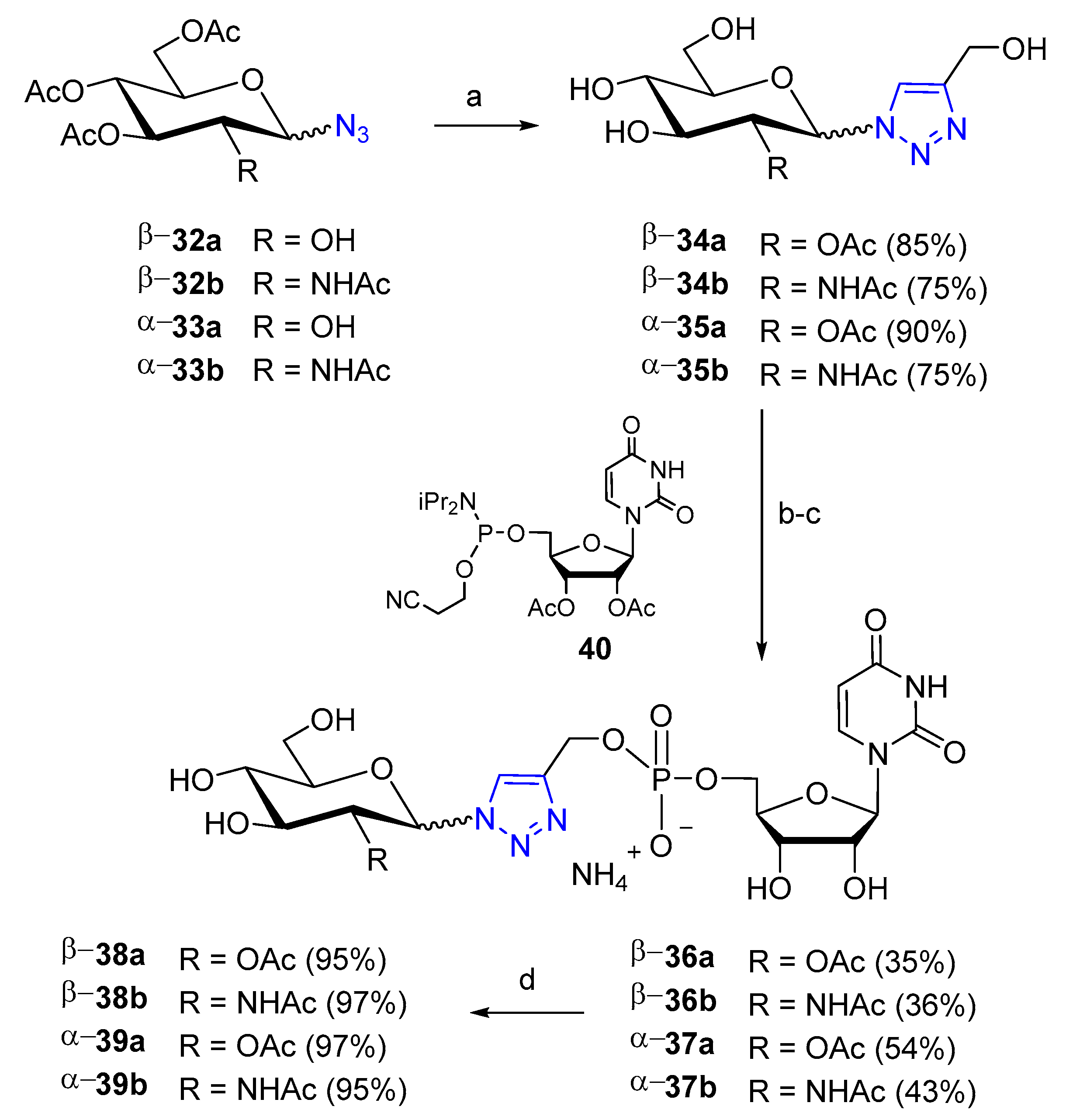
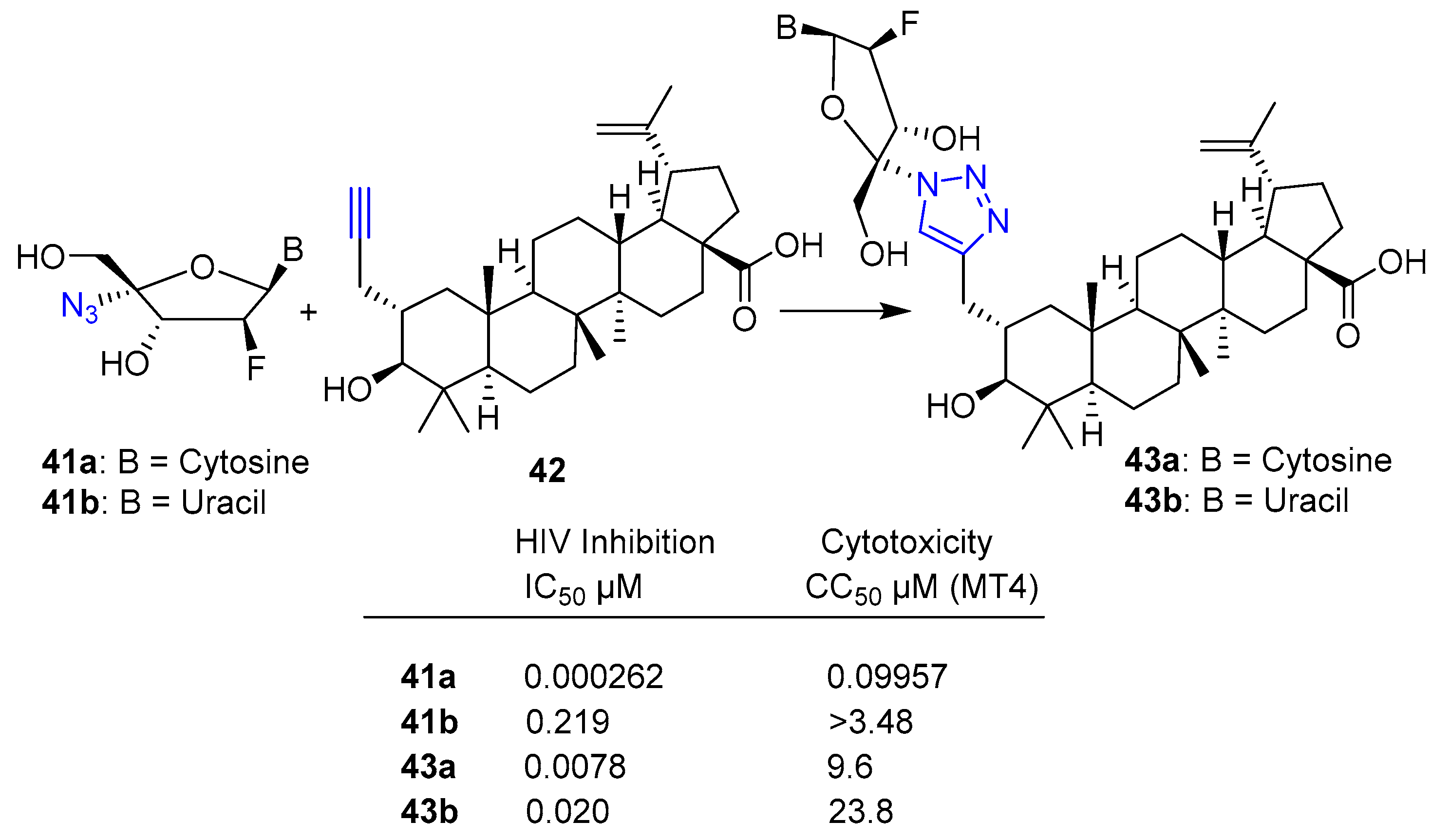
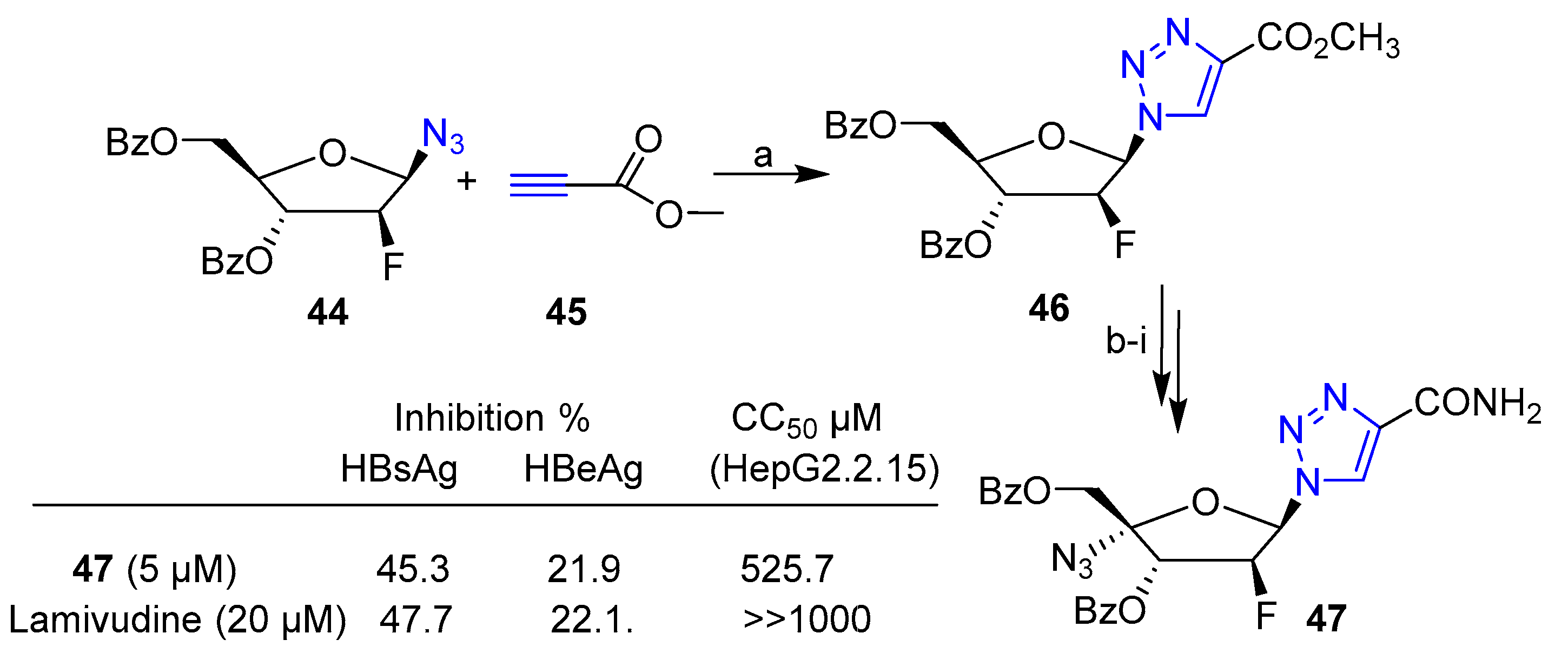
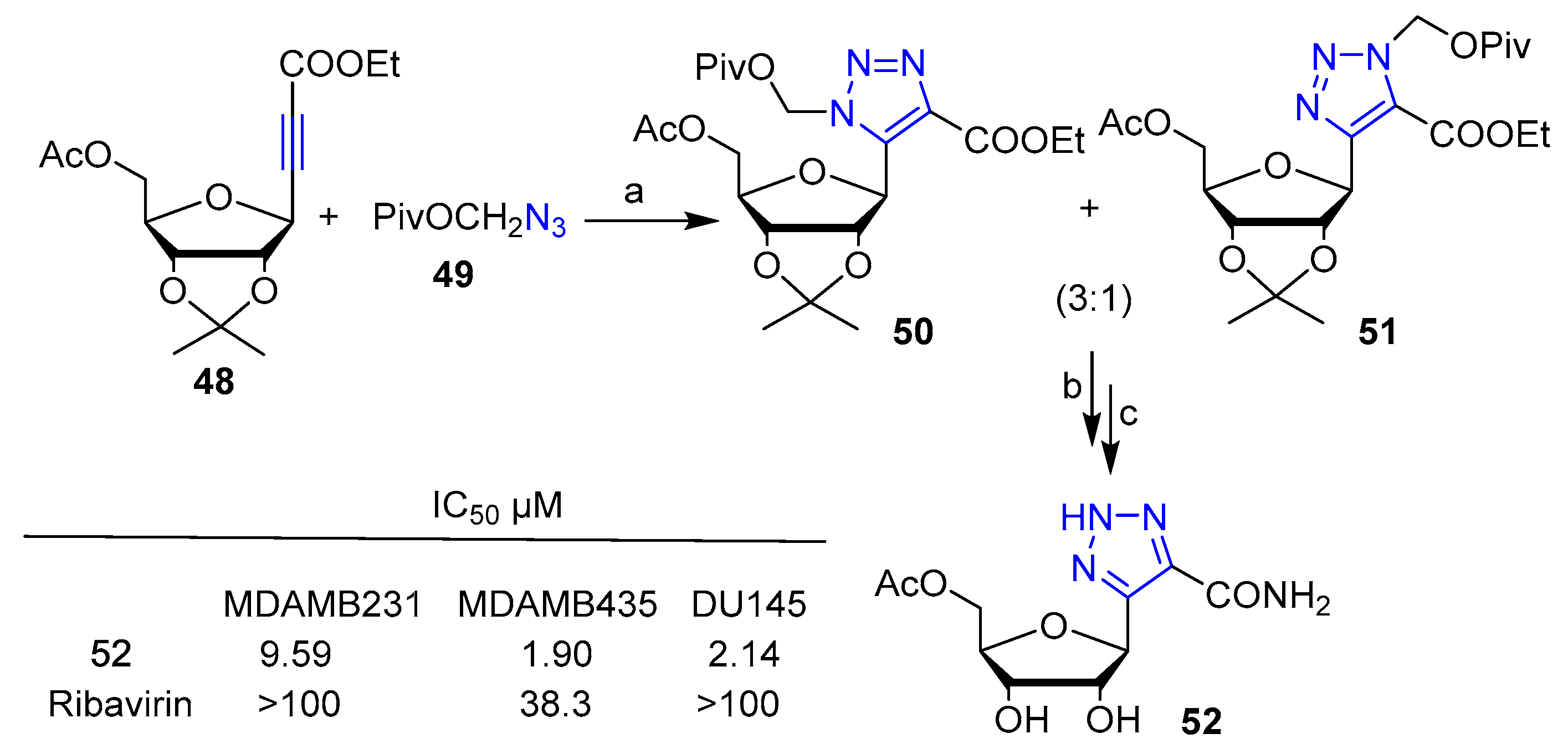
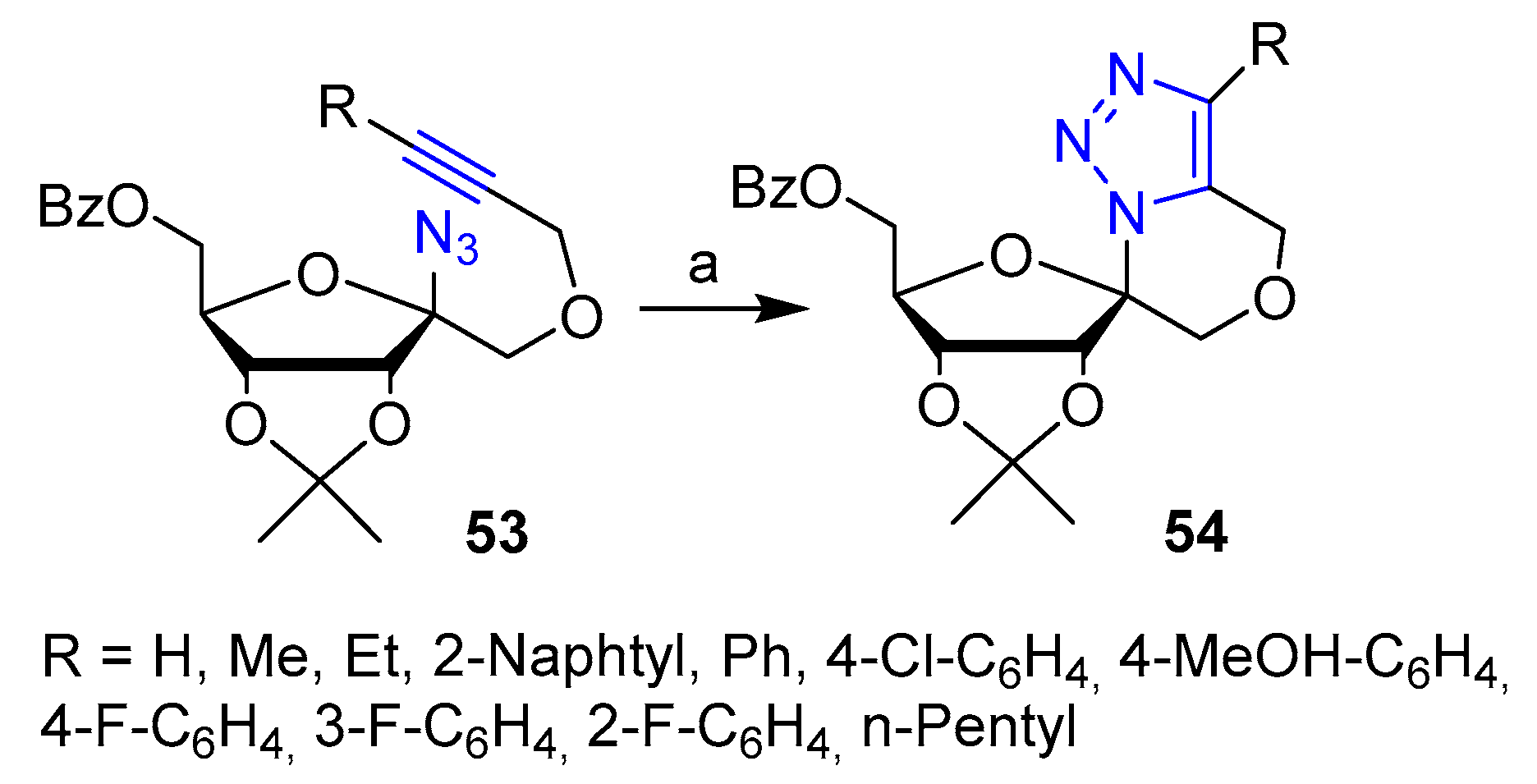

