Optimization of the Biosynthesis of B-Ring Ortho-Hydroxy Lated Flavonoids Using the 4-Hydroxyphenylacetate 3-Hydroxylase Complex (HpaBC) of Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Media, Bacterial Strains and Vectors
2.3. Construction of the HpaB and HpaC Expression Plasmids
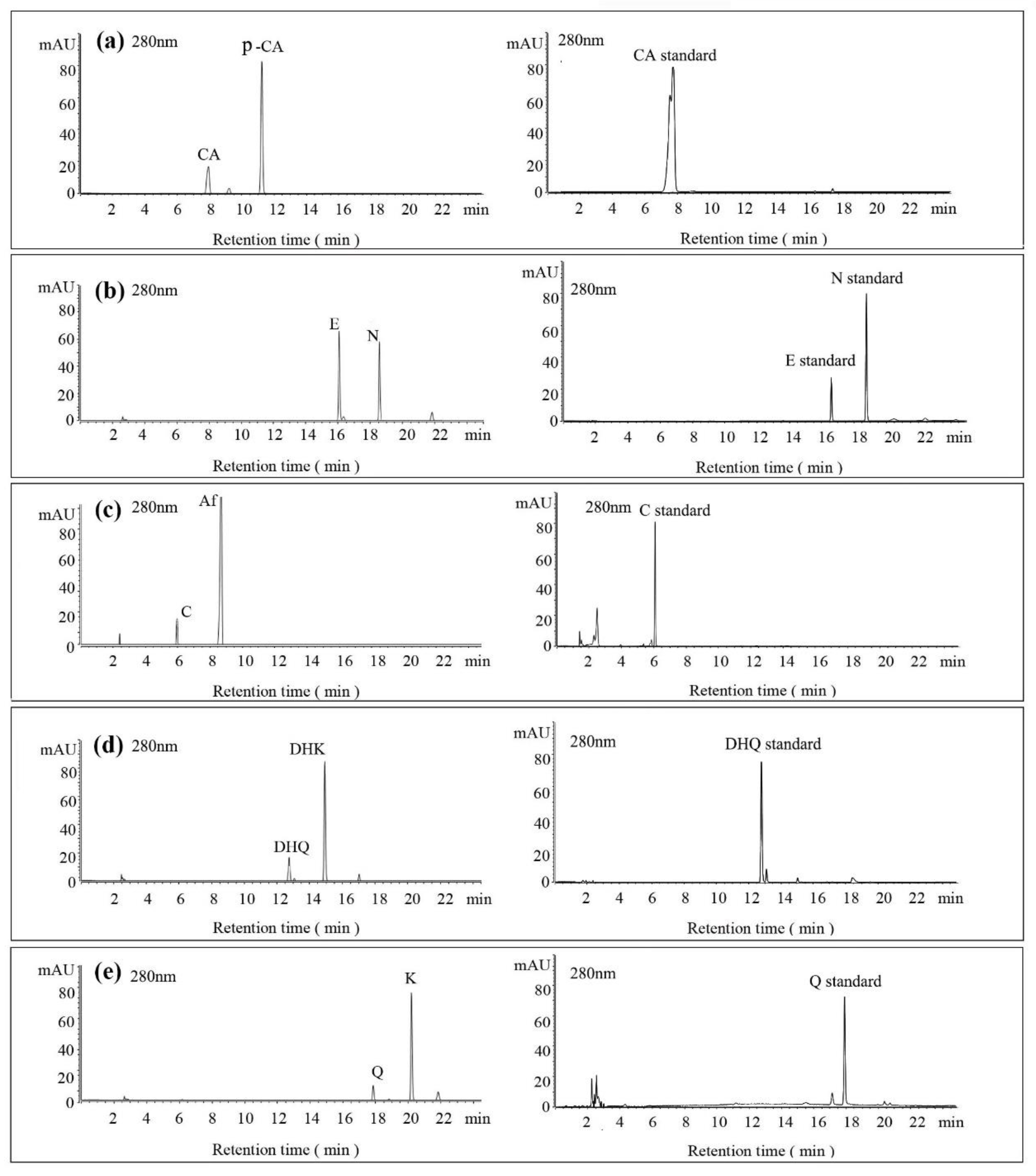
2.4. HPLC and LC-MS Analysis
2.5. Statistical Analysis
3. Results
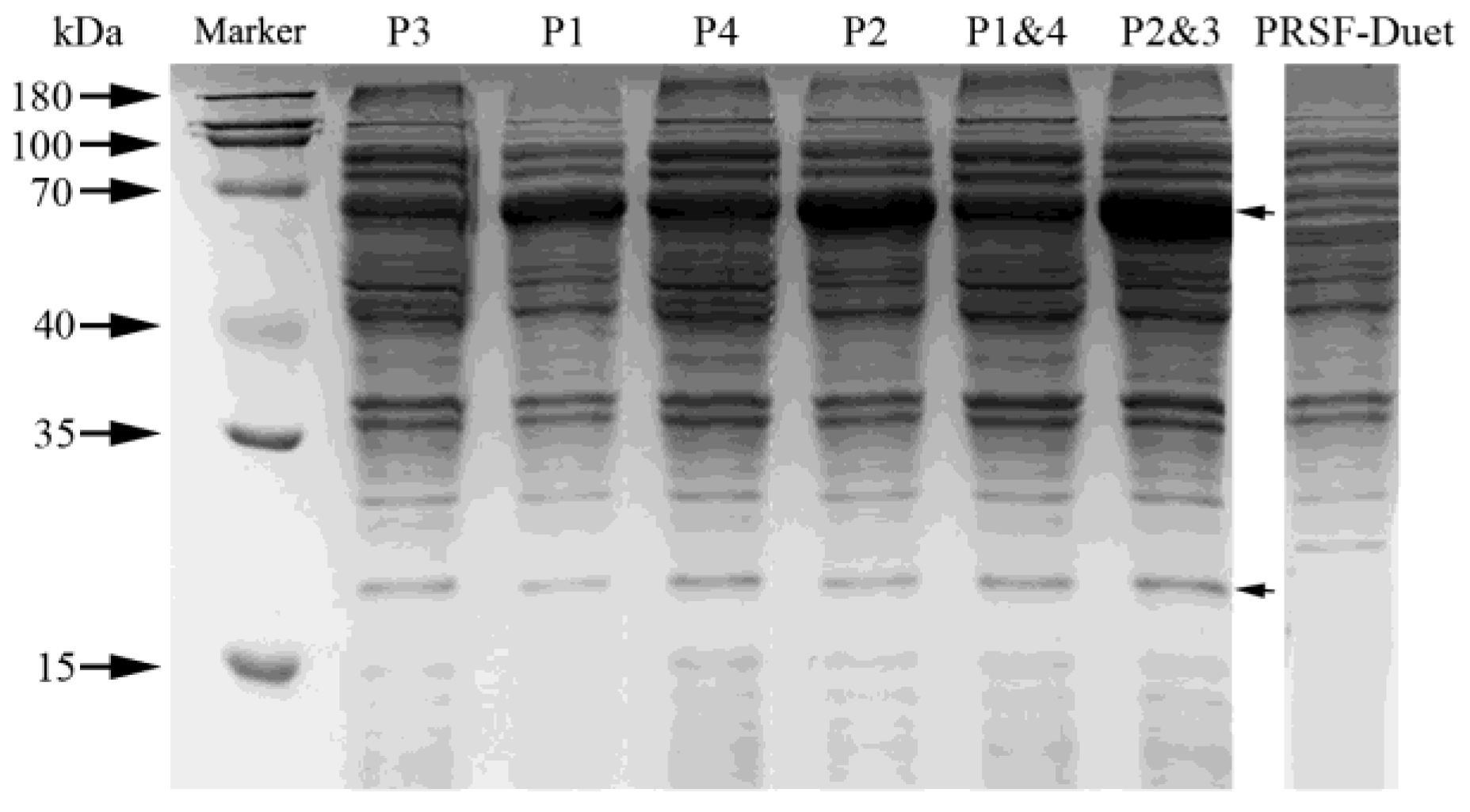
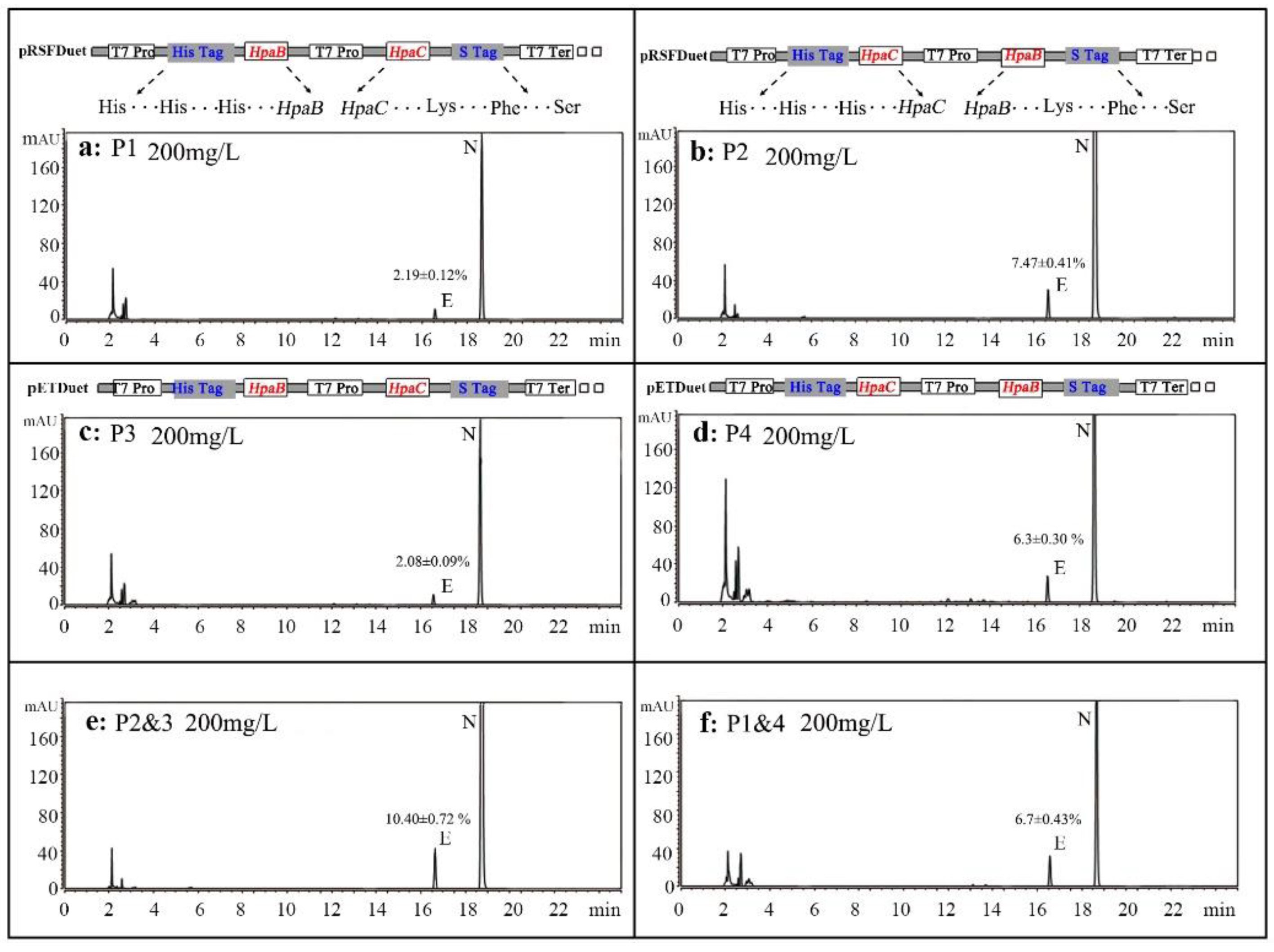
3.1. Expression of HpaB and HpaC in E. coli
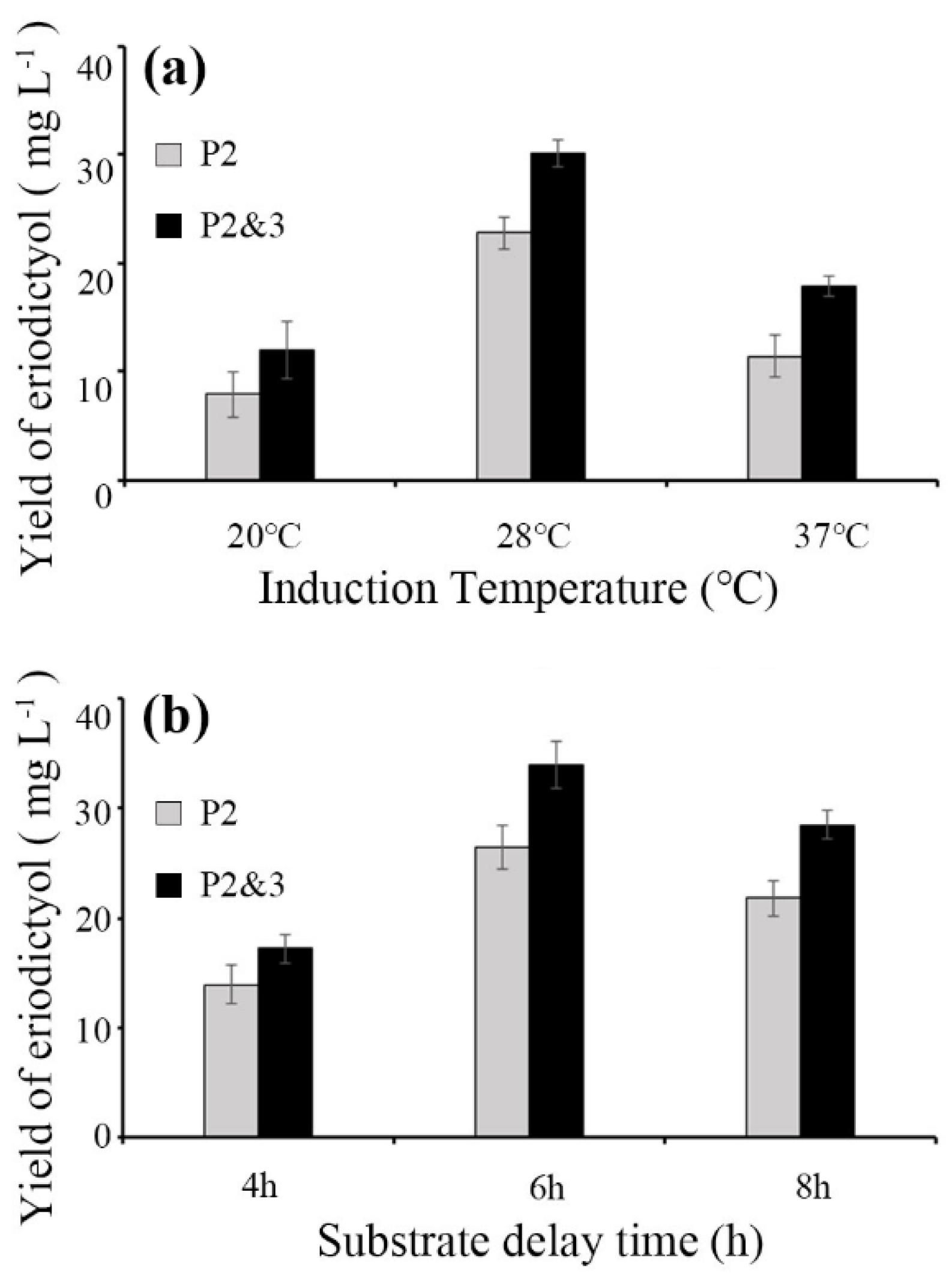
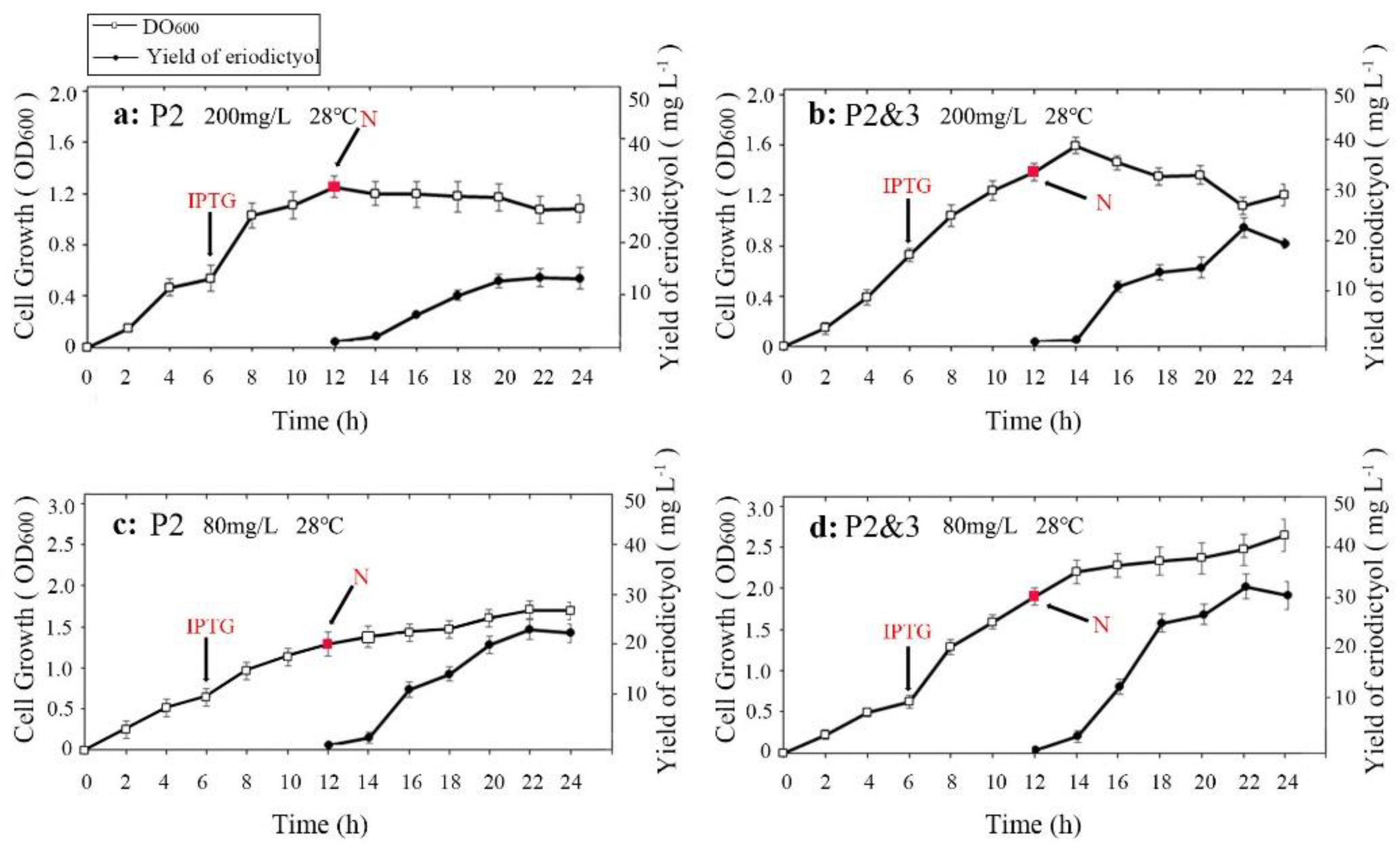
3.2. Optimization of the Induction Temperature and Substrate Delay Time
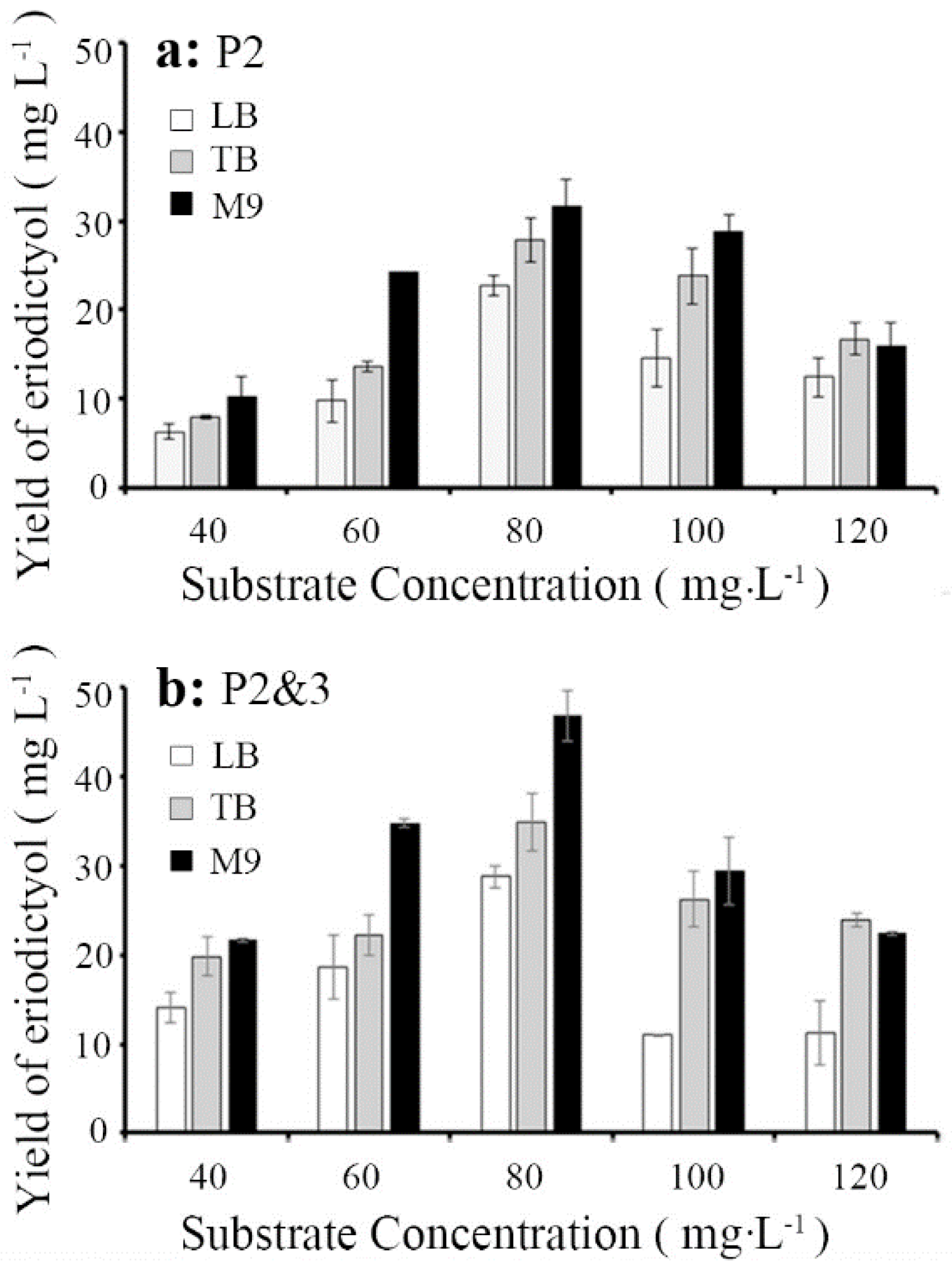
3.3. Optimization of the Substrate Concentration and Medium to Improve Catalytic Efficiency
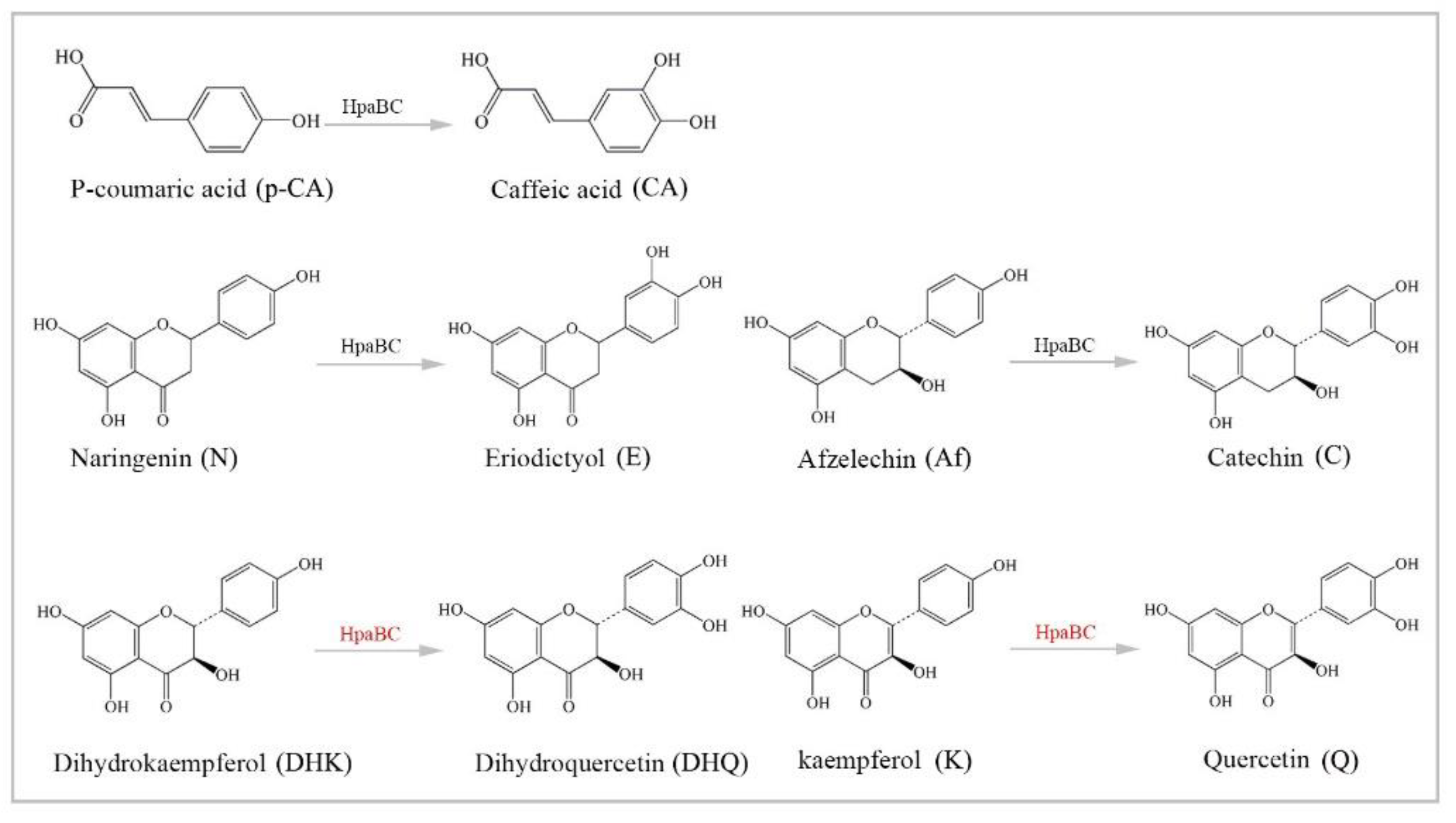
3.4. Substrate Diversity Analysis of the HpaBC Complex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Chen, A.Y.; Chen, Y.C. