Targeting Multiple Signal Transduction Pathways of SARS-CoV-2: Approaches to COVID-19 Therapeutic Candidates
Abstract
1. Introduction
2. COVID-19: Genetics and Structure
3. Clinical Features of COVID-19 Disease
4. SARS-CoV-2 Infection
4.1. ACE2
4.2. TMPRSS2
4.3. Glucose-Regulated Protein 78 (GRP78)
4.4. The Cluster of Differentiation 147 (CD147)
4.5. Dipeptidyl Peptidase (DPP4)
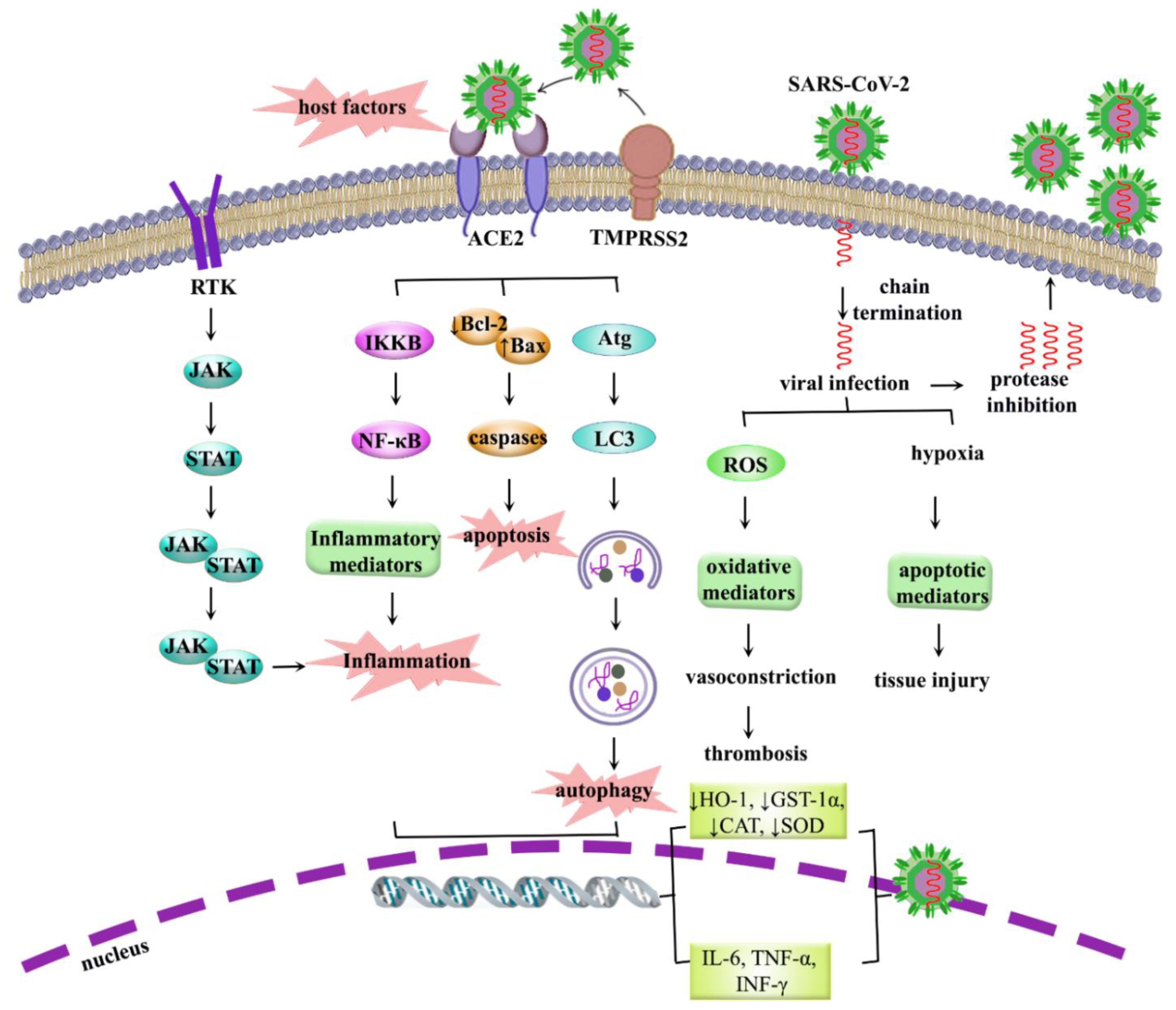
5. COVID-19: Pathogenesis, Dysregulated Pathways and Beyond
5.1. Role of Inflammation in COVID-19
5.2. Role of Oxidative Stress in COVID-19
5.3. Role of Apoptosis in COVID-19
5.4. Role of Autophagy in COVID-19
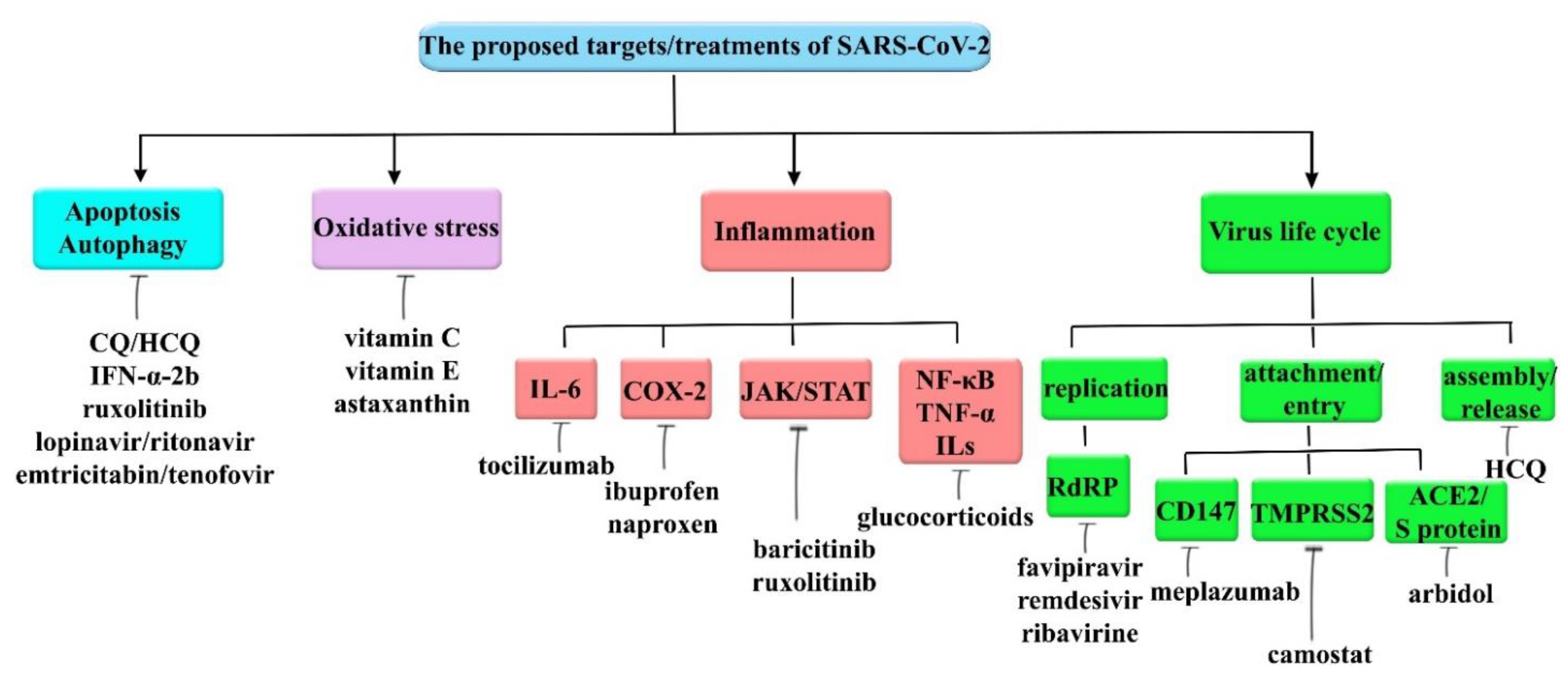
6. Therapeutic Interventions for COVID-19
6.1. Targeting Autophagy and Apoptosis
6.2. Targeting Oxidative Stress
6.3. Targeting SARS-CoV-2 Invasion
6.4. Targeting Inflammation
6.5. Miscellaneous Agents
7. Importance of Phytochemicals in Combating COVID-19
8. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chen, X.; Cai, Y.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; Zhang, Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef]
- Peeri, N.C.; Shrestha, N.; Rahman, M.S.; Zaki, R.; Tan, Z.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.; Haque, U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? Int. J. Epidemiol. 2020, 49, 717–726. [Google Scholar] [CrossRef]
- Shen, K.; Yang, Y.; Wang, T.; Zhao, D.; Jiang, Y.; Jin, R.; Zheng, Y.; Xu, B.; Xie, Z.; Lin, L. Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: Experts’ consensus statement. World J. Pediatr. 2020, 16, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Buchholz, U.; Kühne, A.; Blümel, B. State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. PLoS Curr. 2013, 5. [Google Scholar]
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef]
- Abedi, F.; Rezaee, R.; Karimi, G. Plausibility of therapeutic effects of Rho kinase inhibitors against Severe Acute Respiratory Syndrome Coronavirus 2 (COVID-19). Pharmacol. Res. 2020, 156, 104808. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, K.; Li, H.; Liao, M.; Qi, W. The continuous evolution and dissemination of 2019 novel human coronavirus. J. Infect. 2020, 80, 671–693. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Yang, N.; Shen, H.-M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in covid-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Sriram, K.; Insel, P.A. A hypothesis for pathobiology and treatment of COVID-19: The centrality of ACE1/ACE2 imbalance. Br. J. Pharmacol. 2020, 177, 4825–4844. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, evaluation and treatment coronavirus (COVID-19). In Statpearls [internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- AminJafari, A.; Ghasemi, S. The possible of immunotherapy for COVID-19: A systematic review. Int. Immunopharmacol. 2020, 83, 106455. [Google Scholar] [CrossRef] [PubMed]
- Revuelta-Herrero, J.L.; Chamorro-de-Vega, E.; Rodríguez-González, C.G.; Alonso, R.; Herranz-Alonso, A.; Sanjurjo-Sáez, M. Effectiveness, safety, and costs of a treatment switch to dolutegravir plus rilpivirine dual therapy in treatment-experienced HIV patients. Ann. Pharmacother. 2018, 52, 11–18. [Google Scholar] [CrossRef]
- Laforge, M.; Elbim, C.; Frère, C.; Hémadi, M.; Massaad, C.; Nuss, P.; Benoliel, J.-J.; Becker, C. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat. Rev. Immunol. 2020, 20, 515–516. [Google Scholar] [CrossRef]
- Reddy, K.; Rogers, A.J.; McAuley, D.F. Delving beneath the surface of hyperinflammation in COVID-19. Lancet Rheumatol. 2020, 2, e578–e579. [Google Scholar] [CrossRef]
- Jose, R.J.; Manuel, A. COVID-19 cytokine storm: The interplay between inflammation and coagulation. Lancet Respir. Med. 2020. [Google Scholar] [CrossRef]
- Milne, S.; Yang, C.X.; Timens, W.; Bossé, Y.; Sin, D.D. SARS-CoV-2 receptor ACE2 gene expression and RAAS inhibitors. Lancet Respir. Med. 2020, 8, e50–e51. [Google Scholar] [CrossRef]
- Helmy, Y.A.; Fawzy, M.; Elaswad, A.; Sobieh, A.; Kenney, S.P.; Shehata, A.A. The COVID-19 pandemic: A comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020, 9, 1225. [Google Scholar] [CrossRef]
- Jamwal, S.; Gautam, A.; Elsworth, J.; Kumar, M.; Chawla, R.; Kumar, P. An updated insight into the molecular pathogenesis, secondary complications and potential therapeutics of COVID-19 pandemic. Life Sci. 2020, 257, 118105. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27. [Google Scholar] [CrossRef] [PubMed]
- Angeletti, S.; Benvenuto, D.; Bianchi, M.; Giovanetti, M.; Pascarella, S.; Ciccozzi, M. COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020, 92, 584–588. [Google Scholar] [CrossRef]
- Krichel, B.; Falke, S.; Hilgenfeld, R.; Redecke, L.; Uetrecht, C. Processing of the SARS-CoV pp1a/ab nsp7–10 region. Biochem. J. 2020, 477, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Ceraolo, C.; Giorgi, F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020, 92, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Karampoor, S.; Sholeh, M.; Moradi, P.; Ranjbar, R.; Ghasemi, F. A contemporary review on pathogenesis and immunity of COVID-19 infection. Mol. Biol. Rep. 2020, 47, 5365–5376. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the Cytokine Storm’in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Xiang, P.; Xu, X.; Gao, L.; Wang, H.; Xiong, H.; Li, R. First case of 2019 novel coronavirus disease with Encephalitis. ChinaXiv 2020, 2020, 00015. [Google Scholar]
- Giacomelli, A.; Pezzati, L.; Conti, F.; Bernacchia, D.; Siano, M.; Oreni, L.; Rusconi, S.; Gervasoni, C.; Ridolfo, A.L.; Rizzardini, G. Self-reported olfactory and taste disorders in patients with severe acute respiratory coronavirus 2 infection: A cross-sectional study. Clin. Infect. Dis. 2020, 71, 889–890. [Google Scholar] [CrossRef]
- Ryan, W. There Is A New Symptom of Coronavirus, Doctors Say: Sudden Loss of Smell or Taste. 2020. Available online: https://www.usatoday.com/story/news/health/2020/03/24/coronavirus-symptoms-loss-smell-taste/2897385001/ (accessed on 25 March 2020).