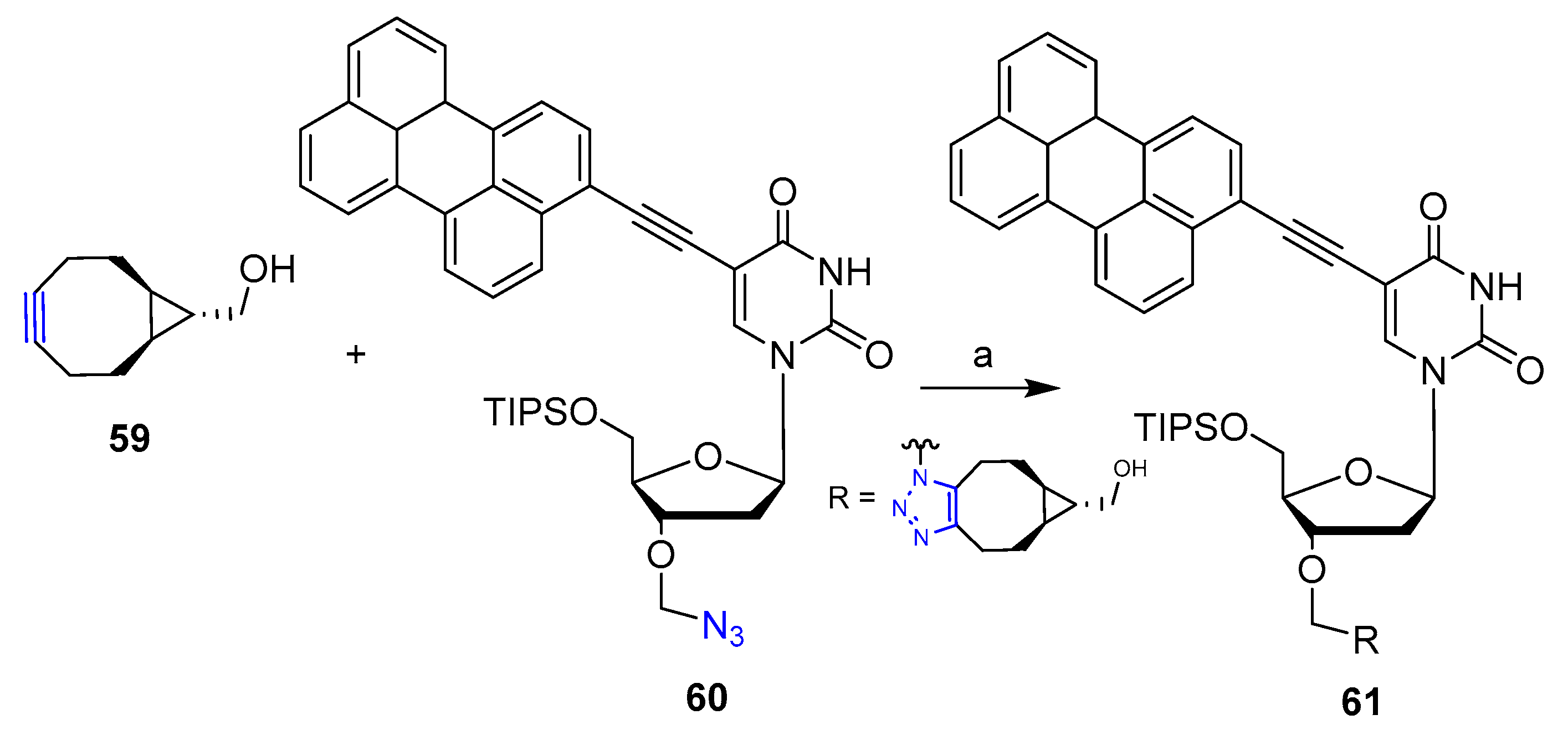
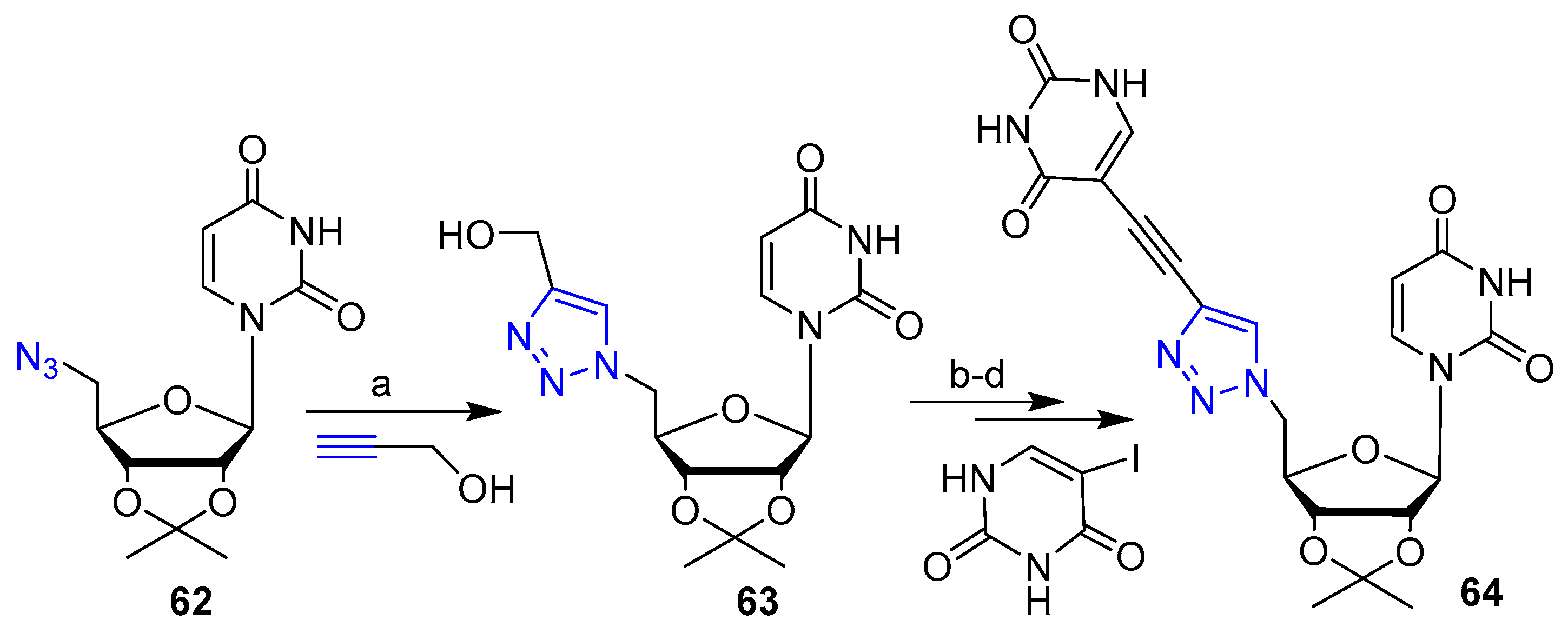
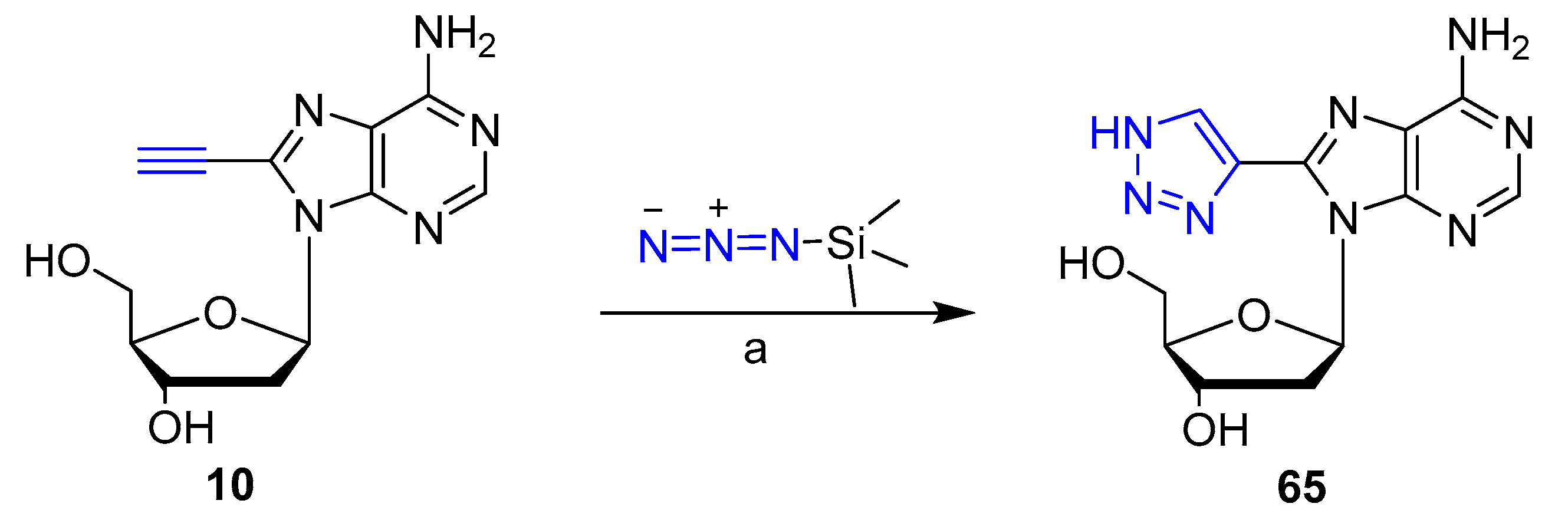



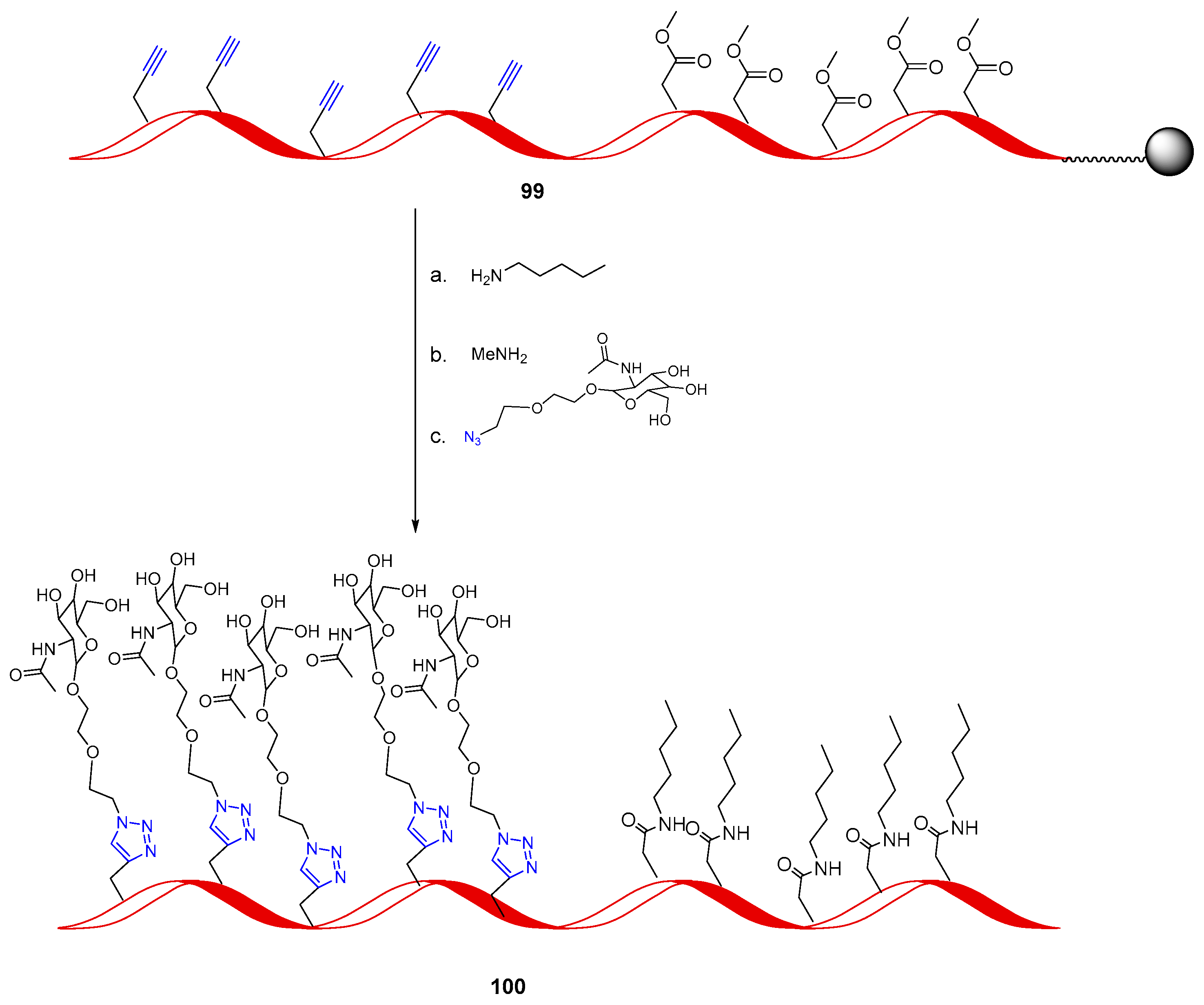
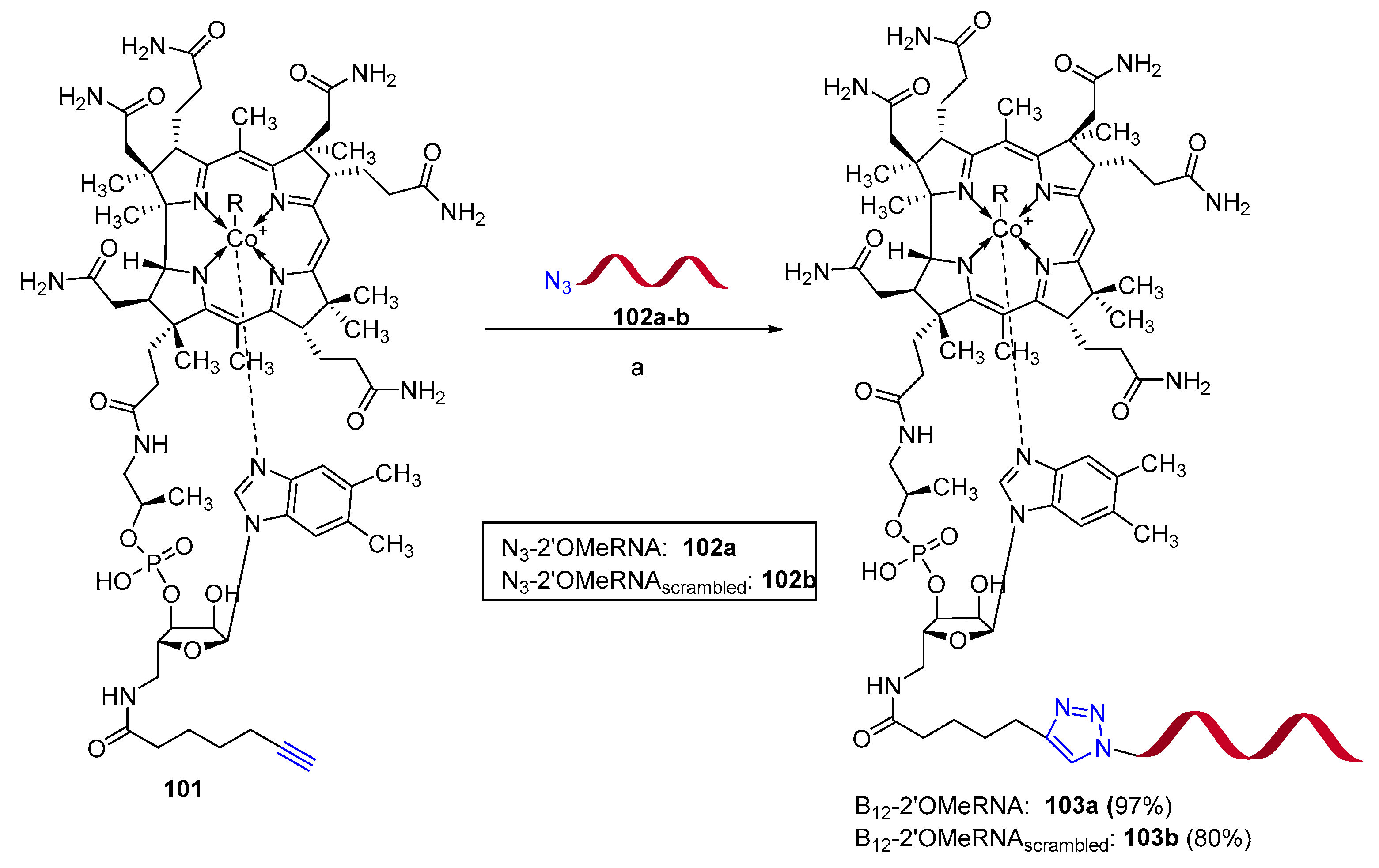