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013, 138, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Hollman, P.C.; Katan, M.B. Absorption, metabolism and health effects of dietary flavonoids in man. Biomed. Pharm. 1997, 51, 305–310. [Google Scholar] [CrossRef]
- Nair, S.; Gupta, R. Dietary antioxidant flavonoids and coronary heart disease. J. Assoc. Phys. India 1996, 44, 699–702. [Google Scholar] [PubMed]
- Edenharder, R.; Keller, G.; Platt, K.L.; Unger, K.K. Isolation and characterization of structurally novel antimutagenic flavonoids from spinach (Spinacia oleracea). J. Agric. Food Chem 2001, 49, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.K.; Raghuram, G.V.; Bhargava, A.; Ahirwar, A.; Samarth, R.; Upadhyaya, R.; Jain, S.K.; Pathak, N. In vitro and in vivo evaluation of the anticarcinogenic and cancer chemopreventive potential of a flavonoid-rich fraction from a traditional Indian herb Selaginella bryopteris. Br. J. Nutr. 2011, 106, 1154–1168. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhao, Y.; Xing, X.; Ma, X.; Sun, X.; Yang, M.; Xiao, X. Antibacterial evaluation of flavonoid compounds against E. coli by microcalorimetry and chemometrics. Appl. Microbiol. Biotechnol. 