- Li, Y.C.; Bai, W.Z.; Hashikawa, T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020, 92, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Koyuncu, O.O.; Hogue, I.B.; Enquist, L.W. Virus infections in the nervous system. Cell Host Microbe 2013, 13, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Fakhri, S.; Piri, S.; Majnooni, M.B.; Farzaei, M.H.; Echeverria, J. Targeting neurological manifestation of coronaviruses by candidate phytochemicals: A mechanistic approach. Front. Pharmacol. 2020, 11, 2291. [Google Scholar]
- Abdennour, L.; Zeghal, C.; Deme, M.; Puybasset, L. Interaction brain-lungs. Ann. Fr. D’anesthesie et de Reanim. 2012, e101–e107. [Google Scholar] [CrossRef]
- Poyiadji, N.; Shahin, G.; Noujaim, D.; Stone, M.; Patel, S.; Griffith, B. COVID-19–associated acute hemorrhagic necrotizing encephalopathy: CT and MRI features. Radiology 2020, 296, E119–E120. [Google Scholar] [CrossRef]
- Steardo, L.; Steardo, L., Jr.; Zorec, R.; Verkhratsky, A. Neuroinfection may contribute to pathophysiology and clinical manifestations of COVID-19. Acta Physiol. 2020, 229, e13473. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Zheng, K.I.; Yan, Q.-Q.; Rios, R.S.; Targher, G.; Byrne, C.D.; Van Poucke, S.; Liu, W.-Y.; Zheng, M.-H. COVID-19 and liver dysfunction: Current insights and emergent therapeutic strategies. J. Clin. Transl. Hepatol. 2020, 8, 18–24. [Google Scholar] [CrossRef]
- Miller, A.J.; Arnold, A.C. The renin–angiotensin system in cardiovascular autonomic control: Recent developments and clinical implications. Clin. Auton. Res. 2019, 29, 231–243. [Google Scholar] [CrossRef]
- Long, B.; Brady, W.J.; Koyfman, A.; Gottlieb, M. Cardiovascular complications in COVID-19. Am. J. Emerg. Med. 2020, 38, 1504–1507. [Google Scholar] [CrossRef]
- Zhu, H.; Rhee, J.-W.; Cheng, P.; Waliany, S.; Chang, A.; Witteles, R.M.; Maecker, H.; Davis, M.M.; Nguyen, P.K.; Wu, S.M. Cardiovascular Complications in Patients with COVID-19: Consequences of Viral Toxicities and Host Immune Response. Curr. Cardiol. Rep. 2020, 22, 32. [Google Scholar] [CrossRef]
- Bandyopadhyay, D.; Akhtar, T.; Hajra, A.; Gupta, M.; Das, A.; Chakraborty, S.; Pal, I.; Patel, N.; Amgai, B.; Ghosh, R.K. COVID-19 pandemic: Cardiovascular complications and future implications. Am. J. Cardiovasc. Drugs 2020, 20, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Villapol, S. Gastrointestinal symptoms associated with COVID-19: Impact on the gut microbiome. Transl. Res. 2020, 226, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Mackett, A.J.; Keevil, V.L. COVID-19 and Gastrointestinal Symptoms—A Case Report. Geriatrics 2020, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.-W.; Da Xu, H.Z.; Zhou, W.; Wang, L.-H.; Cui, X.-G. Identification of a potential mechanism of acute kidney injury during the COVID-19 outbreak: A study based on single-cell transcriptome analysis. Intensive Care Med. 2020, 46, 1114–1116. [Google Scholar] [CrossRef]
- Kissling, S.; Rotman, S.; Gerber, C.; Halfon, M.; Lamoth, F.; Comte, D.; Lhopitallier, L.; Sadallah, S.; Fakhouri, F. Collapsing glomerulopathy in a COVID-19 patient. Kidney Int. 2020, 98, 228–231. [Google Scholar] [CrossRef]
- Durvasula, R.; Wellington, T.; McNamara, E.; Watnick, S. COVID-19 and Kidney Failure in the Acute Care Setting: Our Experience from Seattle. Am. J. Kidney Dis. 2020, 76, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, V.; Fiorentino, M.; Cantaluppi, V.; Gesualdo, L.; Stallone, G.; Ronco, C.; Castellano, G. Acute kidney injury in SARS-CoV-2 infected patients. Crit. Care 2020, 24, 155. [Google Scholar] [CrossRef]
- Pour, P.M.; Fakhri, S.; Asgary, S.; Farzaei, M.H.; Echeverria, J. The signaling pathways, and therapeutic targets of antiviral agents: Focusing on the antiviral approaches and clinical perspectives of anthocyanins in the management of viral diseases. Front. Pharmacol. 2019, 10, 1207. [Google Scholar] [CrossRef]
- Galimberti, S.; Petrini, M.; Baratè, C.; Ricci, F.; Balducci, S.; Grassi, S.; Guerrini, F.; Ciabatti, E.; Mechelli, S.; Di Paolo, A. Tyrosine kinase inhibitors play an antiviral action in patients affected by chronic myeloid leukemia: A possible model supporting their use in the fight against SARS-CoV-2. Front. Oncol. 2020, 10, 1428. [Google Scholar] [CrossRef]
- Miller, R.L.; Meng, T.-C.; Tomai, M.A. The antiviral activity of Toll-like receptor 7 and 7/8 agonists. Drug News Perspect. 2008, 21, 69–87. [Google Scholar] [CrossRef]
- Koumbi, L. Current and future antiviral drug therapies of hepatitis B chronic infection. World J. Hepatol. 2015, 7, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Khan, A.; Ali, A.; Idrees, M. The interplay between viruses and TRIM family proteins. Rev. Med. Virol. 2019, 29, e2028. [Google Scholar] [CrossRef]
- Hakim, M.S.; Spaan, M.; Janssen, H.L.; Boonstra, A. Inhibitory receptor molecules in chronic hepatitis B and C infections: Novel targets for immunotherapy? Rev. Med. Virol. 2014, 24, 125–138. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Krüger, N.; Mueller, M.A.; Drosten, C.; Pöhlmann, S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv 2020. [Google Scholar] [CrossRef]
- Badawi, S.; Ali, B.R. ACE2 Nascence, trafficking, and SARS-CoV-2 pathogenesis: The saga continues. Hum. Genom. 2021, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Haga, S.; Yamamoto, N.; Nakai-Murakami, C.; Osawa, Y.; Tokunaga, K.; Sata, T.; Yamamoto, N.; Sasazuki, T.; Ishizaka, Y. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc. Natl. Acad. Sci. USA 2008, 105, 7809–7814. [Google Scholar] [CrossRef]
- Clarke, N.E.; Turner, A.J. Angiotensin-converting enzyme 2: The first decade. Int. J. Hypertens. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 1–10. [Google Scholar] [CrossRef]
- Koka, V.; Huang, X.R.; Chung, A.C.; Wang, W.; Truong, L.D.; Lan, H.Y. Angiotensin II up-regulates angiotensin I-converting enzyme (ACE), but down-regulates ACE2 via the AT1-ERK/p38 MAP kinase pathway. Am. J. Pathol. 2008, 172, 1174–1183. [Google Scholar] [CrossRef]
- Zhang, R.; Wu, Y.; Zhao, M.; Liu, C.; Zhou, L.; Shen, S.; Liao, S.; Yang, K.; Li, Q.; Wan, H. Role of HIF-1α in the regulation ACE and ACE2 expression in hypoxic human pulmonary artery smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 297, L631–L640. [Google Scholar] [CrossRef]
- Rivellese, F.; Prediletto, E. ACE2 at the centre of COVID-19 from paucisymptomatic infections to severe pneumonia. Autoimmun. Rev. 2020, 19, 102536. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. A novel coronavirus from patients with pneumonia in China, 2019. New Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Dandekar, A.A.; Perlman, S. Immunopathogenesis of coronavirus infections: Implications for SARS. Nat. Rev. Immunol. 2005, 5, 917–927. [Google Scholar] [CrossRef]
- Ziai, S.A.; Rezaei, M.; Fakhrri, S.; Pouriran, R. ACE2: Its potential role and regulation in severe acute respiratory syndrome and COVID-19. J. Cell. Physiol. 2021, 236, 2430–2442. [Google Scholar]
- Li, Y.; Zhou, W.; Yang, L.; You, R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol. Res. 2020, 157, 104833. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir Crit Care Med. 2020, 202, 756–759. [Google Scholar] [CrossRef]
- South, A.M.; Tomlinson, L.; Edmonston, D.; Hiremath, S.; Sparks, M.A. Controversies of renin–angiotensin system inhibition during the COVID-19 pandemic. Nat. Rev. Nephrol. 2020, 16, 305–307. [Google Scholar] [CrossRef]
- Valdes, G.; Neves, L.; Anton, L.; Corthorn, J.; Chacon, C.; Germain, A.; Merrill, D.; Ferrario, C.; Sarao, R.; Penninger, J. Distribution of angiotensin-(1–7) and ACE2 in human placentas of normal and pathological pregnancies. Placenta 2006, 27, 200–207. [Google Scholar] [CrossRef]
- Levy, A.; Yagil, Y.; Bursztyn, M.; Barkalifa, R.; Scharf, S.; Yagil, C. ACE2 expression and activity are enhanced during pregnancy. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 295, R1953–R1961. [Google Scholar] [CrossRef] [PubMed]
- Rees, R.; Feigel, I.; Vickers, A.; Zollman, C.; McGurk, R.; Smith, C. Prevalence of complementary therapy use by women with breast cancer: A population-based survey. Eur. J. Cancer 2000, 36, 1359–1364. [Google Scholar] [CrossRef]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S. Clinical characteristics of coronavirus disease 2019 in China. New Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Fernández-Atucha, A.; Izagirre, A.; Fraile-Bermúdez, A.B.; Kortajarena, M.; Larrinaga, G.; Martinez-Lage, P.; Echevarría, E.; Gil, J. Sex differences in the aging pattern of renin–angiotensin system serum peptidases. Biol. Sex. Differ. 2017, 8, 5. [Google Scholar] [CrossRef]
- Lavrentyev, E.N.; Malik, K.U. High glucose-induced Nox1-derived superoxides downregulate PKC-βII, which subsequently decreases ACE2 expression and ANG (1–7) formation in rat VSMCs. Am. J. Physiol. Heart Circ. Physiol. 2009, 296, H106–H118. [Google Scholar] [CrossRef][Green Version]