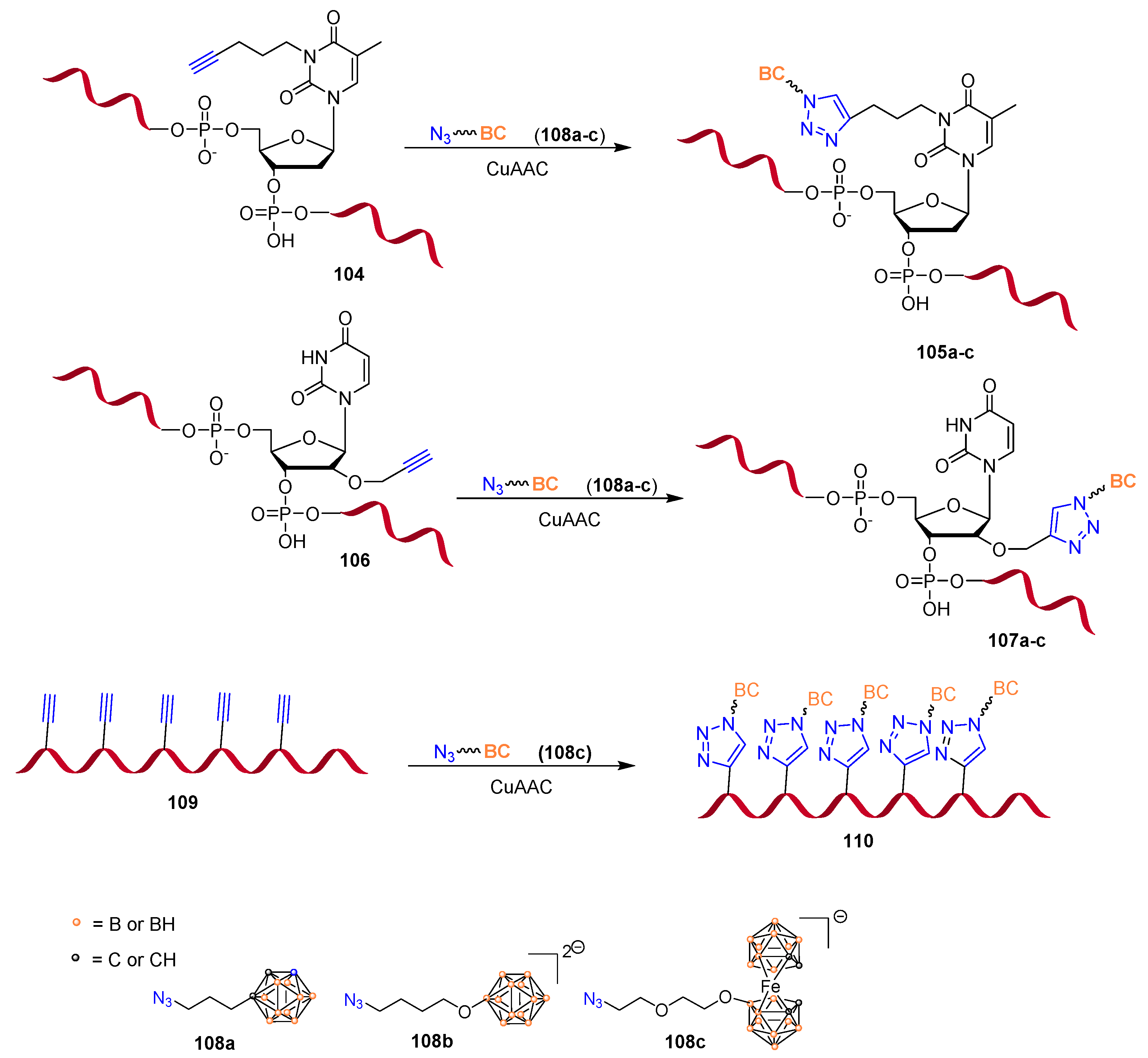
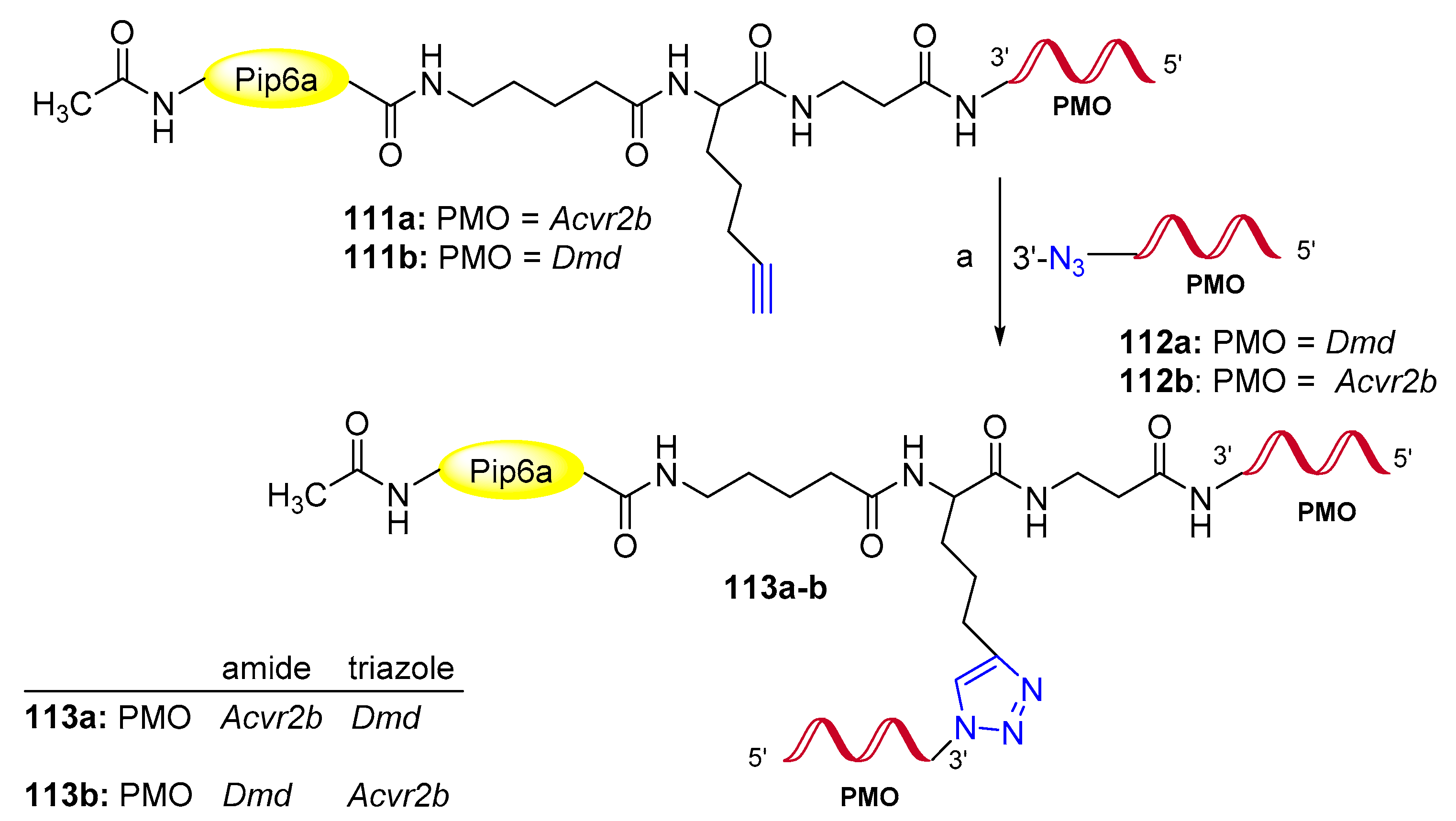


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perrone, D.; Marchesi, E.; Preti, L.; Navacchia, M.L. Modified Nucleosides, Nucleotides and Nucleic Acids via Click Azide-Alkyne Cycloaddition for Pharmacological Applications. Molecules 2021, 26, 3100. https://doi.org/10.3390/molecules26113100
Perrone D, Marchesi E, Preti L, Navacchia ML. Modified Nucleosides, Nucleotides and Nucleic Acids via Click Azide-Alkyne Cycloaddition for Pharmacological Applications. Molecules. 2021; 26(11):3100. https://doi.org/10.3390/molecules26113100
Chicago/Turabian StylePerrone, Daniela, Elena Marchesi, Lorenzo Preti, and Maria Luisa Navacchia. 2021. "Modified Nucleosides, Nucleotides and Nucleic Acids via Click Azide-Alkyne Cycloaddition for Pharmacological Applications" Molecules 26, no. 11: 3100. https://doi.org/10.3390/molecules26113100
APA StylePerrone, D., Marchesi, E., Preti, L., & Navacchia, M. L. (2021). Modified Nucleosides, Nucleotides and Nucleic Acids via Click Azide-Alkyne Cycloaddition for Pharmacological Applications. Molecules, 26(11), 3100. https://doi.org/10.3390/molecules26113100