2015, 99, 6049–6058. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The Science of Flavonoids; Springer: New York, NY, USA, 2006. [Google Scholar]
- Soobrattee, M.A.; Neergheen, V.S.; Luximon-Ramma, A.; Aruoma, O.I.; Bahorun, T. Phenolics as potential antioxidant therapeutic agents: Mechanism and actions. Mutat. Res. 2005, 579, 200–213. [Google Scholar] [CrossRef] [PubMed]
- van Acker, S.A.; van den Berg, D.J.; Tromp, M.N.; Griffioen, D.H.; van Bennekom, W.P.; van der Vijgh, W.J.; Bast, A. Structural aspects of antioxidant activity of flavonoids. Free Radic. Biol. Med. 1996, 20, 331–342. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F. Flower colour and cytochromes P450. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368, 20120432. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant. Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Seitz, C.; Ameres, S.; Forkmann, G. Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase. FEBS Lett. 2007, 581, 3429–3434. [Google Scholar] [CrossRef] [PubMed]
- Seitz, C.; Eder, C.; Deiml, B.; Kellner, S.; Martens, S.; Forkmann, G. Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant. Mol. Biol. 2006, 61, 365–381. [Google Scholar] [CrossRef] [PubMed]
- International Journal of Molecular Sciences. International Journal of Molecular Sciences 2016 Best Paper Award. Natl. Libr. Sci. 2016, 17, 777. [Google Scholar]
- Leonard, E.; Yan, Y.; Koffas, M.A. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab. Eng. 2006, 8, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Prieto, M.A.; Garcia, J.L. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. A two-protein component enzyme. J. Biol. Chem. 1994, 269, 22823–22829. [Google Scholar] [CrossRef]
- Furuya, T.; Kino, K. Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives. Appl. Microbiol. Biotechnol. 2014, 98, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Collins, S.M.; Vernacchio, V.R.; Lachance, D.M.; Koffas, M.A. Optimization of naringenin and p-coumaric acid hydroxylation using the native E. coli hydroxylase complex, HpaBC. Biotechnol. Prog. 