- Chen, K.; Bi, J.; Su, Y.; Chappell, M.C.; Rose, J.C. Sex-specific changes in renal angiotensin-converting enzyme and angiotensin-converting enzyme 2 gene expression and enzyme activity at birth and over the first year of life. Reprod. Sci. 2016, 23, 200–210. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Oakes, J.M.; Fuchs, R.M.; Gardner, J.D.; Lazartigues, E.; Yue, X. Nicotine and the renin-angiotensin system. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018, 315, R895–R906. [Google Scholar] [CrossRef] [PubMed]
- Xudong, X.; Junzhu, C.; Xingxiang, W.; Furong, Z.; Yanrong, L. Age-and gender-related difference of ACE2 expression in rat lung. Life Sci. 2006, 78, 2166–2171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Su, H.; Ma, X.; Xu, X.; Liang, L.; Ma, G.; Shi, L. MiRNA let-7b promotes the development of hypoxic pulmonary hypertension by targeting ACE2. Am. J. Physiol. -Lung Cell. Mol. Physiol. 2019, 316, L547–L557. [Google Scholar] [CrossRef]
- Maruta, H.; He, H. PAK1-blockers: Potential Therapeutics against COVID-19. Med. Drug Discov. 2020, 6, 100039. [Google Scholar] [CrossRef] [PubMed]
- Awad, K.S.; Elinoff, J.M.; Wang, S.; Gairhe, S.; Ferreyra, G.A.; Cai, R.; Sun, J.; Solomon, M.A.; Danner, R.L. Raf/ERK drives the proliferative and invasive phenotype of BMPR2-silenced pulmonary artery endothelial cells. J. Am. J. Physiol. Lung Cell. 2016, 310, L187–L201. [Google Scholar] [CrossRef]
- Maruta, H. Herbal therapeutics that block the oncogenic kinase PAK1: A practical approach towards PAK1-dependent diseases and longevity. Phytother. Res. 2014, 28, 656–672. [Google Scholar] [CrossRef]
- Rico-Mesa, J.S.; White, A.; Anderson, A.S. Outcomes in Patients with COVID-19 Infection Taking ACEI/ARB. Curr. Cardiol. Rep. 2020, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Kam, Y.-W.; Okumura, Y.; Kido, H.; Ng, L.F.; Bruzzone, R.; Altmeyer, R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS ONE 2009, 4, e7870. [Google Scholar] [CrossRef]
- Bittmann, S.; Luchter, E.; Weissenstein, A.; Villalon, G.; Moschüring-Alieva, E. TMPRSS2-inhibitors play a role in cell entry mechanism of COVID-19: An insight into camostat and nafamostat. J. Regen Biol Med. 2020, 2, 1–3. [Google Scholar]
- Thunders, M.; Delahunt, B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J. Clin. Pathol. 2020, 73, 773–776. [Google Scholar] [CrossRef]
- Ragia, G.; Manolopoulos, V.G. Inhibition of SARS-CoV-2 entry through the ACE2/TMPRSS2 pathway: A promising approach for uncovering early COVID-19 drug therapies. Eur. J. Clin. Pharmacol. 2020, 76, 1623–1630. [Google Scholar] [CrossRef]
- Shen, L.W.; Mao, H.J.; Wu, Y.L.; Tanaka, Y.; Zhang, W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie 2017, 142, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhao, J.; Martin, W.; Kallianpur, A.; Chung, M.K.; Jehi, L.; Sharifi, N.; Erzurum, S.; Eng, C.; Cheng, F. New insights into genetic susceptibility of COVID-19: An ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020, 18, 1–8. [Google Scholar] [CrossRef]
- Tanabe, L.M.; List, K. The role of type II transmembrane serine protease-mediated signaling in cancer. FEBS J. 2017, 284, 1421–1436. [Google Scholar] [CrossRef]
- Hardy, B.; Raiter, A. Peptide-binding heat shock protein GRP78 protects cardiomyocytes from hypoxia-induced apoptosis. J. Mol. Med. 2010, 88, 1157–1167. [Google Scholar] [CrossRef]
- Lee, A.S. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 2005, 35, 373–381. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elshahat, M.E.; Elfiky, A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020, 80, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Chan, C.-M.; Zhang, X.; Wang, Y.; Yuan, S.; Zhou, J.; Au-Yeung, R.K.-H.; Sze, K.-H.; Yang, D.; Shuai, H. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018, 293, 11709–11726. [Google Scholar] [CrossRef]
- Balmeh, N.; Mahmoudi, S.; Mohammadi, N.; Karabedianhajiabadi, A. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Inform. Med. Unlocked 2020, 20, 100407. [Google Scholar] [CrossRef]
- Ulrich, H.; Pillat, M.M. CD147 as a target for COVID-19 treatment: Suggested effects of azithromycin and stem cell engagement. Stem Cell Rev. Rep. 2020, 16, 434–440. [Google Scholar] [CrossRef]
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C. Distribution of ACE2, CD147, CD26 and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020, 75, 2829–2845. [Google Scholar] [CrossRef]
- Watanabe, A.; Yoneda, M.; Ikeda, F.; Terao-Muto, Y.; Sato, H.; Kai, C. CD147/EMMPRIN acts as a functional entry receptor for measles virus on epithelial cells. J. Virol. 2010, 84, 4183–4193. [Google Scholar] [CrossRef]
- Zhu, X.; Song, Z.; Zhang, S.; Nanda, A.; Li, G. CD147: A novel modulator of inflammatory and immune disorders. Curr. Med. Chem. 2014, 21, 2138–2145. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sato, Y.; Sasaki, T. Suppression of coronavirus replication by cyclophilin inhibitors. Viruses 2013, 5, 1250–1260. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhou, Y.-S.; Lian, J.-Q.; Zhang, Z.; Du, P.; Gong, L.; Zhang, Y.; Cui, H.-Y.; Geng, J.-J. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. BioRxiv 2020. [Google Scholar] [CrossRef]
- Bian, H.; Zheng, Z.-H.; Wei, D.; Zhang, Z.; Kang, W.-Z.; Hao, C.-Q.; Dong, K.; Kang, W.; Xia, J.-L.; Miao, J.-L. Meplazumab treats COVID-19 pneumonia: An open-labelled, concurrent controlled add-on clinical trial. MedRxiv 2020. [Google Scholar] [CrossRef]
- Klawitter, J.; Klawitter, J.; Schmitz, V.; Brunner, N.; Crunk, A.; Corby, K.; Bendrick-Peart, J.; Leibfritz, D.; Edelstein, C.L.; Thurman, J.M. Low-salt diet and cyclosporine nephrotoxicity: Changes in kidney cell metabolism. J. Proteome Res. 2012, 11, 5135–5144. [Google Scholar] [CrossRef]
- Heinzmann, D.; Noethel, M.; Ungern-Sternberg, S.V.; Mitroulis, I.; Gawaz, M.; Chavakis, T.; May, A.E.; Seizer, P. CD147 is a novel interaction partner of integrin αMβ2 mediating leukocyte and platelet adhesion. Biomolecules 2020, 10, 541. [Google Scholar] [CrossRef] [PubMed]
- Pushkarsky, T.; Yurchenko, V.; Vanpouille, C.; Brichacek, B.; Vaisman, I.; Hatakeyama, S.; Nakayama, K.I.; Sherry, B.; Bukrinsky, M.I. Cell surface expression of CD147/EMMPRIN is regulated by cyclophilin 60. J. Biol. Chem. 2005, 280, 27866–27871. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, V.; Constant, S.; Eisenmesser, E.; Bukrinsky, M. Cyclophilin–CD147 interactions: A new target for anti-inflammatory therapeutics. Clin. Exp. Immunol. 2010, 160, 305–317. [Google Scholar] [CrossRef]
- Tang, W.; Hemler, M.E. Caveolin-1 regulates matrix metalloproteinases-1 induction and CD147/EMMPRIN cell surface clustering. J. Biol. Chem. 2004, 279, 11112–11118. [Google Scholar] [CrossRef]
- Cui, J.; Huang, W.; Wu, B.; Jin, J.; Jing, L.; Shi, W.P.; Liu, Z.Y.; Yuan, L.; Luo, D.; Li, L. N-glycosylation by N-acetylglucosaminyltransferase V enhances the interaction of CD147/basigin with integrin β1 and promotes HCC metastasis. J. Pathol. 2018, 245, 41–52. [Google Scholar] [CrossRef]
- Yu, C.; Lixia, Y.; Ruiwei, G.; Yankun, S.; Jinshan, Y. The Role of FAK in the Secretion of MMP9 after CD147 Stimulation in Macrophages. Int. Heart J. 2018, 59, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Fadini, G.P.; Morieri, M.L.; Longato, E.; Bonora, B.M.; Pinelli, S.; Selmin, E.; Voltan, G.; Falaguasta, D.; Tresso, S.; Costantini, G. Exposure to DPP-4 inhibitors and COVID-19 among people with type 2 diabetes. A case–control study. DiabetesObes. Metab. 2020, 22, 1946–1950. [Google Scholar]
- Lu, G.; Hu, Y.; Wang, Q.; Qi, J.; Gao, F.; Li, Y.; Zhang, Y.; Zhang, W.; Yuan, Y.; Bao, J. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013, 500, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Solerte, S.B.; Di Sabatino, A.; Galli, M.; Fiorina, P. Dipeptidyl peptidase-4 (DPP4) inhibition in COVID-19. Acta Diabetol. 2020, 57, 779–783. [Google Scholar] [CrossRef]
- Strollo, R.; Pozzilli, P. DPP4 inhibition: Preventing SARS-CoV-2 infection and/or progression of COVID-19? Diabetes/Metab. Res. Rev. 2020, 36, e3330. [Google Scholar] [CrossRef] [PubMed]
- Al-Kuraishy, H.M.; Al-Niemi, M.S.; Hussain, N.R.; Al-Gareeb, A.I.; Al-Harchan, N.A.; Al-Kurashi, A.H. The Potential Role of Renin Angiotensin System (RAS) and Dipeptidyl Peptidase-4 (DPP-4) in COVID-19: Navigating the Uncharted. In Selected Chapters from the Renin-Angiotensin System; IntechOpen: London, UK, 2020. [Google Scholar]
- Wagner, L.; Klemann, C.; Stephan, M.; Von Hörsten, S. Unravelling the immunological roles of dipeptidyl peptidase 4 (DPP4) activity and/or structure homologue (DASH) proteins. Clin. Exp. Immunol. 2016, 184, 265–283. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef]
- Klemann, C.; Wagner, L.; Stephan, M.; von Hörsten, S. Cut to the chase: A review of CD26/dipeptidyl peptidase-4′s (DPP4) entanglement in the immune system. Clin. Exp. Immunol. 2016, 185, 1–21. [Google Scholar] [CrossRef]