2016, 32, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, Y.; Li, W.; Zhao, L.; Meng, F.; Wang, Y.; Tan, H.; Yang, H.; Wei, C.; Wan, X. Tissue-Specific, Development-Dependent Phenolic Compounds Accumulation Profile and Gene Expression Pattern in Tea Plant [Camellia sinensis]. PLoS ONE 2013, 8, e62315. [Google Scholar] [CrossRef] [PubMed]
- Sparla, F.; Pupillo, P.; Trost, P. The C-terminal extension of glyceraldehyde-3-phosphate dehydrogenase subunit B acts as an autoinhibitory domain regulated by thioredoxins and nicotinamide adenine dinucleotide. J. Biol. Chem. 2002, 277, 44946–44952. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, H.; Katsuyama, Y.; Danyao, D.; Kahar, P.; Nakamura-Tsuruta, S.; Teramura, H.; Wakai, K.; Yoshihara, K.; Minami, H.; Ogino, C.; et al. Caffeic acid production by simultaneous saccharification and fermentation of kraft pulp using recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 2017, 101, 5279–5290. [Google Scholar] [CrossRef] [PubMed]
| Strains and Plasmids | Relevant Characteristics | Source or Reference |
|---|---|---|
| Plasmids | ||
| pRSFDuet | Double T7 promoter, ColE1 ori. KanR | Novagen |
| pETDuet | Double T7 promoter, ColE1 ori. AmpR | Novagen |
| P1 | pRSFDuet carrying (MCS-1)-HpaB and HpaC (MCS-2) | This study |
| P2 | pRSFDuet carrying (MCS-1)-HpaC and HpaB (MCS-2) | This study |
| P3 | pETDuet carrying (MCS-1)-HpaB and HpaC (MCS-2) | This study |
| P4 | pETDuet carrying (MCS-1)-HpaC and HpaB (MCS-2) | This study |
| Strains | ||
| DH5α | General cloning host | Invitrogen |
| BL21 * (DE3) | Host for flavonoid production and gene clones | Novagen |
| BL21-P1 | General expression strain of pRSFDuet P1 | This study |
| BL21-P2 | General expression strain of pRSFDuet P2 | This study |
| BL21-P3 | General expression strain of pETDuet P3 | This study |
| BL21-P4 | General expression strain of pETDuet P4 | This study |
| BL21-P2&P3 | General co-expression strain of P2 and P3 | This study |
| BL21-P1&P4 | General co-expression strain of P1 and P4 | This study |
| Substrates | Products | Yield (mg·L−1) | Conversion Rate (%) |
|---|---|---|---|
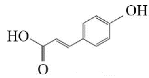 p-Coumaric acid | 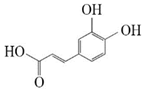 Caffeic acid | 28.91 ± 1.77 | 32.93 ± 2.01 |
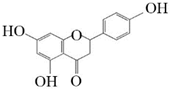 Naringenin |  Eriodictyol | 46.84 ± 2.85 | 57.67 ± 3.36 |
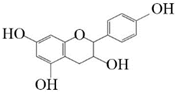 Afzelechin | 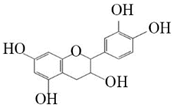 Catechin | 29.81 ± 2.66 | 35.2 ± 3.14 |
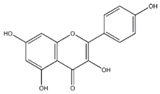 Kaempferol | 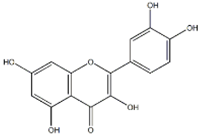 Quercetin | 20.14 ± 0.75 | 23.84 ± 0.88 |
 Dihydrokaempferol | 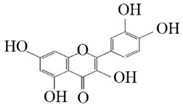 Dihydroquercetin | 20.05 ± 1.48 | 23.74 ± 1.75 |
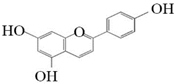 Pelargonidin | 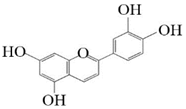 Cyanidin | _ns__ | _ns__ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Ma, X.; Ruan, H.; Chen, Y.; Gao, L.; Lei, T.; Li, Y.; Gui, L.; Guo, L.; Xia, T.; et al. Optimization of the Biosynthesis of B-Ring Ortho-Hydroxy Lated Flavonoids Using the 4-Hydroxyphenylacetate 3-Hydroxylase Complex (HpaBC) of Escherichia coli. Molecules 2021, 26, 2919. https://doi.org/10.3390/molecules26102919