- Iacobellis, G. COVID-19 and diabetes: Can DPP4 inhibition play a role? Diabetes Res. Clin. Pract. 2020, 162. [Google Scholar] [CrossRef] [PubMed]
- Bloomgarden, Z.T. Diabetes and COVID-19. J. Diabetes 2020, 12, 347–348. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.P.; Meng, S.; Wu, Y.-J.; Mao, Y.-P.; Ye, R.-X.; Wang, Q.-Z.; Sun, C.; Sylvia, S.; Rozelle, S.; Raat, H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: A scoping review. Infect. Dis. Poverty 2020, 9, 1–12. [Google Scholar] [CrossRef]
- Filatov, A.; Sharma, P.; Hindi, F.; Espinosa, P.S. Neurological complications of coronavirus disease (COVID-19): Encephalopathy. Cureus 2020, 12, e7352. [Google Scholar] [CrossRef]
- Coyle, J.; Igbinomwanhia, E.; Sanchez-Nadales, A.; Danciu, S.; Chu, C.; Shah, N. A Recovered Case of COVID-19 Myocarditis and ARDS Treated with Corticosteroids, Tocilizumab, and Experimental AT-001. Jacc: Case Rep. 2020, 2, 1331–1336. [Google Scholar] [CrossRef]
- Yarmohammadi, A.; Yarmohammadi, M.; Fakhri, S.; Khan, H. Targeting pivotal inflammatory pathways in COVID-19: A mechanistic review. Eur. J. Pharmacol. 2020, 890, 173620. [Google Scholar] [CrossRef] [PubMed]
- Min, C.-K.; Cheon, S.; Ha, N.-Y.; Sohn, K.M.; Kim, Y.; Aigerim, A.; Shin, H.M.; Choi, J.-Y.; Inn, K.-S.; Kim, J.-H. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci. Rep. 2016, 6, 25359. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.E.; Chambers, R.C. The mercurial nature of neutrophils: Still an enigma in ARDS? Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 306, L217–L230. [Google Scholar] [CrossRef]
- Channappanavar, R.; Perlman, S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. In Seminars in Immunopathology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 529–539. [Google Scholar]
- Cameron, M.J.; Bermejo-Martin, J.F.; Danesh, A.; Muller, M.P.; Kelvin, D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. 2008, 133, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Colafrancesco, S.; Priori, R.; Alessandri, C.; Astorri, E.; Perricone, C.; Blank, M.; Agmon-Levin, N.; Shoenfeld, Y.; Valesini, G. sCD163 in AOSD: A biomarker for macrophage activation related to hyperferritinemia. Immunol. Res. 2014, 60, 177–183. [Google Scholar] [CrossRef]
- Rosário, C.; Zandman-Goddard, G.; Meyron-Holtz, E.G.; D’Cruz, D.P.; Shoenfeld, Y. The hyperferritinemic syndrome: Macrophage activation syndrome, Still’s disease, septic shock and catastrophic antiphospholipid syndrome. BMC Med. 2013, 11, 185. [Google Scholar] [CrossRef]
- Sharif, K.; Vieira Borba, V.; Zandman-Goddard, G.; Shoenfeld, Y. Eppur Si Muove: Ferritin is essential in modulating inflammation. Clin. Exp. Immunol. 2018, 191, 149–150. [Google Scholar] [CrossRef] [PubMed]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef]
- Mahallawi, W.H.; Khabour, O.F.; Zhang, Q.; Makhdoum, H.M.; Suliman, B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 2018, 104, 8–13. [Google Scholar] [CrossRef]
- Wong, C.; Lam, C.; Wu, A.; Ip, W.; Lee, N.; Chan, I.; Lit, L.; Hui, D.; Chan, M.; Chung, S. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004, 136, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, J.M.; Poon, L.L.; Lee, K.C.; Ng, W.F.; Lai, S.T.; Leung, C.Y.; Chu, C.M.; Hui, P.K.; Mak, K.L.; Lim, W. Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003, 361, 1773–1778. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fehr, A.R.; Zheng, J.; Wohlford-Lenane, C.; Abrahante, J.E.; Mack, M.; Sompallae, R.; McCray, P.B.; Meyerholz, D.K.; Perlman, S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019, 129, 3625–3639. [Google Scholar] [CrossRef] [PubMed]
- Kindler, E.; Thiel, V.; Weber, F. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2016; Volume 96, pp. 219–243. [Google Scholar]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef]
- Högner, K.; Wolff, T.; Pleschka, S.; Plog, S.; Gruber, A.D.; Kalinke, U.; Walmrath, H.-D.; Bodner, J.; Gattenlöhner, S.; Lewe-Schlosser, P. Macrophage-expressed IFN-β contributes to apoptotic alveolar epithelial cell injury in severe influenza virus pneumonia. PLoS Pathog. 2013, 9, e1003188. [Google Scholar] [CrossRef]
- Bendickova, K.; Tidu, F.; Fric, J. Calcineurin–NFAT signalling in myeloid leucocytes: New prospects and pitfalls in immunosuppressive therapy. Embo Mol. Med. 2017, 9, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; Yoo, S.-A.; Kim, M.; Kim, W.-U. The Role of Calcium–Calcineurin–NFAT Signaling Pathway in Health and Autoimmune Diseases. Front. Immunol. 2020, 11, 195. [Google Scholar] [CrossRef]
- Islam, A.B.; Khan, M.A.-A.-K. Lung biopsy cells transcriptional landscape from COVID-19 patient stratified lung injury in SARS-CoV-2 infection through impaired pulmonary surfactant metabolism. Sci. Rep. 2020, 10, 19395. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Li, Y.-X.; Jiang, L.-J.; Chen, Q.; Wang, T.; Ye, D.-W. Targeting JAK-STAT Signaling to Control Cytokine Release Syndrome in COVID-19. Trends Pharmacol. Sci. 2020, 41, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Bagca, B.G.; Avci, C.B. The potential of JAK/STAT pathway inhibition by ruxolitinib in the treatment of COVID-19. Cytokine Growth Factor Rev. 2020, 54, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Roche, L.; Mesta, F. Oxidative Stress as Key Player in Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) infection. Arch. Med. Res. 2020, 51, 384–387. [Google Scholar] [CrossRef]
- Camini, F.C.; da Silva, T.F.; da Silva Caetano, C.C.; Almeida, L.T.; Ferraz, A.C.; Vitoreti, V.M.A.; de Mello Silva, B.; de Queiroz Silva, S.; de Magalhães, J.C.; de Brito Magalhães, C.L. Antiviral activity of silymarin against Mayaro virus and protective effect in virus-induced oxidative stress. Antivir. Res. 2018, 158, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.-Y.; Wang, N.; Li, S.; Hong, M.; Wang, X.; Feng, Y. The reactive oxygen species in macrophage polarization: Reflecting its dual role in progression and treatment of human diseases. Oxidative Med. Cell. Longev. 2016, 2016, 2795090. [Google Scholar] [CrossRef]
- Warnatsch, A.; Tsourouktsoglou, T.-D.; Branzk, N.; Wang, Q.; Reincke, S.; Herbst, S.; Gutierrez, M.; Papayannopoulos, V. Reactive oxygen species localization programs inflammation to clear microbes of different size. Immunity 2017, 46, 421–432. [Google Scholar] [CrossRef]
- Wang, J.-Z.; Zhang, R.-Y.; Bai, J. An anti-oxidative therapy for ameliorating cardiac injuries of critically ill COVID-19-infected patients. Int. J. Cardiol. 2020, 312, 137–138. [Google Scholar] [CrossRef] [PubMed]
- Komaravelli, N.; Casola, A. Respiratory viral infections and subversion of cellular antioxidant defenses. J. Pharm. Pharm. 2014, 5, 1000141. [Google Scholar]
- Shukla, K.; Pal, P.B.; Sonowal, H.; Srivastava, S.K.; Ramana, K.V. Aldose reductase inhibitor protects against hyperglycemic stress by activating Nrf2-dependent antioxidant proteins. J. Diabetes Res. 2017, 2017, 6785852. [Google Scholar] [CrossRef]
- Hassan, S.; Jawad, J.; Ahjel, W.; Sing, B.; Sing, J.; Awad, M.; Hadi, R. The Nrf2 Activator (DMF) and Covid-19: Is there a Possible Role. Med. Arch. 2020, 74, 134–138. [Google Scholar] [CrossRef]
- Mao, H.; Tu, W.; Qin, G.; Law, H.K.W.; Sia, S.F.; Chan, P.-L.; Liu, Y.; Lam, K.-T.; Zheng, J.; Peiris, M. Influenza virus directly infects human natural killer cells and induces cell apoptosis. J. Virol. 2009, 83, 9215–9222. [Google Scholar] [CrossRef]
- Tan, Y.-J.; Fielding, B.C.; Goh, P.-Y.; Shen, S.; Tan, T.H.; Lim, S.G.; Hong, W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004, 78, 14043–14047. [Google Scholar] [CrossRef]
- Kvansakul, M. Viral infection and apoptosis. Viruses 2017, 9, 356. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Wong, C.K.; Li, P.; Xie, Y. A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochim. Et Biophys. Acta Gen. Subj. 2008, 1780, 1383–1387. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Fukushi, S.; Saijo, M.; Kurane, I.; Morikawa, S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004, 319, 1228–1234. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Fukushi, S.; Saijo, M.; Kurane, I.; Morikawa, S. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology 2004, 327, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Gharote, M.A. Role of poly (ADP) ribose polymerase-1 inhibition by nicotinamide as a possible additive treatment to modulate host immune response and prevention of cytokine storm in COVID-19. Indian J. Med. Sci. 2020, 72, 25–28. [Google Scholar] [CrossRef]
- Bian, H.; Zhou, Y.; Yu, B.; Shang, D.; Liu, F.; Li, B.; Qi, J. Rho-kinase signaling pathway promotes the expression of PARP to accelerate cardiomyocyte apoptosis in ischemia/reperfusion. Mol. Med. Rep. 2017, 16, 2002–2008. [Google Scholar] [CrossRef]