Wang L, Ma X, Ruan H, Chen Y, Gao L, Lei T, Li Y, Gui L, Guo L, Xia T, et al. Optimization of the Biosynthesis of B-Ring Ortho-Hydroxy Lated Flavonoids Using the 4-Hydroxyphenylacetate 3-Hydroxylase Complex (HpaBC) of Escherichia coli. Molecules. 2021; 26(10):2919. https://doi.org/10.3390/molecules26102919
Chicago/Turabian StyleWang, Longji, Xiubing Ma, Haixiang Ruan, Yang Chen, Liping Gao, Ting Lei, Yan Li, Lin Gui, Lina Guo, Tao Xia, and et al. 2021. "Optimization of the Biosynthesis of B-Ring Ortho-Hydroxy Lated Flavonoids Using the 4-Hydroxyphenylacetate 3-Hydroxylase Complex (HpaBC) of Escherichia coli" Molecules 26, no. 10: 2919. https://doi.org/10.3390/molecules26102919
APA StyleWang, L., Ma, X., Ruan, H., Chen, Y., Gao, L., Lei, T., Li, Y., Gui, L., Guo, L., Xia, T., & Wang, Y. (2021). Optimization of the Biosynthesis of B-Ring Ortho-Hydroxy Lated Flavonoids Using the 4-Hydroxyphenylacetate 3-Hydroxylase Complex (HpaBC) of Escherichia coli. Molecules, 26(10), 2919. https://doi.org/10.3390/molecules26102919