- Zan, J.; Liu, J.; Zhou, J.-W.; Wang, H.-L.; Mo, K.-K.; Yan, Y.; Xu, Y.-B.; Liao, M.; Su, S.; Hu, R.-L. Rabies virus matrix protein induces apoptosis by targeting mitochondria. Exp. Cell Res. 2016, 347, 83–94. [Google Scholar] [CrossRef]
- Chan, C.-M.; Ma, C.-W.; Chan, W.-Y.; Chan, H.Y.E. The SARS-Coronavirus Membrane protein induces apoptosis through modulating the Akt survival pathway. Arch. Biochem. Biophys. 2007, 459, 197–207. [Google Scholar] [CrossRef]
- Lim, H.; Lim, Y.-M.; Kim, K.H.; Jeon, Y.E.; Park, K.; Kim, J.; Hwang, H.-Y.; Lee, D.J.; Pagire, H.; Kwon, H.J. A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef]
- Randhawa, P.K.; Scanlon, K.; Rappaport, J.; Gupta, M.K. Modulation of Autophagy by SARS-CoV-2: A Potential Threat for Cardiovascular System. Front. Physiol. 2020, 11, 1560. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, K.; Chen, C.Z.; Bostwick, R.; Rasmussen, L.; Xu, M.; Pradhan, M.; Tran, B.N.; Zhu, W.; Shamim, K.; Huang, W. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. Biorxiv 2020. [Google Scholar] [CrossRef]
- Bonam, S.R.; Muller, S.; Bayry, J.; Klionsky, D.J. Autophagy as an emerging target for COVID-19: Lessons from an old friend, chloroquine. Autophagy 2020, 1–7. [Google Scholar] [CrossRef]
- Sharma, P.; McAlinden, K.D.; Ghavami, S.; Deshpande, D.A. Chloroquine: Autophagy inhibitor, antimalarial, bitter taste receptor agonist in fight against COVID-19, a reality check? Eur. J. Pharmacol. 2021, 897, 173928. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Bauer, M.A.; Zimmermann, A.; Kainz, K.; Hofer, S.J.; Kroemer, G.; Madeo, F. Digesting the crisis: Autophagy and coronaviruses. Microb. Cell 2020, 7, 119–128. [Google Scholar] [CrossRef]
- Zhang, R.-H.; Zhang, H.-L.; Li, P.-Y.; Gao, J.-P.; Luo, Q.; Liang, T.; Wang, X.-J.; Hao, Y.-Q.; Xu, T.; Li, C.-H. Autophagy is involved in the acute lung injury induced by H9N2 influenza virus. Int. Immunopharmacol. 2019, 74, 105737. [Google Scholar] [CrossRef]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mösbauer, K.; Zellner, A. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Kindrachuk, J.; Ork, B.; Hart, B.J.; Mazur, S.; Holbrook, M.R.; Frieman, M.B.; Traynor, D.; Johnson, R.F.; Dyall, J.; Kuhn, J.H. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 2015, 59, 1088–1099. [Google Scholar] [CrossRef]
- Granato, M.; Romeo, M.A.; Tiano, M.S.; Santarelli, R.; Gonnella, R.; Montani, M.S.G.; Faggioni, A.; Cirone, M. Bortezomib promotes KHSV and EBV lytic cycle by activating JNK and autophagy. Sci. Rep. 2017, 7, 13052. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Yuan, K.; Zhou, L.; Wang, K.; Chen, H.-N.; Lei, Y.; Lan, J.; Pu, Q.; Gao, W.; Zhang, L. PRKAA/AMPK restricts HBV replication through promotion of autophagic degradation. Autophagy 2016, 12, 1507–1520. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yu, W.; Liu, B.; Wang, Y.; Wang, J.; Xia, K.; Liang, C.; Fang, W.; Zhou, C.; Tao, H. Escin induces caspase-dependent apoptosis and autophagy through the ROS/p38 MAPK signalling pathway in human osteosarcoma cells in vitro and in vivo. Cell Death Differ. 2017, 8, e3113. [Google Scholar] [CrossRef] [PubMed]
- García-Pérez, B.E.; González-Rojas, J.A.; Salazar, M.I.; Torres-Torres, C.; Castrejón-Jiménez, N.S. Taming the Autophagy as a Strategy for Treating COVID-19. Cells 2020, 9, 2679. [Google Scholar] [CrossRef]
- Eisenberg-Lerner, A.; Bialik, S.; Simon, H.-U.; Kimchi, A. Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009, 16, 966–975. [Google Scholar] [CrossRef]
- Shojaei, S.; Koleini, N.; Samiei, E.; Aghaei, M.; Cole, L.K.; Alizadeh, J.; Islam, M.I.; Vosoughi, A.R.; Albokashy, M.; Butterfield, Y. Simvastatin increases temozolomide-induced cell death by targeting the fusion of autophagosomes and lysosomes. FEBS J. 2020, 287, 1005–1034. [Google Scholar] [CrossRef]
- Shojaei, S.; Suresh, M.; Klionsky, D.J.; Labouta, H.I.; Ghavami, S. Autophagy and SARS-CoV-2 infection: A possible smart targeting of the autophagy pathway. Virulence 2020, 11, 805–810. [Google Scholar] [CrossRef]
- Kyrmizi, I.; Gresnigt, M.S.; Akoumianaki, T.; Samonis, G.; Sidiropoulos, P.; Boumpas, D.; Netea, M.G.; Van De Veerdonk, F.L.; Kontoyiannis, D.P.; Chamilos, G. Corticosteroids block autophagy protein recruitment in Aspergillus fumigatus phagosomes via targeting dectin-1/Syk kinase signaling. J. Immunol. 2013, 191, 1287–1299. [Google Scholar] [CrossRef]
- Ishida, S.; Akiyama, H.; Umezawa, Y.; Okada, K.; Nogami, A.; Oshikawa, G.; Nagao, T.; Miura, O. Mechanisms for mtorc1 activation and synergistic induction of apoptosis by ruxolitinib and bh3 mimetics or autophagy inhibitors in jak2-v617f-expressing leukemic cells including newly established pvtl-2. Oncotarget 2018, 9, 26834–26851. [Google Scholar] [CrossRef][Green Version]
- Wagener, F.A.; Pickkers, P.; Peterson, S.J.; Immenschuh, S.; Abraham, N.G. Targeting the heme-heme oxygenase system to prevent severe complications following COVID-19 infections. Antioxidants 2020, 9, 540. [Google Scholar] [CrossRef]
- Feyaerts, A.F.; Luyten, W. Vitamin C as prophylaxis and adjunctive medical treatment for COVID-19? Nutrition 2020, 79–80, 110948. [Google Scholar] [CrossRef] [PubMed]
- Colunga Biancatelli, R.M.L.; Berrill, M.; Catravas, J.D.; Marik, P.E. Quercetin and vitamin C: An experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19). Front. Immunol. 2020, 11, 1451. [Google Scholar] [CrossRef]
- Liu, F.; Zhu, Y.; Zhang, J.; Li, Y.; Peng, Z. Intravenous high-dose vitamin C for the treatment of severe COVID-19: Study protocol for a multicentre randomised controlled trial. BMJ Open 2020, 10, e039519. [Google Scholar] [CrossRef]
- Richard, C.; Lemonnier, F.; Thibault, M.; Couturier, M.; Auzepy, P. Vitamin E deficiency and lipoperoxidation during adult respiratory distress syndrome. Crit. Care Med. 1990, 18, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.E.; Guarner-Lans, V.; Soria-Castro, E.; Manzano Pech, L.; Pérez-Torres, I. Is Antioxidant Therapy a Useful Complementary Measure for Covid-19 Treatment? An Algorithm for Its Application. Medicina 2020, 56, 386. [Google Scholar] [CrossRef] [PubMed]
- Fakhri, S.; Nouri, Z.; Moradi, S.Z.; Farzaei, M.H. Astaxanthin, COVID-19 and immune response: Focus on oxidative stress, apoptosis and autophagy. Phytother. Res. 2020, 34, 2790–2792. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA 2020, 323, 1824–1836. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, B.; Li, Q.; Wen, L.; Zhang, R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020, 71, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Yao, Y.; Yeung, M.L.; Deng, W.; Bao, L.; Jia, L.; Li, F.; Xiao, C.; Gao, H.; Yu, P.; et al. Treatment With Lopinavir/Ritonavir or Interferon-beta1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J. Infect. Dis. 2015, 212, 1904–1913. [Google Scholar] [CrossRef] [PubMed]
- Groneberg, D.A.; Poutanen, S.M.; Low, D.E.; Lode, H.; Welte, T.; Zabel, P. Treatment and vaccines for severe acute respiratory syndrome. Lancet Infect. Dis. 2005, 5, 147–155. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. New Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Lu, Z.; Xu, T.; Chen, C.; Yang, G.; Zha, T.; Lu, J.; Xue, Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020, 81, e21–e23. [Google Scholar] [CrossRef]
- Hawman, D.W.; Haddock, E.; Meade-White, K.; Williamson, B.; Hanley, P.W.; Rosenke, K.; Komeno, T.; Furuta, Y.; Gowen, B.B.; Feldmann, H. Favipiravir (T-705) but not ribavirin is effective against two distinct strains of Crimean-Congo hemorrhagic fever virus in mice. Antivir. Res. 2018, 157, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Noguchi, K.; Kimitsuki, K.; Kaimori, R.; Saito, N.; Komeno, T.; Nakajima, N.; Furuta, Y.; Nishizono, A. Reevaluation of the efficacy of favipiravir against rabies virus using in vivo imaging analysis. Antivir. Res. 2019, 172, 104641. [Google Scholar] [CrossRef]
- Fang, Q.-Q.; Huang, W.-J.; Li, X.-Y.; Cheng, Y.-H.; Tan, M.-J.; Liu, J.; Wei, H.-J.; Meng, Y.; Wang, D.-Y. Effectiveness of favipiravir (T-705) against wild-type and oseltamivir-resistant influenza B virus in mice. Virology 2020, 545, 1–9. [Google Scholar] [CrossRef]
- Cai, Q.; Yang, M.; Liu, D.; Chen, J.; Shu, D.; Xia, J.; Liao, X.; Gu, Y.; Cai, Q.; Yang, Y.; et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering 2020, 6, 1192–1198. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q. Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020, 395, 1569–1578. [Google Scholar] [CrossRef]
- Ziaie, S.; Koucheck, M.; Miri, M.; Salarian, S.; Shojaei, S.; Haghighi, M.; Sistanizad, M. Review of therapeutic agents for the treatment of COVID-19. J. Cell. Mol. Anesth. 2020, 5, 32–36. [Google Scholar]
- Yao, X.; Ye, F.; Zhang, M.; Cui, C.; Huang, B.; Niu, P.; Liu, X.; Zhao, L.; Dong, E.; Song, C. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020, 71, 732–739. [Google Scholar] [CrossRef]
- Gautret, P.; Lagier, J.C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020, 56, 105949. [Google Scholar] [CrossRef]
- Cragg, G.M.; Kingston, D.G.; Newman, D.J. Anticancer Agents from Natural Products; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Chen, J.; Liu, D.; Liu, L.; Liu, P.; Xu, Q.; Xia, L.; Ling, Y.; Huang, D.; Song, S.; Zhang, D. A pilot study of hydroxychloroquine in treatment of patients with common coronavirus disease-19 (COVID-19). J. Zhejiang Univ. Med. Sci. 2020, 49, 215–219. [Google Scholar]
- Borba, M.G.S.; Val, F.F.A.; Sampaio, V.S.; Alexandre, M.A.A.; Melo, G.C.; Brito, M.; Mourao, M.P.G.; Brito-Sousa, J.D.; Baia-da-Silva, D.; Guerra, M.V.F.; et al. Effect of High vs Low Doses of Chloroquine Diphosphate as Adjunctive Therapy for Patients Hospitalized With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection: A Randomized Clinical Trial. JAMA Netw. Open 2020, 3, e208857. [Google Scholar] [CrossRef]
- Liu, F.; Li, L.; Xu, M.; Wu, J.; Luo, D.; Zhu, Y.; Li, B.; Song, X.; Zhou, X. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020, 127, 104370. [Google Scholar] [CrossRef] [PubMed]
- Michot, J.-M.; Albiges, L.; Chaput, N.; Saada, V.; Pommeret, F.; Griscelli, F.; Balleyguier, C.; Besse, B.; Marabelle, A.; Netzer, F.; et al. Tocilizumab, an anti-IL6 receptor antibody, to treat Covid-19-related respiratory failure: A case report. Ann. Oncol. 2020, 31, 961–964. [Google Scholar] [CrossRef] [PubMed]
- Hart, B.J.; Dyall, J.; Postnikova, E.; Zhou, H.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G.; Frieman, M.B.; Holbrook, M.R.; Jahrling, P.B.; et al. Interferon-beta and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014, 95, 571–577. [Google Scholar] [CrossRef]
- Sallard, E.; Lescure, F.-X.; Yazdanpanah, Y.; Mentre, F.; Peiffer-Smadja, N. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020, 178, 104791. [Google Scholar] [CrossRef]
- Cantini, F.; Niccoli, L.; Matarrese, D.; Nicastri, E.; Stobbione, P.; Goletti, D. Baricitinib therapy in COVID-19: A pilot study on safety and clinical impact. J. Infect. 2020, 81, 318–356. [Google Scholar] [CrossRef]
- Arabi, Y.M.; Mandourah, Y.; Al-Hameed, F.; Sindi, A.A.; Almekhlafi, G.A.; Hussein, M.A.; Jose, J.; Pinto, R.; Al-Omari, A.; Kharaba, A. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am. J. Respir. Crit. Care Med. 2018, 197, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.-N.; Chen, G.; Sun, J.; Liang, B.-M.; Liang, Z.-A. The effect of corticosteroids on mortality of patients with influenza pneumonia: A systematic review and meta-analysis. Crit. Care 2019, 23, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Russell, C.D.; Millar, J.E.; Baillie, J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020, 395, 473–475. [Google Scholar] [CrossRef]
- Wong, R.S. Disease-modifying effects of long-term and continuous use of nonsteroidal anti-inflammatory drugs (NSAIDs) in spondyloarthritis. Adv. Pharmacol. Sci. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Yousefifard, M.; Zali, A.; Zarghi, A.; Madani Neishaboori, A.; Hosseini, M.; Safari, S. Non-steroidal anti-inflammatory drugs in management of COVID-19; a systematic review on current evidence. Int. J. Clin. Pract. 2020, 74, e13557. [Google Scholar] [CrossRef]
- Russell, B.; Moss, C.; George, G.; Santaolalla, A.; Cope, A.; Papa, S.; Van Hemelrijck, M. Associations between immune-suppressive and stimulating drugs and novel COVID-19—A systematic review of current evidence. Ecancermedicalscience 2020, 14. [Google Scholar] [CrossRef]
- Million, M.; Lagier, J.-C.; Gautret, P.; Colson, P.; Fournier, P.-E.; Amrane, S.; Hocquart, M.; Mailhe, M.; Esteves-Vieira, V.; Doudier, B.; et al. Early treatment of COVID-19 patients with hydroxychloroquine and azithromycin: A retrospective analysis of 1061 cases in Marseille, France. Travel Med. Infect. Dis. 2020, 35, 101738. [Google Scholar] [CrossRef]
- Vouri, S.M.; Thai, T.N.; Winterstein, A.G. An evaluation of co-use of chloroquine or hydroxychloroquine plus azithromycin on cardiac outcomes: A pharmacoepidemiological study to inform use during the COVID19 pandemic. Res. Soc. Adm. Pharm. 2021, 17, 2012–2017. [Google Scholar] [CrossRef]
- Sarayani, A.; Cicali, B.; Henriksen, C.H.; Brown, J.D. Safety signals for QT prolongation or Torsades de Pointes associated with azithromycin with or without chloroquine or hydroxychloroquine. Res. Soc. Adm. Pharm. 2021, 17, 483–486. [Google Scholar] [CrossRef]
- Baron, S.A.; Devaux, C.; Colson, P.; Raoult, D.; Rolain, J.-M. Teicoplanin: An alternative drug for the treatment of COVID-19? Int. J. Antimicrob. Agents 2020, 55, 105944. [Google Scholar] [CrossRef]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Hegarty, P.K.; Kamat, A.M.; Zafirakis, H.; Dinardo, A. BCG vaccination may be protective against Covid-19. Preprint 2020. [Google Scholar] [CrossRef]
- Vyas, N.; Kurian, S.J.; Bagchi, D.; Manu, M.K.; Saravu, K.; Unnikrishnan, M.K.; Mukhopadhyay, C.; Rao, M.; Miraj, S.S. Vitamin D in prevention and treatment of COVID-19: Current perspective and future prospects. J. Am. Coll. Nutr. 2020, 1–14. [Google Scholar] [CrossRef]
- Martineau, A.R.; Forouhi, N.G. Vitamin D for COVID-19: A case to answer? Lancet Diabetes Endocrinol. 2020, 8, 735–736. [Google Scholar] [CrossRef]
- Rahman, A.H.; Branch, A.D. Vitamin D for your patients with chronic hepatitis C? J. Hepatol. 2013, 58, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, X.; Ni, L.; Di, X.; Ma, B.; Niu, S.; Liu, C.; Reiter, R.J. COVID-19: Melatonin as a potential adjuvant treatment. Life Sci. 2020, 250, 117583. [Google Scholar] [CrossRef]
- Moradi, S.Z.; Momtaz, S.; Bayrami, Z.; Farzaei, M.H.; Abdollahi, M. Nanoformulations of Herbal Extracts in Treatment of Neurodegenerative Disorders. Front. Bioeng Biotechnol. 2020, 8, 238. [Google Scholar] [CrossRef]
- Fakhri, S.; Moradi, S.Z.; Farzaei, M.H.; Bishayee, A. Modulation of dysregulated cancer metabolism by plant secondary metabolites: A mechanistic review. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Naithani, R.; Huma, L.C.; Holland, L.E.; Shukla, D.; McCormick, D.L.; Mehta, R.G.; Moriarty, R.M. Antiviral activity of phytochemicals: A comprehensive review. Mini Rev. Med. Chem. 2008, 8, 1106–1133. [Google Scholar] [CrossRef]
- McKee, D.L.; Sternberg, A.; Stange, U.; Laufer, S.; Naujokat, C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020, 157, 104859. [Google Scholar] [CrossRef]
- Majnooni, M.B.; Fakhri, S.; Shokoohinia, Y.; Mohammadi, P.; Gravand, M.M.; Farzaei, M.H.; Echeverria, J. Phytochemicals: Potential therapeutic interventions against coronaviruses-associated lung injury. Front. Pharmacol. 2020, 11, 1744. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ying, Y. The inhibitory effect of curcumin on virus-induced cytokine storm and its potential use in the associated severe pneumonia. Front. Cell Dev. Biol. 2020, 8, 479. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Shi, Y.; Xiao, S.; Li, N.; Zhao, Q.; Zhang, A.; Nan, Y.; Mu, Y.; Sun, Y.; Wu, C. Curcumin is a promising inhibitor of genotype 2 porcine reproductive and respiratory syndrome virus infection. Bmc Vet. Res. 2017, 13, 298. [Google Scholar] [CrossRef]
- Praditya, D.; Kirchhoff, L.; Brüning, J.; Rachmawati, H.; Steinmann, J.; Steinmann, E. Anti-infective properties of the golden spice curcumin. Front. Microbiol. 2019, 10, 912. [Google Scholar] [CrossRef]
- Kannan, S.; Kolandaivel, P. Antiviral potential of natural compounds against influenza virus hemagglutinin. Comput. Biol. Chem. 2017, 71, 207–218. [Google Scholar] [CrossRef]
- Zahedipour, F.; Hosseini, S.A.; Sathyapalan, T.; Majeed, M.; Jamialahmadi, T.; Al-Rasadi, K.; Banach, M.; Sahebkar, A. Potential effects of curcumin in the treatment of COVID-19 infection. Phytother. Res. 2020, 34, 2911–2920. [Google Scholar] [CrossRef]
- Patel, A.; Rajendran, M.; Shah, A.; Patel, H.; Pakala, S.B.; Karyala, P. Virtual screening of curcumin and its analogs against the spike surface glycoprotein of SARS-CoV-2 and SARS-CoV. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef]
- Jena, A.B.; Kanungo, N.; Nayak, V.; Chainy, G.; Dandapat, J. Catechin and Curcumin interact with corona (2019-nCoV/SARS-CoV2) viral S protein and ACE2 of human cell membrane: Insights from Computational study and implication for intervention. Sci. Rep. 2021, 11, 2043. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.-Y.; Han, L.; Shi, X.-l.; Wang, B.-l.; Huang, H.; Wang, X.; Chen, D.-F.; Ju, D.-W.; Feng, M.-Q. Baicalin inhibits autophagy induced by influenza A virus H3N2. Antivir. Res. 2015, 113, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ye, F.; Sun, Q.; Liang, H.; Li, C.; Lu, R.; Huang, B.; Tan, W.; Lai, L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzym. Inhib Med. Chem. 2021, 36, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhang, L.; Xu, Y.; Yang, D.; Yang, S.; Zhang, W.; Wang, J.; Tian, S.; Yang, S.; Yuan, T. The comprehensive study on the therapeutic effects of baicalein for the treatment of COVID-19 in vivo and in vitro. Biochem. Pharmacol. 2021, 183, 114302. [Google Scholar] [CrossRef] [PubMed]
- Gorla, U.S.; Rao, G.K.; Kulandaivelu, U.S.; Alavala, R.R.; Panda, S.P. Lead Finding from Selected Flavonoids with Antiviral (SARS-CoV-2) Potentials against COVID-19: An in-silico Evaluation. Comb. Chem. High. Throughput Screen. 2020. [Google Scholar] [CrossRef]
- Jain, A.S.; Sushma, P.; Dharmashekar, C.; Beelagi, M.S.; Prasad, S.K.; Shivamallu, C.; Prasad, A.; Syed, A.; Marraiki, N.; Prasad, K.S. In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi J. Biol. Sci. 2021, 28, 1040–1051. [Google Scholar] [CrossRef]
- Huseen, N.H.A. Docking Study of Naringin Binding with COVID-19 Main Protease Enzyme. Iraqi J. Pharm. Sci. 2020, 29, 231–238. [Google Scholar]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Clementi, N.; Scagnolari, C.; D’Amore, A.; Palombi, F.; Criscuolo, E.; Frasca, F.; Pierangeli, A.; Mancini, N.; Antonelli, G.; Clementi, M. Naringenin is a powerful inhibitor of SARS-CoV-2 infection in vitro. Pharmacol. Res. 2020, 163, 105255. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Deng, W.; Chen, S. Inactivation of Nf-kb Pathway by Taxifolin Attenuates Sepsis-Induced Acute Lung Injury. Curr. Top. Nutraceutical Res. 2020, 18, 176–182. [Google Scholar]
- Gogoi, N.; Chowdhury, P.; Goswami, A.K.; Das, A.; Chetia, D.; Gogoi, B. Computational guided identification of a citrus flavonoid as potential inhibitor of SARS-CoV-2 main protease. Mol. Divers. 2020, 1–15. [Google Scholar] [CrossRef]
- Verdura, S.; Cuyàs, E.; Llorach-Parés, L.; Pérez-Sánchez, A.; Micol, V.; Nonell-Canals, A.; Joven, J.; Valiente, M.; Sánchez-Martínez, M.; Bosch-Barrera, J. Silibinin is a direct inhibitor of STAT3. Food Chem. Toxicol. 2018, 116, 161–172. [Google Scholar] [CrossRef]
- Bosch-Barrera, J.; Martin-Castillo, B.; Buxó, M.; Brunet, J.; Encinar, J.A.; Menendez, J.A. Silibinin and SARS-CoV-2: Dual targeting of host cytokine storm and virus replication machinery for clinical management of COVID-19 patients. J. Clin. Med. 2020, 9, 1770. [Google Scholar] [CrossRef]
- Yi, L.; Li, Z.; Yuan, K.; Qu, X.; Chen, J.; Wang, G.; Zhang, H.; Luo, H.; Zhu, L.; Jiang, P. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004, 78, 11334–11339. [Google Scholar] [CrossRef]
- Lin, S.-C.; Ho, C.-T.; Chuo, W.-H.; Li, S.; Wang, T.T.; Lin, C.-C. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 2017, 17, 144. [Google Scholar] [CrossRef]
- Yang, M.; Wei, J.; Huang, T.; Lei, L.; Shen, C.; Lai, J.; Yang, M.; Liu, L.; Yang, Y.; Liu, G. Resveratrol inhibits the replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in cultured Vero cells. Phytother. Res. 2020. [Google Scholar] [CrossRef]
- Ho, T.-Y.; Wu, S.-L.; Chen, J.-C.; Li, C.-C.; Hsiang, C.-Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007, 74, 92–101. [Google Scholar] [CrossRef]
- Park, J.-Y.; Jeong, H.J.; Kim, J.H.; Kim, Y.M.; Park, S.-J.; Kim, D.; Park, K.H.; Lee, W.S.; Ryu, Y.B. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012, 35, 2036–2042. [Google Scholar] [CrossRef]
- Qing, Z.-X.; Yang, P.; Tang, Q.; Cheng, P.; Liu, X.-B.; Zheng, Y.-J.; Liu, Y.-S.; Zeng, J.-G. Isoquinoline alkaloids and their antiviral, antibacterial, and antifungal activities and structure-activity relationship. Curr. Org. Chem. 2017, 21, 1920–1934. [Google Scholar] [CrossRef]
- Moradi, M.-T.; Karimi, A.; Rafieian-Kopaei, M.; Fotouhi, F. In vitro antiviral effects of Peganum harmala seed extract and its total alkaloids against Influenza virus. Microb. Pathog. 2017, 110, 42–49. [Google Scholar] [CrossRef]
- Ogunyemi, O.M.; Gyebi, G.A.; Elfiky, A.A.; Afolabi, S.O.; Ogunro, O.B.; Adegunloye, A.P.; Ibrahim, I.M. Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS-Cov-2 RNA-dependent RNA polymerase: An in silico perspective. Antivir. Chem. Chemother. 2020, 28, 2040206620984076. [Google Scholar] [CrossRef]
- Shen, L.; Niu, J.; Wang, C.; Huang, B.; Wang, W.; Zhu, N.; Deng, Y.; Wang, H.; Ye, F.; Cen, S. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019, 93, e00023-19. [Google Scholar] [CrossRef]
- Choy, K.-T.; Wong, A.Y.-L.; Kaewpreedee, P.; Sia, S.F.; Chen, D.; Hui, K.P.Y.; Chu, D.K.W.; Chan, M.C.W.; Cheung, P.P.-H.; Huang, X. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020, 178, 104786. [Google Scholar] [CrossRef]
- Zhang, Y.-N.; Zhang, Q.-Y.; Li, X.-D.; Xiong, J.; Xiao, S.-Q.; Wang, Z.; Zhang, Z.-R.; Deng, C.-L.; Yang, X.-L.; Wei, H.-P. Gemcitabine, lycorine and oxysophoridine inhibit novel coronavirus (SARS-CoV-2) in cell culture. Emerg. Microbes Infect. 2020, 9, 1170–1173. [Google Scholar] [CrossRef]
- Yang, C.-W.; Lee, Y.-Z.; Hsu, H.-Y.; Shih, C.; Chao, Y.-S.; Chang, H.-Y.; Lee, S.-J. Targeting coronaviral replication and cellular JAK2 mediated dominant NF-κB activation for comprehensive and ultimate inhibition of coronaviral activity. Sci. Rep. 2017, 7, 4105. [Google Scholar] [CrossRef]
- Yang, C.-W.; Lee, Y.-Z.; Hsu, H.-Y.; Jan, J.-T.; Lin, Y.-L.; Chang, S.-Y.; Peng, T.-T.; Yang, R.-B.; Liang, J.-J.; Liao, C.-C. Inhibition of SARS-CoV-2 by highly potent broad-spectrum anti-coronaviral tylophorine-based derivatives. Front. Pharmacol. 2020, 11, 2056. [Google Scholar] [CrossRef]
- Kalhori, M.R.; Saadatpour, F.; Arefian, E.; Soleimani, M.; Farzaei, M.H.; Aneva, I.Y.; Echeverría, J. The Potential Therapeutic Effect of RNA Interference and Natural Products on COVID-19: A Review of the Coronaviruses Infection. Front. Pharmacol. 2021, 12, 616993. [Google Scholar] [CrossRef]
- Michaelis, M.; Geiler, J.; Naczk, P.; Sithisarn, P.; Leutz, A.; Doerr, H.W.; Cinatl, J., Jr. Glycyrrhizin exerts antioxidative effects in H5N1 influenza A virus-infected cells and inhibits virus replication and pro-inflammatory gene expression. PLoS ONE 2011, 6, e19705. [Google Scholar] [CrossRef]
- Murck, H. Symptomatic protective action of glycyrrhizin (Licorice) in Covid-19 infection? Front. Immunol. 2020, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Sprague, L.; Lee, J.M.; Hutzen, B.J.; Wang, P.-Y.; Chen, C.-Y.; Conner, J.; Braidwood, L.; Cassady, K.A.; Cripe, T.P. High mobility group box 1 influences HSV1716 spread and acts as an adjuvant to chemotherapy. Viruses 2018, 10, 132. [Google Scholar] [CrossRef]
- Trøseid, M.; Sönnerborg, A.; Nowak, P. High mobility group box protein-1 in HIV-1 infection. Curr. HIV Res. 2011, 9, 6–10. [Google Scholar] [CrossRef]
- Bailly, C.; Vergoten, G. Glycyrrhizin: An alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome? Pharmacol. Ther. 2020, 214, 107618. [Google Scholar] [CrossRef]
- Hassan, M.Z.; Osman, H.; Ali, M.A.; Ahsan, M.J. Therapeutic potential of coumarins as antiviral agents. Eur. J. Med. Chem. 2016, 123, 236–255. [Google Scholar] [CrossRef]
- Lyndem, S.; Sarmah, S.; Das, S.; Roy, A.S. In silico screening of naturally occurring coumarin derivatives for the inhibition of the main protease of SARS-CoV-2. Chem Rxiv. Prepr. 2020. [Google Scholar] [CrossRef]
- Chidambaram, S.K.; Ali, D.; Alarifi, S.; Radhakrishnan, S.; Akbar, I. In silico molecular docking: Evaluation of coumarin based derivatives against SARS-CoV-2. J. Infect. Public Health 2020, 13, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Chidambaram, S.; El-Sheikh, M.A.; Alfarhan, A.H.; Radhakrishnan, S.; Akbar, I. Synthesis of novel coumarin analogues: Investigation of molecular docking interaction of SARS-CoV-2 proteins with natural and synthetic coumarin analogues and their pharmacokinetics studies. Saudi J. Biol. Sci. 2021, 28, 1100–1108. [Google Scholar] [CrossRef]
- Santoyo, S.; Jaime, L.; Plaza, M.; Herrero, M.; Rodriguez-Meizoso, I.; Ibañez, E.; Reglero, G. Antiviral compounds obtained from microalgae commonly used as carotenoid sources. J. Appl. Phycol. 2012, 24, 731–741. [Google Scholar] [CrossRef]
- Vardhan, S.; Sahoo, S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput. Biol. Med. 2020, 124, 103936. [Google Scholar] [CrossRef]
- Yan, Y.Q.; Fu, Y.J.; Wu, S.; Qin, H.Q.; Zhen, X.; Song, B.M.; Weng, Y.S.; Wang, P.C.; Chen, X.Y.; Jiang, Z.Y. Anti-influenza activity of berberine improves prognosis by reducing viral replication in mice. Phytother. Res. 2018, 32, 2560–2567. [Google Scholar] [CrossRef]
- Hussain, T.; Al-Attas, O.S.; Alamery, S.; Ahmed, M.; Odeibat, H.A.; Alrokayan, S. The plant flavonoid, fisetin alleviates cigarette smoke-induced oxidative stress, and inflammation in Wistar rat lungs. J. Food Biochem. 2019, 43, e12962. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Sarkar, C.; El-Kersh, D.M.; Jamaddar, S.; Uddin, S.J.; Shilpi, J.A.; Mubarak, M.S. Natural products and their derivatives against coronavirus: A review of the non-clinical and pre-clinical data. Phytother. Res. 2020, 34, 2471–2492. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.-T.; Hsu, W.-C.; Lin, C.-C. Antiviral natural products and herbal medicines. J. Tradit. Complementary Med. 2014, 4, 24–35. [Google Scholar] [CrossRef] [PubMed]
- ul Qamar, M.T.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants†. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-H.; Wu, K.-L.; Zhang, X.; Deng, S.-Q.; Peng, B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J. Integr. Med. 2020, 18, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.K.; Curtis-Long, M.J.; Lee, K.H.; Kim, D.W.; Ryu, H.W.; Yuk, H.J.; Park, K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorganic Med. Chem. 2013, 21, 3051–3057. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Sarkar, A.; Maulik, U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020, 10, 17699. [Google Scholar] [CrossRef] [PubMed]
- Owis, A.I.; El-Hawary, M.S.; El Amir, D.; Aly, O.M.; Abdelmohsen, U.R.; Kamel, M.S. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020, 10, 19570–19575. [Google Scholar] [CrossRef]
- Rameshkumar, M.R.; Indu, P.; Arunagirinathan, N.; Venkatadri, B.; El-Serehy, H.A.; Ahmad, A. Computational selection of flavonoid compounds as inhibitors against SARS-CoV-2 main protease, RNA-dependent RNA polymerase and spike proteins: A molecular docking study. Saudi J. Biol. Sci. 2021, 28, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zheng, W.; Li, M.; Huang, J.; Bao, S.; Xu, Q.; Ma, Z. Citrus fruits are rich in flavonoids for immunoregulation and potential targeting ACE2. Preprints 2020, 2020020313. [Google Scholar] [CrossRef]
- Maurya, V.K.; Kumar, S.; Prasad, A.K.; Bhatt, M.L.; Saxena, S.K. Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. VirusDisease 2020, 31, 179–193. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, B.; Chen, X.-J.; Xu, D.-Q.; Wang, Y.-X.; Dong, H.-Y.; Ma, S.-R.; Sun, R.-H.; Hui, Y.-P.; Li, Z.-C. Osthole protects lipopolysaccharide-induced acute lung injury in mice by preventing down-regulation of angiotensin-converting enzyme 2. Eur. J. Pharm. Sci. 2013, 48, 819–824. [Google Scholar] [CrossRef]
- Seo, D.J.; Jeon, S.B.; Oh, H.; Lee, B.-H.; Lee, S.-Y.; Oh, S.H.; Jung, J.Y.; Choi, C. Comparison of the antiviral activity of flavonoids against murine norovirus and feline calicivirus. Food Control. 2016, 60, 25–30. [Google Scholar] [CrossRef]
- Wan, L.; Meng, D.; Wang, H.; Wan, S.; Jiang, S.; Huang, S.; Wei, L.; Yu, P. Preventive and therapeutic effects of thymol in a lipopolysaccharide-induced acute lung injury mice model. Inflammation 2018, 41, 183–192. [Google Scholar] [CrossRef]
- Hu, X.; Li, H.; Fu, L.; Liu, F.; Wang, H.; Li, M.; Jiang, C.; Yin, B. The protective effect of hyperin on LPS-induced acute lung injury in mice. Microb. Pathog. 2019, 127, 116–120. [Google Scholar] [CrossRef]
- Lowe, H.I.; Toyang, N.J.; McLaughlin, W. Potential of cannabidiol for the treatment of viral hepatitis. Pharmacogn. Res. 2017, 9, 116–118. [Google Scholar]
- Gani, M.A.; Nurhan, A.D.; Maulana, S.; Siswodihardjo, S.; Shinta, D.W.; Khotib, J. Structure-based virtual screening of bioactive compounds from Indonesian medical plants against severe acute respiratory syndrome coronavirus-2. J. Adv. Pharm. Technol. Res. 2021, 12, 120. [Google Scholar]
- Khan, A.; Heng, W.; Wang, Y.; Qiu, J.; Wei, X.; Peng, S.; Saleem, S.; Khan, M.; Ali, S.S.; Wei, D.-Q. In silico and in vitro evaluation of kaempferol as a potential inhibitor of the SARS-CoV-2 main protease (3CLpro). Phytother. Res 2021. [Google Scholar] [CrossRef]
- Laksmiani, N.P.L.; Larasanty, L.P.F.; Santika, A.A.G.J.; Prayoga, P.A.A.; Dewi, A.A.I.K.; Dewi, N.P.A.K. Active Compounds Activity from the Medicinal Plants against SARS-CoV-2 using in Silico Assay. Biomed. Pharmacol. J. 2020, 13, 873–881. [Google Scholar] [CrossRef]
- Milenkovic, D.; Ruskovska, T.; Rodriguez-Mateos, A.; Heiss, C. Polyphenols could prevent SARS-CoV-2 infection by modulating the expression of miRNAs in the host cells. Aging Dis. 2021. [Google Scholar] [CrossRef]
- Pandey, A.K.; Verma, S. An in-silico evaluation of dietary components for structural inhibition of SARS-Cov-2 main protease. J. Biomol. Struct. Dyn. 2020, 1–7. [Google Scholar] [CrossRef]
| Phytochemical | Compound | Study Type | Mechanism of Antiviral Activity | References |
|---|---|---|---|---|
| Alkaloid | 10′-hydrox-yusambarensine | In silico | ↓RdRp | [263] |
| Berberine | In vitro, In silico | Antiviral effect, ↓ACE2, spike protein and increased Nrf2, HO-1 ↓TGF-β1, ROS | [281] | |
| Cryptospirolepine | In silico | ↓RdRp | [263] | |
| Emetin | In vitro | ↓Viral entry ↓MERS-CoV S-mediated infection, ↓SARS-CoV-2 replication | [264,265] | |
| Lycorine | In vivo In vitro | ↓Spread and replication of HCoV-OC43, ↓SARS-CoV-2 replication | [264,266] | |
| In vitro | ↓Different species of CoV | [283] | ||
| Oxysophoridine | In vitro | ↓SARS-CoV-2 replication | [266,298] | |
| Strychnopentamine | In silico | ↓RdRp | [263] | |
| Tetrandrine | In vitro | ↓HCoV-OC43-infected | [283] | |
| Tylophorine | In vitro | ↓JAK2, ↓NF-κB, ↓inflammation, ↓replication | [267,268] | |
| Anthocyanin | Malvidin | In vitro | ↓Bax/Bcl-2, Caspase-3, IL-1β, TNF-α | [50] |
| Cannabinoid | Cannabidiol | In vitro | ↓MPO, TNF-α, IL-6 | [297] |
| Coumarin | Inophyllum A | In silico | ↓Mpro, ↓replication | [278] |
| Methylgalbanate | In silico | ↓Mpro, ↓replication | [276] | |
| Osthole | In vitro | ↓IL-6, TNF-α, ↑ACE2 and Ang1–7 | [293] | |
| Toddacoumaquinone | In silico | ↓Mpro, ↓replication | [277] | |
| Diarylheptanoid | Hirsutenone | In vitro | ↓PLpro, ↓replication | [260] |
| Flavonoid | Baicalein | In vitro In vivo | ↓3CLpro ↓Vero E6 cells damage, ↓lesions of lung tissue, ↓replication, ↓IL-1β, ↓TNF-α, ↓inflammation | [245,246] |
| Biochanin A | In silico | ↓spike glycoprotein | [247] | |
| Kaempferol | In vitro In silico | ↓3CLpro, ↓replication | [299] | |
| Luteolin | In vitro In silico | ↓Viral entry ↓SARS-CoV infection ↓TNF-α, IL-1β, IL-6, IL-18, NF-κB | [256,300] | |
| Naringenin | In vitro In silico | ↓TPC2, ↓viral infection ↓TNF-α, IL-1β, IL-6, IL-18, NF-κB | [251,300] | |
| Naringin | In silico | ↓Mpro, ↓replication | [249] | |
| In silico | ↓Spike glycoprotein | [248] | ||
| Silibinin | In silico | ↓RdRp | [255] | |
| Silymarin | In silico | ↓ACE2 ↓IL-6, IL-1β, TNF-α, p46-p54, p42, p38, p44, NF-κB, and JNK. | [247] | |
| Taxifolin | In silico | ↓Mpro | [253] | |
| Flavonoid | Cyanidin | In silico | ↓ACE2 and RdRp | [290] |
| Kazinol A | In vitro | ↓SARS-CoV 3CLpro and PLpro | [283] | |
| Narcissin | In silico | Bind to ACE2 | [289] | |
| Tomentin A-E | In silico | ↓PLpro in COVID-19 | [287] | |
| Flavone | Baicalin | In silico | ↓TMPRSS2 and lead to inhibition of COVID-19 | [204] |
| Chrysin | In silico | ↓ACE2 and decline neurological manifestation in COVID-19 | [288] | |
| Flavonol | Fisetin | In vitro, In silico | ↓ACE2, ↓TNF-α, IL-6, IL-1β, ↑Nrf2, GPx, SOD | [282] |
| Hesperetin | In vitro | ↓ACE2 and reduce neurological sign in COVID-19 | [291] | |
| Hesperetin | In vitro | ↓ACE2 and reduce neurological sign in COVID-19 | [291] | |
| Hyperin | In vitro | ↓TNF-α, IL-6, IL-1β, NF-κB | [296] | |
| Isoflavone | Daidzein | In vitro | ↓TLR4, MyD88, NF-κB, MPO, IL-6, TNF-α | [294] |
| Polyphenol | Catechin | In silico | ↓Spike protein, ↓viral entry, ↓ACE2 | [243] |
| Curcumin | In silico | ↓spike protein, ↓viral entry, ↓ACE2 ↓TNF-α, IL-1β, IL-6, IL-18, NF-κB, COX-2 | [242,243,301] | |
| Ellagic acid | In vitro | ↓Mpro, ↓replication | [302] | |
| Resveratrol | In vitro | ↓SARS-CoV-2 infection. | [258,301] | |
| Sinigrin | In vitro | ↓SARS-CoV 3CLpro | [284] | |
| Terpenoid | Carvacrol | In silico | ↓Spike protein | [292] |
| Geraniol | In vitro | ↓Spike protein, ↓TNF-α, IL-1β, IL-6, iNOS, COX-2 | [292] | |
| Limonin | In silico | ↓ACE2, 3CLpro, PLpro, RdRp and spike protein | [280] | |
| Thymol | In vitro | ↓NF-κB, IL-6, TNF-α, IL-1β, ↑SOD | [295] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fakhri, S.; Nouri, Z.; Moradi, S.Z.; Akkol, E.K.; Piri, S.; Sobarzo-Sánchez, E.; Farzaei, M.H.; Echeverría, J. Targeting Multiple Signal Transduction Pathways of SARS-CoV-2: Approaches to COVID-19 Therapeutic Candidates. Molecules 2021, 26, 2917. https://doi.org/10.3390/molecules26102917
Fakhri S, Nouri Z, Moradi SZ, Akkol EK, Piri S, Sobarzo-Sánchez E, Farzaei MH, Echeverría J. Targeting Multiple Signal Transduction Pathways of SARS-CoV-2: Approaches to COVID-19 Therapeutic Candidates. Molecules. 2021; 26(10):2917. https://doi.org/10.3390/molecules26102917
Chicago/Turabian StyleFakhri, Sajad, Zeinab Nouri, Seyed Zachariah Moradi, Esra Küpeli Akkol, Sana Piri, Eduardo Sobarzo-Sánchez, Mohammad Hosein Farzaei, and Javier Echeverría. 2021. "Targeting Multiple Signal Transduction Pathways of SARS-CoV-2: Approaches to COVID-19 Therapeutic Candidates" Molecules 26, no. 10: 2917. https://doi.org/10.3390/molecules26102917
APA StyleFakhri, S., Nouri, Z., Moradi, S. Z., Akkol, E. K., Piri, S., Sobarzo-Sánchez, E., Farzaei, M. H., & Echeverría, J. (2021). Targeting Multiple Signal Transduction Pathways of SARS-CoV-2: Approaches to COVID-19 Therapeutic Candidates. Molecules, 26(10), 2917. https://doi.org/10.3390/molecules26102917










